Abstract
Dysregulated transcription, translation, and protein degradation are common features of cancer cells, regardless of specific genetic profiles. Several clinical anticancer agents take advantage of this characteristic vulnerability, and interfere with the processes of transcription and translation, or inhibit protein degradation. However, traditional assays that follow the process of protein production and removal require multi-step processing, and are not easily amenable to high-throughput screening (HTS). The use of recombinant fluorescent proteins provides a convenient solution to this problem, and moreover, photoconvertable fluorescent proteins allow for ratiometric detection of both new protein production and removal of existing proteins. Here, the photoconvertable protein Dendra2 is used in the development of in-cell assays of protein production and degradation that are optimized and validated for high-throughput screening. Conversion from the green to red emissive form can be achieved using a high intensity light emitting diode (LED) array, producing a stable pool of the red fluorescent form of Dendra2. This allows for rates of protein production or removal to be quantified in a plate reader or by fluorescence microscopy, providing a means to measure the potencies of inhibitors that affect these key processes.
Introduction
The ability to follow the protein life-cycle in living cells from translation through degradation is commonly achieved through the use of several different multi-step methods.1–3 Most of these require samples to be manipulated at each time point to be studied, either through cell lysis or fixation for microscopy. In this context, the protein life-cycle is analyzed through bulk effects, or alternatively, by stitching together data from different groups of cells probed at each time point. More convenient and informative approaches are needed in order to follow these key cellular processes on both the individual cell and population levels.
Fluorescent proteins such as Green Fluorescent Protein (GFP) have been shown to be useful tools to monitor proteins in living cells, where individual cells or the same group of cells can be measured over the complete time-frame of the experiment. The drawback to using fluorescent proteins, however, is that they are continuously being synthesized and degraded, so quantitatively probing these specific processes individually is not possible. One alternative is to use a ratiometric approach so that changes in protein level can be assessed. This is possible using proteins that can be triggered by external stimuli to change their properties.4 Such systems would allow for a single genetically encoded protein to provide both an internal control and a real-time report of protein levels.
A number of light activated fluorescent proteins, characterized by changes in spectral properties induced by exposure to high energy light, have been developed for cell-based imaging applications. These include proteins such as photoactivatable GFP5 and mCherry,6 as well as photoconvertible proteins such as Kaede,7 EosFP,8, 9 and Dendra2.10 These switchable spectroscopic features, combined with molecular biology techniques, allow fusions of fluorescent proteins to be used for spatial tracking of individual proteins, or to study interactions between biomolecules, degradation of proteins, and for super resolution microscopy.11–13
Dendra2 exhibits GFP like fluorescence, with excitation at 488 nm and green emission centered at 507 nm when synthesized and folded. This protein, however, undergoes irreversible conversion to a red emissive species upon exposure to 405 nm light.14, 15 The mechanism of photoconversion is due to covalent changes in the protein backbone, which result in a chromophore containing an imidazolinone conjugated to an imidazole ring.16, 17 The new chromophore exhibits an emission centered at 573 nm. Both forms of the protein are monomeric, nontoxic, and not prone to aggregation. The maturation time (the t1/2, or half-life for the immature state) is only 38 minutes.11 The green emitting species is continually synthesized by the cell, while the red emitting population is only generated upon light exposure. Thus, the green emission increases over time, and the red emission remains constant, or decreases as a result of degradation. This allows Dendra2 to serve as a ratiometric detector of protein production or degradation by comparing the emission in the green and red region of the spectrum.
Several medically important drugs have been developed where their mechanism of action involves the disruption of either protein production or degradation. For example, the platin family of compounds, which are used to treat several types of solid tumor cancers, are thought to interfere with nucleic acid function through covalent adduct formation with nucleic acids.18 This has been shown to involve both DNA and RNA,19 with associated impacts on the DNA damage response, and transcription and translation. The proteasome inhibitor, Velcade (bortezomib), blocks protein degradation through inhibition of the 20S proteasome, and has shown remarkable efficacy in the treatment of multiple myeloma and mantle cell lymphoma. Many compounds that target the ribosome are essential antibiotics, and inhibition of translation is an increasingly active area in cancer research.20 Accordingly, early identification of compounds that modulate different aspects of the protein lifecycle is important in medicinal chemistry efforts. Depending on the type of target protein to be modulated by small-molecule compounds, this assay could be used to rapidly identify lead scaffolds from a compound library, or to determine possible off-target toxicities.
The field of bioinorganic medicinal chemistry has developed many cytotoxic agents with potential as chemotherapeutics. One challenge, however, is that the mechanism of action for inorganic agents can be particularly hard to identify. Many compounds have been shown to interact with DNA in several in vitro assays,21, 22 but it remains to be determined if the behavior is the same in live cells. Alternatively, many compounds work through as-yet unknown mechanisms. As the field progresses towards developing metallo-drugs, a rapid assay to follow protein production and/or degradation in cells would assist SAR efforts.
Here we have developed a high-throughput plate reader assay that utilizes an affordable commercial LED flood array to rapidly photoconvert Dendra2 in 96- and 384-well plates. We demonstrate the utility of this method to follow either protein production or protein degradation using Dendra2 or a fusion of Dendra2 with IκBα (nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha), respectively, and validated the assays using several small molecules known to affect these processes.
Materials and methods
Equipment
A Loctite Flood Array emitting 405 nm light with 144 individually reflectorized LEDs (Item #1167593; regulated with a LED Flood System Controller, Item # 1359255) was used to provide uniform illumination for photoconversion of Dendra2 over large surfaces, as in multiwall plates. A SpectraFluor Plus plate reader (Tecan) was used for the data collection of cells expressing soluble Dendra2, while an Omega Bio-Tek plate reader was used for the IκBα fusion protein studies.
Cell lines
HEK-TREX cells were maintained in DMEM media supplemented with 10% FBS, 50 U/ml Penicillin, 50 μg/ml Streptomycin, and 7.5 μg/ml of blasticidin. A matrigel solution was used to coat the surface of all dishes used in this study. To prepare the matrigel solution, 5 ml of matrigel was thawed on ice and then added to 500 ml of DMEM media. Dishes were incubated with the matrigel at 37 °C for a minimum of 10 min, the media removed, followed by the addition of cells.
Tet responsive Dendra2 and IκBα-Dendra2 cell lines
Dendra2 was amplified by PCR with primers containing KpnI and NotI restriction sites and ligated into the pcDNA4-TO vector. The ligation mix was transformed into DH5α chemically competent cells and selected on LB agar plates containing 100 μg/ml carbenicillin. Colonies were grown in LB with 100 μg/ml ampicillin, followed by plasmid isolation. The presence of Dendra2 was verified by restriction analysis followed by sequencing. For the creation of the IκBα-Dendra2 fusion construct, IκBα (provided by Dr. Tom Huxford, San Diego State University) was used after sequence verification and gene amplification by PCR with primers containing the HindIII and KpnI restriction sites. The pcDNA-4TO Dendra2 vector was digested with HindIII and KpnI and ligated in the presence of IκBα, placing IκBα at the 5’ end of Dendra2. After transformation into DH5α cells, plasmid isolation and sequence analysis was used to confirm the presence of pcDNA4-TO-IκBα-Dendra2.
Cell lines were generated using lipofectamine transfection followed by antibiotic selection. HEK-TREX cells were plated at 250,000 cells per well on a matrigel coated 6-well dish, and allowed to adhere overnight. For transfection, 1 μg of plasmid and 2 μl’s of Lipofectamine 2000 was added to the cell in opti-MEM and allowed to incubate with the cells for 8 hrs. The media was aspirated and growth media added. For the Dendra2 cell line, a stable pool of cells was used. For IκBα-Dendra2, the stable pool was sorted by flow cytometry into separate pools based on the intensity of the Dendra2 emission after 18 hrs in the presence of 1 μg/ml tetracycline.
Microscopy studies
Stable cell lines under the control of the Tet repressor protein were plated in 8 well matrigel coated glass bottom dishes at 30,000 cells per well and allowed to adhere overnight in the presence of 1 μg/ml tetracycline. The cells were then placed in L-15 media. Photoconversion and imaging was performed on an Olympus IX81 equipped with 405, 488 and 561 nm lasers. The intensity of the 405 nm light was 1.5 W/cm2.
Exposure of the cells to 30 sec of 405 nm light resulted in photoconversion of 50% of the Dendra2 in the cells, as determined by the relative changes in intensity of both the green and red emission. Once the cells were photoconverted, at least 3 regions for each well were collected during each experiment. Samples were excited with both 488 nm and 561 nm excitation (100 mW/cm2) and images were collected every 30 minutes. The plating density was sufficient to result in thousands of cells being imaged for each region. For all images background was subtracting using a sliding paraboloid algorithm in ImageJ. The mean fluorescence intensity was then extracted for each region for all time points in the series. For each data set, the emission intensity was normalized to the first time point. The mean of multiple regions was then plotted with error bars determined from the standard deviation.
HTS screening in 96 and 384 well plates with a 405 nm flood array
The wells of 96 or 384 well plates were coated with matrigel for 30 min at 37 °C. The wells were aspirated, and cells added in DMEM growth media containing 1 μg/ml tetracycline (30,000 cells/well for 96 and 12,000 cells/well for 384 well plates). After 18 hrs, the media was removed from the plates and L-15 media containing 1 μg/ml tetracycline added to each well and the plates returned to the incubator.
Compounds were serially diluted in L-15 media with 1 μg/ml tetracycline and transferred to the cells. To photoconvert Dendra2, a 405 nm LED flood array was set up with the lamp set at a distance of 15 cm from the plate. The output from the array was measured at 400 mW of power (509 mW/cm2), 2.5× the output from the 405 nm laser on the microscope.
After a 1 hr incubation with compound, the plates were exposed to the 405 nm light source for 1 min, and then placed in a SpectraFluor Plus plate reader thermostated at 37 °C. The plates were read in kinetic mode, with readings taken every 30 min for a period of 15 hrs. For each time point, two readings were taken. The first scan was performed with 485 nm excitation and 530 nm emission, while the second scan was with a 535 nm excitation and 595 emission. Band pass filters were used with cutoffs of +/− 20 nm, except for the 595 nm emission filter, which had a cutoff of +/− 25 nm.
Results
Production of newly synthesized Dendra2 measured in real-time
The utilization of the tetracycline on-off system allowed for the controlled expression of proteins in mammalian cells, but does not discriminate between transcription and translation. Due to the time requirements for transcription and then translation to produce sufficient quantities of protein for fluorescent detection, protein production is generally not observed at time periods of less than 8 hrs. To reduce this lag time, tetracycline was used to induce protein expression for 18 hrs, followed by subsequent photoconversion of Dendra2. This ensured that a sufficiently large pool of protein was available for immediate fluorescent detection, and also allowed for sensitive detection of rapid modulators of protein production, which could be added immediately prior to or following photoconversion.
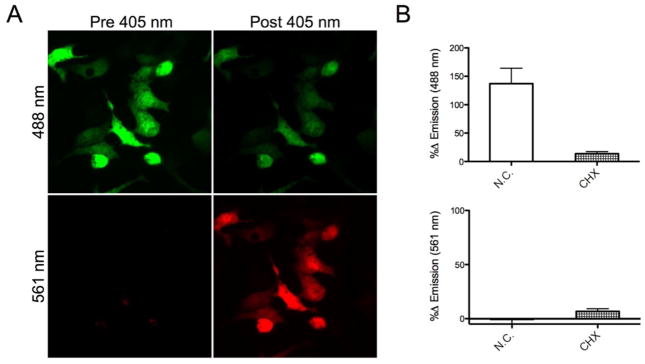
The HEK293 cell line was created using the Tet-on plasmid, where Dendra2 expression remained silent until the addition of 1 μg/ml of tetracycline induced transcription. For microscopy, glass bottom dishes were coated with matrigel solution to adhere the cells to bottom of the dish. The cells were then added in media containing tetracycline and allowed to adhere for 18 hrs. After that time, the cells were exposed to a 405 nm laser for 30 sec with a 4× objective to induce conversion of a population of the pool of Dendra2 from the green to the red emissive form (Figure 1A). This objective was chosen as it allowed for the photo-conversion of a larger population of cells in each field of view for improved statistical analysis. The population of newly synthesized Dendra2 (green emission) was observed over 8 hrs with images taken every 30 min. To verify that the increased emission was due to newly synthesized protein, experiments were performed with the translation inhibitor, cycloheximide (CHX). Addition of CHX suppressed the production of new, green-emissive Dendra2 by 91% (Figure 1B, top). In contrast, the photoconverted, red-emissive dendra2 showed neither a significant increase nor decrease under these conditions, as only a 5% change in emission was observed (Figure 1B, bottom).
Figure 1.
Image of Dendra2 expression in HEK cells. A) Dendra2 before and after exposure to 30 sec of 405 nm light shows photoconversion, where green emission was detected with excitation at 488 nm and red emission was detected with excitation at 561 nm. B) Production of newly translated Dendra2 was measured 8 hrs after treatment with 10 μM CHX.
LED photo-conversion coupled to a fluorescence plate reader for high-throughput screening
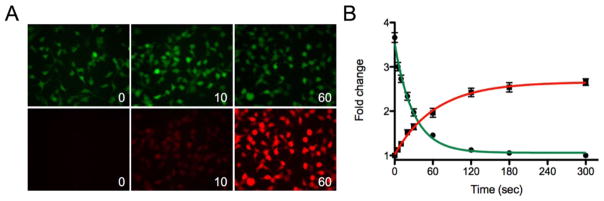
While Dendra2 production and degradation could be followed using a microscope with a programmable stage, the time needed to activate each well within a 96- or 384-microwell plate with 405 nm light and to program the coordinates for each well was deemed to be prohibitive for screening compound libraries. For example, illumination of each well of a 96-well plate would take a minimum of 60 min, and measurements would require a computer capable of storing large image files, in addition to software for data analysis. Thus, to expand this assay to a fluorescence-based 96-well plate reader format, photoconversion using a 405 nm LED flood array was investigated. It was found that illumination with this system was sufficient to convert 50% of Dendra2 from green to red emission within 60 sec. This allowed for screening of multiple plates as an endpoint assay or even in kinetic mode, based on the plate reader configuration.
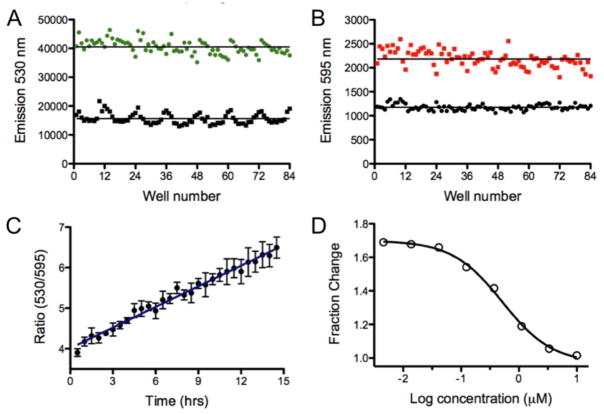
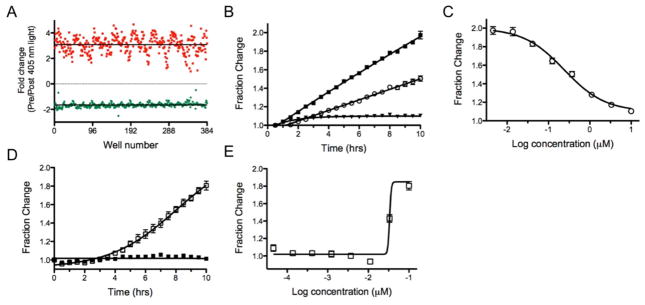
Figure 2 shows the time course for the photoconversion of Dendra2 expressing HEK cells in a 96-well plate. Photoconversion was observed within as little as 10 sec, with 15 +/− 4% of the maximum signal change. The maximum photoconversion was reached within 120 sec, and corresponds to a decrease in signal in the green of 69 +/− 1.5%. Further exposure to the 405 nm light source did not convert the remaining fraction of Dendra2. The change in fluorescence for Dendra2 pre- and post-exposure to 405 nm light is shown in Figure 3; as anticipated, the green emission was reduced and the red emission was increased upon photoconversion. The change in signal was similar across the plate, but some variance in signal intensity was observed at the columns towards the left and right edges of the plate which correlated with variations in the cell number plated for each column (see Figure SI3 and associated discussion).
Figure 2.
Photoconversion of Dendra2 in 96 well plates with an LED flood array. A) Time dependent conversion of HEK cells expressing Dendra2 from green to red upon exposure to 405 nm light for 0 to 60 sec. B) Dendra2 photoconversion reaches a maximum after a 2 min exposure.
Figure 3.
Well-to-well variability before and after Dendra2 photoconversion in a 96-well plate. A) Emission of Dendra2 at 530 nm before (green) and after (black) the cells were exposed to 1 min of 405 nm LED flood array. B) Emission of Dendra2 at 595 nm before (black) and after (red) 1 min of 405 nm light. C) Ratiometric detection of newly synthesized Dendra2 measured over 15 hrs. D) Dose response for cycloheximide plotted as the fractional change in the 530 nm emission as a function of concentration (data was collected in triplicate).
To follow protein production in kinetic mode, Dendra2 expressing cells were photoconverted for 1 min followed by dual reading of the green and red emission every 30 min over a period of 15 hrs. As shown in Figure SI1A, the emission at 530 nm increased linearly over this period, while the emission at 595 nm was constant (Figure SI1B). The ratio of the two emissions is shown in Figure 3C, and demonstrates the linear increase in emission with time, with a 4-fold change in signal. The Dendra2 595 nm signal thus provides an internal control during the time frame of the assay, and could be used as a ratiometric control.
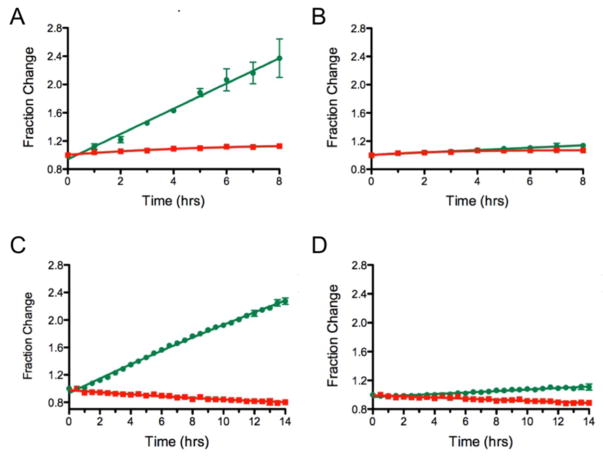
To validate Dendra2 as an assay for quantifying protein translation, cycloheximide was added to the cells and the production of newly synthesized Dendra2 was measured over time in dose response (Figure 3D). After a 1 hr incubation with the cells, Dendra2 was photoconverted. The data was collected both by microscopy and with a plate reader in order to compare both methods. As shown in Figure 4A and 4C, the green emission increases linearly with time in the absence of compound, while the red emission is stable. In contrast, in Figure 4B and 4D, cycloheximide completely blocked translation of Dendra2 at 10 μM. Both instrumentation set-ups showed similar responses and demonstrated excellent agreement in the fraction change in emission and the kinetics. Thus, Dendra2 emission can be used to follow protein production and detect compounds that inhibit this process.
Figure 4.
Comparison of detection methods. A) Dendra2 production over 8 hrs as measured by live cell imaging shows a >2-fold increase in newly synthesized Dendra2 (green filled circles) without loss of photoconverted Dendra2 (red filled squares). B) Addition of 10 μM CHX inhibits the production of newly synthesized Dendra2 as measured by live cell imaging. C) Dendra2 production measured in 96 well plates over a period of 14 hrs in a fluorescence plate reader. D) Addition of 10 μM CHX inhibits Dendra2 production as measured with a fluorescence plate reader.
IκBα-Dendra2 fusion to measure effect of compounds on protein degradation
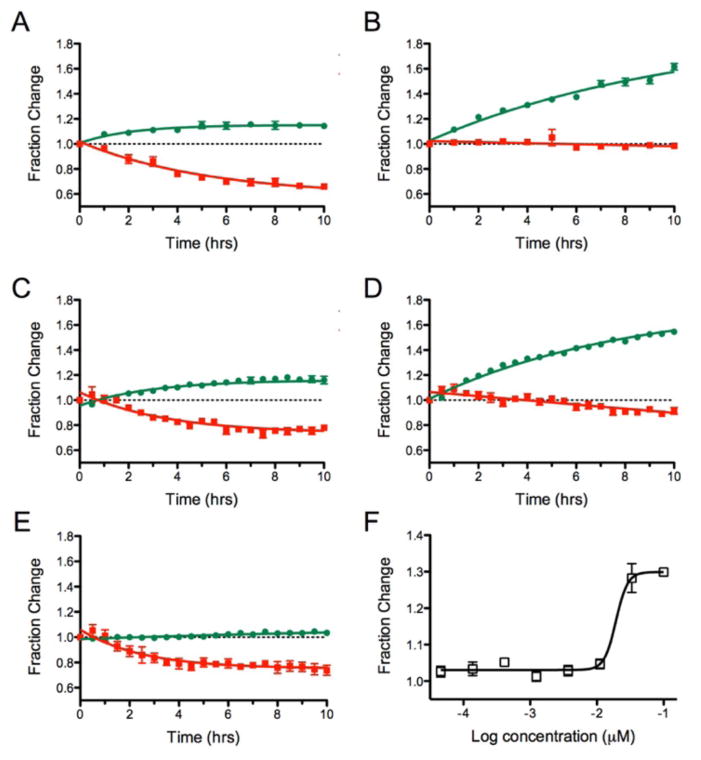
IκBα is an inhibitor of the transcription factor NF-κB. In cells it is continuously expressed but also continuously degraded.23 It has previously been shown that the addition of Dendra2 as a fusion to IκBα produces a fluorescent form of IκBα that recapitulates the degradation rate seen for IκBα alone.24 Since the half-life for this protein is on the order of minutes, we explored the application of this fusion protein to follow the effect of compounds that inhibit protein degradation. The proteasome inhibitors, MG132 and Velcade were dosed for 1 hr prior to photoconversion, and the emission followed at 530 and 595 nm.
As shown in Figure 5A, the amount of new protein that can be observed reaches a plateau after 3 hrs in the absence of any compound. This was not surprising, as the short half-life of IκBα would result in the competitive degradation of both the newly translated, green, protein and post-translated, photoconverted, red, protein. As expected, the red emissive form of the protein continually decreased.
Figure 5.
Comparison of results for IκBα-Dendra2 by live cell imaging versus a multi-well plate reader. A) Change in IκBα-Dendra2 over time after exposure to 1 min of 405 nm light as measured by live cell imaging. B) Addition of 0.1 μM Velcade followed by imaging. C) Change in IκBα-Dendra2 over time after exposure to 1 min of 405 nm light followed by change in protein emission over time measured in a plate reader. D) Addition of 0.1 μM Velcade; E) addition of 10 μM CHX. F) Dose response of Velcade with fraction change from the 530 nm emission plotted against concentration.
In the presence of increasing concentrations of proteasome inhibitors, a block in the degradation of the red IκBα-Dendra2 was seen (Figure 5B). There was an associated steady increase in the newly synthesized IκBα-Dendra2. Full inhibition of protein degradation was readily observed by microscopy. Similar results were observed in a 96 well plate format where the emission signals for each well were measured over time (Figure 5C). Data collection with the plate reader following the addition of the proteaseome inhibitor, Velcade, showed a slight decrease in the red IκBα-Dendra2 during the time course of the experiment (Figure 5D). Both detection approaches showed the same fractional change in the newly translated IκBα-Dendra2 protein in the presence and absence of 0.1 μM Velcade. Translation inhibitors could also be studied in this assay, where only the green form of the IκBα-Dendra2 was affected. (Figure 5E).
To further extend the utility of this assay, the kinetic data was collected in dose response to determine if dose-response curves could be generated. Both CHX and Velcade were tested. As shown in Figure SI2, the IC50 value for Dendra2 production with CHX in the HEK-Dendra2 cell line was 0.52+/−0.08 μM. Velcade gave an IC50 of 0.02+/−0.0013 μM in the IκBα-Dendra2 cell line (Figure 5F). The IC50 value for CHX was also determined in the IκBα-Dendra2 cell line, giving a value of 0.41+/−0.07 μM, in close agreement with the HEK-Dendra2 cell line. Thus, IC50 values can be obtained for both inhibitors of protein production and degradation.
Extension of the assays to high-throughput screening in 384-well plates
To validate the assay in a high density format, 384-well low volume plates were seeded with the HEK-Dendra2 cells followed by the addition of tetracycline for 18 hrs. A maximal signal change between wells containing tetracycline and those without was found when 12,000 cells per well were used. Photoconversion resulted in an average decrease in green emission of 1.66 fold, with an increase in red emission of 3.06 fold (Figure 6A). Addition of CHX to the cells resulted in a dose dependent decrease in the production of Dendra2; analysis of the signal change after 10 hrs incubation resulted in an IC50 value of 0.24+/−0.06 μM. This value is 2-fold more potent than determined in 96-well (Figure 6B, C). Similar results for Velcade were seen with the IκBα-Dendra2 cell line, where an IC50 of 0.033+/−0.001 μM was determined, compared to 0.02 μM when assayed in 96-well plates (Figure 6D, E). While the data is in good agreement, care must be taken when comparing values from 96- vs. 384-well plates.
Figure 6.
Translation and proteasome inhibition assays in 384-well format. A) Well-to-well variability in emission of Dendra2 at 595 nm (red) and 530 nm (green). B) Fractional change in the emission signal at 530 nm for CHX inhibition of protein production with the HEK-Dendra2 cell line (filled squares = NC; open circles = 0.3 μM, filled triangles = 10 μM); C) Dose response for CHX inhibition of Dendra2 production at 10 hr time point. D) Velcade inhibition of IκBα-Dendra2 degradation (filled squares = NC; open squares = 0.1 μM Velcade). Fraction change is shown for the 530 nm emission signal. E) Dose response for Velcade inhibition of IκBα-Dendra2 degradation at 10 hr time point.
Discussion
Since the discovery of photoconvertable proteins, only limited studies have sought to employ them for high-throughput screening. Rather, development of methods using Dendra2 have focused on detection of the protein by microscopy. Expanding the utility of Dendra2 to other assay formats has been challenging due to the requirement of a 405 nm laser to induce the photoreaction of the chromophore. While photoconversion has been shown to occur at wavelengths up to 450 nm, the length of time required to photoconvert is prohibitively long. This has limited the application of Dendra2 and other photosensitive proteins for HTS.
The use of a high intensity LED array allows for the rapid illumination of an entire plate of any format required for screening. We used this to convert cells in a single well, and up to 384-well formats. Live cells expressing the protein are then amenable to any microscopy study or protein quantification using standard plate readers. The rapid quantification of fluorescence intensity in plate readers allows for high-throughput screening of compounds that inhibit protein production or protein degradation, with data that can be generated as kinetic time courses or as an end point assay. This provides complementary data on large populations of cells that can be compared to the high resolution information obtained on single cells or small collections of cells via microscopy.
One issue that is concerning with using Dendra2 to follow protein degradation is the inefficiency of multiwell plate readers to detect red emission. In the microscope the signal intensities for the green and red emissions were equivalent. The plate reader did exhibit the same fold change, with around a 10-fold decrease in signal intensity for the red form of Dendra2. The difference in sensitivity for the plate reader for the green and red detection is evident in the Z’ values for the two different detection wavelengths. The Z’ reflects the number of standard deviations an individual reading is from the mean reading. The Z’ is 0.51 for detection of green emission, and is 0.33 for the red emission; thus, the assay is in the excellent range in the green channel and marginal in the red in the plate reader and both values are better using the microscope. Optimized filter sets or plate readers with greater sensitivity in the red of the spectrum would allow for improved sensitivity for the red form of the Dendra2.
While the 2-fold change in signal may appear limiting, the assay is actually quite robust, as a change in Dendra2 of greater than 10% can be reliably detected. The data can be used to discriminate this signal change using a three standard deviation analysis from the no compound control.25 Thus, changes in protein level of more than 10% can be detected with confidence, as shown in Figure SI4.
These in-cell assays for global protein production and destruction provide important tools for the discovery of agents that target these processes. The assays also allow for detection of undesired off-target effects for compounds not anticipated to interfere with proteostasis. Furthermore, in vitro systems are available for detecting inhibition of transcription and translation (using the minimum required biological components),2, 22 or direct inhibition of the proteasome, but the cellular environment is an intrinsically more complex environment, and multiple biological factors play a role in proteostasis. The combination of these new in-cell assays with existing in vitro assays may allow for validation or exclusion of specific targets in the complex machinery regulating protein levels.
Acknowledgments
This work was supported by the National Institutes of Health (1R01GM107586). A.F. was supported by by NIDA T32 Research Fellowship (NIH DA016176). The authors are grateful to Dr. Tom Huxford (San Diego State University) for the gift of the plasmid containing the gene for IκBα.
References
- 1.Capece MC, Kornberg GL, Petrov A, et al. A simple real-time assay for in vitro translation. RNA. 2015;21:296–305. doi: 10.1261/rna.047159.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rosenblum G, Chen C, Kaur J, et al. Real-time assay for testing components of protein synthesis. Nucleic Acids Res. 2012;40:e88. doi: 10.1093/nar/gks232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rosenblum G, Cooperman BS. Engine out of the chassis: cell-free protein synthesis and its uses. FEBS Lett. 2014;588:261–268. doi: 10.1016/j.febslet.2013.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lukyanov KA, Chudakov DM, Lukyanov S, et al. Innovation: Photoactivatable fluorescent proteins. Nat Rev Mol Cell Biol. 2005;6:885–891. doi: 10.1038/nrm1741. [DOI] [PubMed] [Google Scholar]
- 5.Patterson GH, Lippincott-Schwartz J. A photoactivatable GFP for selective photolabeling of proteins and cells. Science. 2002;297:1873–1877. doi: 10.1126/science.1074952. [DOI] [PubMed] [Google Scholar]
- 6.Subach FV, Patterson GH, Manley S, et al. Photoactivatable mCherry for high-resolution two-color fluorescence microscopy. Nat Methods. 2009;6:153–159. doi: 10.1038/nmeth.1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ando R, Hama H, Yamamoto-Hino M, et al. An optical marker based on the UV-induced green-to-red photoconversion of a fluorescent protein. Proc Natl Acad Sci U S A. 2002;99:12651–12656. doi: 10.1073/pnas.202320599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang M, Chang H, Zhang Y, et al. Rational design of true monomeric and bright photoactivatable fluorescent proteins. Nat Methods. 2012;9:727–729. doi: 10.1038/nmeth.2021. [DOI] [PubMed] [Google Scholar]
- 9.Wiedenmann J, Ivanchenko S, Oswald F, et al. EosFP, a fluorescent marker protein with UV-inducible green-to-red fluorescence conversion. Proc Natl Acad Sci U S A. 2004;101:15905–15910. doi: 10.1073/pnas.0403668101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gurskaya NG, Verkhusha VV, Shcheglov AS, et al. Engineering of a monomeric green-to-red photoactivatable fluorescent protein induced by blue light. Nat Biotechnol. 2006;24:461–465. doi: 10.1038/nbt1191. [DOI] [PubMed] [Google Scholar]
- 11.Wang S, Moffitt JR, Dempsey GT, et al. Characterization and development of photoactivatable fluorescent proteins for single-molecule-based superresolution imaging. Proc Natl Acad Sci U S A. 2014;111:8452–8457. doi: 10.1073/pnas.1406593111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McKinney SA, Murphy CS, Hazelwood KL, et al. A bright and photostable photoconvertible fluorescent protein. Nat Methods. 2009;6:131–133. doi: 10.1038/nmeth.1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Betzig E, Patterson GH, Sougrat R, et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 14.Chudakov DM, Lukyanov S, Lukyanov KA. Using photoactivatable fluorescent protein Dendra2 to track protein movement. Biotechniques. 2007;42:553, 555, 557. doi: 10.2144/000112470. [DOI] [PubMed] [Google Scholar]
- 15.Chudakov DM, Lukyanov S, Lukyanov KA. Tracking intracellular protein movements using photoswitchable fluorescent proteins PS-CFP2 and Dendra2. Nat Protoc. 2007;2:2024–2032. doi: 10.1038/nprot.2007.291. [DOI] [PubMed] [Google Scholar]
- 16.Mizuno H, Mal TK, Tong KI, et al. Photo-induced peptide cleavage in the green-to-red conversion of a fluorescent protein. Mol Cell. 2003;12:1051–1058. doi: 10.1016/s1097-2765(03)00393-9. [DOI] [PubMed] [Google Scholar]
- 17.Hoi H, Shaner NC, Davidson MW, et al. A monomeric photoconvertible fluorescent protein for imaging of dynamic protein localization. J Mol Biol. 2010;401:776–791. doi: 10.1016/j.jmb.2010.06.056. [DOI] [PubMed] [Google Scholar]
- 18.Cohen SM, Lippard SJ. Cisplatin: From DNA damage to cancer chemotherapy. Progress in Nucleic Acid Research and Molecular Biology. 2001;67:93–130. doi: 10.1016/s0079-6603(01)67026-0. [DOI] [PubMed] [Google Scholar]
- 19.Hostetter AA, Osborn MF, DeRose VJ. RNA-Pt adducts following cisplatin treatment of Saccharomyces cerevisiae. ACS Chem Biol. 2012;7:218–225. doi: 10.1021/cb200279p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bhat M, Robichaud N, Hulea L, et al. Targeting the translation machinery in cancer. Nat Rev Drug Discov. 2015;14:261–278. doi: 10.1038/nrd4505. [DOI] [PubMed] [Google Scholar]
- 21.Wang J, Newman J, Jr, Higgins SL, et al. Red-light-induced inhibition of DNA replication and amplification by PCR with an Os/Rh supramolecule. Angew Chem Int Ed Engl. 2013;52:1262–1265. doi: 10.1002/anie.201207083. [DOI] [PubMed] [Google Scholar]
- 22.Heidary DK, Glazer EC. A light-activated metal complex targets both DNA and RNA in a fluorescent in vitro transcription and translation assay. Chembiochem. 2014;15:507–511. doi: 10.1002/cbic.201300681. [DOI] [PubMed] [Google Scholar]
- 23.Henkel T, Machleidt T, Alkalay I, et al. Rapid proteolysis of I kappa B-alpha is necessary for activation of transcription factor NF-kappa B. Nature. 1993;365:182–185. doi: 10.1038/365182a0. [DOI] [PubMed] [Google Scholar]
- 24.Zhang L, Gurskaya NG, Merzlyak EM, et al. Method for real-time monitoring of protein degradation at the single cell level. Biotechniques. 2007;42:446, 448, 450. doi: 10.2144/000112453. [DOI] [PubMed] [Google Scholar]
- 25.Malo N, Hanley JA, Cerquozzi S, et al. Statistical practice in high-throughput screening data analysis. Nat Biotechnol. 2006;24:167–175. doi: 10.1038/nbt1186. [DOI] [PubMed] [Google Scholar]