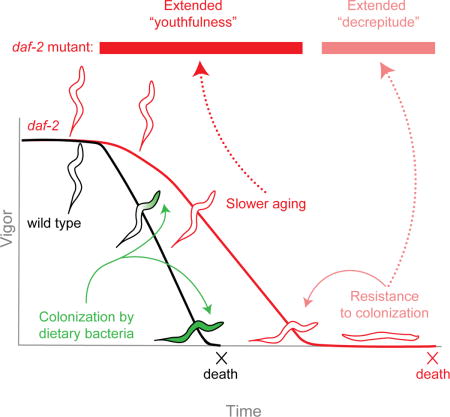
Summary
The goal of aging research is to extend healthy, active life. For decades, C. elegans daf-2 insulin/IGF-1 receptor mutants have served as a model for extended lifespan and youthfulness. However, a recent report suggested that their longevity is associated with an undesirable phenotype: a disproportionately long period of decrepitude at the end of life. In a human population, such an outcome would be a burden to society, bringing into question the relevance of daf-2 mutants as a model for life extension. However, here we report that, following an extended period of movement, daf-2 mutants survive longer in a decrepit state because of a beneficial trait: they are resistant to colonization of the digestive tract by dietary bacteria, a condition that leads to premature death in wild type and prevents their manifestation of decrepitude. If bacterial colonization is prevented, daf-2 mutants lead chronologically and proportionately healthier lives relative to wild type.
Graphical abstract
Introduction
Aging research aims not simply to increase lifespan but rather to increase the duration of healthy, disability-free life, or healthspan. In theory, lifespan can be extended by decreasing the rate of aging, postponing its onset, or eliminating a cause of mortality in old individuals. These mechanisms would impact the durations of healthspan and age-related deterioration differently.
Caenorhabditis elegans has been an invaluable experimental organism for discovery and characterization of conserved pathways that extend lifespan. In particular, reduced signaling through the stress and nutrient-sensing insulin/IGF-1 pathway was first shown to double the lifespan of C. elegans (Kenyon et al., 1993) and later found to increase longevity of other species, including mammals (Bartke, 2008). Moreover, polymorphisms in IGF-1-pathway genes and low plasma IGF-1 levels are associated with extreme longevity in humans (Kenyon, 2010; Milman et al., 2014; Suh et al., 2008; Willcox et al., 2008). C. elegans with partial loss-of-function mutations in daf-2, the C. elegans insulin/IGF-1-receptor gene, not only live longer but also maintain more youthful characteristics, such as active movement (Huang et al., 2004; Kenyon et al., 1993), neuronal function (Liu et al., 2013) and memory (Kauffman et al., 2010), indicating an extension of healthspan as well as lifespan. However, a recent study followed functional ability of daf-2 mutants and found that the daf-2 healthspan, though chronologically longer than that of wild type, did not scale with lifespan, resulting in a disproportionately extended period of age-related decrepitude (Bansal et al., 2014). This report was disconcerting because such an outcome would be undesirable in a human society, where population aging has already massively increased healthcare costs (Centers for Medicare & Medicaid Services, 2015; Meara et al., 2004), and also brought into question the validity of C. elegans as a model organism to study healthy life extension.
In this study, we set out to accomplish three goals: to undertake a quantitative large-scale analysis to corroborate the reported disproportionately extended end-of-life decrepitude in a daf-2 mutant; to determine whether this phenotype could be due to behavioral particularities of the specific daf-2 allele that was examined; and, if not, to elucidate the cause of this apparently undesirable phenotype.
Results and Discussion
Food limitation causes wild-type C. elegans juveniles to arrest development and enter the quiescent state of dauer diapause. Loss-of-function daf-2 mutants become dauers even in the presence of food. The partial loss-of-function daf-2(e1370) mutant grows to adulthood and is very long-lived, but displays mild dauer-like quiescence, particularly when shifted to high temperature (Gems et al., 1998). The mild quiescence of this mutant is likely due to altered sensory perception caused by its elevated expression of the odorant receptor gene odr-10 (Hahm et al., 2015). Importantly, dauer-like quiescence is not necessary for daf-2 mutants to live long, because other daf-2 mutants, such as daf-2(e1368) mutants, live long but do not exhibit quiescence (Arantes-Oliveira et al., 2003; Ewald et al., 2014; Gems et al., 1998). Thus, we wondered whether the disproportionately long late-life immobility reported for the daf-2(e1370) mutant (Bansal et al., 2015) could result from the intrinsic movement defects of this particular mutant. To this end, we compared daf-2(e1370) to daf-2(e1368) mutants.
First, we used the Multi-Worm Tracker computer-vision system (Swierczek et al., 2011) to carefully measure locomotory speed and behaviors of these two daf-2 mutants during young adulthood (Figure 1A). In the lab, C. elegans are typically observed responding to stimulation; for example, to the jolt of their culture dishes landing on the stage of a dissecting microscope (Figure 1A). When measured after delivering a mechanical stimulus in a controlled manner, stimulated movement speed of daf-2(e1370) mutants was modestly but significantly slower than wild type (22%; Figure 1B). When the speed of normal explorative locomotion was measured in the absence of external stimulation, daf-2(e1370) mutants moved markedly more slowly than wild type (53% slower; Figure 1B). The impairment was particularly pronounced in the backward movement (Figure 1C). In contrast, both stimulated and unstimulated movement speed of the daf-2(e1368) mutant closely resembled that of the wild type (Figure 1B–C).
Figure 1. daf-2(e1370) but not daf-2(e1368) mutants exhibit several behavioral phenotypes during early adulthood.
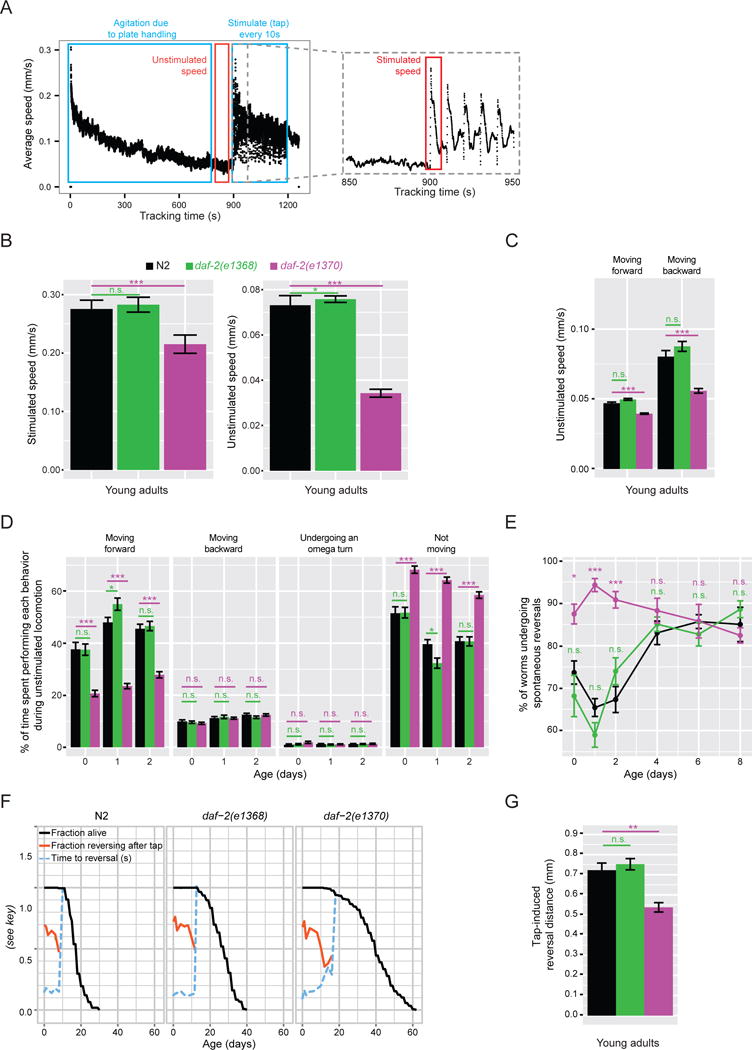
a) Data collection using the Multi-Worm Tracker. As an example, speed of N2 young adults on a single plate over the course of a tracking session is shown: 0–900 s – no stimulation; starting at 900 s, a mechanical tap stimulus was delivered every 10 seconds 30 times. b) Average speed of stimulated and unstimulated locomotion. c) Speed of unstimulated locomotion in forward and backward directions. d) Time spent by each animal performing each of the four stereotypical behaviors and e) fraction of the population spontaneously reversing movement direction during unstimulated locomotion (measured over a 60 s window). f) Fraction of animals responding (i.e. reversing) and average response time to a mechanical tap stimulus (the first tap). A reversal following the tap cannot be interpreted as a response to the tap if the time to a reversal is equal to the time between spontaneous reversals. Once this happens, tap response curves are no longer shown (i.e. in older animals). g) Distance traveled backward following a tap stimulus before resuming forward locomotion. Age – adult age (0 – young adult). n > 200. * − p < 0.05; ** − p < 0.005; *** − p < 0.0005; n.s. – not significant. Figure S1A–C shows behavioral phenotypes in the absence of FUDR. Figure S2E–F shows young adult behavior in additional daf-2 mutants.
The daf-2(e1370) mutant displayed a number of additional behavioral phenotypes. It spent less time moving forward and more time taking pauses during explorative locomotion (Figure 1D). During young adulthood, but not after day 4, daf-2(e1370) animals spontaneously reversed direction of locomotion more frequently than did wild type (Figure 1E). Furthermore, while the probability of initiating an escape response following a tap-stimulus, and the amount of time it took to react, were normal (Figure 1F), the magnitude of the response (i.e. distance traveled during the reversal) was reduced (Figure 1G). All of these behaviors were unaffected in the daf-2(e1368) mutant (Figure 1D–G). Together, these results support and extend previous reports that daf-2(e1370) mutants have movement impairments as young adults, suggesting a mild dauer-like phenotype, and show that these early life impairments are completely absent in the daf-2(e1368) mutant. In contrast, these animals exhibited completely normal behaviors, yet still lived long.
To test the hypothesis that the early-life dauer-like phenotypes are related to the extension of late-life immobility, we measured the behavior of daf-2(e1370) and daf-2(e1368) mutants throughout life. Both mutants had extended lifespans (Figure 2A) and had a greater average movement speed than wild type starting at day 9 of adulthood, as wild-type movement declined (Figure 2B, S1D). Because MWT only detects animals that can move at least half a body length within a 1 min interval, any live but immobile animals would not be taken into account for speed calculation, resulting in average speed of a population with immobile animals to be overestimated. Thus, we quantified the fraction of live animals that were able to move (i.e. were detectable) at a given age. This analysis also showed that the two daf-2 mutants preserved movement ability longer than wild type (Figure 2C). Thus, both daf-2 mutants gain additional healthy days of life, indicating a slower rate of behavioral aging.
Figure 2. daf-2 mutants maintain vigor longer but have an extended period of immobility at the end of life.
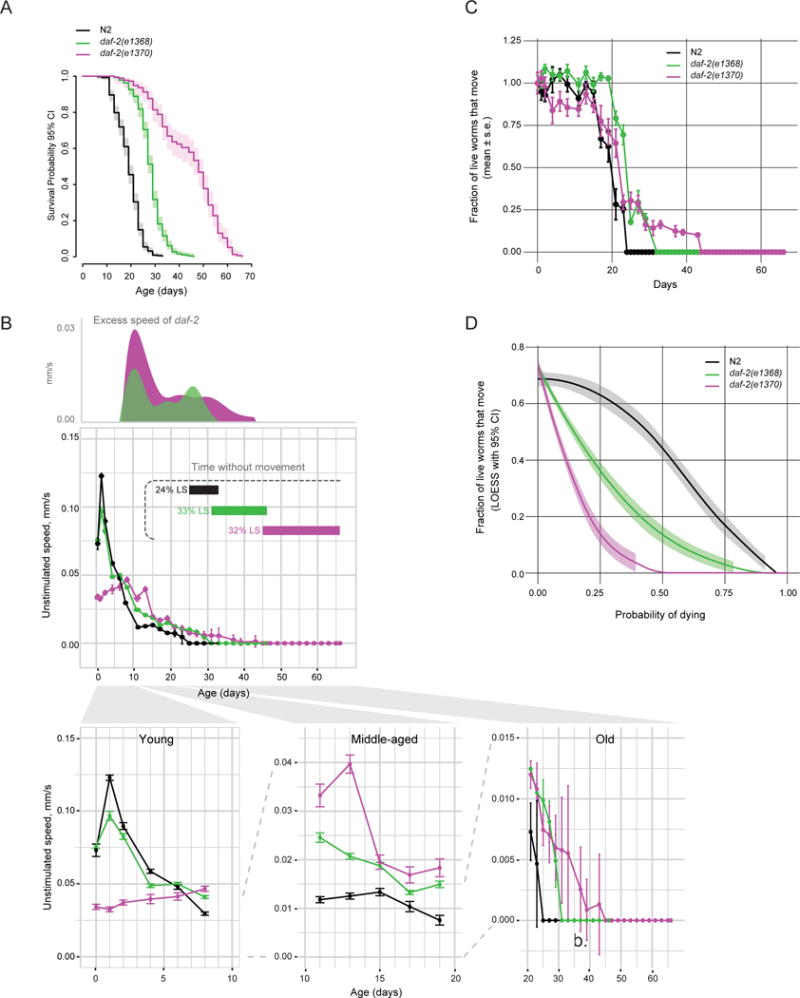
a) Survival analysis of daf-2(e1368) and daf-2(e1370) mutants under standard growth conditions. b) Unstimulated movement speed of detected animals as a function of adult age. Excess speed of daf-2 mutants is calculated by subtracting wild-type speed from mutant speed. Locomotion of young, middle-aged and old animals is also plotted separately below. Horizontal bars indicate duration of life and proportion of maximal lifespan without any detectable movement in the population. See Figure S1D for analysis of stimulated movement. n > 200 on day 0. c) Fraction of live worms exhibiting detectable movement at a given age. The data are averages of 8 independent replicate plates with 30–50 worms per plate on day 0. d) Same as (c) but plotted versus survival probability (i.e. normalized to lifespan). If every animal exhibited movement right until death regardless of whether it died early or late (i.e. there were no late-life decrepitude), this plot would be a straight horizontal line. Local polynomial regression (LOESS) fit with a 95% confidence interval is shown. Figure S1E–F shows stimulated and unstimulated speed scaled to maximal lifespan. Figure S2A–E shows early and late-life movement phenotypes in additional daf-2 mutants. Table S1 shows survival statistics for all lifespan experiments.
However, we were surprised to find that following this extended period of motility, both mutants spent a relatively long time (that is, a greater proportion of maximal lifespan relative to wild type) without any spontaneous or stimulated movement (Figure 2B, S1D). Accordingly, in the daf-2(e1370) population, and to a lesser extent in the daf-2(e1368) population, a larger fraction of live animals were immobile at a given survival probability (i.e. proportion of lifespan; Figure 2D). For example, with half of the population still alive, 42% of the live wild type exhibited movement, whereas only about 15% of daf-2(e1368) and 0% of daf-2(e1370) did. Together, these data show that although they have an extended chronological healthspan relative to wild type, daf-2 mutants end up spending a greater fraction of life in a state of decrepitude (which we define as a severely impaired spontaneous and stimulated movement ability that follows a gradual age-dependent behavioral decline). This extended period of decline at the end of life results in daf-2 mutants’ appearing less healthy if compared to wild type at a corresponding percentile of maximal lifespan (Figure S1E–F), consistent with what had been reported for daf-2(e1370) (Bansal et al., 2015). Because disproportionate extension of late-life decrepitude was observed in both daf-2(e1368) and daf-2(e1370) mutants, it can be genetically uncoupled from the early-life slow movement phenotypes seen in daf-2(e1370) mutants, arguing against a causal connection. Consistent with this, we analyzed two additional daf-2 perturbations and found no correlation between vigor in young adults and the duration of decrepitude at the end of life (Figure S2).
In summary, we found that two very different daf-2 mutants both remain active longer and age more slowly than wild type, at least through mid-life, but then go on to stay alive but decrepit for a long time. We wanted to understand what might cause this extended decrepitude. Theoretically, eliminating a cause of death that kills relatively young individuals would result in a population’s growing older and frailer. We wondered whether resistance to bacterial toxicity might play a role. daf-2 mutants are resistant to a wide variety of environmental stresses, including bacterial pathogens such as Staphylococcus aureus and Pseudomonas aeruginosa (Evans et al., 2008; Garsin et al., 2003). Bacteria are the natural food source of C. elegans in the wild (Samuel et al., 2016), and in the lab, they are fed an O-antigen-negative strain of Escherichia coli called OP50 that is considered to be non-pathogenic. However, wild-type worms fed dead OP50 have been shown to live longer than worms fed live OP50 (Garigan et al., 2002; Gems and Riddle, 2000). Therefore, we hypothesized that pathogenicity of this E. coli strain is a significant cause of death in wild-type C. elegans and that daf-2 mutants are more resistant to this pathogenicity. If so, the still-alive daf-2 mutants might continue to age, falling into a state of decrepitude. This situation would extend the overall proportion of life spent in a decrepit state.
To test this hypothesis, we first asked whether the E. coli food source is a risk factor for death in C. elegans that is reduced in daf-2 mutants. To visualize bacteria in the animal, we imaged GFP-expressing OP50 E. coli over the course of life. We found that bacteria accumulate in old wild-type worms (Figure 3A–B). In particular, we observed an age-dependent increase in bacterial density in the terminal bulb of the pharynx and in the proximal intestine (100 μm from the pharyngeal-intestinal valve). Every dead wild-type worm in our study displayed this pattern of colonization (Figure 3A–B). During mid-life, we observed a mixture of colonized and non-colonized worms. We found that worms that did not display detectable colonization during mid-life (day 9 of adulthood) lived significantly longer than worms that did (Figure 3C). In fact, in these experiments, we never observed a worm dying prior to colonization (an affliction that eventually affects all worms). This association suggested that bacterial colonization, or something tightly linked to colonization, kills the animal, which is consistent with our previously-published results (Garigan et al., 2002). To address causality more directly, we “cured” colonized worms of bacteria with the antibiotic gentamicin on day 9, and found that curing significantly increased their lifespans (Figure 3D). Gentamicin effectively penetrated inside the animal (Figure 3E–F) and did not affect lifespan on its own (Figure 3G). In addition, gentamicin-killed bacteria did not extend lifespan via dietary restriction (Figure S3A–C). Together, these data show that colonization is strongly associated with death in wild-type C. elegans and that reducing bacterial load in colonized animals significantly improves their survival.
Figure 3. Colonization by the E. coli food source is a risk factor for death in C. elegans.
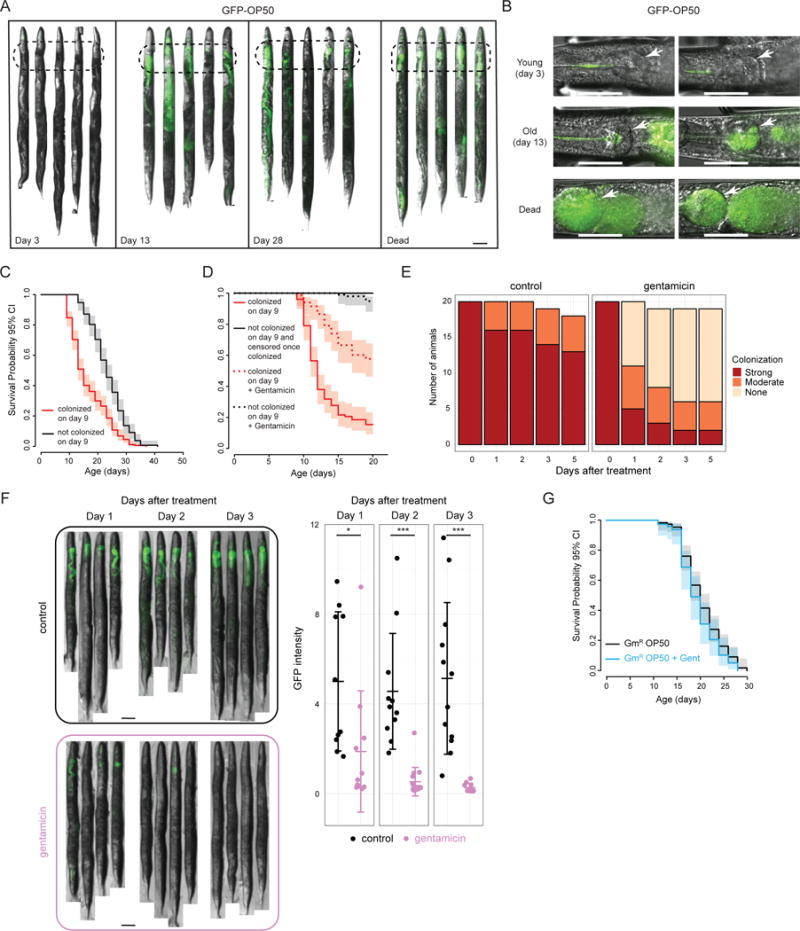
(a–b) Representative images of GFP-labeled OP50 E. coli accumulating in the terminal bulb and anterior intestine (dashed ovals) of wild-type worms with age. For quantification using > 15 animals, see Figure 4B,C. Worms were grown on GFP-OP50 starting at L1. Images of dead worms were taken at a lower exposure to avoid saturation. Arrows point to the terminal bulb of the pharynx. Scale bar = 100 μm. c) Survival of wild-type animals that did or did not display colonization of the terminal bulb and anterior intestine on day 9 of adulthood. d) Worms that displayed colonization on day 9 were treated with gentamicin starting on day 9 and for the remainder of their lifespan. Solid black curve – population of worms that did not display any colonization on day 9 and from which any live worm that developed colonization was physically removed. Survival curves end when there were no more animals left in this group, since all wild-type worms eventually developed colonization. Note that there were no deaths in worms without colonization. e) Gentamicin effectively reduces bacterial load in colonized worms. Worms colonized by GFP-OP50 on day 9 were isolated and split into two groups. Experimental animals were transferred to GFP-OP50 lawns treated with gentamicin. Because dead OP50 do not make GFP, to monitor clearance of colonizing bacteria that could normally occur in these worms, control animals were transferred to vehicle-treated plates seeded with non-GFP-expressing OP50. Twenty worms were singled out from each treatment group and if alive, scored in the days following treatment as strongly colonized (same as on day 9), moderately colonized (less than on day 9) or not colonized (not detectable by eye). f) Animals were treated as in (e), but >10 animals were picked randomly from gentamicin-treated and untreated plates and imaged. Representative images as well as mean GFP intensity per worm (mean ± s.d.) are shown. Scale bar = 100 μm. * − p < 0.05; *** − p < 0.0005. g) C. elegans lifespan is not affected by life-long gentamicin treatment when grown on gentamicin-resistant OP50 E. coli. Figure S3A–C shows that gentamicin-killed OP50 does not induce dietary restriction.
We then measured bacterial colonization of a daf-2 mutant, focusing initially on the daf-2(e1368) mutant. As shown in Figure 4A–B, colonization of the upper digestive tract was delayed and never reached the same maximum as in wild type. daf-2(e1368) animals had reduced colonization relative to both age-matched wild type, and to wild type with the same survival probability (Figure 4C), demonstrating that they spend both a larger amount of time and a larger proportion of life with less colonization. This finding is consistent with the idea that resistance to colonization allows daf-2 mutants to survive into old age. Why bacterial colonization occurs in old C. elegans and how exactly it causes death remains unknown. [Interestingly, the kinetics of colonization look biphasic, so there could be more than one reason (see Figure 4 legend).] Decreased immune function with age (Youngman et al., 2011) could contribute to bacterial accumulation and proliferation, and daf-2 mutants have higher expression of some antimicrobial genes (Murphy et al., 2003) that curtail the rate of bacterial proliferation in the intestine (Hahm et al., 2011). One possibility for why bacterial accumulation kills worms is that metabolites produced by live metabolically-active bacteria reach toxic levels when bacterial density increases inside the animal. Because the intestine becomes permeable with age (Gelino et al., 2016), another possibility is that worms die when bacteria leak out of the lumen and enter the body cavity. Although we did not observe any bacteria within tissues of live old worms (data not shown), it is possible that death occurs immediately after bacteria invade.
Figure 4. daf-2 mutants resist bacterial colonization.
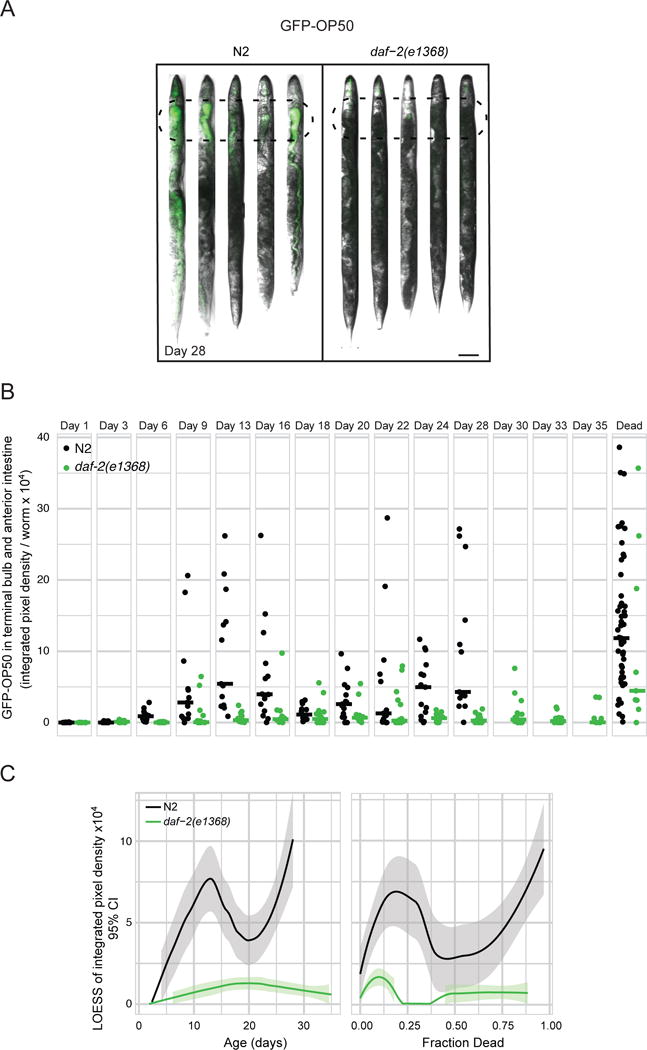
a) Representative images and b) quantification of GFP-OP50 accumulation in the terminal bulb and anterior intestine (100 μm from the terminal bulb; dashed ovals) in N2 and daf-2(e1368) with age. Scale bar = 100 μm. Each dot represents a worm, and horizontal bars represent medians. n > 15. The extent of colonization appears to be biphasic. This could be due to bacteria that accumulate in relatively young adults being cleared by the animal, or due to a subpopulation of highly-colonized worms dying, while the remaining, less colonized, worms go on to develop further colonization later in life. We favor the second idea, given that there is little clearance of colonizing bacteria in day 9 animals (Figure 3E) and given the wide distribution of colonization severities in day 9–16 animals. c) Local regression of data for live worms in (c) plotted as a function of age (days of adulthood) or lifespan. Note that the daf-2 curve ends when 88% of animals are dead rather than 100%. Figure S5 shows colonization in daf-2(e1370).
If reduced risk of death due to bacterial colonization allows daf-2 mutants to live long enough to become decrepit, then eliminating bacterial colonization as a cause of “premature” death should allow wild-type worms, too, to live long enough to enter a state of end-of-life decrepitude. To test this hypothesis, we fed wild-type animals OP50 bacteria killed by gentamicin from the time of hatching. Using killed bacteria as a food source extended wild-type lifespan by 40% (Figure 5A). The amount of time (and proportion of lifespan) spent without detectable movement increased from 8 days (24%) on live OP50 (see Figure 2B) to 16 days (37%) on dead OP50 in wild-type worms (Figure 5B). Importantly, feeding dead bacteria did not change the rate of behavioral aging; it specifically extended the period of infirmity (Figure S3D–F). The likelihood of exhibiting movement at a given survival probability was significantly reduced by a diet of dead OP50 in wild-type worms (Figure 5C). In this regard, wild-type animals fed dead OP50 more closely resembled daf-2(e1368) mutants fed the standard diet of live OP50 than wild type fed the standard diet. Therefore, eliminating bacterial colonization as a cause of death in wild-type worms phenocopied the extended period of decrepitude seen in daf-2(e1368) mutants. Reducing food pathogenicity in a different way, by feeding worms less pathogenic soil bacteria, Bacillus subtilis, also increased lifespan and the proportion of life spent in a decrepit state, although the increase in the latter was smaller than that achieved by dead OP50 (Figure S4A–C). The conclusion that feeding worms non-pathogenic bacteria makes wild-type healthspan more similar to daf-2 healthspan was supported by a number of additional approaches to quantifying healthspan (Figure S4D–F).
Figure 5. The extended period of decrepitude in daf-2 mutants can be attributed to a reduced risk of death by bacterial colonization.
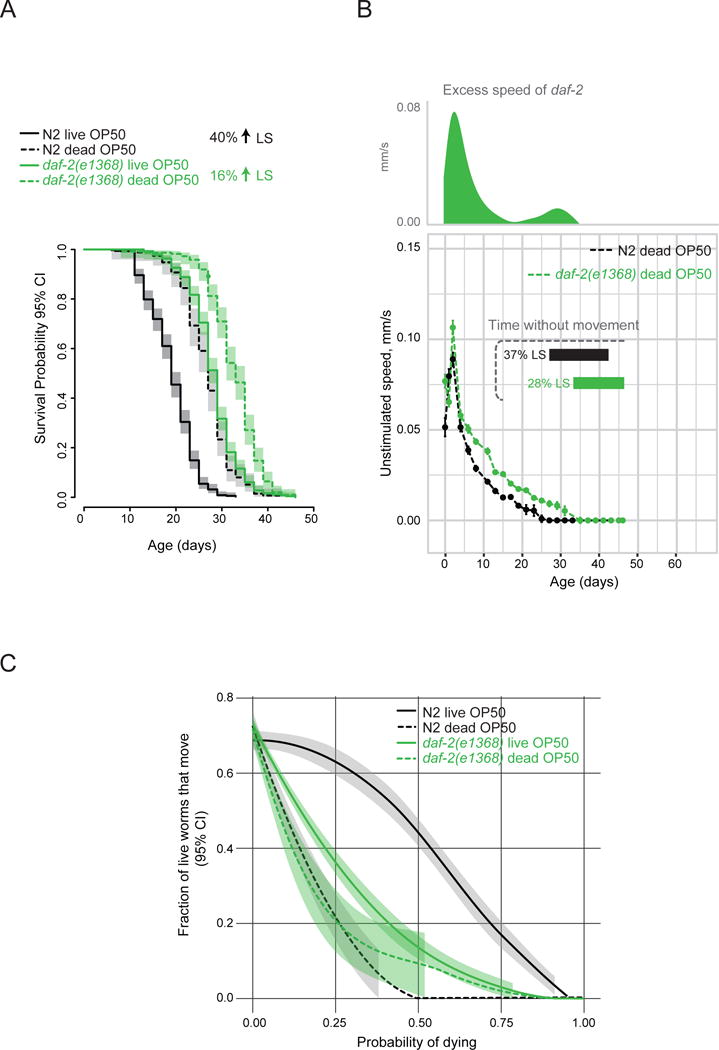
a) Lifespans of wild-type and daf-2(e1368) mutants grown on gentamicin-killed OP50 E. coli from hatching. This diet did not affect the amount of time it took the animals to reach adulthood (data not shown). b) Unstimulated movement speed of wild-type and daf-2(e1368) animals when grown on dead OP50. Excess speed of daf-2 mutants was calculated by subtracting wild-type speed from mutant speed. Horizontal bars indicate duration of life and proportion of maximal lifespan without any detectable movement in the population. n > 200. c) Fraction of live worms exhibiting detectable movement at a given survival probability (i.e. normalized to lifespan). Local polynomial regression (LOESS) fit of data from 8 independent replicate plates with 30–50 worms per plate on day 0 and a 95% confidence interval are shown. Figure S3D–F shows rates of behavioral changes on live and dead bacteria. See Figure S4 for additional quantification of healthspan. Figure S5 shows effect of dead bacteria in daf-2(e1370).
A diet of dead bacteria also extended the daf-2(e1368)-mutant lifespan, but by a smaller proportion (16% extension; Figure 5A), with the mutant still living longer than wild type. Furthermore, when fed dead bacteria, the speed of daf-2-mutant locomotion still declined more slowly than did that of wild type fed dead bacteria (Figure 5B). These two results show that daf-2 mutants model aging/longevity per se, and not simply increased immunity/stress resistance. The healthspan of daf-2(e1368) mutants was largely unaffected by dead bacteria. There was a slight decrease in the fraction of live daf-2(e1368) mutant animals exhibiting movement at a given proportion of lifespan in response to dead bacteria (albeit a much smaller one than in the wild type; Figure 5C), suggesting that the modest lifespan extension achieved by dead bacteria in daf-2(e1368) mutants is likely due to elimination of residual susceptibility of these mutants to colonization. Together these findings provide an explanation for the disproportionately-extended period of decrepitude of daf-2 mutants. Moreover, they indicate that daf-2 mutants are not intrinsically unhealthy; instead they survive a wave of mortality that kills wild type, which allows their subsequent manifestation of decrepitude, likely as an extension of the aging process.
Interestingly, a recent study that looked at variability of individual healthspans within a wild-type C. elegans population (Zhang et al., 2016) found that individuals that die early, die looking relatively youthful, whereas individuals that live longer spend a greater proportion of life in a state of decline. Both colonization severity (see Figure 4B) and pathogen resistance (Sanchez-Blanco and Kim, 2011) vary substantially in wild-type animals (likely due to stochastic processes). Therefore, this finding about the healthspans of individual wild-type animals is consistent with greater pathogen resistance allowing some worms to survive long enough to enter decrepitude.
We also examined bacterial colonization in the daf-2(e1370) mutant, which is longer-lived than daf-2(e1368). This mutant had an even greater delay and reduction in the maximal level of bacterial colonization. Even very late in life (day 55), GFP-OP50 bacteria were barely detectable in these animals (Figure S5A–C). Feeding dead bacteria to daf-2(e1370) mutants did not further extend their lifespan, but, rather unexpectedly, shortened it slightly (by 10%) (Figure S5D). We do not know the basis for this phenomenon. One possible explanation is that there may be a beneficial role of live bacteria in lifespan extension (for example, bacteria could provide an essential signaling molecule to old animals), but this beneficial effect is normally preceded by the detrimental effect of the bacteria’s establishing colonization and killing susceptible animals. An example of a bacterially-derived pro-longevity molecule is nitric oxide, which is produced by B.subtilis (Gusarov et al., 2013) (note, however, that this molecule is not produced by OP50 E.coli). The daf-2(e1370) mutant is highly resistant to bacterial colonization [more so than daf-2(e1368)], so death associated with colonization is not a hazard to these animals, allowing the beneficial effect of live bacteria to become limiting to lifespan. In other words, to be extremely long-lived [like daf-2(e1370) mutants], C. elegans may need live bacteria, but only if they don’t colonize and kill them prematurely. Consistent with this idea, feeding live but less pathogenic B. subtilis did not shorten the lifespan of daf-2(e1370) (Figure S4A). It should also be noted that although dead bacteria shortened the lifespan of daf-2(e1370) mutants, their lifespan was still longer than the lifespan of wild-type or daf-2(e1368) animals fed either live or dead bacteria (Figure S5D).
In summary, we find that the level of bacterial colonization predicts wild-type lifespan. The extent of colonization is significantly greater in wild type than in daf-2 mutants, and eliminating colonization in wild-type animals allows them to avoid an early death and instead remain alive for a longer time in a decrepit, aged, state, just like daf-2 mutants. Therefore, we conclude that a beneficial trait (resistance to bacterial colonization) can explain the extended end-of-life frailty of daf-2 mutants. Surviving the hazard from bacterial colonization allows these mutants to grow biologically older and more decrepit than end-of-life wild type animals. Importantly, if colonization is eliminated as a risk factor for death [i.e. if daf-2(e1368) and wild-type worms are both fed dead bacteria], daf-2 mutants still exhibit a gain in the number of “healthy” days but they do not exhibit a disproportionately long period of decrepitude relative to wild type, because extended end-of-life decrepitude can now be manifested in the wild type as well. This leads to a situation in which daf-2(e1368) mutants appear at least as, or more, vigorous than wild type at every day and percentile of life (Figure 5B and Figure S4G).
Together these findings support the argument that C. elegans daf-2 mutants are valuable for studying healthy lifespan extension. daf-2 mutants live longer due to a two-part mechanism: a slower rate of aging (leading to extension of healthspan) and an increased ability to resist death due to bacterial colonization (leading to extension of decrepitude). Other studies showing that it is possible to increase lifespan of daf-2 mutants even further (up to 6-fold), strongly suggest that daf-2 mutations can affect lifespan far beyond what is possible by simply delaying death in old worms by suppressing pathogenesis (Arantes-Oliveira et al., 2003; Ewald et al., 2014). More generally, the results presented here show how the healthspan of an organism can be affected in opposite ways at different times of life by an intervention that both decreases the rate of aging and also mitigates a disease that kills old individuals. This is important to keep in mind when seeking to develop interventions that act by different demographic mechanisms to increase human lifespan.
Experimental Procedures
C. elegans strains and maintenance
Animals were maintained at 20°C under standard conditions. See Supplemental Information for additional details.
Lifespan assays
2′fluoro-5′deoxyuridine (FUDR, Sigma 50 μM) was added during late L4 stage to prevent progeny hatching. FUDR did not affect lifespan or movement (Figure S1A–C). Adults were scored manually as dead or alive at least every other day. A worm was considered alive if it moved spontaneously or, in cases where it wasn’t moving, if it responded to a light touch on the head with a platinum wire. Animals that crawled off the plates, had progeny that hatched internally, burrowed or ruptured were censored and included in the analysis until the time of censorship.
Behavioral measurements
Movement was recorded using the Multi-Worm Tracker (MWT) computer vision system and analyzed using Choreography (v 1.3.1 build 1063), as described previously (Swierczek et al., 2011). Specifically, MWT was run using default settings save for a minimal object size of 100 pixels and a background adaptation rate “alpha” of 7 (resulting in 4× slower adaptation rate than usual) and Choreography was run using −p 0.02652 −M 1 −t 30 – shadowless options. With these parameters, an animal had to have moved at least half a body length anytime within a 1 min time interval to be detected. After detection, tracking continued even if the animal was still. Recordings lasted for 1260 s (21 min): animals were allowed to calm down from being handled (800 s), after which unstimulated behaviors were measured. Starting at 900 s, a mechanical stimulus was delivered using a solenoid tapper every 10 seconds for a total of 30 taps with 10 second intervals (Swierczek et al., 2011). Animals responded less to each subsequent tap due to habituation. Tap response behaviors and stimulated movement speed were measured following the first tap. Tracking was performed using 8 separate plates per condition, starting with 20–50 young adults per plate. Animals were tracked every day for the first three days of adulthood and every other day thereafter for the remainder of lifespan. Lifespan assays were done by hand on the same worms observed with the MWT.
Statistical analyses
All data were analyzed in R (v 3.2.1). Behavioral parameters were extracted using Choreography (Swierczek et al., 2011). Differences between groups were compared using Student’s unpaired t-test. Kaplan-Meier estimates of survival curves were calculated using survival (v 2.38–3) and rms (v 4.5–0) R packages and differences were tested using log-rank test. All experiments were performed independently at least two times. Data are presented as means ± SEM, unless noted otherwise. Values of p < 0.05 were considered statistically significant. The number of animals used in each experiment is indicated in the figure legends.
Supplementary Material
Acknowledgments
We thank Adam Freund, David Botstein and Jeff Settleman for comments on the manuscript and Antoine Roux and T. Richard Parenteau for technical assistance. We thank Kenyon lab members for helpful discussions and the Calico microscopy team for support with imaging. This project was supported at UCSF by the NIH R01 AG011816 to C.K., and then by Calico LLC. All authors are now employees of Calico Life Sciences LLC.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Additional experimental procedures are included in the Supplemental Material
Author Contributions
K.P. and C.K. conceived the experiments and wrote the manuscript. K.P. performed experiments. C.K. secured funding. R.A.K provided expertise and feedback.
References
- Arantes-Oliveira N, Berman JR, Kenyon C. Healthy animals with extreme longevity. Science. 2003;302:611. doi: 10.1126/science.1089169. [DOI] [PubMed] [Google Scholar]
- Bansal A, Kwon ES, Conte DJ, Liu H, Gilchrist MJ, MacNeil LT, Tissenbaum HA. Transcriptional regulation of Caenorhabditis elegans FOXO/DAF-16 modulates lifespan. Longevity & healthspan. 2014;3:5. doi: 10.1186/2046-2395-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal A, Zhu LJ, Yen K, Tissenbaum HA. Uncoupling lifespan and healthspan in Caenorhabditis elegans longevity mutants. Proceedings of the National Academy of Sciences of the United States of America. 2015 doi: 10.1073/pnas.1412192112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartke A. Insulin and aging. Cell cycle. 2008;7:3338–3343. doi: 10.4161/cc.7.21.7012. [DOI] [PubMed] [Google Scholar]
- Centers for Medicare & Medicaid Services. NHE fact sheet. Baltimore: p. 2015. [Google Scholar]
- Evans EA, Chen WC, Tan MW. The DAF-2 insulin-like signaling pathway independently regulates aging and immunity in C. elegans. Aging cell. 2008;7:879–893. doi: 10.1111/j.1474-9726.2008.00435.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewald CY, Landis JN, Abate JP, Murphy CT, Blackwell TK. Dauer-independent insulin/IGF-1-signalling implicates collagen remodelling in longevity. Nature. 2014 doi: 10.1038/nature14021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garigan D, Hsu AL, Fraser AG, Kamath RS, Ahringer J, Kenyon C. Genetic analysis of tissue aging in Caenorhabditis elegans: a role for heat-shock factor and bacterial proliferation. Genetics. 2002;161:1101–1112. doi: 10.1093/genetics/161.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garsin DA, Villanueva JM, Begun J, Kim DH, Sifri CD, Calderwood SB, Ruvkun G, Ausubel FM. Long-lived C. elegans daf-2 mutants are resistant to bacterial pathogens. Science. 2003;300:1921. doi: 10.1126/science.1080147. [DOI] [PubMed] [Google Scholar]
- Gelino S, Chang JT, Kumsta C, She X, Davis A, Nguyen C, Panowski S, Hansen M. Intestinal Autophagy Improves Healthspan and Longevity in C. elegans during Dietary Restriction. PLoS genetics. 2016;12:e1006135. doi: 10.1371/journal.pgen.1006135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gems D, Riddle DL. Genetic, Behavioral and Environmental Determinants of Male Longevity in Caenorhabditis elegans. Genetics. 2000;154:1597–1610. doi: 10.1073/pnas.96.13.7394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gems D, Sutton AJ, Sundermeyer ML, Albert PS, King KV, Edgley ML, Larsen PL, Riddle DL. Two pleiotropic classes of daf-2 mutation affect larval arrest, adult behavior, reproduction and longevity in Caenorhabditis elegans. Genetics. 1998;150:129–155. doi: 10.1093/genetics/150.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusarov I, Gautier L, Smolentseva O, Shamovsky I, Eremina S, Mironov A, Nudler E. Bacterial nitric oxide extends the lifespan of C. elegans. Cell. 2013;152:818–830. doi: 10.1016/j.cell.2012.12.043. [DOI] [PubMed] [Google Scholar]
- Hahm JH, Kim S, DiLoreto R, Shi C, Lee SJV, Murphy CT, Nam HG. C. elegans maximum velocity correlates with healthspan and is maintained in worms with an insulin receptor mutation. Nature communications. 2015;6:1–7. doi: 10.1038/ncomms9919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahm JH, Kim S, Paik YK. GPA-9 is a novel regulator of innate immunity against Escherichia coli foods in adult Caenorhabditis elegans. Aging cell. 2011;10:208–219. doi: 10.1111/j.1474-9726.2010.00655.x. [DOI] [PubMed] [Google Scholar]
- Huang C, Xiong C, Kornfeld K. Measurements of age-related changes of physiological processes that predict lifespan of Caenorhabditis elegans. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:8084–8089. doi: 10.1073/pnas.0400848101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauffman AL, Ashraf JM, Corces-Zimmerman MR, Landis JN, Murphy CT. Insulin signaling and dietary restriction differentially influence the decline of learning and memory with age. PLoS biology. 2010;8:e1000372. doi: 10.1371/journal.pbio.1000372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. A C. elegans mutant that lives twice as long as wild type. Nature. 1993:1–4. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- Kenyon CJ. The genetics of ageing. Nature. 2010;464:504–512. doi: 10.1038/nature08980. [DOI] [PubMed] [Google Scholar]
- Liu J, Zhang B, Lei H, Feng Z, Liu J, Hsu AL, Xu XZS. Functional aging in the nervous system contributes to age-dependent motor activity decline in C. elegans. Cell metabolism. 2013;18:392–402. doi: 10.1016/j.cmet.2013.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meara E, White C, Cutler DM. Trends In Medical Spending By Age, 1963–2000. Health Affairs. 2004;23:176–183. doi: 10.1377/hlthaff.23.4.176. [DOI] [PubMed] [Google Scholar]
- Milman S, Atzmon G, Huffman DM, Wan J, Crandall JP, Cohen P, Barzilai N. Low insulin-like growth factor-1 level predicts survival in humans with exceptional longevity. Aging cell. 2014;13:769–771. doi: 10.1111/acel.12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy CT, McCarroll SA, Bargmann CI, Fraser A, Kamath RS, Ahringer J, Li H, Kenyon C. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature. 2003;424:277–283. doi: 10.1038/nature01789. [DOI] [PubMed] [Google Scholar]
- Samuel BS, Rowedder H, Braendle C, Félix M-A, Ruvkun G. Caenorhabditis elegansresponses to bacteria from its natural habitats. Proceedings of the National Academy of Sciences of the United States of America. 2016:201607183–14. doi: 10.1073/pnas.1607183113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Blanco A, Kim SK. Variable pathogenicity determines individual lifespan in Caenorhabditis elegans. PLoS genetics. 2011;7:e1002047. doi: 10.1371/journal.pgen.1002047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh Y, Atzmon G, Cho MO, Hwang D, Liu B, Leahy DJ, Barzilai N, Cohen P. Functionally significant insulin-like growth factor I receptor mutations in centenarians. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:3438–3442. doi: 10.1073/pnas.0705467105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swierczek NA, Giles AC, Rankin CH, Kerr RA. High-throughput behavioral analysis in C. elegans. Nature methods. 2011;8:592–598. doi: 10.1038/nmeth.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willcox BJ, Donlon TA, He Q, Chen R, Grove JS, Yano K, Masaki KH, Willcox DC, Rodriguez B, Curb JD. FOXO3A genotype is strongly associated with human longevity. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:13987–13992. doi: 10.1073/pnas.0801030105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngman MJ, Rogers ZN, Kim DH. A Decline in p38 MAPK Signaling Underlies Immunosenescence in Caenorhabditis elegans. PLoS genetics. 2011;7:e1002082. doi: 10.1371/journal.pgen.1002082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang WB, Sinha DB, Pittman WE, Hvatum E, Stroustrup N, pincus Z. Extended Twilight among Isogenic C. elegans Causes a Disproportionate Scaling between Lifespan and Health. Cell Systems. 2016;3:333–345.e4. doi: 10.1016/j.cels.2016.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


