Abstract
Cryptosporidiosis and giardiasis are recognized as significant enteric diseases due to their long-term health effects in humans and their economic impact in agriculture and medical care. Molecular analysis is essential to identify species and genotypes causing these infectious diseases and provides a potential tool for monitoring. This study uses information on species and genetic variants to gain insights into the geographical distribution and spatial patterns of Cryptosporidium and Giardia parasites. Here, we describe the population heterogeneity of genotypic groups within Cryptosporidium and Giardia present in New Zealand using gp60 and gdh markers to compare the observed variation with other countries around the globe. Four species of Cryptosporidium (C. hominis, C. parvum, C. cuniculus and C. erinacei) and one species of Giardia (G. intestinalis) were identified. These species have been reported worldwide and there are not unique Cryptosporidium gp60 subtype families and Giardia gdh assemblages in New Zealand, most likely due to high gene flow of historical and current human activity (travel and trade) and persistence of large host population sizes. The global analysis revealed that genetic variants of these pathogens are widely distributed. However, genetic variation is underestimated by data biases (e.g. neglected submission of sequences to genetic databases) and low sampling. New genotypes are likely to be discovered as sampling efforts increase according to accumulation prediction analyses, especially for C. parvum. Our study highlights the need for greater sampling and archiving of genotypes globally to allow comparative analyses that help understand the population dynamics of these protozoan parasites. Overall our study represents a comprehensive overview for exploring local and global protozoan genotype diversity and advances our understanding of the importance for surveillance and potential risk associated with these infectious diseases.
Author summary
Infectious diseases threaten the health and well-being of wildlife, livestock and human populations and contribute to significant economic impact in agriculture and medical care. Cryptosporidium and Giardia are enteric protozoan pathogens that cause diarrhea and nutritional disorders on a global level. Using molecular analysis and a review framework we showed that species and genetic variants within genera Cryptosporidium and Giardia (including two species recently infecting humans) found in an island system are not different from other parts of the world. This similarity is likely due to high gene flow of historical and current human activity (travel and trade) and persistence of large host population sizes, such as cattle and people. We also show that, although species and genotypes are widely distributed, new variants will arise when sampling effort increase and their dispersal will be facilitated by human activity. These findings suggest that geographical distribution of species and genotypes within Cryptosporidium and Giardia parasites may yield important clues for designing effective surveillance strategies and identification of factors driving within and cross species transmission.
Introduction
Infectious diseases are the major leading causes of death and disability worldwide [1]. Classical public health and sanitation measures have long served to minimize dissemination and human exposure to many pathogens that are spread by routes such as contaminated water or via vectors [2] but the recent Ebola and Zika outbreaks have highlighted the need for a global framework to reduce the risk and mitigate health crises [3–5]. Determining genetic diversity and the distribution of microbes can guide more efficient interventions that decrease the incidence of infectious diseases. Understanding patterns of geographical distribution and genetic variation is essential to monitor pathogen evolution and adaptation, design effective surveillance strategies, identify factors driving within and cross species transmission, and ultimately, reduce the burden of disease [6–8].
Gastroenteritis is a well-known public health threat and a significant burden of disease that requires improved monitoring [2, 9]. A substantial number of gastroenteritis cases around the world are caused by the parasitic protozoans Cryptosporidium and Giardia [10–14]. These organisms are recognized as major causes of parasite-induced diarrhoea in humans and other animals that can be spread through various means, but transmitted mainly by interactions between humans and animals and by contaminated water or food [14]. Most human cryptosporidiosis cases worldwide are caused by Cryptosporidium parvum and C. hominis [15], although other species in domesticated and wild animals can infect humans and cause high morbidity and mortality in children, travellers and immunocompromised individuals [11, 16–21]. It has been suggested that Cryptosporidium can be found in 1% of stools of immunocompetent hosts from high-income countries and 5–10% of hosts from low-resource settings but recent molecular studies suggest an underestimation of the reported frequency of infection and the global burden of the disease [16, 22]. Giardia is a common enteric parasite that infects a wide range of mammals, including domestic animals (livestock, dogs and cats), birds and amphibians. In humans, G. intestinalis (synonyms G. lamblia and G. duodenalis) is the most common intestinal protozoan parasite, frequently reported in association with water- and food-borne outbreaks [15, 23, 24], affecting about 280 million people worldwide [25] with some 500,000 new symptomatic cases reported each year in developing countries only [26].
Species within each of these two protozoan genera are morphologically indistinguishable and their taxonomic and genotype differentiation is mainly based on molecular characterization [14, 27–29]. Multilocus sequence typing has been developed for identification and genotyping of Cryptosporidium and Giardia but the most commonly employed markers in epidemiological investigations and population studies are the cell surface glycoprotein (gp60) and glutamate dehydrogenase (gdh), respectively [30, 31]. The gp60 and gdh genes have been readily adopted as a key component for molecular epidemiological investigations because of their high reliability in the characterization of genotypes, standardization in genotype nomenclature and detection of variants within populations [15]. Although not all Cryptosporidium species can be detected with the gp60 typing tool [23, 32–34] a high variety of subtype families have been identified based on this gene, whilst several assemblages can be determined using gdh for Giardia.
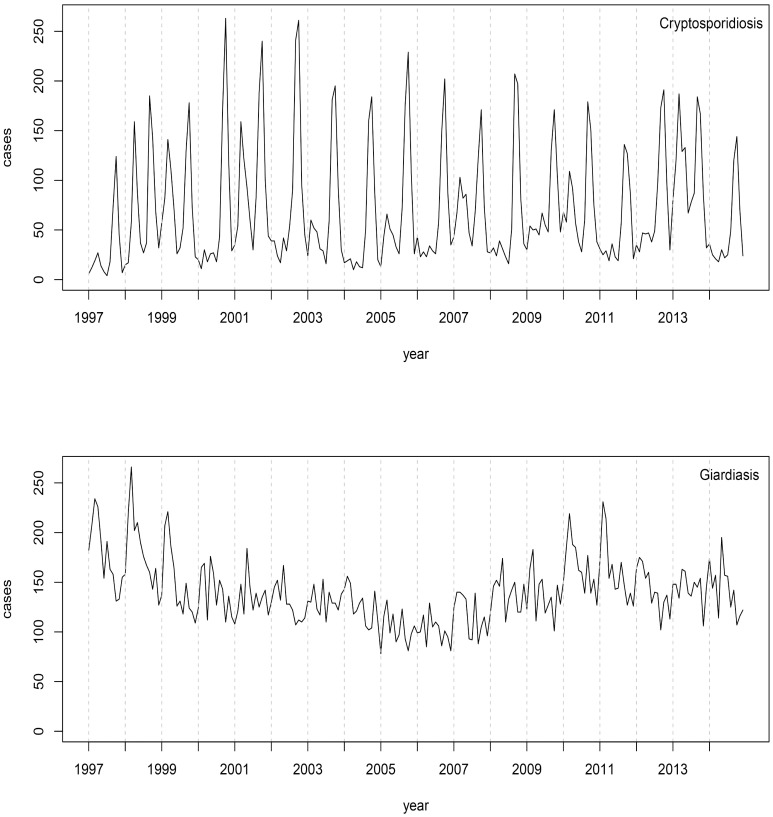
Recent studies have revealed that some genotypes are genetically diverse, host restricted and comprise a zoonotic or anthroponotic reservoir. For instance, C. parvum subtype families IIc and IIe are considered anthroponotic and IIa is predominant in humans and other animals worldwide [35] whilst G. intestinalis genotypes A and B are the only assemblages found in humans [23]. Additionally, the prevalence of some genotypes of Cryptosporidium and Giardia are unevenly distributed in different regions of the world [15, 36] with major occurrences of novel and rare variants in developing countries compared to developed countries [15, 30]. However, both diseases have been reported with a high incidence rate in New Zealand [15, 37]. The peak of cryptosporidiosis cases each year coincides with the cattle calving season [38, 39], while giardiasis incidence is more evenly distributed throughout the year (Fig 1). The annual number of notifications reported in New Zealand range between 500 and 700 and 1600 and 1800 for cryptosporidiosis and giardiasis, respectively [40]. Here, we reported the species and genotypes of Cryptosporidium and Giardia in New Zealand using gp60 and gdh markers during a long-term monitoring study (2009–2015). New Zealand has a recent and limited domestic animal movement [41] and it is an excellent island system example to compare genetic diversity against continental situations. Genotypes for species isolated from New Zealand were contrasted with those recovered from other countries to understand their spatial distribution and similarity. Specifically, we determined the genetic variants of gp60 subtype families in Cryptosporidium and gdh assemblages in Giardia to address the following questions: What are the subtype families of Cryptosporidium and assemblages of Giardia in New Zealand detected with gp60 and gdh genetic markers? How is the genetic variation of these parasites distributed in New Zealand compared to the rest of the world?
Fig 1. Trends in cryptosporidiosis and giardiasis notifications in New Zealand.
Source: New Zealand Public Health Observatory (http://www.nzpho.org.nz/NotifiableDisease.aspx).
Materials and methods
Sampling
We performed an observational study of giardiasis and cryptosporidiosis in New Zealand. Both diseases are notifiable in New Zealand; therefore, suspected cases are confirmed through accredited laboratories. Fresh feacal samples were screened from symptomatic humans in New Zealand between 2009 and 2015 using either fluorescence microscopy or ELISA analysis in three national diagnostic laboratories, from both the North and South Islands of New Zealand. Positive stool samples were sent to the Hopkirk Research Institute, Massey University, New Zealand for further molecular typing analyses (see below). Human samples were submitted anonymously and no information about the patients was available. Our human sample covers 10% (range 4–19%) notified cryptosporidiosis and 13% (2–32%) notified giardiasis cases from 2009 to 2015 in New Zealand. A microscopy-based screen was employed for feacal samples of other sources that were obtained from farms, zoos, animal hospitals and urban wildlife, on an ad hoc basis, and as part of other studies [e.g., 42]. Stools were deposited in 2 ml screw cap tubes and stored at 4°C in the laboratory until DNA extraction.
Molecular techniques
DNA extractions from stool samples were performed using the Isolate fecal DNA (Bioline) or QIAamp DNA Stool (Qiagen) kits following the manufacturer’s instructions and carried out at the Hopkirk Research Institute. DNA extraction required disruption of the oocyst/cysts using a beadbeater (Tissue Lyser II, Qiagen) at 30 Hz for 5 min. DNA quantification was measured using the Qubit fluorometer. A fragment of the gp60 and gdh genes were PCR amplified using a combination of external and internal primers (S1 Table) for Cryptosporidium [43, 44] and Giardia [31], respectively. Both strands of the amplification products were sequenced using Big Dye Terminator version 3.1 reagents and an ABI 3730XL automated DNA sequencer (Applied Biosystems, Foster City, California, USA).
Data analysis
DNA sequences analysis
Consensus sequences were assembled from forward and reverse reads and edited manually using Geneious v.10.1.3 [45]. The sequences derived were used to identify species and genotypes of Cryptosporidium and Giardia by aligning to sequence entries in nucleotide databases using the program BLAST (http://www.ncbi.nlm.nih.gov/blast/) and checked by their corresponding genotype [e.g., 46].
Worldwide distribution of subtype families and assemblages
We extracted a dataset of available gp60 and gdh sequences from GenBank for the same species of Cryptosporidium and Giardia found in New Zealand. The search strategy used consist of one search string (species name) combined by the Boolean operator “AND” for gene name (gp60 or gdh). To check for duplicate sequences, accession numbers were compared between datasets from different countries. All results were verified and cleaned for further analyses (final searches were conducted on February 20, 2016). Sequences of gp60 and gdh for each specific country were allocated to the corresponding subtype families/assemblages following the source information from GenBank when available. In cases where this information was not available in GenBank we either aligned all sequences using SATé-II v.2.2.7 [47] or used BLAST software (http://www.ncbi.nlm.nih.gov/blast/) to determine the corresponding genotype. We complement this information with data from papers published in scientific journals (last search conducted on June 15, 2017) using the PubMed search engine via Geneious v.10.1.3. The search strategy for Cryptosporidium included the keywords gp60, gp40/15, gp15, pgp60, 60 kDa, 60 kilodalton glycoprotein, glycoprotein, genotype, subtype family, and cryptosporid*, whilst the search strategy for Giardia included the keywords gdh, glutamate dehydrogenase, assemblage, and giard* [30]. Geographical distributions were visualized as maps using ggplot2 [48] and rworldmap [49] packages in RStudio v.0.98.1091.
Extrapolation and similarity of parasite genotypes
We used a proportional similarity index (PSI) in the R package spaa [50] to calculate niche overlap (co-occurrence) patterns among countries according to the type and number of genetic variants reported. Values close to 1 indicate very similar countries in their genotype diversity, whereas values close to 0 indicate countries with dissimilar genotypes. Next, we used the “specaccum” function in the R package vegan [51] to generate genotypes (cf. species) accumulation curves [52] by plotting the total number of different subtype families/assemblages against the number of all countries reporting data on GenBank combined and all countries in the world (~200). Rarefaction was used to produce smooth mean parasite genotypes variability and calculate 95% upper and lower bounds.
Pairwise FST and an analysis of molecular variance (AMOVA) were performed in Arlequin v.3.5.2.2 [53] using the sequence data obtained from GenBank to recognize the genetic differentiation between pairs of populations and the level of population genetic structure, respectively. Gblocks v.0.91b [54] was used to remove nonconserved positions adjacent to gaps and nucleotide repeats in the protein coding-gene gp60.
Accession numbers
DNA sequences have been submitted to GenBank: accession numbers KY123918–KY124121. Note that only unique haplotypes were submitted but similar haplotypes found in different hosts were repeatedly submitted (see S2 Table).
Results
Molecular identification of species and genotypes
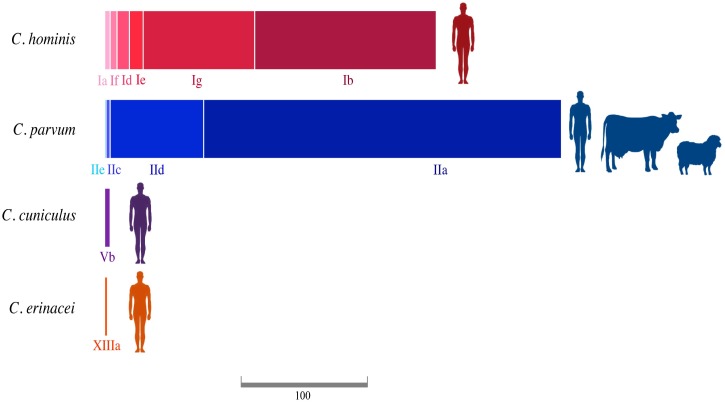
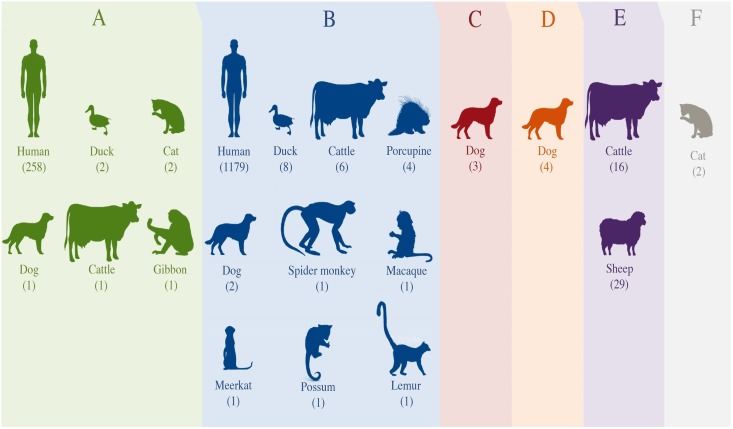
A total of 579 sequences were obtained for Cryptosporidium and 1523 for Giardia (S3 Table). The sequences presented > 98% identity and E-value of 0.0 with BLAST searching against sequences in GenBank, confirming a reliable identification of species and genotypes. Cryptosporidium species detected in New Zealand using the gp60 marker were C. parvum, C. hominis, C. cuniculus and C. erinacei. Our molecular analysis identified 332 samples with the presence of C. parvum, 241 C. hominis, four C. cuniculus and two C. erinacei. Samples of C. parvum were collected from humans (86.4%), cattle (13%) and sheep (0.6%), whilst the remaining species were collected only from humans (Fig 2 and S3 Table). Subtype families found in C. parvum were IIa, IIc, IId and IIe (Fig 2). The most common subtype families were IIa (78.3%) and IId (20.5%). The subtype families in C. hominis most frequently identified were Ib (54.8%) and Ig (33.6%). Other subtype families found in our study were Ia, Id, Ie and If, which represented the remaining 11.6% (Fig 2 and S3 Table). Cryptosporidium cuniculus and C. erinacei were assigned to subfamilies Vb and XIIIa, respectively (Fig 2 and S3 Table). Assemblages from A to F of G. intestinalis are presented in New Zealand but assemblage B is notably dominant (79%) and found in humans, domestic, wild and zoo animals (Fig 3 and S3 Table).
Fig 2. Subtype families and hosts of Cryptosporidium species found in this study.
The size of the squares is proportional to the number of samples identified for each subtype family. The scale bar corresponds to 100 samples. See S3 Table for details.
Fig 3. Range of hosts and assemblages (A to F) of Giardia intestinalis found in New Zealand.
The number of samples by hosts identified within each assemblage is found in brackets.
Global geographic distribution of gp60 subtype families and gdh assemblages
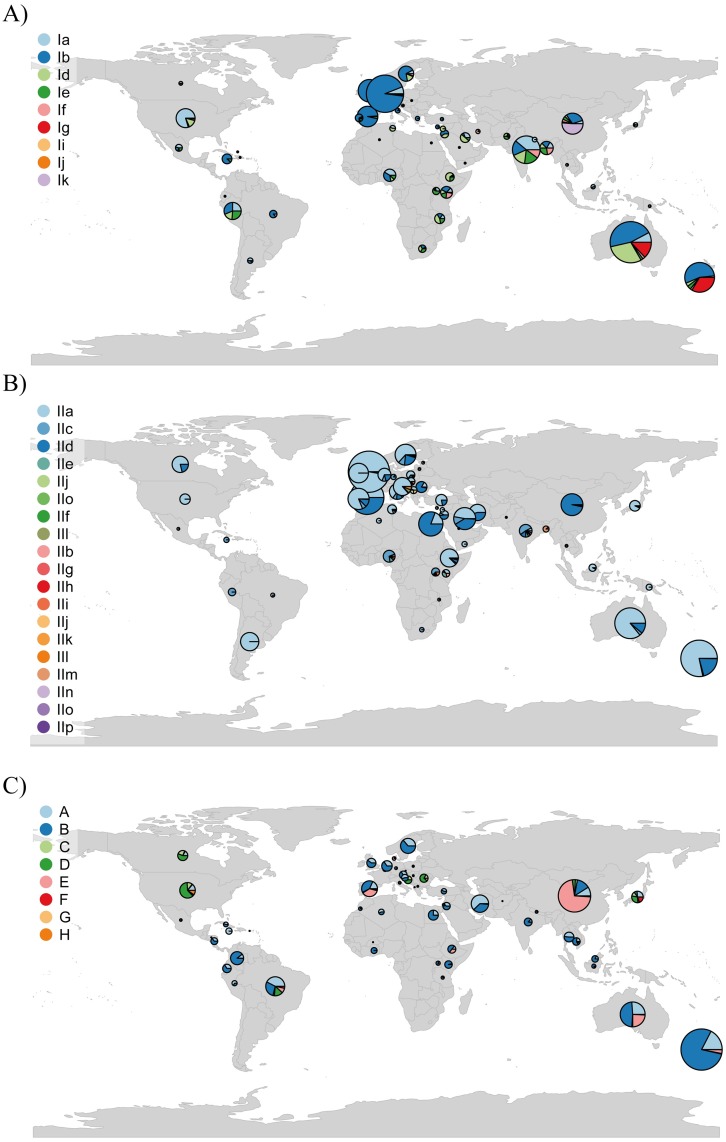
Cryptosporidium parvum and C. hominis were found across all continents (Fig 4a and 4b and S4 Table) but Australia and the United Kingdom accounted for the majority of data for both pathogens (S4 Table). Ten subtype families of C. hominis (Ia-Ik) are found around the world whilst 16 have been identified for C. parvum (IIa-IIp). Cryptosporidium hominis subtype families Ia, Ib, Id and Ie are the most abundant worldwide (S4 Table). Regardless of the high diversity of subtype families in C. parvum, IIa and IId are the most abundant. A more restricted geographical distribution is reported for C. cuniculus and C. erinacei, with data from Australia, China, Czech Republic, Poland, the United Kingdom, Algeria and Germany (S4 Table). Both of the current recognized genotypes for C. cuniculus (Va and Vb) were reported in China and the United Kingdom but Vb has been additionally found in Australia, Czech Republic and Poland.
Fig 4. Global records of Cryptosporidium and Giardia genotypes data.
(a) and (b) Distribution of gp60 subtype families in C. hominis and C. parvum, respectively. (c) Distribution of gdh assemblages in Giardia intestinalis. The size of the pies is proportional to the frequency of data obtained from publications and GenBank data. Data for New Zealand includes this study.
We extracted 4348 records corresponding to G. intestinalis assemblages from 64 countries spanning all continents except Antarctica (Fig 4c and S4 Table). However, the majority of available data were retrieved from China, Australia and Brazil. Assemblages A and B represent more than 60% of the reported data worldwide (S4 Table). There are two additional assemblages (G and H) that are not found in New Zealand. Assemblage G is reported in Australia, China and Spain whilst assemblage H is found in Colombia and USA.
Countries similarity and global genotypes accumulation curves
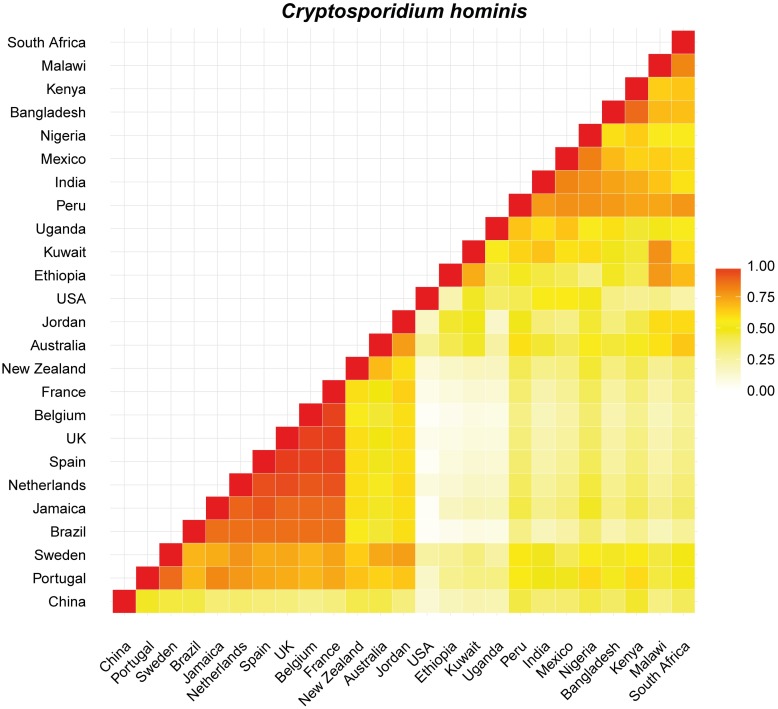
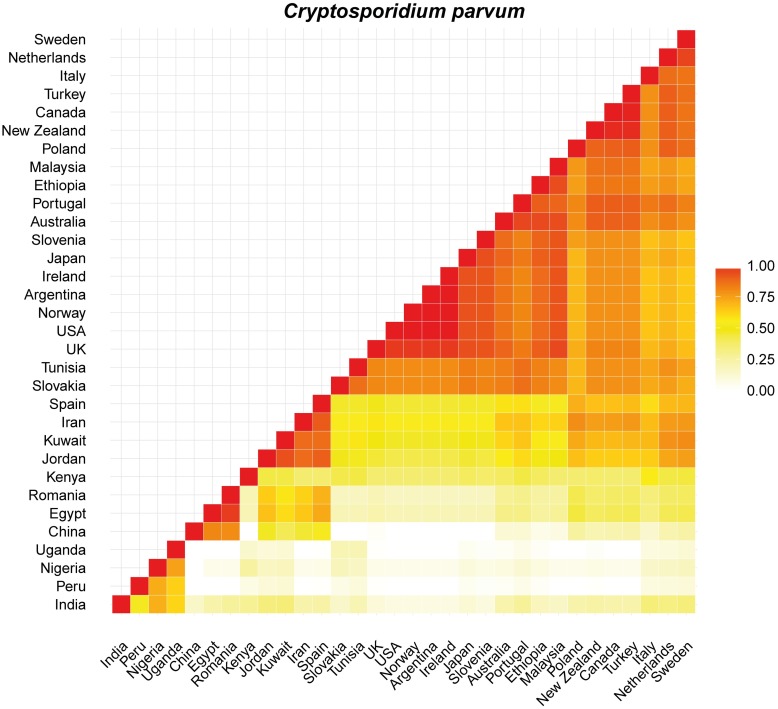
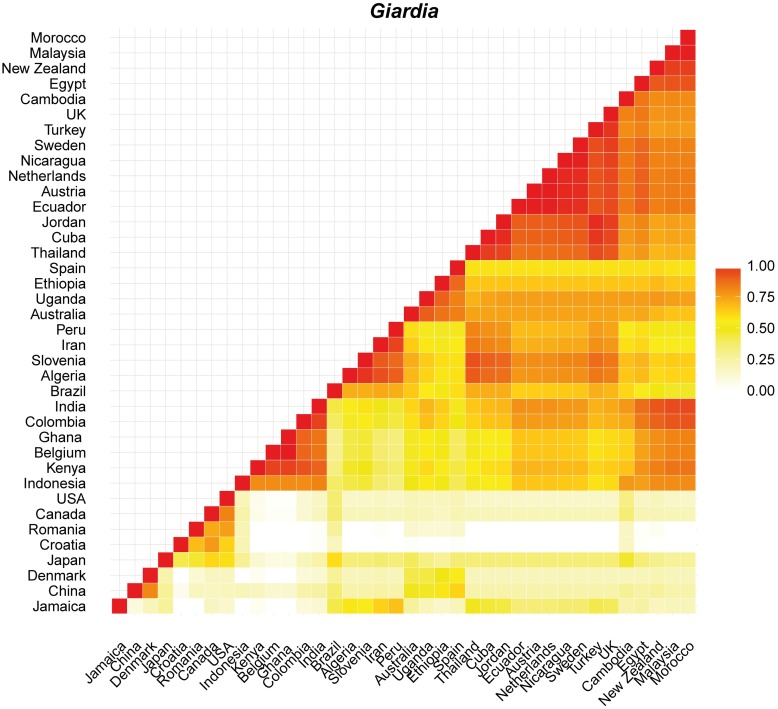
Proportional similarity analyses were carried out only between countries with more than 10 records retrieved. In general, C. hominis showed intermediate to high values (> 0.5) of similarity for most country comparisons with New Zealand visually similar to Australia, Portugal, Sweden and the United Kingdom (Figs 5 and S1). Cryptosporidium parvum showed high similarity (> 0.75) among most country comparisons with exception of comparisons against Uganda, Nigeria, China, Peru and India (Figs 6 and S2). In this case, New Zealand is most like countries from Europe, including the Netherlands, Sweden, the United Kingdom and Portugal and countries from other hemispheres such as Canada and Australia. High values of similarity were also observed for comparisons of G. intestinalis between countries spanning different continents but Denmark, Croatia, Romania, Canada and USA were visually dissimilar (< 0.5) (Figs 7 and S3). New Zealand is similar to many countries from across the world, from Europe to Asia, Africa and Australia (Fig 7).
Fig 5. Plot showing the proportional similarity of C. hominis genotypes between pairs of countries.
Highly similar countries are showed in darker colours.
Fig 6. Plot showing the proportional similarity of C. parvum genotypes between pairs of countries.
Highly similar countries are showed in darker colours.
Fig 7. Plot showing the proportional similarity of G. intestinalis genotypes between pairs of countries.
Highly similar countries are showed in darker colours.
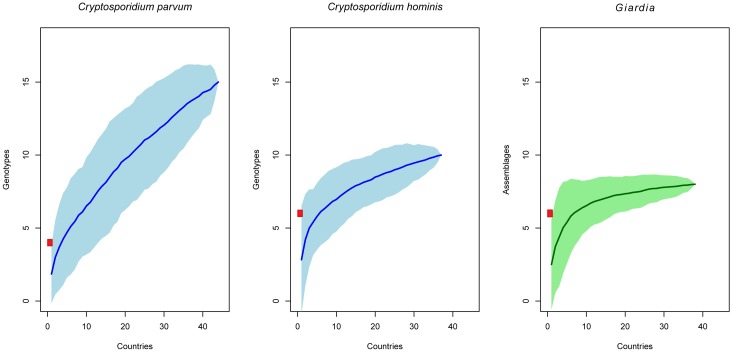
Fig 8 shows the cumulative number of genotypes for all countries combined. Accumulation curves for G. intestinalis and C. hominis were starting to show some downward curvature below 10 genotypes, which indicates a declining rate of parasite genetic type discovery, but only G. intestinalis approached an asymptote. The number of genetic types found in New Zealand for these two species is close to the plateau area of the curves (Fig 8). Interestingly, C. parvum show a different pattern with an upward curvature which indicates that many more genetic types than those currently recorded will be found if sampling in other countries is performed. However, this species may never attain an asymptote (Figs 8 and S4) indicating that other genotypes will be likely detected worldwide as well as in New Zealand. Rarefaction analysis using only countries with more than 10 records did not show differences of these trends (S5 Fig).
Fig 8. Genotype accumulation curves of C. parvum (left), C. hominis (center) and G. intestinalis (right) estimated by the number of countries with reported data.
For each curve the lighter shaded region shows 95% confidence intervals and the red square correspond to the number of genotypes for each species found in New Zealand.
Population structure analyses were carried out between countries with more than 10 sequences available in GenBank. Sequences that do not overlap at least 50% of the total length of the dataset were discarded. Gaps and nucleotide repeats were not considered for pairwise FST and AMOVA analyses. Large and significant FST values are an indication of population subdivision and/or low gene flow between populations. There was not significant (p < 0.0001) population differentiation of comparisons among Nigeria, Mexico, Kenya and USA for C. hominis. New Zealand showed high significant pairwise FST when compared with other countries with exception of comparisons among Nigeria, Mexico, USA and Australia (S5 Table). A lack of population differentiation for C. parvum was evident between most comparisons of estimated pairwise FST (S5 Table). We did not find structured populations among closely geographical European countries (e.g. United Kingdom, Sweden, Spain, Slovenia and Poland) as well as other countries spanning around the world, with exception of most comparisons among Egypt, India and China (S5 Table). New Zealand showed no population differentiation with Australia, Argentina, USA, Sweden, Poland, Slovenia and Turkey. Giardia intestinalis also revealed no population subdivision among most countries including those from different continents, with exception of most comparisons among Ethiopia, Egypt, Denmark, Croatia, Romania and Colombia. New Zealand presented no population subdivision with countries from different latitudes including Norway, Malaysia, Uganda, Sweden, Indonesia, Japan, Germany, Australia, among others (S5 Table). However, there was substantial polymorphism within populations (> 80%) found in the AMOVA which result in low but significant proportions of the total variation partitioned among countries (14%, 19% and 16%, p < 0.0001 for C. hominis, C. parvum and G. intestinalis, respectively).
Discussion
Our relatively large sample size and genotyping diversity of Cryptosporidium and Giardia from New Zealand allowed us to perform comparative analyses with available worldwide data. All species and genetic variants reported in New Zealand of either Cryptosporidium or Giardia are found in several countries and this homogenization suggests that their populations are well established and mixed throughout the globe [55, 56]. Some cases of structure relating to geography have been indicated by previous studies [e.g., 57, 58] but they also show sharing alleles among different populations around the world. We also found evidence of geographically-associated differentiation with high FST values that were generally confirmed by low PSI values, despite large confidence intervals (S1, S2 and S3 Figs). The evidence of structure is likely a consequence of limitations in the estimators due to an uneven number of samples, high within-population variation and reporting bias [59]. The use of a single and high variable locus and non-contemporaneous samples also affect the probability of a reliable estimation of the genetic differentiation from sequence data [60, 61]. Furthermore, the population structure of these species, going from clonal to panmictic, is influenced by diverse factors such as transmission dynamics and host-specificity [57, 62].
The global distribution of species and genotypes within Cryptosporidium and Giardia might represent the geographic range shifts of domesticated animals, historical movement of livestock and transmission facilitated by human travel and non-native fauna [35, 63, 64]. This is likely the case of the rare C. cuniculus and C. erinacei causing human disease in New Zealand. Cryptosporidium cuniculus has recently expanded its known host range from rabbits to humans and kangaroos [65] and it is a human pathogen of public health importance related to waterborne outbreaks in the United Kingdom [66]. The presence of this species and a similar genotype in New Zealand and few other countries may suggest a dispersal event caused by human factors. Most recently, C. erinacei has been identified as a cause of gastroenteritis in immunocompetent man in the Czech Republic, having previously only been identified in hedgehogs and horses [20]. Though we have no data on the origin of the infection, finding that humans in New Zealand have been infected with C. erinacei could be an indication of cross-species transmission events from either domestic or wildlife hosts, introduced pets or non-native mammals such as possums, hedgehogs, rabbits and horses. Specific future studies could include active surveillance of human, domestic and wild animal populations and longitudinal studies with advanced molecular methods [16]. More systematic sampling of local and introduced fauna along with whole genome sequences over short timescales will allow us to identify major functional loci involved in host specificity, proliferation and host shifting. This information will provide insights into the zoonotic potential of different genotypes, host range, cross-species transmission and risks for host shifts to humans [67].
We acknowledge that Cryptosporidium species diversity will be underestimated because most species within the genus fail to amplify with standard primers used for the gp60 marker [68, 69]. For instance, C. andersoni and C. bovis have been previously identified in cows in New Zealand using the 18S rRNA gene markers [70]. Moreover, our approaches are dependent on sequences submission, accurate taxonomy and complementary information in GenBank. Some genotypes have been recognized as species or wrongly submitted to a different species. Cryptosporidium erinacei was previously described as C. sp. hedgehog [20] with the single XIIIa subtype family but a subtype family VIIa was also proposed (S4 Table) [71]. Likewise, the subtype family IIf in C. parvum has recently been classified as C. tyzzeri [72]. This is also the case for G. intestinalis. Several assemblages of G. intestinalis are genetically isolated lineages and have been nominated as different species [73]. Some genotypes in C. parvum and G. intestinalis might be named as new species when more data become available.
Sequences from public databases are an extremely valuable source of information [74]. Genotypes and abundance in different countries may not be totally represented because some researchers avoid submitting multiple sequences if the same genotype is already found within the sequence data collection of GenBank [74]. The potential bias caused by this practice does not help to capture a substantial part of the genetic diversity globally. This information is fundamental for analyses of the evolution and movement of the parasites, the possible origin and patterns of dispersal of these organisms or their genotypes, prioritization on potential hazard reduction to susceptible hosts and control to prevent spread between regions. For example, C. parvum haplotypes IIg, IIh, IIi are reported in Uganda, IIm in Bangladesh, and IIn in India; C. hominis Ii is found in Asia; and G. intestinalis assemblage G in Australia. Given current reporting practices, it is unclear if these genotypes are unique to these regions deserving greater attention or if they are widely distributed but simply unreported on GenBank. Using our literature review framework (S1 References), we found that subtype families IIg, IIi and IIm were also reported in India, Ii in USA and assemblage G in China and Spain. We predict that unreported sequences from different countries will decrease the geographical population structure, however, it will be dependent on whether researchers do not report sequences because of 100% identity or because they are the same genotype. Another alternative source of information to measure the population genetic variation of these parasites is ZoopNet [75, 76] but this database is not publicly available. A previous study using ZoopNet found high genetic diversity in C. hominis and C. parvum from Australia and the United Kingdom, respectively [75]. Estimations of the genetic diversity in this study align with our results and the similarity between these nations and New Zealand.
It has been demonstrated that mixed infections (different alleles per isolate) within a host can produce recombinant genotypes that comprise alleles inherited from each parental line [77, 78]. These recombinant events are associated to the high probability of finding new genotypes (S4 Fig). Cryptosporidium parvum displayed a large genotypic variation as a result of gene combinations and host range shift [67]. Similarly, genetic recombination has been shown to occur commonly in C. hominis and G. intestinalis in developing countries [59, 78]. This is an important issue for public health risks because genetic recombination is a driving force for the emergence of virulent subtypes [79]. The distribution of these species in humans might vary among geographical areas and socioeconomic conditions [68] and interestingly, high genetic diversity and new genotypes have been particularly found in children or unexpected hosts in rural settings from developing countries [e.g., 80, 81–85]. Unfortunately, the data reported from developing countries is low with limited information from Africa, Latin America and populous countries such as China and India [30].
Comprehensive surveillance strategies will require efforts at multiple scales to help decrease the significant burden caused by Cryptosporidium and Giardia. Understanding of the transmission pathways of these parasitic protozoans is imperative. The large number of zoonotic cases highlights the importance of rates of contact and host densities. For example, local spread of Cryptosporidium and Giardia in New Zealand has been facilitated by cattle densities in the catchment of drinking water particularly within the most populous and agricultural regions [37, 39, 42, 86]. It is uncertain in our study if this is the transmission pathway, particularly for Giardia which shows less seasonality in case notification rates (Fig 1), but a substantial number of diarrheal cases around the world suggest that infections among humans has an increasing trend caused by waterborne transmission [e.g., 24, 87]. Animal exposure, seasonal patterns and contaminated food are other key factors for transmission in certain geographical settings and more detailed analysis will be need to identify the pathways of human cryptosporidiosis and giardiasis.
Supporting information
Highly similar countries are showed in darker colours and 95% lower (above) and upper (below) bootstrapped confidence limits in numbers within each box.
(TIF)
Highly similar countries are showed in darker colours and 95% lower (above) and upper (below) bootstrapped confidence limits in numbers within each box.
(TIF)
Highly similar countries are showed in darker colours and 95% lower (above) and upper (below) bootstrapped confidence limits in numbers within each box.
(TIF)
(TIF)
For each curve the lighter shaded region shows 95% confidence intervals and the red square correspond to the number of genotypes for each species found in New Zealand.
(TIF)
(DOCX)
(DOCX)
(DOCX)
Details for the literature used on this data are found in the accompanying pdf document.
(XLSX)
Highly significant values (p < 0.0001) are shown in red.
(XLSX)
(PDF)
Acknowledgments
We are grateful to the zoos, vet clinics, farms, animal hospitals and all diagnostic medical laboratories around New Zealand that kindly provided samples necessary for this study. Elaine Moriarty and Brent Gilpin from ESR, Alex Grinberg from IVABS, mEpiLab members and four anonymous reviewers provided helpful comments that greatly improved this manuscript.
Data Availability
All sequences are available from the GenBank database (accession numbers KY123918-KY124121).
Funding Statement
This research was funded by the New Zealand Ministry of Health. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Morens DM, Folkers GK, Fauci AS. The challenge of emerging and re-emerging infectious diseases. Nature. 2004;430(6996):242–9. 10.1038/nature02759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morse SS. Factors in the emergence of infectious diseases. Emerging Infectious Diseases. 1995;1(1):7–15. 10.3201/eid0101.950102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Currie J, Grenfell B, Farrar J. Beyond Ebola. Science. 2016;351(6275):815–6. 10.1126/science.aad8521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Espinal M, Aldighieri S, John RS, Becerra-Posada F, Etienne C. International health regulations, Ebola, and emerging infectious diseases in Latin America and the Caribbean. American Journal of Public Health. 2015;106(2):279–82. 10.2105/AJPH.2015.302969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Woolhouse MEJ, Rambaut A, Kellam P. Lessons from Ebola: Improving infectious disease surveillance to inform outbreak management. Science Translational Medicine. 2015;7(307):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heymann D, L., Rodier G. Global Surveillance, National Surveillance, and SARS. Emerging Infectious Diseases. 2004;10(2):173–5. 10.3201/eid1002.031038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilson ME. Travel and the emergence of infectious diseases. Emerging Infectious Diseases. 1995;1(2):39–46. 10.3201/eid0102.952001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weiss RA, McMichael AJ. Social and environmental risk factors in the emergence of infectious diseases. Nature Medicine. 2004;10(12):S70—S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones KE, Patel NG, Levy MA, Storeygard A, Balk D, Gittleman JL, et al. Global trends in emerging infectious diseases. Nature. 2008;451(7181):990–3. http://www.nature.com/nature/journal/v451/n7181/suppinfo/nature06536_S1.html. 10.1038/nature06536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mead PS, Slutsker L, Dietz V, McCaig LF, Bresee JS, Shapiro C, et al. Food-related illness and death in the United States. Emerging Infectious Diseases. 1999;5(5):607–25. 10.3201/eid0505.990502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Enserink R, van den Wijngaard C, Bruijning-Verhagen P, van Asten L, Mughini-Gras L, Duizer E, et al. Gastroenteritis attributable to 16 enteropathogens in children attending day care: Significant effects of Rotavirus, Norovirus, Astrovirus, Cryptosporidium and Giardia. The Pediatric Infectious Disease Journal. 2015;34(1):5–10. 10.1097/INF.0000000000000472 [DOI] [PubMed] [Google Scholar]
- 12.Ng-Hublin JSY, Hargrave D, Combs B, Ryan U. Investigation of a swimming pool-associated cryptosporidiosis outbreak in the Kimberley region of Western Australia. Epidemiology & Infection. 2015;143(05):1037–41. 10.1017/S095026881400106X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gertler M, Dürr M, Renner P, Poppert S, Askar M, Breidenbach J, et al. Outbreak of cryptosporidium hominis following river flooding in the city of Halle (Saale), Germany, August 2013. BMC Infectious Diseases. 2015;15(1):1–10. 10.1186/s12879-015-0807-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Savioli L, Smith H, Thompson A. Giardia and Cryptosporidium join the ‘Neglected Diseases Initiative’. Trends in Parasitology. 2006;22(5):203–8. 10.1016/j.pt.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 15.Cacciò SM, Thompson RCA, McLauchlin J, Smith HV. Unravelling Cryptosporidium and Giardia epidemiology. Trends in Parasitology. 2005;21(9):430–7. 10.1016/j.pt.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 16.Checkley W, White AC Jr, Jaganath D, Arrowood MJ, Chalmers RM, Chen X-M, et al. A review of the global burden, novel diagnostics, therapeutics, and vaccine targets for cryptosporidium. The Lancet Infectious Diseases. 2015;15(1):85–94. 10.1016/S1473-3099(14)70772-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rossignol J-F. Cryptosporidium and Giardia: Treatment options and prospects for new drugs. Experimental Parasitology. 2010;124(1):45–53. 10.1016/j.exppara.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 18.Broglia A, Weitzel T, Harms G, Cacció SM, Nöckler K. Molecular typing of Giardia duodenalis isolates from German travellers. Parasitol Res. 2013;112(10):3449–56. 10.1007/s00436-013-3524-y [DOI] [PubMed] [Google Scholar]
- 19.Sallon S, Deckelbaum RJ, Schmid II, Harlap S, Baras M, Spira DT. Cryptosporidium, malnutrition, and chronic diarrhea in children. American Journal of Diseases of Children. 1988;142(3):312–5. 10.1001/archpedi.1988.02150030086027 [DOI] [PubMed] [Google Scholar]
- 20.Kváč M, Saková K, Kvĕtoňová D, Kicia M, Wesołowska M, McEvoy J, et al. Gastroenteritis caused by the Cryptosporidium hedgehog genotype in an immunocompetent man. Journal of Clinical Microbiology. 2014;52(1):347–9. 10.1128/JCM.02456-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kotloff KL, Nataro JP, Blackwelder WC, Nasrin D, Farag TH, Panchalingam S, et al. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. The Lancet. 2013;382(9888):209–22. 10.1016/S0140-6736(13)60844-2 [DOI] [PubMed] [Google Scholar]
- 22.Lima A, Samie A, Guerrant R. Cryptosporidiosis In: Guerrant R, Walker D, Weller P, editors. Tropical Infectious Diseases. Philadelphia, USA: Elsevier-Churchill Livingstone; 2011. p. 640–63. [Google Scholar]
- 23.Feng Y, Xiao L. Zoonotic potential and molecular epidemiology of Giardia species and giardiasis. Clinical Microbiology Reviews. 2011;24(1):110–40. 10.1128/CMR.00033-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baldursson S, Karanis P. Waterborne transmission of protozoan parasites: Review of worldwide outbreaks—An update 2004–2010. Water Research. 2011;45(20):6603–14. 10.1016/j.watres.2011.10.013. [DOI] [PubMed] [Google Scholar]
- 25.Lane S, Lloyd D. Current trends in research into the waterborne parasite Giardia. Critical Reviews in Microbiology. 2002;28(2):123–47. 10.1080/1040-840291046713 [DOI] [PubMed] [Google Scholar]
- 26.WHO. Fighting Disease Fostering Development. Geneva: 1996. [Google Scholar]
- 27.Xiao L, Escalante L, Yang C, Sulaiman I, Escalante AA, Montali RJ, et al. Phylogenetic analysis of Cryptosporidium parasites based on the small-subunit rRNA gene locus. Applied and Environmental Microbiology. 1999;65(4):1578–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fayer R. Taxonomy and species delimitation in Cryptosporidium. Experimental Parasitology. 2010;124(1):90–7. 10.1016/j.exppara.2009.03.005. [DOI] [PubMed] [Google Scholar]
- 29.McDonnell PA, Upcroft J, Upcroft P, Buret A. Morphological identification markers for distinguishing avian from mammalian Giardia species—do they need reconsideration? European Journal of Protistology. 2001;37(3):273–80. 10.1078/0932-4739-00818. [DOI] [Google Scholar]
- 30.Jex AR, Gasser RB. Genetic richness and diversity in Cryptosporidium hominis and C. parvum reveals major knowledge gaps and a need for the application of “next generation” technologies—Research review. Biotechnology Advances. 2010;28(1):17–26. 10.1016/j.biotechadv.2009.08.003. [DOI] [PubMed] [Google Scholar]
- 31.Read CM, Monis PT, Andrew Thompson RC. Discrimination of all genotypes of Giardia duodenalis at the glutamate dehydrogenase locus using PCR-RFLP. Infection, Genetics and Evolution. 2004;4(2):125–30. 10.1016/j.meegid.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 32.Jex AR, Smith HV, Monis PT, Campbell BE, Gasser RB. Cryptosporidium—Biotechnological advances in the detection, diagnosis and analysis of genetic variation. Biotechnology Advances. 2008;26(4):304–17. 10.1016/j.biotechadv.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 33.Strong WB, Gut J, Nelson RG. Cloning and sequence analysis of a highly polymorphic Cryptosporidium parvum gene encoding a 60-kilodalton glycoprotein and characterization of its 15- and 45-kilodalton zoite surface antigen products. Infection and Immunity. 2000;68(7):4117–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Monis PT, Mayrhofer G, Andrews RH, Homan WL, Limper L, Ey PL. Molecular genetic analysis of Giardia intestinalis isolates at the glutamate dehydrogenase locus. Parasitology. 1996;112(01):1–12. 10.1017/S0031182000065021 [DOI] [PubMed] [Google Scholar]
- 35.Wang R, Zhang L, Axen C, Bjorkman C, Jian F, Amer S, et al. Cryptosporidium parvum IId family: clonal population and dispersal from Western Asia to other geographical regions. Scientific Reports. 2014;4(4208):srep04208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Putignani L, Menichella D. Global distribution, public health and clinical impact of the Protozoan pathogen Cryptosporidium. Interdisciplinary Perspectives on Infectious Diseases. 2010;2010:39 10.1155/2010/753512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Snel SJ, Baker MG, Kamalesh V, French N, Learmonth J. A tale of two parasites: the comparative epidemiology of cryptosporidiosis and giardiasis. Epidemiology & Infection. 2009;137(11):1641–50. 10.1017/S0950268809002465 [DOI] [PubMed] [Google Scholar]
- 38.Lal A, Ikeda T, French N, Baker MG, Hales S. Climate variability, weather and enteric disease incidence in New Zealand: Time series analysis. PLoS ONE. 2013;8(12):e83484 10.1371/journal.pone.0083484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Learmonth JIM, Ionas G, Pita A, Cowie R. Seasonal shift in Cryptosporidium parvum transmission cycles in New Zealand. Journal of Eukaryotic Microbiology. 2001;48:34–5. 10.1111/j.1550-7408.2001.tb00444.x [DOI] [PubMed] [Google Scholar]
- 40.ESR. Surveillance report: Notifiable diseases in New Zealand Annual report 2014. Porirua, New Zealand: 2015.
- 41.Binney BM, Biggs PJ, Carter PE, Holland BM, French NP. Quantification of historical livestock importation into New Zealand 1860–1979. New Zealand Veterinary Journal. 2014;62(6):309–14. 10.1080/00480169.2014.914861 [DOI] [PubMed] [Google Scholar]
- 42.Al Mawly J, Grinberg A, Velathanthiri N, French N. Cross sectional study of prevalence, genetic diversity and zoonotic potential of Cryptosporidium parvum cycling in New Zealand dairy farms. Parasites & Vectors. 2015;8:240 10.1186/s13071-015-0855-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Glaberman S, Moore JE, Lowery CJ, Chalmers RM, Sulaiman I, Elwin K, et al. Three Drinking-Water–Associated Cryptosporidiosis Outbreaks, Northern Ireland. Emerging Infectious Diseases. 2002;8(6):631–3. 10.3201/eid0806.010368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Waldron LS, Ferrari BC, Power ML. Glycoprotein 60 diversity in C. hominis and C. parvum causing human cryptosporidiosis in NSW, Australia. Experimental Parasitology. 2009;122(2):124–7. 10.1016/j.exppara.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 45.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28(12):1647–9. 10.1093/bioinformatics/bts199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xiao L. Molecular epidemiology of cryptosporidiosis: An update. Experimental Parasitology. 2010;124(1):80–9. 10.1016/j.exppara.2009.03.018. [DOI] [PubMed] [Google Scholar]
- 47.Liu K, Warnow TJ, Holder MT, Nelesen SM, Yu J, Stamatakis AP, et al. SATé-II: Very fast and accurate simultaneous estimation of multiple sequence alignments and phylogenetic trees. Systematic Biology. 2012;61(1):90–106. 10.1093/sysbio/syr095 [DOI] [PubMed] [Google Scholar]
- 48.Wickham H. ggplot2: Elegant graphics for data analysis. New York: Springer-Verlag; 2009. [Google Scholar]
- 49.South A. rworldmap: A new R package for mapping global data. The R Journal. 2011;3(1):35–43. [Google Scholar]
- 50.Zhang J, Ding Q, Huang J. spaa: Species association analysis. R package version 0.2. 1; 2013.
- 51.Dixon P. VEGAN, a package of R functions for community ecology. Journal of Vegetation Science. 2003;14(6):927–30. 10.1111/j.1654-1103.2003.tb02228.x [DOI] [Google Scholar]
- 52.Dove ADM, Cribb TH. Species accumulation curves and their applications in parasite ecology. Trends in Parasitology. 2006;22(12):568–74. 10.1016/j.pt.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 53.Excoffier L, Laval G, Schneider S. Arlequin (version 3.0): An integrated software package for population genetics data analysis. Evolutionary Bioinformatics Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- 54.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Molecular Biology and Evolution. 2000;17(4):540–52. [DOI] [PubMed] [Google Scholar]
- 55.Widmer G. Meta-analysis of a polymorphic surface glycoprotein of the parasitic protozoa Cryptosporidium parvum and Cryptosporidium hominis. Epidemiology & Infection. 2009;137(12):1800–8. 10.1017/S0950268809990215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Choy SH, Mahdy MAK, Al-Mekhlafi HM, Low VL, Surin J. Population expansion and gene flow in Giardia duodenalis as revealed by triosephosphate isomerase gene. Parasites & Vectors. 2015;8(1):1–10. 10.1186/s13071-015-1084-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tanrıverdi S, Grinberg A, Chalmers RM, Hunter PR, Petrovic Z, Akiyoshi DE, et al. Inferences about the global population structures of Cryptosporidium parvum and Cryptosporidium hominis. Applied and Environmental Microbiology. 2008;74(23):7227–34. 10.1128/AEM.01576-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cacciò SM, de Waele V, Widmer G. Geographical segregation of Cryptosporidium parvum multilocus genotypes in Europe. Infection, Genetics and Evolution. 2015;31(0):245–9. 10.1016/j.meegid.2015.02.008. [DOI] [PubMed] [Google Scholar]
- 59.Widmer G, Sullivan S. Genomics and population biology of Cryptosporidium species. Parasite Immunology. 2012;34(2–3):61–71. 10.1111/j.1365-3024.2011.01301.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Meirmans PG, Hedrick PW. Assessing population structure: FST and related measures. Molecular Ecology Resources. 2011;11(1):5–18. 10.1111/j.1755-0998.2010.02927.x [DOI] [PubMed] [Google Scholar]
- 61.Meirmans PG. Using the AMOVA framework to estimate a standardized genetic differentiation measure. Evolution. 2006;60(11):2399–402. [PubMed] [Google Scholar]
- 62.Fayer R, Xiao L. Cryptosporidium and Cryptosporidiosis, Second Edition: CRC Press; 2008. [Google Scholar]
- 63.Siripattanapipong S, Leelayoova S, Mungthin M, Thompson RCA, Boontanom P, Saksirisampant W, et al. Clonal diversity of the glutamate dehydrogenase gene in Giardia duodenalis from Thai Isolates: evidence of genetic exchange or Mixed Infections? BMC Microbiology. 2011;11:206- 10.1186/1471-2180-11-206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Heyworth MF. Giardia duodenalis genetic assemblages and hosts. Parasite. 2016;23:13–7. 10.1051/parasite/2016013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Koehler AV, Whipp MJ, Haydon SR, Gasser RB. Cryptosporidium cuniculus—new records in human and kangaroo in Australia. Parasites & Vectors. 2014;7:492 10.1186/s13071-014-0492-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chalmers R, Robinson G, Elwin K, Hadfield S, Xiao L, Ryan U, et al. Cryptosporidium sp. rabbit genotype, a newly identified human pathogen. Emerging Infectious Diseases. 2009;15(5):829–30. 10.3201/eid1505.081419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Garcia-R JC, Hayman DTS. Origin of a major infectious disease in vertebrates: The timing of Cryptosporidium evolution and its hosts. Parasitology. 2016;143(13):1683–90. 10.1017/S0031182016001323 [DOI] [PubMed] [Google Scholar]
- 68.Ryan U, Fayer R, Xiao L. Cryptosporidium species in humans and animals: current understanding and research needs. Parasitology. 2014;141(13):1667–85. 10.1017/S0031182014001085 [DOI] [PubMed] [Google Scholar]
- 69.Li N, Xiao L, Alderisio K, Elwin K, Cebelinski E, Chalmers R, et al. Subtyping Cryptosporidium ubiquitum,a Zoonotic Pathogen Emerging in Humans. Emerging Infectious Diseases. 2014;20(2):217–24. 10.3201/eid2002.121797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shrestha RD, Grinberg A, Dukkipati VSR, Pleydell EJ, Prattley DJ, French NP. Infections with multiple Cryptosporidium species and new genetic variants in young dairy calves on a farm located within a drinking water catchment area in New Zealand. Veterinary Parasitology. 2014;202(3–4):287–91. 10.1016/j.vetpar.2014.03.034. [DOI] [PubMed] [Google Scholar]
- 71.Dyachenko V, Kuhnert Y, Schmaeschke R, Etzold M, Pantchev N, Daugschies A. Occurrence and molecular characterization of Cryptosporidium spp. genotypes in European hedgehogs (Erinaceus europaeus L.) in Germany. Parasitology. 2010;137(02):205–16. 10.1017/S0031182009991089 [DOI] [PubMed] [Google Scholar]
- 72.Ren X, Zhao J, Zhang L, Ning C, Jian F, Wang R, et al. Cryptosporidium tyzzeri n. sp. (Apicomplexa: Cryptosporidiidae) in domestic mice (Mus musculus). Experimental Parasitology. 2012;130(3):274–81. 10.1016/j.exppara.2011.07.012. [DOI] [PubMed] [Google Scholar]
- 73.Xu F, Jerlstrom-Hultqvist J, Andersson JO. Genome-wide analyses of recombination suggest that Giardia intestinalis assemblages represent different species. Mol Biol Evol. 2012;29 10.1093/molbev/mss107 [DOI] [PubMed] [Google Scholar]
- 74.Slapeta J. Centenary of the genus Cryptosporidium: from morphological to molecular species identification In: Ortega-Pierres MG, Caccio S, Fayer R, Mank T, Smith H, Thompson RCA, editors. Giardia and Crytosporidium: CABI Publishing; 2009. p. 31–50. [Google Scholar]
- 75.Takumi K, Cacciò SM, Giessen Jvd, Xiao L, Sprong H. Hypothesis: Cryptosporidium genetic diversity mirrors national disease notification rate. Parasites & Vectors. 2015;8(308):1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sprong H, Cacciò SM, van der Giessen JWB, on behalf of the Zn, partners. Identification of Zoonotic Genotypes of Giardia duodenalis. PLoS Negl Trop Dis. 2009;3(12):e558 10.1371/journal.pntd.0000558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Feng X, Rich SM, Tzipori S, Widmer G. Experimental evidence for genetic recombination in the opportunistic pathogen Cryptosporidium parvum. Molecular and Biochemical Parasitology. 2002;119(1):55–62. 10.1016/S0166-6851(01)00393-0. [DOI] [PubMed] [Google Scholar]
- 78.Kosuwin R, Putaporntip C, Pattanawong U, Jongwutiwes S. Clonal diversity in Giardia duodenalis isolates from Thailand: evidences for intragenic recombination and purifying selection at the beta giardin locus. Gene. 2010;449:1–8. 10.1016/j.gene.2009.09.010 [DOI] [PubMed] [Google Scholar]
- 79.Feng Y, Torres E, Li N, Wang L, Bowman D, Xiao L. Population genetic characterisation of dominant Cryptosporidium parvum subtype IIaA15G2R1. International Journal for Parasitology. 2013;43(14):1141–7. 10.1016/j.ijpara.2013.09.002. [DOI] [PubMed] [Google Scholar]
- 80.Hira KG, Mackay MR, Hempstead AD, Ahmed S, Karim MM, O'Connor RM, et al. Genetic diversity of Cryptosporidium spp. from Bangladeshi children. Journal of Clinical Microbiology. 2011;49(6):2307–10. 10.1128/JCM.00164-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ajjampur SSR, Liakath FB, Kannan A, Rajendran P, Sarkar R, Moses PD, et al. Multisite study of cryptosporidiosis in children with diarrhea in India. Journal of Clinical Microbiology. 2010;48(6):2075–81. 10.1128/JCM.02509-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ramírez JD, Heredia RD, Hernández C, León CM, Moncada LI, Reyes P, et al. Molecular diagnosis and genotype analysis of Giardia duodenalis in asymptomatic children from a rural area in central Colombia. Infection, Genetics and Evolution. 2015;32:208–13. 10.1016/j.meegid.2015.03.015. [DOI] [PubMed] [Google Scholar]
- 83.Morse T D, Nichols R A B, Grimason A M, Campbell B M, Tembo K C, Smith H V. Incidence of cryptosporidiosis species in paediatric patients in Malawi. Epidemiology and Infection. 2007;135(8):1307–15. 10.1017/S0950268806007758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Abe N, Matsubayashi M, Kimata I, Iseki M. Subgenotype analysis of Cryptosporidium parvum isolates from humans and animals in Japan using the 60-kDa glycoprotein gene sequences. Parasitol Res. 2006;99(3):303–5. 10.1007/s00436-006-0140-0 [DOI] [PubMed] [Google Scholar]
- 85.Li W, Kiulia NM, Mwenda JM, Nyachieo A, Taylor MB, Zhang X, et al. Cyclospora papionis, Cryptosporidium hominis, and human-pathogenic Enterocytozoon bieneusi in captive baboons in Kenya. Journal of Clinical Microbiology. 2011;49(12):4326–9. 10.1128/JCM.05051-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lal A, Dobbins T, Bagheri N, Baker MG, French NP, Hales S. Cryptosporidiosis risk in New Zealand children under 5 years old is greatest in areas with high dairy cattle densities. EcoHealth. 2016;13(4):652–60. 10.1007/s10393-016-1187-8 [DOI] [PubMed] [Google Scholar]
- 87.Zahedi A, Monis P, Aucote S, King B, Paparini A, Jian F, et al. Zoonotic Cryptosporidium species in animals inhabiting Sydney water catchments. PLOS ONE. 2016;11(12):e0168169 10.1371/journal.pone.0168169 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Highly similar countries are showed in darker colours and 95% lower (above) and upper (below) bootstrapped confidence limits in numbers within each box.
(TIF)
Highly similar countries are showed in darker colours and 95% lower (above) and upper (below) bootstrapped confidence limits in numbers within each box.
(TIF)
Highly similar countries are showed in darker colours and 95% lower (above) and upper (below) bootstrapped confidence limits in numbers within each box.
(TIF)
(TIF)
For each curve the lighter shaded region shows 95% confidence intervals and the red square correspond to the number of genotypes for each species found in New Zealand.
(TIF)
(DOCX)
(DOCX)
(DOCX)
Details for the literature used on this data are found in the accompanying pdf document.
(XLSX)
Highly significant values (p < 0.0001) are shown in red.
(XLSX)
(PDF)
Data Availability Statement
All sequences are available from the GenBank database (accession numbers KY123918-KY124121).