Abstract
Glioma stem cells (GSCs) have a central role in glioblastoma (GBM) development and chemo/radiation resistance, and their elimination is critical for the development of efficient therapeutic strategies. Recently, we showed that lysine demethylase KDM1A is overexpressed in GBM. In the present study, we determined whether KDM1A modulates GSCs stemness and differentiation and tested the utility of two novel KDM1A-specific inhibitors (NCL-1 and NCD-38) to promote differentiation and apoptosis of GSCs. The efficacy of KDM1A targeting drugs was tested on purified GSCs isolated from established and patient-derived GBMs using both in vitro assays and in vivo orthotopic preclinical models. Our results suggested that KDM1A is highly expressed in GSCs and knockdown of KDM1A using shRNA-reduced GSCs stemness and induced the differentiation. Pharmacological inhibition of KDM1A using NCL-1 and NCD-38 significantly reduced the cell viability, neurosphere formation and induced apoptosis of GSCs with little effect on differentiated cells. In preclinical studies using orthotopic models, NCL-1 and NCD-38 significantly reduced GSCs-driven tumor progression and improved mice survival. RNA-sequencing analysis showed that KDM1A inhibitors modulate several pathways related to stemness, differentiation and apoptosis. Mechanistic studies showed that KDM1A inhibitors induce activation of the unfolded protein response (UPR) pathway. These results strongly suggest that selective targeting of KDM1A using NCL-1 and NCD-38 is a promising therapeutic strategy for elimination of GSCs.
INTRODUCTION
Glioblastoma (GBM) are the most common and deadliest tumors of the central nervous system. Despite recent advances in multimodal therapies, patients with GBM have poor prognosis due to tumor recurrence and resistance to therapy.1 Median survival is < 15 months and the 5-year survival rate after diagnosis is 5%.2 The standard therapy for GBM is surgery followed by radiation with adjuvant chemotherapy.
GBM are highly infiltrative and mutable in nature, exhibit extensive cellular heterogeneity.3 Emerging studies are implicating cancer stem cells (CSCs) in tumor initiation, progression, and therapy resistance.4–6 GBM display hierarchical organization with a subpopulation of undifferentiated and self-renewing glioma stem cells (GSCs) at the apex.3,7,8 GSCs have characteristics such as self-renewal, multi-lineage differentiation ability and express various neural stem cell markers such as nestin, CD133 and olig2.7–9 Although GSCs comprise only a small portion of the tumor, GSCs are highly tumorigenic, sustain the tumor growth and recapitulate the cellular heterogeneity and hierarchy of the original tumor. Recent studies demonstrated that GSCs promote tumor angio-genesis, immune evasion and possess high DNA repair capacity that often contribute to tumor relapse and therapy resistance.10–12 Eradication of GSCs is critical for the development of efficient therapeutic strategies,13 and several strategies of targeting GSCs are currently being developed.14 A potential therapeutic strategy for GBM would be to use forced differentiation and apoptosis of GSCs.15
GBM development is a multistep process that results from aberrant genetic alterations.16 In addition to genetic alterations, epigenetic changes have a pivotal role in GBM development.17,18 Histone methylation is a dynamic process regulated by histone methylases and demethylases, and alterations in histone methylation have a vital role in neoplastic processes.19,20 The lysine-specific demethylase-1 (KDM1A, LSD1, AOF2) was the first demethylase discovered. It demethylates both mono- and dimethylated lysine residue-4 specifically on histone H321 and also on lysine-9 of histone H3 in an AR-22 and ESR1-23 dependent manner. KDM1A regulates gene expression programs by changing the epigenetic histone marks at the gene promoters.20 KDM1A overexpression has been associated with various cancers including neuroblastoma,24 colon cancer,25 breast cancer,26 ovarian cancer,27 bladder cancer,28 prostate cancer,29 hepatocellular cancer27,30 and glioma.31 Recent studies demonstrated that KDM1A is essential to maintain the undifferentiated state of human embryonic stem cells32 and regulates neural stem cell proliferation and differentiation.33 KDM1A is essential for the oncogenic potential of MLL-AF9 leukemia stem cells34 and its inhibition resulted in selective inhibition of pluripotent stem cell proliferation.35 However, little is known about the functional significance of KDM1A signaling in GSCs and whether KDM1A inhibitors have clinical utility in eradicating GSCs.
KDM1A-mediated demethylation process involves flavin adenine dinucleotide-dependent enzymatic oxidation. The mono-amine oxidase inhibitors such as tranylcypromine, pargyline, clorgyline and polyamine derivatives are known to inhibit the KDM1A enzyme activity. However, their selectivity for KDM1A is very low and requires higher concentrations to inhibit the KDM1A activity,36 which causes side effects and limits their use as potential therapeutic agents. We recently developed a novel KDM1A-specific inhibitor NCL-1 (N-[(1S)-3-[3-(trans-2-Aminocyclo-propyl)phenoxy]-1-(benzylcarbamoyl)propyl] benzamide)37,38 that has potent inhibitory activity on various cancer cells.31,38,39 Further, we developed another potent KDM1A inhibitor called NCD-38 (2-(N-4-phenylbenzenecarbonyl)amino-6-(trans-2-phenyl-cyclopropane-1-amino-N-(3-chlorobenzyl)hexaneamide trifluoroacetate) based on a novel concept of direct delivery of phenylcyclopropylamine to the KDM1A active site.40 Here, we describe the therapeutic utility of two novel KDM1A inhibitors NCL-1 and NCD-38 on GSCs using in vitro and in vivo models. Our results demonstrate that KDM1A is highly expressed in GSCs and inhibition of KDM1A reduces the stemness, induces differentiation and promotes apoptosis of GSCs. Mechanistic studies showed that KDM1A inhibitors activate the unfolded protein response (UPR) pathway. Further, KDM1A inhibition reduces tumor progression and improves mice survival.
RESULTS
KDM1A is highly expressed in GSCs compared with differentiated cells
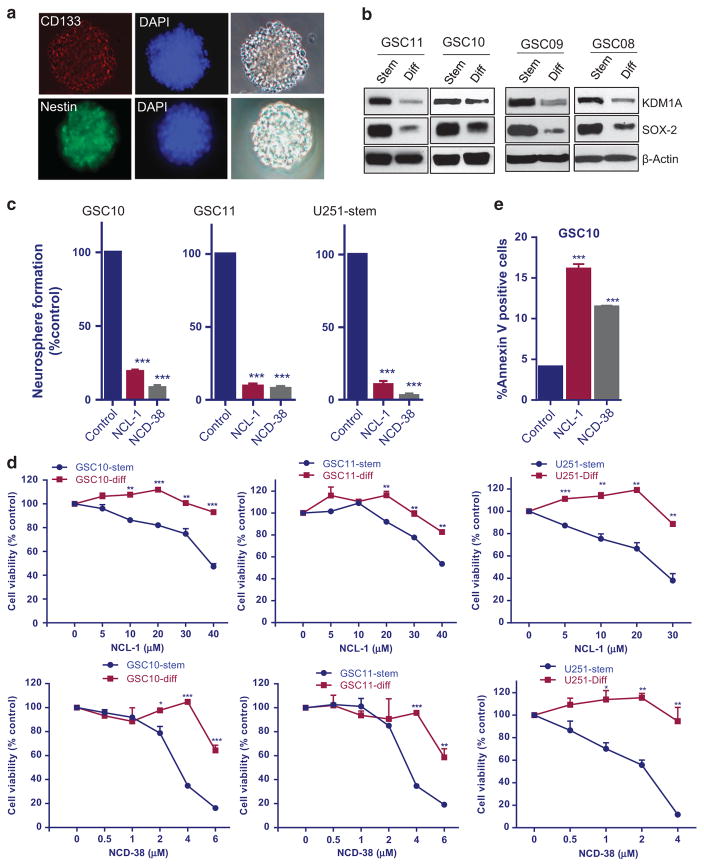
To determine the significance of KDM1A in GSCs stemness, GSCs were isolated using established stem cell marker CD133 from primary GBM specimens (GBM08, GBM09, GBM10 and GBM11). The expression of stemness markers CD133 and nestin in purified GSCs was confirmed with immunofluorescence analysis (Figure 1a). We then examined whether differentiation of GSCs promote alterations in KDM1A expression. To induce the differentiation process, GSCs were cultured in the presence of 10% FBS for 7 days. Western blot analysis showed that KDM1A expression was high in GSCs and KDM1A expression was substantially downregulated in differentiated cells (Figure 1b). As GSCs express stem cell marker SOX2, the decrease in SOX2 levels was used to confirm the process of differentiation of GSCs.
Figure 1.
KDM1A expression was reduced in differentiated GSCs. (a) GSC10 were subjected to immunofluorescence staining to detect the presence of the stemness markers CD133 and nestin. (b) GSCs were isolated from patient-derived GBM using established stem cell marker CD133 and differentiation was induced in the presence of 10% FBS for 7 days. KDM1A expression was examined in stem and differentiated GSCs using Western blotting. (c) GSCs (GSC10, GSC11 and U251-GSCs) were isolated from patient-derived GBM (GBM10, GBM11) and U251-GBM cells, respectively, and treated with KDM1A inhibitors NCL-1 and NCD-38 for 7 days, and the number of neurospheres were counted. (d) Undifferentiated and differentiated GSCs were treated with vehicle, NCL-1 or NCD-38 for 72 h, and the cell viability rates were measured by CellTiter-Glo assays. All the results were representative of three independent experiments. (e) Replicate cultures of GSC10 were treated with vehicle, NCL-1 or NCD-38 for 48 h followed by Annexin V-FITC and propidium iodide (PI) staining for 15 min. The Annexin V–positive apoptotic populations were determined by using flow cytometry. Data are represented as mean ± s.e. *P < 0.05, **P < 0.01, ***P < 0.001 based on the Student’s t-test.
KDM1A inhibition reduced the neurosphere formation, cell viability and induced apoptosis of GSCs
As KDM1A is highly expressed in GSCs, we next examined the effect of the most potent KDM1A inhibitors NCL-1 and NCD-38 on neurosphere formation. The design and synthesis of novel KDM1A inhibitors NCL-1 and NCD-38 was described in our previous studies.37,40 GSCs (U251-GSCs, GSC10 and GSC11) were treated with either vehicle or KDM1A inhibitors for 7 days, and the number of newly formed neurospheres were counted. As shown in Figure 1c, pharmacological inhibition of KDM1A significantly reduced the neurosphere formation of GSCs. To determine whether KDM1A inhibitors affect the cell viability of GSCs (U251-GSCs, GSC10 and GSC11), CellTiter-Glo viability assays were performed on GSCs and differentiated GSCs. NCL-1 and NCD-38 significantly reduced the viability of GSCs in a dose-dependent manner but exhibited minimal effect on the viability of the differentiated cells at the tested doses, suggesting the selectivity of KDM1A inhibitors on GSCs (Figure 1d). These results demonstrate that KDM1A inhibitors reduced the viability of GSCs, which express high levels of KDM1A, but these inhibitors have less effect on differentiated cells that have low levels of KDM1A. To further demonstrate the specific requirement of KDM1A for NCL-1 and NCD-38 actions, U251-KDM1A knockout cells were generated using CRISPR/Cas9 technology and the effect of KDM1A inhibitors on the cell viability was determined. As shown in Supplementary Figure 1, NCL-1 and NCD-38 showed minimal effect on the cell viability of U251-KDM1A knockout cells compared with control, confirming the specificity of KDM1A inhibitors. To further examine the effect of KDM1A inhibitors on apoptosis, GSC10 were treated with vehicle or NCL-1 or NCD-38, and apoptosis was measured by Annexin V/PI staining. As shown in Figure 1e, NCL-1 and NCD-38 treatment significantly induced apoptosis of GSCs.
KDM1A inhibitors reduced the stemness and induced the differentiation of GSCs
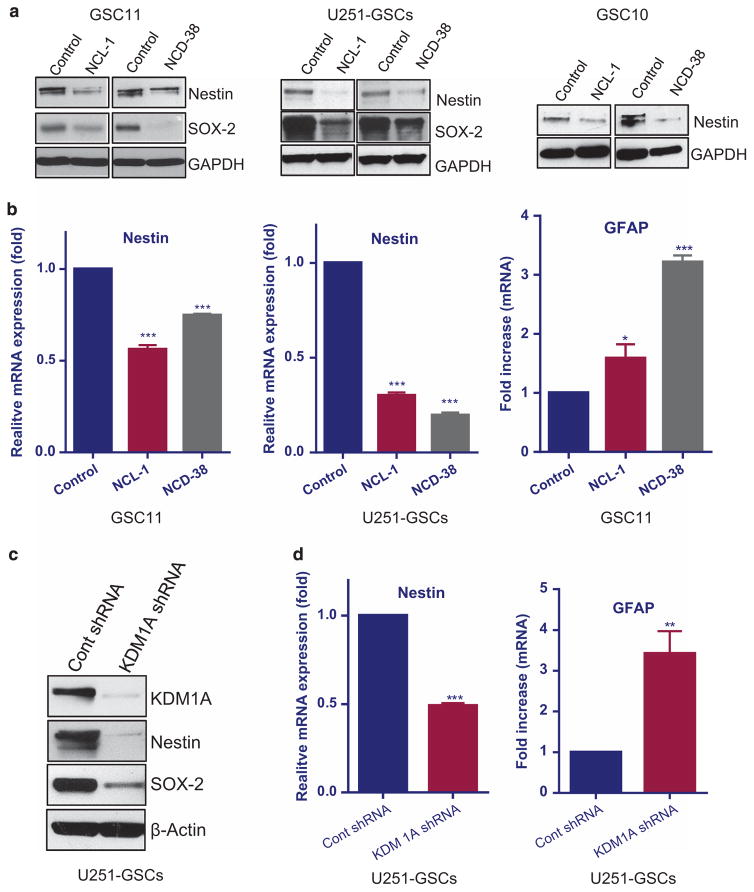
To determine the effect of KDM1A inhibition on stemness and differentiation, GSCs isolated from U251-GBM cells, GSC10 and GSC11 were treated with NCL-1 and NCD-38 for 5 days and analyzed for expression of stemness markers (nestin and sox2) and astrocytic differentiation marker GFAP. Western blotting and qRT-PCR analysis demonstrated that NCL-1 and NCD-38 significantly reduced the expression of nestin and sox2 and induced GFAP (Figures 2a and b). To further validate these findings, we have examined whether KDM1A knockdown also alters stemness of GSCs. CD133-positive cells separated from U251-GBM cells were infected with control or KDM1A-specific short hairpin RNA (shRNA) lentiviral vectors. Western blot and qRT-PCR analysis showed that KDM1A knockdown significantly reduced the expression of nestin and sox2 and induced the expression of GFAP (Figures 2c and d). These results suggest that KDM1A inhibition reduced the stemness and induced the differentiation of GSCs.
Figure 2.
KDM1A inhibition reduced the expression of stemness genes and increased the expression of differentiation genes. Replicate cultures of GSC10, GSC11 and U251-GSCs were treated with vehicle or the KDM1A inhibitors NCL-1 (20 μM) or NCD-38 (2 μM) for 5 days. (a) The expression levels of stem cell markers nestin and sox-2 were tested using Western blotting. (b) The expression levels of nestin and differentiation marker GFAP was examined by RT-qPCR. (c) Replicate cultures of U251-GSCs were infected with control shRNA and KDM1A-shRNA lentiviral plasmids and the expression levels of nestin and sox-2 was examined by Western blotting. (d) mRNA expression of nestin and GFAP was tested in control shRNA and KDM1A-shRNA GSCs using qRT-PCR. All the results were representative of three independent experiments. Data are represented as mean ±s.e. *P < 0.05, **P < 0.01, ***P < 0.001 based on the Student’s t-test.
Analysis of KDM1A inhibition-mediated global transcriptional changes in GSCs
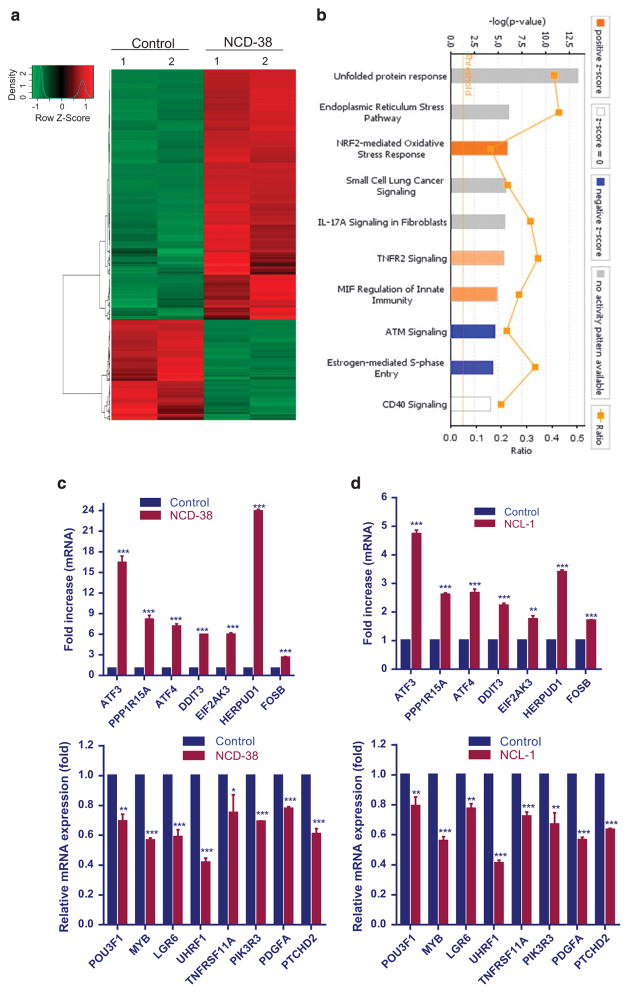
To examine the global changes in the gene expression following KDM1A inhibition on GSCs, we performed global transcriptome analysis. GSC10 were treated with either vehicle or NCD-38 for 24 h, and the isolated RNA was subjected to RNA-seq analysis. Differentially expressed genes with two fold change in expression (P < 0.01) were chosen for analysis. Overall, 1320 genes were differentially expressed in NCD-38-treated GSCs: 943 genes were upregulated and 377 genes were downregulated. A representative heat map is shown in Figure 3a. The complete list of differentially expressed genes is given in Supplementary Table 2. To further examine the biological significance of the differentially expressed genes, ingenuity pathway analysis (IPA) was performed. The top 10 canonical pathways modulated by NCD-38 are shown in Figure 3b. We found that the top three pathways that were modulated by NCD-38 were UPR, endoplasmic reticulum stress pathway and NRF2-mediated oxidative stress response. Further, the top networks that were differentially expressed after NCD-38 treatment were related to cellular assembly and organization, cellular compromise, DNA replication, recombination and repair (Supplementary Figures 2A and 3). Analysis of molecular and cellular functions of the differentially expressed genes revealed that they are involved in cell death and survival, cellular compromise, cellular function and maintenance (Supplementary Figure 2B).
Figure 3.
Analysis of global transcriptome changes modulated by KDM1A inhibition. Total RNA was isolated from GSC10 cells that were treated with either vehicle or NCD-38 for 24 h using RNeasy mini kit, and the quality of RNA was tested using a Bio-analyzer (Agilent Technologies, Santa Clara, CA, USA) and subjected to RNA-sequencing. (a) Heat map of differentially expressed genes between vehicle and NCD-38 is shown. (b) Differentially expressed genes were subjected to pathway analysis using IPA software (Qiagen, Valencia, CA, USA) and the top 10 canonical pathways are shown. (c, d) GSC10 were treated with vehicle or the KDM1A inhibitors NCL-1 (20 μM) and NCD-38 (2 μM) for 24 h, and the differentially expressed genes that were upregulated or downregulated following KDM1A inhibitor treatment were validated using RT-qPCR. All RT-qPCR experiments were representative of three independent experiments. Data are represented as mean ±s.e. *P < 0.05, **P < 0.01, ***P < 0.001 based on the Student’s t-test.
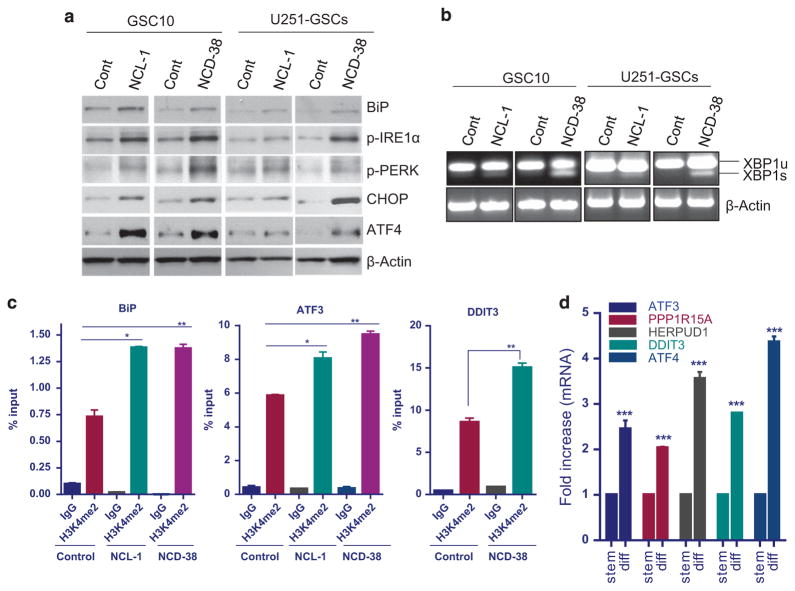
KDM1A inhibition induced the expression of UPR pathway genes The UPR pathway is induced in cells to alleviate the endoplasmic reticulum stress that is caused by unfolded or misfolded proteins. Several studies demonstrated that the induction of UPR induces apoptosis of various cancer cells.41,42 Emerging studies also showed the involvement of UPR in the differentiation of stem cells.43,44 The PERK-eIF-2α axis of UPR, activates the transcription factor ATF4,45 which further transactivate expression of several genes, such as ATF3, DDIT3 (CHOP) and PPP1R15A.41 Our RNA-seq analysis of NCD-38-treated GSC10 revealed that several genes of the PERK branch of UPR pathway, such as EIF2AK3, ATF4, ATF3, DDIT3 and PPP1R15A were significantly upregulated in the KDM1A inhibitor-treated group when compared with control. Further RNA-sequencing analysis also revealed that the IRE1 axis genes such as IRE1, XBP1 and ERO1LB and ATF6 target genes such as GRP78 (BiP, HSPA5), HERPUD1, HYOU1, and PDIA4 were upregulated following NCD-38 treatment compared with control. We independently validated the regulation of some of the UPR genes using GSCs that were treated with NCL-1 and NCD-38. RT-qPCR analysis revealed that several genes of the UPR pathway were upregulated significantly in both NCL-1- and NCD-38-treated GSCs (Figures 3c and d). Further, several other genes that involved in GBM stemness and oncogenesis such as POU3F1, PDGFA, MYB, LGR6, PIK3R3, PTCHD2, UHRF1 and TNFRSF11A were significantly downregulated following KDM1A inhibition (Figures 3c and d). The chaperone GRP78/BiP is one of the major targets of UPR and its transcriptional activation was used as an indicator of onset of UPR.46,47 To examine whether KDM1A inhibitors induces the expression of BiP, GSCs were treated with vehicle or NCL-1 or NCD-38 and its expression was determined by western blotting. Tunicamycin treatment was used as a positive control to validate the detected bands by specific antibodies (Supplementary Figure 4C). As shown in Figure 4a, both NCL-1 and NCD-38 increased the expression of BiP compared with control. Activation of UPR results in autophosphorylation of PERK and IRE1α and further activation of downstream targets.41,42 We then determined KDM1A inhibition increases the expression of UPR components by western blot analysis. Our results demonstrated that KDM1A inhibitors increased the phosphorylation of IRE1α and PERK and the expression of downstream molecules such as ATF4 and CHOP in GSCs (Figure 4a). Activated IRE1α process the intron from XBP1 mRNA there by generating spliced XBP1, which encodes an active transcription factor. In consistent with phosphorylation of IRE1-α , our results demonstrated that KDM1A inhibitors NCL-1 and NCD-38 induces XBP1 splicing in GSCs (Figure 4b). To examine whether NCL-1 and NCD-38 inhibition of the KDM1A demethylase function affects the alteration in epigenetic modifications at the endoplasmic reticulum (ER) stress response element (ERSE) on BiP promoter and ATF4 response element on the ATF3 and DDIT3 promoters, the status of the H3K4me2 marks were examined in GSCs that are treated with control or KDM1A inhibitors. Chromatin immunoprecipitation (ChIP) assay results revealed significant enrichment of H3K4me2 at the promoters of the UPR stress genes BiP, ATF3 and DDIT3 in U251-GSCs following treatment with KDM1A inhibitors (Figure 4c). To confirm the induction of UPR during differentiation of GSCs, the expression of UPR genes were examined in undifferentiated and differentiated GSCs. As shown Figure 4d, the expression of UPR components ATF3, ATF4, PPP1R15A, HERPUD1 and DDIT3 was significantly increased in differentiated cells than in undifferentiated GSCs. These results suggest that KDM1A inhibitors significantly induce the UPR stress genes by modifying epigenetic marks at target gene promoters, leading to the differentiation and apoptosis of GSCs.
Figure 4.
KDM1A inhibition leads to activation of UPR. (a) Replicate cultures of GSC10 and U251-GSCs were treated with NCL-1 (20 μM) and NCD-38 (2 μM) for 72 h and the expression levels of BiP, p-IRE1α, p-PERK, CHOP and ATF4 were determined using western blotting. (b) Replicate cultures of GSC10 and U251-GSCs were treated with NCL-1 (20 μM) and NCD-38 (2 μM) for 24 h and Xbp1 splicing was determined by RT-PCR. (c) Replicate cultures of U251-GSCs were treated with vehicle, NCL-1 (20 μM) or NCD-38 (2 μM) for 24 h, and the status of the activation marker H3K4me2 at the promoters of BiP, ATF3 and DDIT3 was determined by chromatin immunoprecipitation. (d) Replicate cultures of GSC10 were subjected to serum induced differentiation for 7 days and the expression of UPR pathway genes were determined in stem and differentiated cells using RT-qPCR. All experiments were representative of two independent experiments. Data are represented as mean ±s.e. *P < 0.05, **P < 0.01, ***P < 0.001 based on the Student’s t-test.
KDM1A inhibition reduced the in vivo tumor progression and increased the survival of tumor-bearing mice
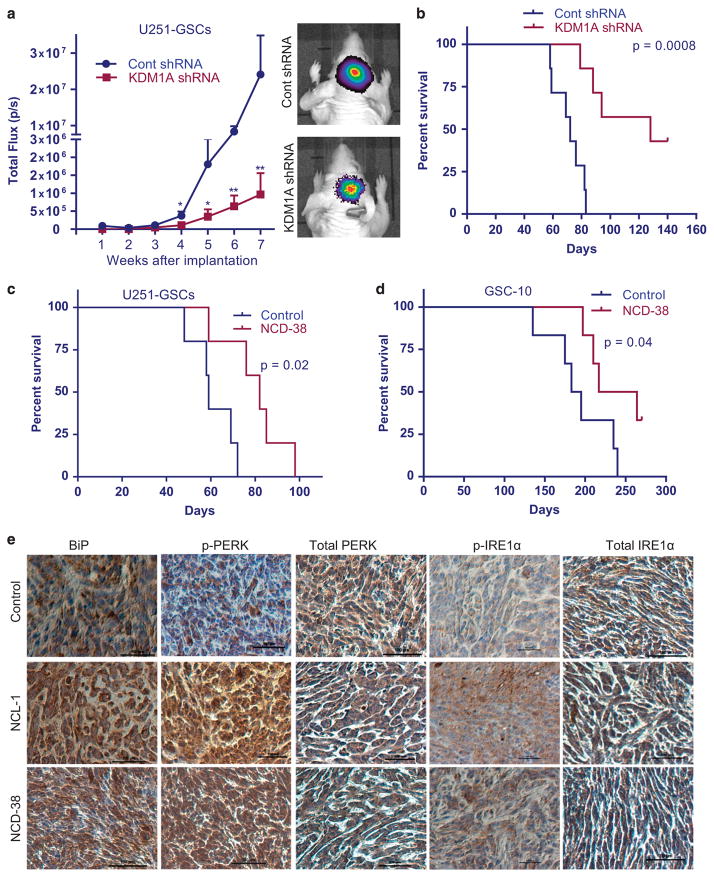
As our in vitro results demonstrated that KDM1A is essential for GSCs maintenance and survival, we evaluated whether KDM1A inhibition in vivo will have therapeutic benefit. To determine whether knockdown of KDM1A suppresses the GSCs-mediated tumor progression, GFP-luciferase-labeled control shRNA and KDM1A-shRNA U251-GSCs were implanted intracranially in immunocompromised mice, and tumor growth in terms of luciferase intensity was measured weekly using Xenogen in vivo imaging systems (IVIS) (PerkinElmer, Waltham, MA, USA). KDM1A knockdown significantly reduced the U251-GSCs-driven tumor progression when compared with progression in the control mice (Figure 5a). Compared with the survival of control mice, KDM1A knockdown significantly increased overall survival of the mice (Figure 5b). To determine the therapeutic benefit of KDM1A inhibitors on GSCs tumor growth, GSCs were implanted into the mouse brains. After tumors were established, the mice were randomized into a control group, which received vehicle, and a treatment group, which received NCL-1 or NCD-38 through oral gavage. Compared with the vehicle, NCL-1 or NCD-38 treatment significantly reduced the in vivo tumor growth of U251-GSCs (Supplementary Figures 4A and B). Further, NCD-38 treatment significantly increased the overall survival of U251-GSCs and GSC10 tumorbearing mice compared with controls (Figures 5c and d). To examine whether KDM1A inhibitors induces UPR activation in vivo, the expression of BiP and phosphorylation of PERK and IRE1α was tested immunohistochemically on tumor sections. NCL-1 and NCD-38 treatment significantly increased the expression of BiP, p-PERK and p-IRE1α compared with control suggesting the activation of UPR in KDM1A inhibitor-treated tumors (Figure 5e). Further, to examine whether KDM1A inhibitors caused any toxicity, we have analyzed various organs histopathologically. We did not observe any damage to spleen, liver, heart and kidney in NCL-1 and NCD-38 treatment groups compared with controls (Supplementary Figure 5A). In addition, we also determined the expression of proliferation marker Ki67 in the spleen and no significant differences in the number of Ki67 positive cells in control and KDM1A inhibitor-treated group were observed (Supplementary Figure 5B).
Figure 5.
KDM1A inhibition reduced GSCs tumor growth and increased mice survival. (a) U251-GSCs stably expressing GFP-luciferase were transduced with control shRNA and KDM1A-shRNA and implanted intracranially into the right cerebrum of nude mice. Tumor growth was monitored using luciferase intensity and was measured using Xenogen IVIS imaging every week (n =7). (a) Representative images of tumor-bearing mice are shown. (b) Survival of the mice was determined using Kaplan–Meier analysis (n =7). (c, d) U251-GSCs (n =5) and GSC10 (n =6) were implanted orthotopically into the brain and after the tumors established, mice were treated with vehicle or NCD-38 (5 mg/kg body weight/day). Survival of the mice was determined using Kaplan–Meier survival analysis. GSC10 implanted mice were treated with NCD-38 for 75 days. (e) Tumor sections collected from control or NCL-1-, or NCD-38-treated U251-GSCs groups were processed and subjected to immunohistochemical staining for BiP, p-IRE1α, total IRE1α, p-PERK and total PERK as described in ‘Materials and methods’ section. Data are represented as mean ±s.e. *P < 0.05, **P < 0.01 based on Mann–Whitney t-test.
KDM1A inhibition reduced stemness and induced differentiation and apoptosis in vivo
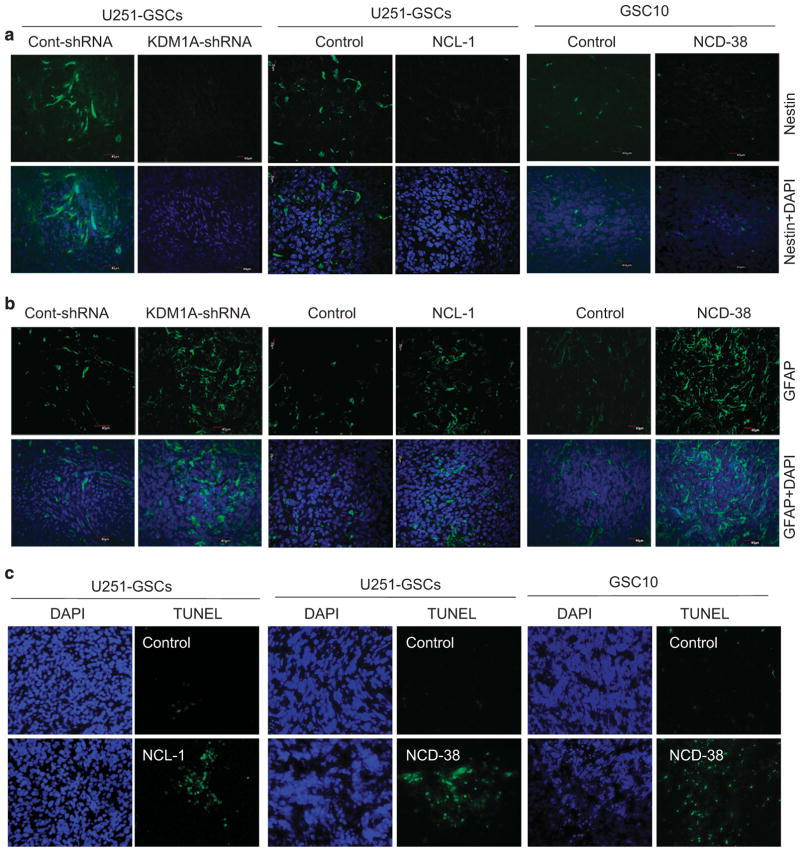
To examine the effect of inhibition of KDM1A on stemness and differentiation in vivo, tumor sections derived from control shRNA and KDM1A-shRNA U251-GSCs tumors and vehicle- or KDM1A inhibitors-treated U251-GSCs and GSC10 tumors were analyzed using immunofluorescence. KDM1A knockdown or its pharmacological inhibition significantly reduced the expression of nestin in tumors (Figure 6a). Further, the expression of GFAP was significantly higher in KDM1A knockdown and KDM1A inhibitor-treated tumors than in control tumors (Figure 6b). To determine whether KDM1A inhibition induces apoptosis in vivo, tumor sections from vehicle or KDM1A inhibitor-treated U251-GSCs and GSC10 tumors were analyzed by using the TUNEL assay. The number of apoptotic cells was significantly higher in KDM1A inhibitor-treated tumors than in control tumors (Figure 6c). These results suggest that KDM1A inhibition decreased the stemness of GSCs and concomitantly induced their differentiation and apoptosis. Further to determine whether KDM1A inhibitors affect the tumor growth by altering epigenetic modifications, the status of global H3K4 and H3K9 methylation were examined in tumors. Immunohistochemical analysis showed that the expression of global H3K4me2 was substantially increased and H3K9me2 was substantially decreased in NCD-38-treated tumors than in control tumors (Supplementary Figure 6).
Figure 6.
KDM1A inhibitors reduced stemness and induced differentiation of GSCs in vivo. Tumor sections collected from control or KDM1shRNA-, NCL-1- or NCD-38-treated groups were processed and subjected to immunofluorescence staining for nestin (a) and GFAP (b) and TUNEL assay (c) as described. DAPI was used to visualize the nuclei.
DISCUSSION
The lysine demethylase KDM1A is overexpressed in several human malignancies including GBM,24,27,28,31,48 and emerging studies suggest that KDM1A is essential for stem cell maintenance and survival.32–34 As epigenetic changes are reversible, selective targeting of KDM1A is of high importance and could be a viable therapy. In the present study, we examined the potent novel KDM1A-specific inhibitors NCL-1 and NCD-38 as potential therapeutic agents to target GSCs. Our results suggest that (1) KDM1A is highly expressed in GSCs; (2) inhibition of KDM1A reduce GSCs stemness and induce the differentiation and apoptosis; (3) KDM1A inhibitors induce the UPR pathway activation and (4) KDM1A inhibitors decrease GSCs-driven tumor progression and improve mice survival. Thus, these results suggest that therapeutic targeting of GSCs with potent KDM1A inhibitors could be a potential therapy for GBM.
In our previous study, we found that KDM1A is overexpressed in glioma and correlated with the histological malignancy, and that pharmacological inhibition of KDM1A resulted in an antitumor effect in a subcutaneous GBM model. Recent studies demonstrated that KDM1A and HDAC inhibitors have synergistic activity on GBM,49 and that KDM1A is essential for maintenance of the undifferentiated state of human embryonic stem cells,32 and regulates neural stem cell proliferation and differentiation.50 Further, KDM1A regulates the stemness and chemoresistance of liver cancer initiating cells.51 In this study, we found that KDM1A is highly expressed in undifferentiated GSCs and that KDM1A inhibitors had more potency in reducing cell viability of the undifferentiated GSCs than the differentiated cells. Further, KDM1A knockdown or pharmacological inhibition reduced the sphere formation ability and reduced the expression of nestin and sox2, and increased GFAP. Earlier studies demonstrated that inhibition of KDM1A promoted the osteogenic differentiation of human adipose-derived stem cells52 and blockade of KDM1A induced the differentiation of leukemia stem cells,53,54 further support the our conclusion that KDM1A blockade induces GSCs differentiation.
Monoamine oxidase (MAO) inhibitors were used as the first small molecular inhibitors of KDM1A because of the homology of MAOs amino acid sequence with catalytic domain of KDM1A,55 and the use of them significantly affects the cell proliferation of various cancer cells in vitro and in vivo.24,29,31 Given the importance of the MAO role in psychiatric and neurological disorders, use of non-selective KDM1A inhibitors in higher doses may cause side effects that limit their use as therapeutic agents.36 Therefore, specific inhibitors are required to selectively target KDM1A. We developed the potent KDM1A inhibitor NCL-1 that efficiently inhibits KDM1A but not type A and type B MAOs.38 NCL-1 inhibited KDM1A demethylase activity and decreased cell proliferation in prostate cancer, glioma and breast cancer cell lines31,56 with the IC50 range of 10–50 μM depending on cell type. Recently, we synthesized a series of novel KDM1A inhibitors based on the concept of direct delivery of phenylcyclopropylamine directly to the KDM1A active site.40 The compound 1t (NCD-38) showed higher KDM1A inhibitory activity and cancer cell growth inhibitory activity without affecting MAOA and MOAB. In our studies, the compound NCD-38 significantly inhibited the cell viability of GSCs with an IC50 range of 2–3 μM. More importantly, our in vivo studies demonstrated that KDM1A inhibitors NCL-1 and NCD-38 did not show any toxicities in various organs.
UPR is triggered in cells in response to stress in the endoplasmic reticulum caused by misfolded or unfolded protein accumulation in the ER due to altered cellular redox status, calcium homeostasis and increased synthesis of secretory proteins. The UPR stress is initiated by three transmembrane sensor proteins PERK, IRE1 and ATF6.42,57 The PERK directly phosphorylates the alpha subunit of eukaryotic initiation factor-2 α (eIF-2α ), leading to attenuation of general translation initiation and phosphorylated eIF-2α selectively translates UPR dependent genes including the transcription factor ATF4.45 ATF4 transactivates expression of several genes, such as C/EBP homologous protein (CHOP), GADD34 and ATF3.41 In addition, PERK also phosphorylates NRF2, which promotes the induction of antioxidant genes.58 An activated IRE1 splices the XBP1 mRNA, which encodes potent active transcription factor and ATF6 undergoes proteolytic cleavage in Golgi and the cleaved nuclear form translocate to nucleus. The transcription factors activated by all three UPR transducers cooperate to activate the UPR target genes. Our RNA-sequencing and IPA analysis demonstrated that the top three canonical pathways modulated by KDM1A inhibitor treatment include UPR, endoplasmic reticulum stress pathway and NRF2- mediated oxidative stress response. Validation studies confirmed many of the genes in UPR pathway were upregulated following KDM1A inhibitor treatment in GSCs. Most importantly, KDM1A inhibition resulted in increased active methylation mark H3K4me2 at the promoters of BiP, ATF3 and DDIT3, further suggesting a role of KDM1A-mediated epigenetic modifications in GSCs function. Ability of KDM1A inhibitors (1) to increase phosphorylation of PERK and IRE1α , (2) to increase splicing of XBP1 (3) to increase expression of GRP78, ATF4 and CHOP and (4) to activate multiple ATF6 target genes such as GRP78, HERPUD1, HYOU1, PDIA4, ERO1LB suggest that KDM1A inhibitors have potential to activate all major branches of UPR pathway. RNAseq data also showed alteration of genes in immune pathway such as IL17A and CD40. As KDM1A also known to have an important role in the regulation of multiple pathways via its interactions with NF-κB and C/EBPβ, induction of KDM1A suppressed genes is expected with treatment of KDM1A inhibitors. Even through this represent an important observation, future studies are needed to test the biological significance of their activation and is beyond the scope our present study.
UPR activation alleviates ER stress. If ER stress remains unresolved, apoptosis is induced.41 Recent studies also implicated ER stress in osteogenic differentiation.59 UPR has an important role in the stem cell differentiation in intestinal epithelium, and induction of ER stress causes the loss of stemness in a PERK-eIF-2α-dependent manner43 and UPR-induced differentiation of colon CSCs enhances response to chemotherapy in vitro and in vivo.60 Activation of UPR results in the differentiation of definitive endoderm cells from mouse ESCs and inhibition of UPR prevents the specific differentiation.61 UPR components Xbp1 and Hrd1 restrict the proliferative activity of ISCs in the intestinal epithelium.62 In addition, ER stress is also implicated in the embryonic development of CNS,44 and all three branches of UPR were induced during neuronal differentiation of rat bone marrow stromal cells and mouse embryonic stem cells.63 In consistent with these studies, our results suggest that UPR activation by KDM1A inhibitors may contribute to decrease in stemness and induction of differentiation and apoptosis of GSCs. We also observed activation of UPR (increased expression of ATF3, ATF4, HERPUD1, EIF2AK3 and DDIT3) when GSCs are allowed to differentiate with the addition of serum. However, this differentiation did not promoted apoptosis, possibly due to simultaneous activation of survival pathways by serum. However, KDM1A inhibitors mediated differentiation of GSCs promoted both UPR activation and apoptosis, suggesting KDM1A inhibitor action is complex and may involve activation/blockage of multiple pathways.
It has been recently shown that partial silencing of KDM1A activates H3K4me3 at MYC locus resulting in increased MYC expression and tumorigenicity, in contrast efficient knockdown of KDM1A induces cell death in vitro.64 However, other recent studies demonstrated that KDM1A is overexpressed in GSCs compared with normal progenitor cells. Further mRNA and protein levels are reduced in serum induced differentiated glioblastoma cells (DGCs) and synthetic KDM1A inhibitor S2101 reduced viability of stem-like tumor propagating cells (TPCs) with no effect on DGCs in vitro.49,65 In consistent with these studies, we also observed that serum induced differentiation resulted reduction of KDM1A levels in GSCs derived from several primary tumors. KDM1A inhibitors NCL-1 and NCD-38 significantly reduced the viability of GSCs with little effect on differentiated cells. Further, our studies provided first in vivo proof that both NCL-1 and NCD-38 also reduce in vivo tumor progression by GSCs.
In conclusion, our study demonstrated that KDM1A inhibition using novel KDM1A-specific inhibitors reduced the stemness and induced the differentiation and apoptosis of GSCs in vitro and in in vivo by inducing the activation of UPR. NCL-1 and NCD-38 reduced the in vivo tumor growth in intracranial models and extended mice survival. These findings suggest that selective targeting of KDM1A using NCL-1 and NCD-38 may be a promising therapeutic strategy for GSCs.
MATERIALS AND METHODS
Cell culture, reagents and generation KDM1A-shRNA cells
Human GBM cell line U251 was obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA). Cells were maintained in DMEM supplemented with 10% fetal bovine serum (Sigma Chemical, St Louis, MO, USA). U251 cells were passaged in our laboratory for fewer than 6 months after receipt or resuscitation. Neurobasal medium and B27 serum-free supplement were obtained from Invitrogen (Carlsbad, CA, USA). KDM1A antibody (Cat # A300-215A) was purchased from Bethyl Laboratories (Montgomery, TX, USA). The nestin (Cat # 4760), sox-2 (Cat # 3579), GFAP (Cat # 12389), CHOP (Cat # 2895), ATF4 (Cat # 11815), BiP (Cat # 3177) and GAPDH (Cat # 8884) antibodies were obtained from Cell Signaling Technology (Beverly, MA, USA), p-PERK (Cat # sc-32577), nestin (Cat # sc-377380), PERK (Cat # sc-377400), IRE1α (Cat # sc-20790) antibodies were obtained from Santa Cruz Biotechnology (Dallas, TX, USA), p-IRE1-α (Cat # NB-100-2323) and IgG isotype control (Cat # NBP2-24891) antibodies were obtained from Novus Biologicals (Littleton, CO, USA) and Ki67 antibody (Cat # ab1667) was purchased from Abcam (Cambridge, MA, USA). KDM1A-specific shRNA lentiviral plasmids, β-actin (Cat # A2066) and all secondary antibodies were purchased from Sigma Chemical. H3K4me2 (Cat # 39141), H3K9me2 (Cat # 07-441) antibodies were purchased from Active Motif (Carlsbad, CA, USA) and Millipore (Billerica, MA, USA), respectively. GSCs stably expressing KDM1A-shRNA were generated by infecting cells with human-specific KDM1A-shRNA lentivirus, and stable clones were selected with puromycin (1 μg/ml). Lentiviral particles expressing non-targeted shRNA were used to generate control cells. CD133 antibody was obtained from Miltenyi Biotech and EGF, bFGF and LIF were purchased from BioLegend (San Diego, CA, USA).
Isolation of GSCs
Patient-derived primary GBM cells were isolated from discarded specimens obtained from patients undergoing surgery using IRB approved protocol and no clinical linkers or codes for the specimens were accessible to any research personnel or the PI. Patients provided informed consent for surgery and use of their tissues for research. Briefly, fresh tumor tissue was collected intraoperatively, dispersed into single cells, and cultured briefly in a neurobasal media for expansion and purification followed by intracranial injection in nude mice. The resulting tumors were characterized for consistency by histology with the primary tumor, growth attributes including time to animal demise, and for gene expression profiling for the molecular subtype. Primary GBM lines GBM08, GBM09, GBM10 and GBM11 were cultured in neurobasal medium supplemented with B27 serum-free supplement, EGF (20 ng/ml), bFGF (20 ng/ml), LIF (10 ng/ml) and heparin (5 μg/ml) as described previously.66 GSCs were enriched using established stem cells marker CD133 from primary GBMs and U251-GBM cells by labeling with PE-conjugated CD133 antibody and sorting by flow cytometry. Although other putative stem cell markers were used previously to enrich GSCs including SSEA-1,67 CD4468 and integrin α 669 and L1CAM,70 CD133 has been shown to be a widely used marker of self-renewing and tumorigenic GBM cells,6,71–73 therefore, we have used this marker in our studies. To induce the differentiation, CD133-postive GSCs were cultured in 10% FBS for 7 days. GSCs isolated from GBM08, GBM09, GBM10 and GBM11 were called as GSC08, GSC09, GSC10 and GSC11, respectively.
Neurosphere and cell viability assays
For neurosphere assays, single cell suspensions of GSCs were seeded in 24-well plates (100 cells per well) in triplicates and treated with vehicle, NCL-1 (20 μM) or NCD-38 (2 μM) for 7 days. Then, newly formed neurospheres were counted in each well and the values were plotted on a histogram. The effect of KDM1A inhibitors on the viability of GSCs was determined using CellTiter-Glo Luminescent Cell Viability Assay (Promega, Madison, WI, USA) according to the manufacturer’s instructions. Undifferentiated GSCs and differentiated GSCs were seeded in 96-well, fiat, clear-bottom, opaque-wall microplates and treated with vehicle, NCL-1 or NCD-38 for 72 h. The total ATP content as an estimate of total number of viable cells was measured on an automatic Fluoroskan Luminometer (Thermo Scientific, Waltham, MA, USA).
Annexin V apoptosis assay
The effect of the KDM1A inhibitors NCL-1 and NCD-38 on GSCs apoptosis was analyzed using the Annexin V/PI kit as per the manufacturer’s instructions (BioLegend). Briefly, GSCs were treated with either vehicle, NCL-1 (25 μM) or NCD-38 (2.5 μM) for 48 h and harvested cells at a density of 2.5 × 106 cells/ml in Annexin V binding buffer. Then, 100 μl of the cell suspension was incubated with Annexin V-FITC and PI for 15 min at room temperature in the dark. Annexin V binding buffer (400 μl) was then added to each sample and stained cells were analyzed using flow cytometry.
Western blotting
Whole cell lysates were prepared from GSCs using RIPA buffer containing protease and phosphatase inhibitors (Sigma Chemical). Total proteins (50 μg) were mixed with SDS sample buffer and subjected to SDS–PAGE, and the resolved proteins were transferred onto nitrocellulose membranes. Blots were then blocked with 5% non-fat dry milk powder for 1 h at room temperature and incubated with primary antibodies over night at 4 °C followed by incubation with secondary antibody for 1 h at room temperature. Blots were developed using the ECL kit (Thermo Scientific, Waltham, MA, USA).
RNA-sequencing and qRT-PCR
GSC10 were treated with either vehicle or NCD-38 (2 μM) for 24 h, and total RNA was isolated using RNAesy mini kit according to the manufacturer’s instructions (Qiagen, Valencia, CA, USA). The RNA purity was determined using Agilent 2100 BioAnalyzer (Agilent Technologies, Santa Clara, CA, USA). Illumina TruSeq RNA Sample preparation was conducted according to the manufacturer's instructions, and the samples were run on an Illumina HiSeq 2000 (Illumina, Inc., San Diego, CA, USA) in duplicates. RNA-sequencing was performed as described previously.66 Differential expression analysis was performed by DEseq, and significant genes with a twofold change with P < 0.01 were chosen for analysis. The interpretation of biological significance of differentially expressed genes was determined with IPA. To validate the selected genes RNA was subjected to reverse transcription reactions using SuperScript III First Strand kit (Invitrogen), according to the manufacturer’s protocol. Real-time PCR was performed with SYBR Green (Thermo Scientific) on an Illumina RT-PCR system. Data were normalized to β-actin and the difference in fold change was calculated using delta-delta-CT method. Primer sequences are provided in the Supplementary Table 1. The RNAseq data was deposited to GEO data base with GEO accession# GSE84603.
XBP1 mRNA splicing assay
GSC10 and U251-GSCs were treated with either vehicle or NCL-1 (20 μM) or NCD-38 (2 μM) for 24 h, and total RNA was isolated using RNAesy mini kit using manufacturer’s protocol. First-strand cDNA was synthesized using SuperScript III First Strand kit (Invitrogen). Unspliced and spliced Xbp1 cDNA fragments were amplified by PCR using the following primers, XBP1u/XBP1s-F-5′-CCTGGTTGCTGAAGAGGAGG-3′, XBP1u/XBP1s-R-5′-CCA TGGGGAGATGTTCTGGAG-3′. Unspliced and spliced fragments were separated by 3% agarose gel and stained with ethidium bromide.
Immunohistochemistry and TUNEL assay
Immunohistochemical studies were performed as described previously.74 Tumor sections were incubated overnight with H3K4me2, H3K9me2, p-IRE1α , p-PERK and BiP primary antibodies followed by secondary antibody incubation for 45 min at room temperature and positivity was visualized by using the DAB substrate and counterstained with haematoxylin (Vector Lab, Burlingame, CA, USA). Antibody specificity was validated using isotype control primary antibody and also by omitting the primary antibody (Supplementary Figure 4D). For the immunofluorescence studies, tissue sections were blocked in 5% normal goat serum followed by incubation with primary antibodies and fluorochrome-conjugated secondary antibody (Alexa Fluor 488). DAPI was used to visualize the nuclei and images were captured using confocal microscopy. TUNEL assays were performed using In situ Cell Death Detection Kit, Fluorescein (Roche, Indianapolis, IN, USA) utilizing manufacturer’s protocol as described previously.31
Chromatin immunoprecipitation assay
U251-GSCs were treated with vehicle, NCL-1 (20 μM) or NCD-38 (2 μM) for 24 h, and a ChIP assay was performed using Pierce Magnetic ChIP Kit using manufacturer’s protocol (Thermo Scientific). Briefly, cells were cross-linked using 1% formaldehyde, and the extracted chromatin was subjected to immunoprecipitation using H3K4me2 or isotype-specific control IgG antibodies overnight at 4 °C. Extracted DNA was then subjected to real-time PCR using ERSE (ER stress response elements) of the grp78 promoter F-5′-CGGAGCAGTGACGTTTAT-3′, R-5′-GTCGCCT ACTCGGCTTAT-3′ and ATF3, DDIT3 specific ATF4 response element primers: ATF3-F-5′- (−690) CAGATCTGGCTTGGGTGTTT-3′, ATF3-R-5′- (−470) GGTGGAGGAGTGTTTGCATT-3′; DDIT3-F-5′- (−174) ACAGCTTAGATGGGA GGCTG-3′, DDIT3-R-5′- (+25) AAAACATGGTTGAACAGCAAAA-3′.
In vivo orthotopic tumor model
All animal experiments were performed after obtaining IACUC approval, and by using UTHSCSA institution’s protocol guidelines. Male mice (athymic nude mice, homozygous) of 8 weeks’ old were purchased from Charles River (Wilmington, MO, USA). GSCs were labeled with GFP-Luciferase reporter and CD133-positive GSC10 (1 × 104) and U251-GSCs (5 × 104) were injected orthotopically into the mouse right cerebrum. Once the tumor is established, mice were randomized into control or treatment groups based on tumor volume. Investigators were not blinded in the animal studies. A sample size of mice/treatment was derived using information from previous studies and with the assumption of equal variance for the two groups and two-sided significance level 0.05. The control group received vehicle (30% Captisol) and the treatment group received NCL-1 (10 mg/kg body weight/day) or NCD-38 (5 mg/kg body weight/day) orally in 30% Captisol. Tumor growth was measured weekly using the Xenogen IVIS system. After treatment, the mice were euthanized, and brains were collected and processed for histological studies. For KDM1A knockdown experiments, GFP-Luciferase labeled U251-GSCs were infected with control shRNA or KDM1A-shRNA lentivirus and selected with puromycin (1 μg/ml) before injection. Mouse survival was determined using Kaplan–Meier survival curves and log-rank test using GraphPad Prism 6 software (GraphPad Software, San Diego, CA, USA).
Statistical analyses
Statistical differences between groups were analyzed with unpaired Student’s t-test using GraphPad Prism 6 software (GraphPad Software). All in vitro assays were performed in triplicate and repeated at least three times. RNA-seq data were analyzed using IPA software. Log-rank (Kaplan–Meier) test was analyzed using GraphPad Prism. All the data represented in plots are shown as means ± s.e. A value of P < 0.05 was considered as statistically significant.
Supplementary Material
Acknowledgments
This study was supported by the 2014 AACR-AMGEN Inc. fellowship in clinical/translational cancer research (Grant Number: 14-40-11-SARE) and CPRIT training grant (RP140105) awarded to GRS and NCI Grant CA178499 (RKV and AB).
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Supplementary Information accompanies this paper on the Oncogene website (http://www.nature.com/onc)
References
- 1.Johnson DR, O'Neill BP. Glioblastoma survival in the United States before and during the temozolomide era. J Neurooncol. 2012;107:359–364. doi: 10.1007/s11060-011-0749-4. [DOI] [PubMed] [Google Scholar]
- 2.Ostrom QT, Gittleman H, Liao P, Rouse C, Chen Y, Dowling J, et al. CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2007–2011. Neuro Oncol. 2014;16:iv1–63. doi: 10.1093/neuonc/nou223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonavia R, Inda MM, Cavenee WK, Furnari FB. Heterogeneity maintenance in glioblastoma: a social network. Cancer Res. 2011;71:4055–4060. doi: 10.1158/0008-5472.CAN-11-0153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu G, Yuan X, Zeng Z, Tunici P, Ng H, Abdulkadir IR, et al. Analysis of gene expression and chemoresistance of CD133+ cancer stem cells in glioblastoma. Mol Cancer. 2006;5:67. doi: 10.1186/1476-4598-5-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee Y, Kim KH, Kim DG, Cho HJ, Kim Y, Rheey J, et al. FoxM1 promotes stemness and radio-resistance of glioblastoma by regulating the master stem cell regulator Sox2. PLoS One. 2015;10:e0137703. doi: 10.1371/journal.pone.0137703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 7.Magee JA, Piskounova E, Morrison SJ. Cancer stem cells: impact, heterogeneity, and uncertainty. Cancer Cell. 2012;20(21):283–296. doi: 10.1016/j.ccr.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Venere M, Fine HA, Dirks PB, Rich JN. Cancer stem cells in gliomas: identifying and understanding the apex cell in cancer's hierarchy. Glia. 2011;59:1148–1154. doi: 10.1002/glia.21185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Galli R, Binda E, Orfanelli U, Cipelletti B, Gritti A, De VS, et al. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res. 2004;64:7011–7021. doi: 10.1158/0008-5472.CAN-04-1364. [DOI] [PubMed] [Google Scholar]
- 10.Mukherjee B, McEllin B, Camacho CV, Tomimatsu N, Sirasanagandala S, Nanne-paga S, et al. EGFRvIII and DNA double-strand break repair: a molecular mechanism for radioresistance in glioblastoma. Cancer Res. 2009;69:4252–4259. doi: 10.1158/0008-5472.CAN-08-4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sato A, Sunayama J, Matsuda K, Seino S, Suzuki K, Watanabe E, et al. MEK-ERK signaling dictates DNA-repair gene MGMT expression and temozolomide resistance of stem-like glioblastoma cells via the MDM2-p53 axis. Stem Cells. 2011;29:1942–1951. doi: 10.1002/stem.753. [DOI] [PubMed] [Google Scholar]
- 12.Bao S, Wu Q, Sathornsumetee S, Hao Y, Li Z, Hjelmeland AB, et al. Stem cell-like glioma cells promote tumor angiogenesis through vascular endothelial growth factor. Cancer Res. 2006;66:7843–7848. doi: 10.1158/0008-5472.CAN-06-1010. [DOI] [PubMed] [Google Scholar]
- 13.Vescovi AL, Galli R, Reynolds BA. Brain tumour stem cells. Nat Rev Cancer. 2006;6:425–436. doi: 10.1038/nrc1889. [DOI] [PubMed] [Google Scholar]
- 14.Khan IS, Ehtesham M. Targeting glioblastoma cancer stem cells: the next great hope? Neurosurg Focus. 2014;37:E7. doi: 10.3171/2014.9.focus14509. [DOI] [PubMed] [Google Scholar]
- 15.Piccirillo SG, Reynolds BA, Zanetti N, Lamorte G, Binda E, Broggi G, et al. Bone morphogenetic proteins inhibit the tumorigenic potential of human brain tumour-initiating cells. Nature. 2006;444:761–765. doi: 10.1038/nature05349. [DOI] [PubMed] [Google Scholar]
- 16.Crespo I, Vital AL, Gonzalez-Tablas M, Patino MC, Otero A, Lopes MC, et al. Molecular and genomic alterations in glioblastoma multiforme. Am J Pathol. 2015;185:1820–1833. doi: 10.1016/j.ajpath.2015.02.023. [DOI] [PubMed] [Google Scholar]
- 17.Noushmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, Berman BP, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17:510–522. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kondo Y, Katsushima K, Ohka F, Natsume A, Shinjo K. Epigenetic dysregulation in glioma. Cancer Sci. 2014;105:363–369. doi: 10.1111/cas.12379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol. 2005;6:838–849. doi: 10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- 20.Hojfeldt JW, Agger K, Helin K. Histone lysine demethylases as targets for antic-ancer therapy. Nat Rev Drug Discov. 2013;12:917–930. doi: 10.1038/nrd4154. [DOI] [PubMed] [Google Scholar]
- 21.Lan F, Nottke AC, Shi Y. Mechanisms involved in the regulation of histone lysine demethylases. Curr Opin Cell Biol. 2008;20:316–325. doi: 10.1016/j.ceb.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Metzger E, Wissmann M, Yin N, Muller JM, Schneider R, Peters AH, et al. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437:436–439. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- 23.Nair SS, Nair BC, Cortez V, Chakravarty D, Metzger E, Schule R, et al. PELP1 is a reader of histone H3 methylation that facilitates oestrogen receptor-alpha target gene activation by regulating lysine demethylase 1 specificity. EMBO Rep. 2010;11:438–444. doi: 10.1038/embor.2010.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schulte JH, Lim S, Schramm A, Friedrichs N, Koster J, Versteeg R, et al. Lysine-specific demethylase 1 is strongly expressed in poorly differentiated neuro-blastoma: implications for therapy. Cancer Res. 2009;69:2065–2071. doi: 10.1158/0008-5472.CAN-08-1735. [DOI] [PubMed] [Google Scholar]
- 25.Ding J, Zhang ZM, Xia Y, Liao GQ, Pan Y, Liu S, et al. LSD1-mediated epigenetic modification contributes to proliferation and metastasis of colon cancer. Br J Cancer. 2013;109:994–1003. doi: 10.1038/bjc.2013.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lim S, Janzer A, Becker A, Zimmer A, Schule R, Buettner R, et al. Lysine-specific demethylase 1 (LSD1) is highly expressed in ER-negative breast cancers and a biomarker predicting aggressive biology. Carcinogenesis. 2010;31:512–520. doi: 10.1093/carcin/bgp324. [DOI] [PubMed] [Google Scholar]
- 27.Shao G, Wang J, Li Y, Liu X, Xie X, Wan X, et al. Lysine-specific demethylase 1 mediates epidermal growth factor signaling to promote cell migration in ovarian cancer cells. Sci Rep. 2015;5:15344. doi: 10.1038/srep15344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hayami S, Kelly JD, Cho HS, Yoshimatsu M, Unoki M, Tsunoda T, et al. Over-expression of LSD1 contributes to human carcinogenesis through chromatin regulation in various cancers. Int J Cancer. 2011;128:574–586. doi: 10.1002/ijc.25349. [DOI] [PubMed] [Google Scholar]
- 29.Wang M, Liu X, Guo J, Weng X, Jiang G, Wang Z, et al. Inhibition of LSD1 by Pargyline inhibited process of EMT and delayed progression of prostate cancer in vivo. Biochem Biophys Res Commun. 2015;467:310–315. doi: 10.1016/j.bbrc.2015.09.164. [DOI] [PubMed] [Google Scholar]
- 30.Zhao ZK, Yu HF, Wang DR, Dong P, Chen L, Wu WG, et al. Overexpression of lysine specific demethylase 1 predicts worse prognosis in primary hepatocellular carcinoma patients. World J Gastroenterol. 2012;18:6651–6656. doi: 10.3748/wjg.v18.i45.6651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sareddy GR, Nair BC, Krishnan SK, Gonugunta VK, Zhang QG, Suzuki T, et al. KDM1 is a novel therapeutic target for the treatment of gliomas. Oncotarget. 2013;4:18–28. doi: 10.18632/oncotarget.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Adamo A, Sese B, Boue S, Castano J, Paramonov I, Barrero MJ, et al. LSD1 regulates the balance between self-renewal and differentiation in human embryonic stem cells. Nat Cell Biol. 2011;13:652–659. doi: 10.1038/ncb2246. [DOI] [PubMed] [Google Scholar]
- 33.Sun G, Alzayady K, Stewart R, Ye P, Yang S, Li W, et al. Histone demethylase LSD1 regulates neural stem cell proliferation. Mol Cell Biol. 2010;30:1997–2005. doi: 10.1128/MCB.01116-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harris WJ, Huang X, Lynch JT, Spencer GJ, Hitchin JR, Li Y, et al. The histone demethylase KDM1A sustains the oncogenic potential of MLL-AF9 leukemia stem cells. Cancer Cell. 2012;21:473–487. doi: 10.1016/j.ccr.2012.03.014. [DOI] [PubMed] [Google Scholar]
- 35.Wang J, Lu F, Ren Q, Sun H, Xu Z, Lan R, et al. Novel histone demethylase LSD1 inhibitors selectively target cancer cells with pluripotent stem cell properties. Cancer Res. 2011;71:7238–7249. doi: 10.1158/0008-5472.CAN-11-0896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang M, Culhane JC, Szewczuk LM, Jalili P, Ball HL, Machius M, et al. Structural basis for the inhibition of the LSD1 histone demethylase by the antidepressant trans-2-phenylcyclopropylamine. Biochemistry. 2007;46:8058–8065. doi: 10.1021/bi700664y. [DOI] [PubMed] [Google Scholar]
- 37.Ogasawara D, Suzuki T, Mino K, Ueda R, Khan MN, Matsubara T, et al. Synthesis and biological activity of optically active NCL-1, a lysine-specific demethylase 1 selective inhibitor. Bioorg Med Chem. 2011;19:3702–3708. doi: 10.1016/j.bmc.2010.12.024. [DOI] [PubMed] [Google Scholar]
- 38.Ueda R, Suzuki T, Mino K, Tsumoto H, Nakagawa H, Hasegawa M, et al. Identification of cell-active lysine specific demethylase 1-selective inhibitors. J Am Chem Soc. 2009;131:17536–17537. doi: 10.1021/ja907055q. [DOI] [PubMed] [Google Scholar]
- 39.Pajtler KW, Weingarten C, Thor T, Kunkele A, Heukamp LC, Buttner R, et al. The KDM1A histone demethylase is a promising new target for the epigenetic therapy of medulloblastoma. Acta Neuropathol Commun. 2013;1:19. doi: 10.1186/2051-5960-1-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ogasawara D, Itoh Y, Tsumoto H, Kakizawa T, Mino K, Fukuhara K, et al. Lysine-specific demethylase 1-selective inactivators: protein-targeted drug delivery mechanism. Angew Chem Int Ed Engl. 2013;52:8620–8624. doi: 10.1002/anie.201303999. [DOI] [PubMed] [Google Scholar]
- 41.Sano R, Reed JC. ER stress-induced cell death mechanisms. Biochim Biophys Acta. 2013;1833:3460–3470. doi: 10.1016/j.bbamcr.2013.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoshida H. ER stress and diseases. FEBS J. 2007;274:630–658. doi: 10.1111/j.1742-4658.2007.05639.x. [DOI] [PubMed] [Google Scholar]
- 43.Heijmans J, van Lidth de Jeude JF, Koo BK, Rosekrans SL, Wielenga MC, van de Wetering M, et al. ER stress causes rapid loss of intestinal epithelial stemness through activation of the unfolded protein response. Cell Rep. 2013;3:1128–1139. doi: 10.1016/j.celrep.2013.02.031. [DOI] [PubMed] [Google Scholar]
- 44.Zhang X, Szabo E, Michalak M, Opas M. Endoplasmic reticulum stress during the embryonic development of the central nervous system in the mouse. Int J Dev Neurosci. 2007;25:455–463. doi: 10.1016/j.ijdevneu.2007.08.007. [DOI] [PubMed] [Google Scholar]
- 45.Saito A, Ochiai K, Kondo S, Tsumagari K, Murakami T, Cavener DR, et al. Endoplasmic reticulum stress response mediated by the PERK-eIF2(alpha)-ATF4 pathway is involved in osteoblast differentiation induced by BMP2. J Biol Chem. 2011;286:4809–4818. doi: 10.1074/jbc.M110.152900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Baumeister P, Luo S, Skarnes WC, Sui G, Seto E, Shi Y, et al. Endoplasmic reticulum stress induction of the Grp78/BiP promoter: activating mechanisms mediated by YY1 and its interactive chromatin modifiers. Mol Cell Biol. 2005;25:4529–4540. doi: 10.1128/MCB.25.11.4529-4540.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kaufman RJ. Orchestrating the unfolded protein response in health and disease. J Clin Invest. 2002;110:1389–1398. doi: 10.1172/JCI16886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao X, Huang Q, Jin Y. Gold nanorod delivery of LSD1 siRNA induces human mesenchymal stem cell differentiation. Mater Sci Eng C Mater Biol Appl. 2015;54:142–149. doi: 10.1016/j.msec.2015.05.013. [DOI] [PubMed] [Google Scholar]
- 49.Singh MM, Manton CA, Bhat KP, Tsai WW, Aldape K, Barton MC, et al. Inhibition of LSD1 sensitizes glioblastoma cells to histone deacetylase inhibitors. NeuroOncol. 2011;13:894–903. doi: 10.1093/neuonc/nor049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Amente S, Lania L, Majello B. The histone LSD1 demethylase in stemness and cancer transcription programs. Biochim Biophys Acta. 2013;1829:981–986. doi: 10.1016/j.bbagrm.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 51.Lei ZJ, Wang J, Xiao HL, Guo Y, Wang T, Li Q, et al. Lysine-specific demethylase 1 promotes the stemness and chemoresistance of Lgr5(+) liver cancer initiating cells by suppressing negative regulators of beta-catenin signaling. Oncogene. 2015;34:3188–3198. doi: 10.1038/onc.2015.129. [DOI] [PubMed] [Google Scholar]
- 52.Ge W, Liu Y, Chen T, Zhang X, Lv L, Jin C, et al. The epigenetic promotion of osteogenic differentiation of human adipose-derived stem cells by the genetic and chemical blockade of histone demethylase LSD1. Biomaterials. 2014;35:6015–6025. doi: 10.1016/j.biomaterials.2014.04.055. [DOI] [PubMed] [Google Scholar]
- 53.Fiskus W, Sharma S, Shah B, Portier BP, Devaraj SG, Liu K, et al. Highly effective combination of LSD1 (KDM1A) antagonist and pan-histone deacetylase inhibitor against human AML cells. Leukemia. 2014;28:2155–2164. doi: 10.1038/leu.2014.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schenk T, Chen WC, Gollner S, Howell L, Jin L, Hebestreit K, et al. Inhibition of the LSD1 (KDM1A) demethylase reactivates the all-trans-retinoic acid differentiation pathway in acute myeloid leukemia. Nat Med. 2012;18:605–611. doi: 10.1038/nm.2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee MG, Wynder C, Schmidt DM, McCafferty DG, Shiekhattar R. Histone H3 lysine 4 demethylation is a target of nonselective antidepressive medications. Chem Biol. 2006;13:563–567. doi: 10.1016/j.chembiol.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 56.Cortez V, Mann M, Tekmal S, Suzuki T, Miyata N, Rodriguez-Aguayo C, et al. Targeting the PELP1–KDM1 axis as a potential therapeutic strategy for breast cancer. Breast Cancer Res. 2012;14:R108. doi: 10.1186/bcr3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Diaz-Villanueva JF, Diaz-Molina R, Garcia-Gonzalez V. Protein folding and mechanisms of proteostasis. Int J Mol Sci. 2015;16:17193–17230. doi: 10.3390/ijms160817193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cullinan SB, Diehl JA. PERK-dependent activation of Nrf2 contributes to redox homeostasis and cell survival following endoplasmic reticulum stress. J Biol Chem. 2004;279:20108–20117. doi: 10.1074/jbc.M314219200. [DOI] [PubMed] [Google Scholar]
- 59.Pereira RC, Delany AM, Canalis E. CCAAT/enhancer binding protein homologous protein (DDIT3) induces osteoblastic cell differentiation. Endocrinology. 2004;145:1952–1960. doi: 10.1210/en.2003-0868. [DOI] [PubMed] [Google Scholar]
- 60.Wielenga MC, Colak S, Heijmans J, van Lidth de Jeude JF, Rodermond HM, Paton JC, et al. ER-stress-induced differentiation sensitizes colon cancer stem cells to chemotherapy. Cell Rep. 2015;13:490–494. doi: 10.1016/j.celrep.2015.09.016. [DOI] [PubMed] [Google Scholar]
- 61.Xu H, Tsang KS, Wang Y, Chan JC, Xu G, Gao WQ. Unfolded protein response is required for the definitive endodermal specification of mouse embryonic stem cells via Smad2 and beta-catenin signaling. J Biol Chem. 2014;289:26290–26301. doi: 10.1074/jbc.M114.572560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang L, Zeng X, Ryoo HD, Jasper H. Integration of UPRER and oxidative stress signaling in the control of intestinal stem cell proliferation. PLoS Genet. 2014;10:e1004568. doi: 10.1371/journal.pgen.1004568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cho YM, Jang YS, Jang YM, Chung SM, Kim HS, Lee JH, et al. Induction of unfolded protein response during neuronal induction of rat bone marrow stromal cells and mouse embryonic stem cells. Exp Mol Med. 2009;41:440–452. doi: 10.3858/emm.2009.41.6.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kozono D, Li J, Nitta M, Sampetrean O, Gonda D, Kushwaha DS, et al. Dynamic epigenetic regulation of glioblastoma tumorigenicity through LSD1 modulation of MYC expression. Proc Natl Acad Sci USA. 2015;112:E4055–E4064. doi: 10.1073/pnas.1501967112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Suva ML, Rheinbay E, Gillespie SM, Patel AP, Wakimoto H, Rabkin SD, et al. Reconstructing and reprogramming the tumor-propagating potential of glioblastoma stem-like cells. Cell. 2014;157:580–594. doi: 10.1016/j.cell.2014.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sareddy GR, Li X, Liu J, Viswanadhapalli S, Garcia L, Gruslova A, et al. Selective estrogen receptor beta agonist LY500307 as a novel therapeutic agent for glioblastoma. Sci Rep. 2016;6:24185. doi: 10.1038/srep24185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Son MJ, Woolard K, Nam DH, Lee J, Fine HA. SSEA-1 is an enrichment marker for tumor-initiating cells in human glioblastoma. Cell Stem Cell. 2009;4:440–452. doi: 10.1016/j.stem.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Anido J, Saez-Borderias A, Gonzalez-Junca A, Rodon L, Folch G, Carmona MA, et al. TGF-beta receptor inhibitors target the CD44(high)/Id1(high) glioma-initiating cell population in human glioblastoma. Cancer Cell. 2010;18:655–668. doi: 10.1016/j.ccr.2010.10.023. [DOI] [PubMed] [Google Scholar]
- 69.Lathia JD, Gallagher J, Heddleston JM, Wang J, Eyler CE, Macswords J, et al. Integrin alpha 6 regulates glioblastoma stem cells. Cell Stem Cell. 2010;6:421–432. doi: 10.1016/j.stem.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bao S, Wu Q, Li Z, Sathornsumetee S, Wang H, McLendon RE, et al. Targeting cancer stem cells through L1CAM suppresses glioma growth. Cancer Res. 2008;68:6043–6048. doi: 10.1158/0008-5472.CAN-08-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bai Y, Lathia JD, Zhang P, Flavahan W, Rich JN, Mattson MP. Molecular targeting of TRF2 suppresses the growth and tumorigenesis of glioblastoma stem cells. Glia. 2014;62:1687–1698. doi: 10.1002/glia.22708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, et al. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 73.Xie Q, Wu Q, Mack SC, Yang K, Kim L, Hubert CG, et al. CDC20 maintains tumor initiating cells. Oncotarget. 2015;6:13241–13254. doi: 10.18632/oncotarget.3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sareddy GR, Nair BC, Gonugunta VK, Zhang QG, Brenner A, Brann DW, et al. Therapeutic significance of estrogen receptor beta agonists in gliomas. Mol Cancer Ther. 2012;11:1174–1182. doi: 10.1158/1535-7163.MCT-11-0960. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.