Abstract
BACKGROUND & AIMS
The role of tobacco smoke in the etiology of inflammatory bowel disease (IBD) is unclear. We investigated interactions between genes and smoking (gene–smoking interactions) that affect risk for Crohn’s disease (CD) and ulcerative colitis (UC) in a case-only study of patients and in mouse models of IBD.
METHODS
We used 55 immunochip-wide data sets that included 19,735 IBD cases (10,856 CD cases and 8879 UC cases) of known smoking status. We performed 3 meta-analyses each for CD, UC, and IBD (CD and UC combined), comparing data for never vs ever smokers, never vs current smokers, and never vs former smokers. We studied the effects of exposure to cigarette smoke in Il10−/− and Nod2−/− mice, as well as in Balb/c mice without disruption of these genes (wild-type mice). Mice were exposed to the smoke of 5 cigarettes per day, 5 days a week, for 8 weeks, in a ventilated smoking chamber, or ambient air (controls). Intestines were collected and analyzed histologically and by reverse transcription PCR.
RESULTS
We identified 64 single nucleotide polymorphisms (SNPs) for which the association between the SNP and IBD were modified by smoking behavior (meta-analysis Wald test P<5.0×10−5; heterogeneity Cochrane Q test P>.05). Twenty of these variants were located within the HLA region at 6p21. Analysis of classical HLA alleles (imputed from SNP genotypes) revealed an interaction with smoking. We replicated the interaction of a variant in NOD2 with current smoking in relation to the risk for CD (frameshift variant fs1007insC; rs5743293). We identified 2 variants in the same genomic region (rs2270368 and rs17221417) that interact with smoking in relation to CD risk. Approximately 45% of the SNPs that interact with smoking were in close vicinity (≤1 Mb) to SNPs previously associated with IBD; many were located near or within genes that regulate mucosal barrier function and immune tolerance. Smoking modified the disease risk of some variants in opposite directions for CD vs UC. Exposure of IL10-deficient mice to cigarette smoke accelerated development of colitis and increased expression of interferon gamma in the small intestine, compared to wild-type mice exposed to smoke. NOD2-deficient mice exposed to cigarette smoke developed ileitis, characterized by increased expression of interferon gamma, compared to wildtype mice exposed to smoke.
CONCLUSION
In an analysis of 55 immunochip-wide data sets, we identified 64 SNPs whose association with risk for IBD is modified by tobacco smoking. Gene–smoking interactions were confirmed in mice with disruption of Il10 and Nod2—variants of these genes have been associated with risk for IBD. Our findings from mice and humans revealed that the effects of smoking on risk for IBD depend on genetic variants.
Keywords: animal model, nicotine, inflammation, gene–environment interaction
Inflammatory bowel diseases (IBD), including Crohn’s disease (CD; MIM 266600) and ulcerative colitis (UC; MIM 191390), are chronic lifelong illnesses of early onset that seriously impede the quality of life of patients and their families. IBD is affecting more than 2.5 million people in Europe (~0.5%) and is becoming increasingly frequent in Asia and in developing countries.1 The etiology of IBD involves both genetic and environmental factors, but the biological mechanisms of IBD development are still poorly understood. In particular, little is known about the possible role of gene–environment interaction (G×E) in IBD. In consequence, despite the many genotype-phenotype associations that have been identified in past genome-wide association studies (GWAS), more than 70% of the heritability of IBD is still unaccounted for.1–3
Smoking is the only well-established environmental risk factor for IBD.4–6 Early case-control studies revealed an increased risk for both CD and UC in former smokers whereas current smoking seems to predispose to CD, but to protect against UC.6,7 This differential effect on risk was recently confirmed in a large prospective study of 229,111 women from the US Nurses’ Health Study6 where the CD hazard ratio was found to be 1.35 for former and 1.90 for current smokers, using never smokers as a reference. By contrast, the UC hazard ratio was equal to 1.56 for former, but 0.86 for current smokers. However, with a 95% confidence interval ranging from 0.61 to 1.20, the apparent UC protective effect of current smoking was not statistically significant.
The etiological role of smoking in IBD is not yet fully understood mainly because of the complex chemical composition of tobacco smoke.8 Many candidate mechanisms appear worth consideration, including epigenetic changes that alter gene expression relevant to the innate and adaptive immune responses.8 Smoking also induces compositional changes of the gut microbiota, which provides a plausible link to disease etiology as well.9,10 Other possible mechanisms involve the post-translational modification of key proteins by constituents of tobacco smoke that activates the immune response and induces inflammation. For example, smoking has been found to induce citrullination of various proteins.11 Citrullination affects the 3-dimensional structure of proteins in such a way that the latter may unfold and interior domains become exposed that can subsequently act as antigens. In rheumatoid arthritis, for example, smoking has been identified as an environmental trigger of anti-citrulline immunity in individuals with particular HLA-DRB1 ‘shared epitope’ alleles, a mechanism that might also explain why UC risk stays high even decades after smoking cessation.6,12
G×E studies are one way to unravel the biological mechanisms of disease development. As yet, however, only few studies of interactions between genes and smoking (gene–smoking interactions) have been conducted in the context of IBD.13–15 One of these studies reported a statistically significant interaction between NOD2 gene variant 1007fs, predisposing to CD, and both ever and current smoking.13 Two other small studies observed a significantly higher risk of CD for smokers among GG homozygotes for SNP rs2241880 in the ATG16L1 gene and among CC (wild-type) homozygotes for SNP rs1343151 in the IL23R gene.14,15
So far, gene–smoking interactions in IBD have not been investigated at a genome-wide level. Using the genotype data available from the International Inflammatory Bowel Disease Genetics Consortium (IIBDGC), we therefore investigated whether the relative IBD risk of smokers is modified by any of the genetic variants included on the Illumina Immunochip itself, or by variants in the HLA region that can be imputed from Immunochip data using publicly available databases. For that purpose, we adopted a two-tiered approach, including the verification in control individuals of the gene–smoking independence assumption implicit to the case-only design (stage I), followed by a case-only analysis to identify gene–smoking interactions (stage II). The epidemiological findings were complemented by functional studies of mice deficient for two of the genes identified as potential G×E partners and which encode for interleukin-10 (referred as Il10) and nucleotide-binding oligomerization domain 2 protein (referred as Nod2).
Materials and Methods
IBD Immunochip Dataset
All DNA samples used in the present study were collected through the IIBDGC and originated from 48 sites in 17 countries in Europe, North America and Australia.2 Genotyping with the Immunochip custom genotyping array (Illumina) was performed in 34 batches in 11 different centers, as described elsewhere.2 After quality control,16 genotype data for a total of 132,890 SNPs with minor allele frequency >1% were tested for an interaction with smoking. For SNPs identified as potential G×E partners, additional quality control was carried out by visual inspection of the corresponding cluster plots.
Only samples with known smoking status were included in our study. We confined our meta-analyses to those IIBDGC centers that provided at least 10 samples with either CD or UC in each of the three smoking categories (never, current or former), which yielded a total of 19,735 cases (10,856 CD, 8879 UC; Table 1). Ten of the participating centers also had genotype data from controls available (n=8143), 60% of which (n=4887) were of known smoking status.
Table 1.
Overview of case-only and control data used for SNP-smoking interaction meta-analyses
| Study center | Crohn’s disease (CD) | Ulcerative colitis (UC) | Controls | ||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| Cases | Percentage Current Smokers | Percentage Former Smokers | Cases | Percentage Current Smokers | Percentage Former Smokers | ||
| USA, Los Angeles | 1451 | 11.3 | 8.3 | 791 | 7.4 | 16.6 | NA |
| Italy, Florence | 1068 | 37.3 | 13.3 | 765 | 12.4 | 26.3 | NA |
| Belgium, Leuven | 908 | 37.4 | 7.5 | 516 | 21.7 | 29.8 | 340 |
| Germany, Kiel | 714 | 31.4 | 16.1 | 692 | 12.9 | 28.0 | 2490 |
| UK, Newcastle | 655 | 22.9 | 25.8 | 553 | 6.5 | 30.2 | NA |
| UK, Exeter | 428 | 38.3 | 19.6 | 663 | 15.7 | 38.5 | NA |
| USA, Pittsburgh | 620 | 28.4 | 8.1 | 449 | 7.8 | 19.1 | 312 |
| Australia, Brisbane | 435 | 44.4 | 7.8 | 447 | 19.9 | 28.6 | 528 |
| New Zealand, Christchurch | 435 | 25.5 | 23.4 | 425 | 13.2 | 37.4 | NA |
| UK, Edinburgh | 339 | 24.2 | 33.3 | 399 | 9.8 | 43.6 | NA |
| Belgium, Liege | 349 | 50.1 | 5.4 | 255 | 13.7 | 18.0 | 122 |
| Canada, Torontoa | 298 | 15.8 | 7.7 | 294 | 9.2 | 18.4 | 112 |
| Canada, Montreal | 293 | 30.0 | 8.9 | 202 | 13.9 | 35.1 | 258 |
| UK, Cambridge | 348 | 31.9 | 16.7 | 133 | 9.0 | 24.1 | NA |
| Sweden, Örebro | 293 | 33.8 | 16.4 | 151 | 16.6 | 29.1 | NA |
| Lithuania | 112 | 22.3 | 19.6 | 297 | 12.8 | 26.9 | NA |
| UK, Torbay | 111 | 31.5 | 36.9 | 294 | 6.5 | 39.1 | NA |
| UK, Oxford | 125 | 28.8 | 19.2 | 257 | 9.3 | 39.3 | NA |
| UK, London | 362 | 31.5 | 26.0 | NA | NA | NA | NA |
| Canada, Torontob | 195 | 26.2 | 9.2 | 148 | 7.4 | 26.4 | NA |
| Australia, Fremantle | 178 | 24.7 | 27.5 | 165 | 12.7 | 35.8 | NA |
| USA, Yale | 182 | 17.0 | 11.5 | 149 | 10.1 | 16.8 | 322 |
| UK, Dundee | 108 | 42.6 | 17.6 | 216 | 18.5 | 38.4 | NA |
| USA, Chicago | 155 | 23.2 | 9.0 | 128 | 8.6 | 19.5 | 92 |
| Norway | NA | NA | NA | 272 | 11.0 | 31.6 | NA |
| Sweden, Karolinska | 210 | 23.3 | 32.9 | NA | NA | NA | NA |
| Belgium, Brussels | 192 | 40.6 | 15.1 | NA | NA | NA | NA |
| Germany, Munich | NA | NA | NA | 168 | 11.3 | 24.4 | 311 |
| Netherlands, Groningen | 152 | 42.1 | 14.5 | NA | NA | NA | NA |
| Australia, Adelaide | 92 | 33.7 | 15.2 | 50 | 22.0 | 20.0 | NA |
| Australia, Townsville | 48 | 31.2 | 39.6 | NA | NA | NA | NA |
|
| |||||||
| Total | 10,856 | 8879 | 4887 | ||||
NA: data not available.
National Institute of Diabetes, Digestive and Kidney Diseases (NIDDK) IBD genetics consortium
University of Toronto
To study the specific role of the HLA region, we used imputed classical HLA alleles for the 19,735 cases of interest from a previous IIBDGC study16 (see Supplementary Methods).
Animal Studies
All animal studies were approved by the local investigational review board (AF 16/20090) in an accredited establishment at the Institute Pasteur de Lille (N° B59-108) according to governmental guidelines N°86/609/CEE. Age- and gender-matched Nod2-deficient (Nod2−/−), Interleukin 10-deficient (Il10−/−) and control Balb/c mice (without disruption of these genes; wild-type mice) had free access to a standard laboratory chow diet in a half-day light cycle exposure and temperature-controlled environment. 3R4F research cigarettes were purchased from the University of Kentucky. Eight to ten week-old mice were exposed to the smoke of 5 cigarettes per day, 5 days a week, for 8 weeks, in a ventilated smoking chamber (InExpose® System, Emka, Scireq, Canada). The control group was exposed to ambient air.
Formalin-fixed, paraffin-embedded colon specimens were blindly scored for inflammation by two investigators (see Supplementary Methods). Relative mRNA levels were determined in colon samples according to standard methods using Actb as an internal reference gene (see Supplementary Methods).
Statistical Analysis
All statistical analyses of the human data were performed with either PLINK17 or the R software (v. 3.2.1), as appropriate. The statistical significance of pairwise SNP-smoking interactions was assessed by logistic regression analysis as implemented in PLINK,17 following a case-only approach.18 We employed an additive allelic model of the genotype-phenotype relationship and encoded individual SNP genotypes (G) by allele counts. Genotypes were treated as predictor variables whereas the binary smoking status (E, see below) was treated as the response variable, i.e.
| (1) |
Following Piegorsch et al,18 we do not include any additional predictor variables, such as age or sex, into the model.
Any significant association between G and E that occurs in cases points towards G×E at the population level, provided that the two assumptions underlying the case-only design are met, namely that (i) the disease is sufficiently rare (i.e. prevalence <5%) and (ii) G and E are uncorrelated in the general population.18 Note that the case-only approach does not involve any further assumptions. The case-only paradigm is exemplified in Supplementary Table 10 for SNPs rs17221417 and rs2270368 from the NOD2 gene region. Conceptually, multiplicative interaction between G and E is defined as the extent to which the true joint effect of G and E differs from the product of the two individual effects. From a case-only study, the genotypic odds ratio (OR) for exposure, i.e. the odds of E given the presence of G divided by the odds of E given the absence of G, can be derived by taking the antilog of the θ estimate (equation 1). One premise of the case-only design is that this OR can be interpreted as the multiplicative interaction between G and E on disease risk.
We performed a two-tiered G×E study separately for CD, UC and IBD (i.e. CD and UC combined). In stage I, the validity of the G-E independence assumption underlying the case-only design was assessed for all 132,890 SNPs. To this end, the logistic regression model of equation 1 was fitted to the available control data. In total, 15,196 SNPs were removed because of a nominally significant violation of the G-E independence assumption in controls (meta-analysis p<0.05) for at least one of the three smoking contrasts never vs ever, never vs current or never vs former, leaving 117,694 SNPs for stage II.
In stage II, case-only analyses of gene–smoking interaction were carried out for all 117,694 SNPs and for 11,248 variants in the HLA region separately for each participating center. Population stratification correction was performed in individual study centers, following a recently proposed genomic control-based approach for case-only studies19 (see Supplementary Methods).
The case-only analyses were carried out in triplicate, each time considering one of the three smoking contrasts never vs ever, never vs current and never vs former (Table 1). For meta-analysis, fixed- and random-effect models were fitted to the results using PLINK. A SNP was considered worth further consideration if the meta-analysis gene–smoking interaction (Wald) test yielded p<5.0×10−5 and the heterogeneity (Cochrane Q) test yielded p>0.05. Note that these criteria were not meant to control the family-wise error rate, i.e. define a threshold for genome-wide statistical significance. In recognition of many previous human studies, and particularly our own mouse data (see below), the present study was not geared towards disproving a genome-wide lack of gene–smoking interaction for IBD (e.g. the ‘global null hypothesis’) but rather served to identify the strongest candidate genes for G×E. Thus, the significance thresholds employed here served as sensible filters to prioritize nominally significant findings. We also calculated the false discovery rate20 (FDR) for each SNP to control the estimated proportion of false positive results among the identified potential G×E. We adopted 0.1 as a common threshold for the FDR in subsequent considerations. At the chosen significance level of α=5.0×10−5, the meta-analysis of the never vs ever IBD cohorts (n=19,735) had approximately 90% power21 to detect even a small G×E effect (OR=1.15), assuming a risk allele frequency of 0.15, a smoking frequency of 0.4, a genetic OR of 1.2 (as observed, on average, for IBD1) and a smoking OR of 1.4.6 Expectedly,21–23 a case-control analysis including all cases plus the available 4887 controls with known smoking status would have yielded dramatically smaller power of only 3%, assuming the same interaction effect and leaving all other parameters unchanged. Since smoking rates differed between centers, we also investigated, for each of the 64 SNPs identified as potential G×E partners, the relationship between the center-specific interaction ORs and smoking rates, using Spearman correlation coefficient.
To assess whether a given region harbored multiple independent gene–smoking interactions, regions with more than one SNP with p<5.0×10−5 were scrutinized further. All analyses were repeated including into the respective statistical model the SNP with the smallest gene–smoking interaction p value within a given 1 Mb region (henceforth called the ‘top SNP’) as a mandatory predictor. SNPs with a nominally significant Wald test (p<0.05) in the conditional analysis were deemed independent gene–smoking interaction partners.
To assess whether the gene–smoking interactions identified in our study overlapped or coincided with previously reported IBD associations,1,24 pair-wise linkage disequilibrium was estimated in the available control samples (n=8143) irrespective of whether smoking information was also available or not. To this end, r2 was computed in each center between pairs of SNPs no more than 1 Mb apart, where one showed an interaction in our study and one had been identified as a genetic main effect in a previous GWAS,1,24 followed by the calculation of a sample size-weighted average of the center-wise r2 values.
In order to identify SNPs that show interaction with smoking in opposite direction in CD and UD, we searched for SNPs with a meta-analysis gene–smoking interaction (Wald test) p<0.01 and a heterogeneity (Cochrane Q test) p>0.05 for which the case-only G×E OR for one and the same risk allele was reversed between CD and UC (i.e. OR<1 in CD and OR>1 in UC, or vice versa).
We illustrated the validity of the case-only approach by performing case-control analyses of CD for two selected SNPs from the NOD2 gene region (see Supplementary Methods). These calculations naturally had to be confined to the centers that provided controls with smoking information. We also carried out stratified analyses of the genetic main effects of the two SNPs in never smokers and current smokers. We confined the genome-wide analysis to a case-only approach because this has much higher power than a case-control approach, as was noted above.
The mice data were analyzed statistically using a Kruskal-Wallis test or two-way ANOVA as implemented in GraphPad Prism 5 Version 5.02. Statistical significance was defined as p<0.05; measurements were summarized as mean ± standard error of the mean.
Functional annotation of the interacting SNPs and gene prioritization, pathway and tissue/cell type enrichment analysis, and regional linkage disequilibrium plots and annotation of association boundaries were performed as described in the supplementary methods section using publicly accessible databases.
Results
SNP-Smoking Interaction
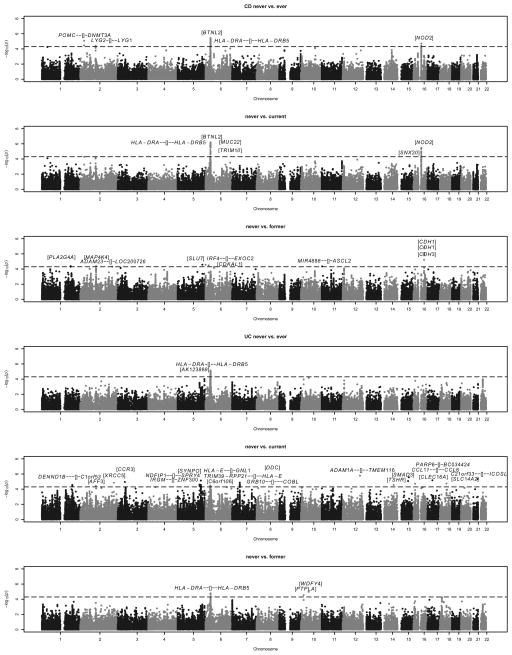
Three Immunochip-wide meta-analyses of the interaction between smoking behavior (contrasts never vs ever, never vs current and never vs former) and genotype (117,694 SNPs complying with the G-E independence assumption in controls, minor allele frequency >1%) were performed separately for CD, UC and IBD (i.e. CD and UC combined). Manhattan plots of the meta-analyses results highlighted several potentially interacting loci (Figure 1 and Supplementary Figure 1). Study-wide λ values, calculated to adjust for potential population stratification, were found to be small to moderate, with a maximum of 1.15 obtained in the “USA, Los Angeles” CD cohort (Supplementary Table 1 and Supplementary Figure 2).
Figure 1. Manhattan plots of six Immunochip-wide meta-analyses highlighting potentially smoking-interacting loci for CD and UC.
Panels 1–3 (from top) refer to three different smoking contrasts for CD: never vs ever, never vs current and never vs former. Similarly, panels 4–6 (from top) refer to three different smoking contrasts for UC. The horizontal dashed line indicates the threshold (p=5.0×10−5) for suggestive evidence of gene–smoking interaction. All SNPs included in the analyses complied with the G-E independence assumption underlying the case-only design, leaving 117,694 SNPs for case-only meta-analysis. Top SNPs from Table 2 and the gene context (see Table 2 legend for gene context definition) are shown above the suggestive threshold line (in bold). Manhattan plots for IBD (i.e. CD and UC combined) are provided as Supplementary Figure 1.
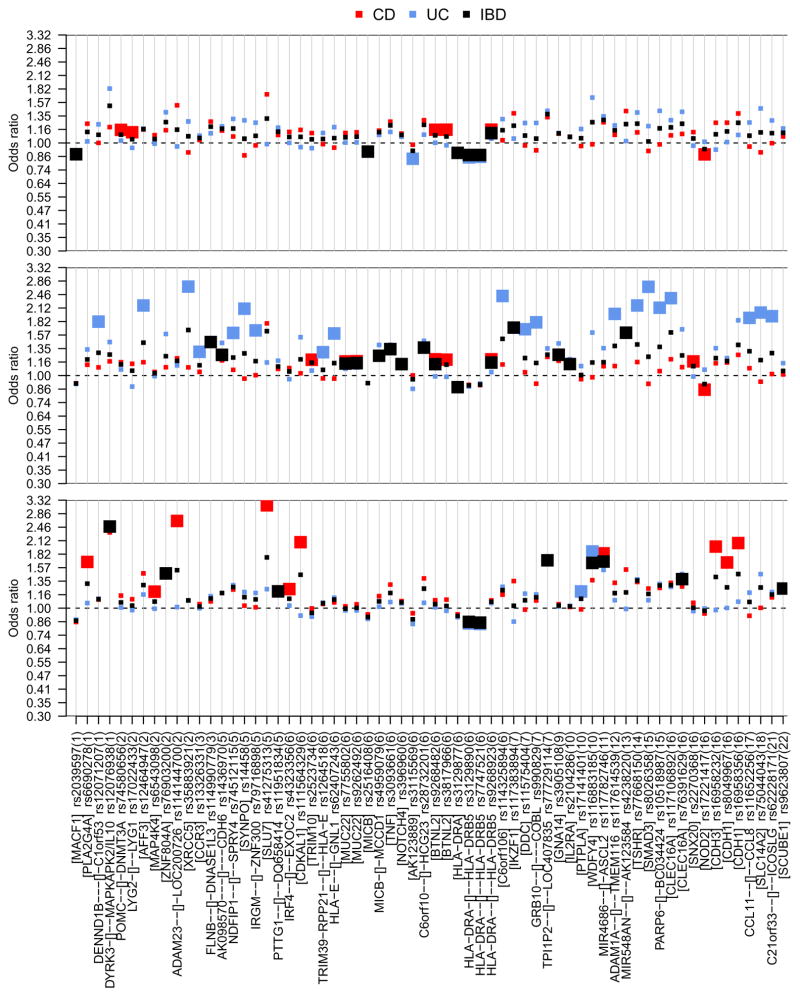
With the never vs ever contrast, 46 interacting SNPs for CD (in 5 genomic regions), 53 interacting SNPs for UC (in 1 genomic region) and 65 interacting SNPs for IBD (in 3 genomic regions) initially fulfilled our filter criteria (Supplementary Table 2). When conditioning upon the genotypes of the region-specific top SNPs, 1 (CD), 1 (UC) and 3 (IBD) additional SNPs were found to exhibit residual SNP-smoking interaction of nominal significance (Supplementary Table 3), thereby indicating potentially independent interaction signals from the same genomic regions. Overall, a total of 6 SNPs (5 top, 1 additional independent), 2 SNPs (1, 1) and 6 SNPs (3, 3) were thus identified as interacting with smoking for CD, UC and IBD risk, respectively, in the never vs ever smoker meta-analyses. Similarly, meta-analyses with the never vs current and never vs former smoker contrasts identified an additional 18 interacting SNPs (13 top, 5 additional independent) for CD, 24 SNPs (21, 3) for UC and 23 SNPs (18, 5) for IBD (Supplementary Tables 4–7). In summary, considering at least one of the three smoking contrasts and after adjustment for possible population stratification, 19 SNPs for CD, 25 SNPs for UC and 25 SNPs for IBD were identified as interacting with smoking according to our filter criteria (Wald test p<5.0×10−5, Cochrane Q test p>0.05). Since two SNPs (rs9268923, rs117782746) were overlapping between CD and IBD and three SNPs (rs3129890, rs7747521, rs116883185) were overlapping between UC and IBD, the total number of unique SNPs was 64 (Figure 2, Table 2; for regional linkage disequilibrium plots, see Supplementary Figures 3–11). Some 52 of these 64 SNPs yielded a false discovery rate (FDR) below 0.1. Interestingly, the largest number of interacting markers was identified with the never vs current smoker contrast for UC (middle panel in Figure 2), but with the never vs former smoker contrast for CD (bottom panel). No correlation became apparent for any SNP between the center-specific smoking rates and the center-specific interaction ORs (Supplementary Figure 12). For the NOD2 risk locus, we replicated the interaction between tobacco smoke exposure and the frameshift polymorphism fs1007insC (rs5743293; p=4.5×10−3 for never vs current smoker contrast in CD) and identified two suggestive independent SNP-smoking interactions in the same gene region (rs2270368, p=2.9×10−5, never vs current; rs17221417, p=3.3×10−6; never vs current) by means of conditional analysis (Supplementary Table 8).
Figure 2. Visualization of SNPs identified as interacting with smoking (.
Table 2).
The odds ratio is shown for all markers with suggestive gene–smoking interaction (p<5.0×10−5); gene context (see Table 2 legend for gene context definition) is provided with each rs-number and chromosome numbers are given in parentheses. The three panels refer to different smoking contrasts, namely never vs ever (top), never vs current (middle) and never vs former (bottom). The square color refers to the IBD type (CD: red, UC: blue, IBD: black), large squares mark meta-analysis (Wald test) p values <5.0×10−5.
Table 2.
SNPs with a suggestive gene–smoking interaction (meta-analysis Wald test p<5.0×10−5 and heterogeneity Cochrane Q test ph>0.05) for at least one smoking contrast (never vs ever, never vs current, or never vs former). All listed SNPs complied with the G-E independence assumption (p≥0.05) in healthy controls
| Chr:Pos | Gene context | Alleles (A/B) | SNP | never vs ever (6052 cases vs 4804 cases) | never vs current (6052 cases vs 3177 cases) | never vs former (6052 cases vs 1627 cases) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR [95% CI] | p | FDR | ph | NS | OR [95% CI] | p | FDR | ph | NS | OR [95% CI] | p | FDR | ph | NS | ||||
| CD | ||||||||||||||||||
| 1:186946010 | [PLA2G4A] | A/G | rs6690278 | 1.24 [1.03, 1.50] | 0.026 | 1.00 | 0.49 | 28 | 1.13 [0.90, 1.41] | 0.284 | 0.985 | 0.72 | 28 | 1.67 [1.31, 2.14] | 4.0 × 10−5 | 0.324 | 0.96 | 28 |
| 2:25451789 | POMC--[]-DNMT3A | A/G | rs74580656 | 1.16 [1.08, 1.23] | 8.3 × 10−6 | 0.051 | 0.10 | 29 | 1.16 [1.08, 1.25] | 5.1×10−5 | 0.105 | 0.23 | 29 | 1.15 [1.05, 1.26] | 3.2×10−3 | 0.610 | 0.17 | 29 |
| 2:99873658 | LYG2-[]--LYG1 | A/T | rs17022433 | 1.13 [1.06, 1.19] | 3.9 × 10−5 | 0.115 | 0.90 | 29 | 1.14 [1.07, 1.21] | 5.8×10−5 | 0.112 | 0.81 | 29 | 1.10 [1.02, 1.20] | 0.018 | 0.742 | 0.20 | 29 |
| 2:102444956 | [MAP4K4] | G/A | rs6543098 | 1.09 [1.03, 1.16] | 5.5×10−3 | 0.895 | 0.44 | 29 | 1.04 [0.97, 1.12] | 0.254 | 0.980 | 0.46 | 29 | 1.20[1.10, 1.31] | 4.0 × 10−5 | 0.324 | 0.95 | 29 |
| 2:207500875 | ADAM23--[]-LOC200726 | A/G | rs114144700 | 1.52 [1.00, 2.31] | 0.050 | 1.00 | 0.95 | 10 | 1.21 [0.76, 1.93] | 0.419 | 0.989 | 1.00 | 9 | 2.64 [1.65, 4.21] | 4.8 × 10−5 | 0.324 | 0.91 | 13 |
| 5:159840935 | [SLU7] | A/T | rs41275313 | 1.72 [1.17, 2.52] | 5.8×10−3 | 0.900 | 0.98 | 16 | 1.79 [1.11, 2.87] | 0.017 | 0.879 | 0.96 | 13 | 3.14 [1.84, 5.35] | 2.6 × 10−5 | 0.324 | 0.98 | 12 |
| 6:451373 | IRF4--[]--EXOC2 | A/G | rs4323356 | 1.13 [1.05, 1.21] | 5.2×10−4 | 0.466 | 0.89 | 29 | 1.09 [1.01, 1.18] | 0.037 | 0.904 | 0.62 | 29 | 1.24 [1.12, 1.36] | 2.6 × 10−5 | 0.324 | 0.75 | 29 |
| 6:20607724 | [CDKAL1] | G/A | rs111564329 | 1.16 [0.89, 1.51] | 0.287 | 1.00 | 0.57 | 25 | 1.03 [0.71, 1.49] | 0.894 | 0.999 | 0.48 | 18 | 2.08 [1.47, 2.94] | 3.2 × 10−5 | 0.324 | 0.57 | 23 |
| 6:30129676 | [TRIM10] | C/A | rs2523734 | 1.12 [1.03, 1.20] | 4.9×10−3 | 0.863 | 0.92 | 29 | 1.19 [1.10, 1.30] | *4.9 × 10−5 | 0.105 | 0.84 | 29 | 1.00 [0.89, 1.12] | 0.964 | 0.999 | 0.82 | 29 |
| 6:30982209 | [MUC22] | G/A | rs7755802 | 1.12 [1.06, 1.18] | 1.3×10−4 | 0.229 | 0.57 | 29 | 1.17 [1.10, 1.25] | 1.4 × 10−6 | 0.007 | 0.21 | 29 | 1.02 [0.94, 1.11] | 0.622 | 0.965 | 0.43 | 29 |
| 6:32367777 | [BTNL2] | T/A | † rs9268482 | 1.16 [1.09, 1.23] | 5.4 × 10−6 | 0.049 | 0.39 | 29 | 1.20 [1.12, 1.29] | 6.1 × 10−7 | 0.006 | 0.38 | 29 | 1.09 [1.00, 1.20] | 0.058 | 0.813 | 0.70 | 29 |
| 6:32367847 | [BTNL2] | G/A | † rs3817966 | 1.16 [1.09, 1.23] | 3.6 ×10−6 | 0.049 | 0.56 | 29 | 1.19 [1.11, 1.28] | 7.5 × 10−7 | 0.006 | 0.54 | 29 | 1.10 [1.00, 1.20] | 0.039 | 0.812 | 0.68 | 29 |
| 6:32432835 | HLA-DRA--[]--HLA-DRB5 | A/G | rs9268923 | 1.16 [1.08, 1.24] | *1.2 × 10−5 | 0.060 | 0.83 | 28 | 1.20 [1.11, 1.29] | *1.7 × 10−6 | 0.007 | 0.92 | 28 | 1.09 [0.99, 1.20] | 0.083 | 0.826 | 0.90 | 28 |
| 11:2282206 | MIR4686--[]-ASCL2 | A/C | rs117782746 | 1.26 [1.01, 1.57] | 0.041 | 1.00 | 0.87 | 27 | 1.11 [0.85, 1.44] | 0.437 | 0.989 | 0.92 | 24 | 1.84 [1.37, 2.46] | 4.3 × 10−5 | 0.324 | 0.95 | 27 |
| 16:50714335 | [SNX20] | G/A | rs2270368 | 1.13 [1.06, 1.20] | 3.3×10−4 | 0.399 | 0.06 | 29 | 1.17 [1.09, 1.26] | *2.9 × 10−5 | 0.067 | 0.17 | 29 | 1.06 [0.97, 1.17] | 0.218 | 0.878 | 0.33 | 29 |
| 16:50739582 | [NOD2] | C/G | rs17221417 | 0.88 [0.83, 0.93] | 1.8 × 10−5 | 0.072 | 0.22 | 29 | 0.85 [0.80, 0.91] | 3.3 × 10−6 | 0.011 | 0.12 | 29 | 0.94 [0.87, 1.03] | 0.172 | 0.861 | 0.42 | 29 |
| 16:68681383 | [CDH3] | G/A | rs16958232 | 1.25 [1.00, 1.56] | 0.049 | 1.00 | 0.58 | 26 | 1.15 [0.88, 1.49] | 0.313 | 0.987 | 0.41 | 25 | 1.98 [1.47, 2.67] | *5.9 × 10−6 | 0.175 | 0.63 | 24 |
| 16:68822479 | [CDH1] | A/G | rs8049967 | 1.25 [1.08, 1.45] | 2.7×10−3 | 0.677 | 0.50 | 29 | 1.16 [0.98, 1.37] | 0.093 | 0.947 | 0.76 | 29 | 1.66 [1.36, 2.03] | *7.4 × 10−7 | 0.043 | 0.36 | 29 |
| 16:68824965 | [CDH1] | G/A | rs16958356 | 1.39 [1.14, 1.69] | 1.0×10−3 | 0.547 | 0.92 | 29 | 1.26 [1.00, 1.58] | 0.049 | 0.908 | 0.97 | 27 | 2.06 [1.58, 2.70] | 1.3 × 10−7 | 0.016 | 0.90 | 27 |
| UC | never vs ever (5239 cases vs 3640 cases) | never vs current (5239 cases vs 1080 cases) | never vs former (5239 cases vs 2560 cases) | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1:197865112 | DENND1B---[]-C1orf53 | C/A | rs12071207 | 1.23 [1.01, 1.50] | 0.041 | 0.930 | 0.67 | 26 | 1.82 [1.38, 2.41] | 2.8 × 10−5 | 0.102 | 0.81 | 25 | 1.11 [0.88, 1.40] | 0.380 | 0.970 | 0.53 | 26 |
| 2:100746909 | [AFF3] | A/T | rs12464947 | 1.17 [0.92, 1.49] | 0.195 | 0.979 | 0.85 | 23 | 2.18 [1.50, 3.16] | 3.8 × 10−5 | 0.102 | 0.95 | 20 | 1.16 [0.89, 1.53] | 0.270 | 0.962 | 0.89 | 22 |
| 2:217005480 | [XRCC5] | G/A | rs35883921 | 1.27 [0.93, 1.74] | 0.136 | 0.977 | 0.69 | 17 | 2.69 [1.72, 4.20] | 1.5 × 10−5 | 0.102 | 0.99 | 16 | 1.09 [0.72, 1.65] | 0.688 | 0.983 | 0.78 | 13 |
| 3:46298561 | [CCR3] | A/G | rs13326331 | 1.08 [1.00, 1.17] | 0.045 | 0.943 | 0.91 | 26 | 1.30 [1.16, 1.46] | 1.1 × 10−5 | 0.102 | 0.66 | 26 | 1.00 [0.91, 1.09] | 0.982 | 0.999 | 0.89 | 26 |
| 5:141611379 | NDFIP1--[]--SPRY4 | A/G | rs74512115 | 1.31 [1.12, 1.52] | 4.9×10−4 | 0.264 | 0.16 | 26 | 1.61 [1.29, 2.00] | 2.4 × 10−5 | 0.102 | 0.44 | 26 | 1.29 [1.09, 1.53] | 3.2×10−3 | 0.784 | 0.06 | 26 |
| 5:150038672 | [SYNPO] | G/A | rs14458 | 1.28 [1.02, 1.61] | 0.030 | 0.906 | 0.65 | 25 | 2.10 [1.52, 2.91] | 7.2 × 10−6 | 0.102 | 1.00 | 24 | 1.20 [0.93, 1.54] | 0.164 | 0.962 | 0.86 | 24 |
| 5:150272060 | IRGM--[]-ZNF300 | A/T | rs79716898 | 1.25 [1.06, 1.47] | 7.2×10−3 | 0.780 | 0.26 | 26 | 1.65 [1.31, 2.09] | *2.7 × 10−5 | 0.102 | 0.72 | 26 | 1.18 [0.98, 1.43] | 0.076 | 0.962 | 0.38 | 26 |
| 6:30410206 | TRIM39-RPP21--[]--HLA-E | A/G | rs1264518 | 1.12 [1.03, 1.21] | 9.1×10−3 | 0.812 | 0.11 | 26 | 1.30 [1.14, 1.47] | *4.5 × 10−5 | 0.102 | 0.25 | 26 | 1.06 [0.97, 1.17] | 0.202 | 0.962 | 0.10 | 26 |
| 6:30505000 | HLA-E--[]-GNL1 | G/A | rs62407243 | 1.19 [1.03, 1.39] | 0.022 | 0.892 | 0.92 | 26 | 1.60 [1.28, 1.99] | 3.3 × 10−5 | 0.102 | 0.99 | 26 | 1.12 [0.95, 1.33] | 0.174 | 0.962 | 0.96 | 26 |
| 6:32224139 | [AK123889] | A/G | rs3115569 | 0.84 [0.78, 0.91] | *9.0 × 10−6 | 0.063 | 0.94 | 26 | 0.86 [0.76, 0.97] | 0.017 | 0.397 | 0.77 | 26 | 0.84 [0.77, 0.91] | 7.1×10−5 | 0.307 | 0.91 | 26 |
| 6:32414273 | HLA-DRA-[]--HLA-DRB5 | G/A | rs3129890 | 0.85 [0.79, 0.91] | 7.3 × 10−6 | 0.063 | 0.50 | 26 | 0.88 [0.79, 0.98] | 0.025 | 0.444 | 0.72 | 26 | 0.84 [0.78, 0.91] | 2.6 × 10−5 | 0.302 | 0.44 | 26 |
| 6:32431105 | HLA-DRA--[]--HLA-DRB5 | G/A | rs7747521 | 0.86 [0.80, 0.92] | 2.1 × 10−5 | 0.067 | 0.55 | 26 | 0.92 [0.82, 1.02] | 0.112 | 0.650 | 0.85 | 26 | 0.84 [0.77, 0.91] | 1.7 × 10−5 | 0.302 | 0.46 | 26 |
| 6:34561587 | [C6orf106] | A/T | rs114325894 | 1.30 [0.96, 1.77] | 0.090 | 0.977 | 0.31 | 20 | 2.42 [1.58, 3.70] | 4.4 × 10−5 | 0.102 | 0.99 | 16 | 1.27 [0.88, 1.82] | 0.197 | 0.962 | 0.32 | 19 |
| 7:50565963 | [DDC] | G/A | rs11575404 | 1.25 [1.06, 1.47] | 8.2×10−3 | 0.798 | 0.44 | 26 | 1.67 [1.33, 2.11] | 1.3 × 10−5 | 0.102 | 0.69 | 26 | 1.18 [0.98, 1.41] | 0.088 | 0.962 | 0.63 | 26 |
| 7:50909698 | GRB10--[] COBL | G/A | rs990829 | 1.25 [1.02, 1.52] | 0.027 | 0.892 | 0.29 | 25 | 1.81 [1.37, 2.37] | *2.2 × 10−5 | 0.102 | 0.88 | 24 | 1.16 [0.92, 1.47] | 0.195 | 0.962 | 0.40 | 25 |
| 10:17642806 | [PTPLA] | A/G | rs17141401 | 1.17 [1.08, 1.26] | 1.1×10−4 | 0.155 | 0.67 | 26 | 1.11 [0.98, 1.25] | 0.087 | 0.607 | 0.94 | 26 | 1.20 [1.10, 1.31] | 2.9 × 10−5 | 0.302 | 0.32 | 26 |
| 10:50003599 | [WDFY4] | A/G | rs116883185 | 1.66 [1.29, 2.12] | 6.8×10−5 | 0.137 | 0.81 | 19 | 1.61 [1.09, 2.38] | 0.017 | 0.392 | 0.92 | 19 | 1.88 [1.42, 2.49] | 1.0 × 10−5 | 0.302 | 0.76 | 16 |
| 12:112357984 | ADAM1A--[]--TMEM116 | A/G | rs117614539 | 1.22 [1.00, 1.48] | 0.055 | 0.964 | 0.57 | 24 | 1.98 [1.50, 2.63] | 1.6 × 10−6 | 0.102 | 0.98 | 23 | 1.09 [0.87, 1.38] | 0.454 | 0.973 | 0.42 | 25 |
| 14:81500600 | [TSHR] | A/G | rs77668150 | 1.40 [1.10, 1.78] | 5.6×10−3 | 0.747 | 0.95 | 25 | 2.18 [1.51, 3.13] | 2.7 × 10−5 | 0.102 | 0.96 | 24 | 1.38 [1.06, 1.79] | 0.018 | 0.953 | 1.00 | 25 |
| 15:67444393 | [SMAD3] | A/G | rs8026358 | 1.21 [0.91, 1.60] | 0.197 | 0.979 | 0.97 | 23 | 2.68 [1.74, 4.14] | 8.4 × 10−6 | 0.102 | 0.99 | 19 | 1.09 [0.79, 1.51] | 0.592 | 0.980 | 0.98 | 22 |
| 15:72565787 | PARP6-[]-BC034424 | A/G | rs10518987 | 1.43 [1.14, 1.79] | 2.1×10−3 | 0.579 | 0.76 | 24 | 2.12 [1.55, 2.91] | 2.7 × 10−6 | 0.102 | 0.99 | 23 | 1.31 [1.01, 1.69] | 0.041 | 0.962 | 0.88 | 25 |
| 16:11112283 | [CLEC16A] | G/A | rs117106892 | 1.29 [0.97, 1.70] | 0.077 | 0.977 | 0.50 | 21 | 2.36 [1.59, 3.51] | 1.9 × 10−5 | 0.102 | 0.95 | 21 | 1.27 [0.90, 1.79] | 0.176 | 0.962 | 0.88 | 19 |
| 17:32632378 | CCL11--[]--CCL8 | A/G | rs11652256 | 1.26 [1.03, 1.53] | 0.022 | 0.892 | 0.59 | 26 | 1.90 [1.44, 2.51] | 6.7 × 10−6 | 0.102 | 0.86 | 26 | 1.19 [0.95, 1.49] | 0.129 | 0.962 | 0.92 | 26 |
| 18:42839184 | [SLC14A2] | T/A | rs75044043 | 1.47 [1.19, 1.81] | 3.1×10−4 | 0.221 | 0.59 | 26 | 2.01 [1.46, 2.77] | 1.8 × 10−5 | 0.102 | 0.93 | 25 | 1.46 [1.16, 1.84] | 1.1×10−3 | 0.693 | 0.88 | 26 |
| 21:45597657 | C21orf33--[]--ICOSLG | A/G | rs62228171 | 1.28 [1.04, 1.58] | 0.017 | 0.892 | 0.14 | 24 | 1.93 [1.46, 2.56] | 4.6 × 10−6 | 0.102 | 0.56 | 25 | 1.20 [0.95, 1.51] | 0.136 | 0.962 | 0.47 | 24 |
| IBD | never vs ever (11,291 cases vs 8444 cases) | never vs current(11,291 cases vs 4257 cases) | never vs former (11,291 cases vs 4187 cases) | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1:39749884 | [MACF1] | A/C | rs2039597 | 0.88 [0.83, 0.93] | 1.5 × 10−5 | 0.034 | 0.92 | 55 | 0.92 [0.85, 0.98] | 0.017 | 0.451 | 0.49 | 55 | 0.87 [0.81, 0.94] | 1.8×10−4 | 0.229 | 0.98 | 55 |
| 1:206827199 | DYRK3-[]--MAPKAPK2/IL10 | C/A | rs12076938 | 1.51 [1.10, 2.08] | 0.012 | 0.750 | 0.58 | 17 | 1.26 [0.79, 2.02] | 0.331 | 0.834 | 0.89 | 15 | 2.48 [1.65, 3.72] | 1.2 × 10−5 | 0.090 | 0.87 | 15 |
| 2:185601641 | [ZNF804A] | G/A | rs76903200 | 1.26 [1.09, 1.46] | 2.2×10−3 | 0.519 | 0.90 | 52 | 1.24 [1.02, 1.50] | 0.028 | 0.512 | 0.97 | 48 | 1.47 [1.23, 1.77] | 3.2 × 10−5 | 0.162 | 1.00 | 53 |
| 3:58169958 | FLNB--[]-DNASE1L3 | G/A | rs114990779 | 1.20 [1.04, 1.38] | 0.014 | 0.774 | 0.97 | 53 | 1.45 [1.22, 1.73] | 3.3 × 10−5 | 0.101 | 0.94 | 51 | 1.11 [0.92, 1.35] | 0.270 | 0.841 | 1.00 | 51 |
| 5:30639738 | AK098570 [] CDH6 | C/A | rs1436970 | 1.17 [1.08, 1.28] | 2.5×10−4 | 0.205 | 0.87 | 55 | 1.26 [1.13, 1.40] | 2.0× 10−5 | 0.095 | 0.87 | 55 | 1.18 [1.06, 1.32] | 2.3×10−3 | 0.473 | 0.54 | 55 |
| 5:159880356 | PTTG1--[]--DQ658414 | A/G | rs11951834 | 1.13 [1.06, 1.21] | 3.2×10−4 | 0.223 | 0.02 | 55 | 1.11 [1.01, 1.21] | 0.022 | 0.484 | 0.18 | 55 | 1.21 [1.11, 1.31] | 2.0 × 10−5 | 0.125 | 0.35 | 55 |
| 6:30986015 | [MUC22] | G/A | rs9262492 | 1.07 [1.03, 1.12] | 1.9×10−3 | 0.491 | 0.36 | 55 | 1.15 [1.09, 1.21] | 6.7 × 10−7 | 0.030 | 0.71 | 55 | 1.00 [0.95, 1.06] | 0.902 | 0.988 | 0.20 | 55 |
| 6:31463491 | [MICB] | A/G | rs2516408 | 0.91 [0.87, 0.95] | *2.4 × 10−5 | 0.051 | 0.82 | 55 | 0.92 [0.87, 0.97] | 4.0×10−3 | 0.337 | 0.54 | 55 | 0.91 [0.86, 0.96] | 6.5×10−4 | 0.340 | 0.87 | 55 |
| 6:31488879 | MICB-[]-MCCD1 | A/G | rs4959079 | 1.13 [1.03, 1.23] | 7.8×10−3 | 0.692 | 0.98 | 55 | 1.25 [1.12, 1.39] | *4.5 × 10−5 | 0.101 | 1.00 | 55 | 1.08 [0.96, 1.21] | 0.202 | 0.805 | 0.81 | 55 |
| 6:31543758 | [TNF] | A/G | rs3093661 | 1.22 [1.09, 1.35] | 3.1×10−4 | 0.219 | 0.85 | 55 | 1.34 [1.18, 1.52] | *6.6 × 10−6 | 0.070 | 0.97 | 54 | 1.18 [1.03, 1.36] | 0.016 | 0.585 | 0.97 | 55 |
| 6:32191581 | [NOTCH4] | A/T | rs396960 | 1.09 [1.04, 1.15] | 2.2×10−4 | 0.195 | 0.72 | 55 | 1.14 [1.07, 1.21] | *2.6 × 10−5 | 0.101 | 0.45 | 55 | 1.07 [1.00, 1.13] | 0.039 | 0.647 | 0.47 | 55 |
| 6:32350454 | C6orf10--[]-HCG23 | A/G | rs28732201 | 1.22 [1.09, 1.37] | 8.2×10−4 | 0.339 | 0.30 | 54 | 1.36 [1.18, 1.57] | *1.6 × 10−5 | 0.086 | 0.90 | 52 | 1.24 [1.07, 1.45] | 5.7×10−3 | 0.507 | 0.86 | 53 |
| 6:32408597 | [HLA-DRA] | A/G | rs3129877 | 0.90 [0.85, 0.94] | *3.9 × 10−6 | 0.012 | 0.55 | 55 | 0.88 [0.83, 0.93] | 2.9 × 10−5 | 0.101 | 0.08 | 55 | 0.92 [0.86, 0.97] | 3.8×10−3 | 0.487 | 0.99 | 55 |
| 6:32414273 | HLA-DRA-[]--HLA-DRB5 | G/A | rs3129890 | 0.87 [0.83, 0.92] | 2.9 × 10−8 | 0.001 | 0.10 | 55 | 0.90 [0.84, 0.95] | 3.7×10−4 | 0.192 | 0.43 | 55 | 0.86 [0.81, 0.91] | 7.4 × 10−7 | 0.010 | 0.25 | 55 |
| 6:32431105 | HLA-DRA--[]--HLA-DRB5 | G/A | rs7747521 | 0.87 [0.83, 0.92] | 3.3 × 10−8 | 0.001 | 0.14 | 55 | 0.90 [0.85, 0.96] | 1.2×10−3 | 0.258 | 0.29 | 55 | 0.85 [0.80, 0.91] | 2.3 × 10−7 | 0.006 | 0.55 | 55 |
| 6:32432835 | HLA-DRA--[]--HLA-DRB5 | A/G | rs9268923 | 1.12 [1.06, 1.17] | *3.8 × 10−5 | 0.071 | 0.66 | 52 | 1.15 [1.08, 1.23] | 9.3 × 10−6 | 0.073 | 0.93 | 52 | 1.07 [1.00, 1.15] | 0.041 | 0.654 | 0.72 | 52 |
| 7:50464756 | [IKZF1] | A/C | rs117383894 | 1.21 [0.99, 1.49] | 0.068 | 0.939 | 0.35 | 35 | 1.70 [1.32, 2.19] | 3.7 × 10−5 | 0.101 | 0.82 | 30 | 1.03 [0.75, 1.40] | 0.867 | 0.983 | 0.65 | 31 |
| 7:128729699 | TPI1P2--[]--LOC407835 | C/A | rs77729114 | 1.38 [1.12, 1.69] | 2.2×10−3 | 0.519 | 0.87 | 43 | 1.26 [0.96, 1.65] | 0.093 | 0.643 | 0.98 | 40 | 1.70 [1.34, 2.16] | 1.1 × 10−5 | 0.090 | 0.98 | 46 |
| 9:80226683 | [GNA14] | A/G | rs3905108 | 1.11 [1.02, 1.22] | 0.021 | 0.819 | 0.48 | 55 | 1.26 [1.13, 1.41] | 4.5 × 10−5 | 0.101 | 0.81 | 55 | 1.03 [0.92, 1.16] | 0.626 | 0.947 | 0.90 | 55 |
| 10:6099045 | [IL2RA] | G/A | rs2104286 | 1.07 [1.02, 1.12] | 5.8×10−3 | 0.630 | 0.75 | 55 | 1.14 [1.07, 1.21] | 3.5 × 10−5 | 0.101 | 0.85 | 55 | 1.02 [0.96, 1.08] | 0.509 | 0.926 | 0.63 | 55 |
| 10:50003599 | [WDFY4] | A/G | rs116883185 | 1.26 [1.06, 1.49] | 8.2×10−3 | 0.707 | 0.32 | 41 | 1.16 [0.92, 1.45] | 0.205 | 0.760 | 0.54 | 40 | 1.65 [1.33, 2.05] | 5.6 × 10−6 | 0.051 | 0.95 | 38 |
| 11:2282206 | MIR4686--[]-ASCL2 | A/C | rs117782746 | 1.29 [1.09, 1.53] | 3.1×10−3 | 0.538 | 0.99 | 46 | 1.16 [0.92, 1.46] | 0.207 | 0.761 | 0.99 | 39 | 1.68 [1.36, 2.07] | 1.1 × 10−6 | 0.013 | 1.00 | 46 |
| 13:100108807 | MIR548AN--[]--AK123584 | G/A | rs4238220 | 1.23 [1.03, 1.47] | 0.022 | 0.819 | 0.65 | 48 | 1.61 [1.29, 2.01] | 2.7 × 10−5 | 0.101 | 0.70 | 43 | 1.19 [0.94, 1.52] | 0.154 | 0.776 | 0.96 | 43 |
| 16:11269399 | [CLEC16A] | C/A | rs76391629 | 1.24 [1.10, 1.40] | 6.2×10−4 | 0.300 | 0.35 | 53 | 1.25 [1.07, 1.46] | 4.9×10−3 | 0.344 | 0.69 | 53 | 1.38 [1.19, 1.61] | 3.5 × 10−5 | 0.169 | 0.95 | 52 |
| 22:43704052 | [SCUBE1] | C/A | rs9623807 | 1.12 [1.04, 1.20] | 2.5×10−3 | 0.520 | 0.58 | 55 | 1.05 [0.96, 1.16] | 0.280 | 0.808 | 0.50 | 55 | 1.24 [1.14, 1.36] | 1.6 × 10−6 | 0.018 | 0.93 | 55 |
rs9268482 showed the most significant interaction (p=6.1×10−7) in the never vs current smoker analysis whereas rs3817966 showed the most significant interaction (p=3.6×10−6) in the never vs ever analysis.
Chr:Pos: chromosome number, base-pair position; Gene context: Gene(s) spanning or flanking (<1Mb) the interacting SNP, brackets indicate the position of the SNP, dashes indicate distance to flanking gene (-, >1 kb; --, >10kb; ---, >100kb); Alleles (A/B): minor/major alleles; OR, p: genotypic odds ratio for exposure (see Methods) and p value from a fixed-effects inverse-variance meta-analysis, based upon center-specific Wald tests. Before meta-analysis, p values were individually adjusted for possible population stratification, following a genomic control approach. 95% CI: 95% confidence interval; FDR: false discovery rate calculated by the Benjamini-Hochberg procedure as implemented in the p.adjust tool of R; ph: heterogeneity p value from a Cochrane Q test; NS: number of study centers providing data for meta-analyses of the respective SNP.
Secondary signals with unconditioned p values (see Supplementary Tables 3, 5 and 7 for p values of conditional analyses).
In addition to the candidate variants in Table 2, seven nominally significant interactions (Wald test p<0.01) were found to be of opposite direction for CD and UC (Table 3) which implies that one and the same allele of each of these SNPs increases the risk of CD in smokers, but at the same time protects smokers against UC.
Table 3.
Differential gene–smoking interaction in CD and UC
| Smoking contrast | Chr | SNP | Minor allele | CD | UC | p* | ||||
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| OR [95% CI] | p | ph | OR [95% CI] | p | ph | |||||
| never vs ever | 6 | rs3127599 | A | 1.10 [1.04, 1.17] | 1.5×10−3 | 0.88 | 0.91 [0.85, 0.97] | 7.1×10−3 | 0.19 | 3.6×10−5 |
| 11 | rs117833518 | A | 0.81 [0.70, 0.95] | 8.4×10−3 | 0.49 | 1.33 [1.12, 1.58] | 1.2×10−3 | 0.86 | 3.0×10−5 | |
| 19 | rs2230330 | A | 0.58 [0.38, 0.87] | 8.7×10−3 | 0.76 | 1.84 [1.18, 2.85] | 6.8×10−3 | 0.63 | 1.6×10−4 | |
|
| ||||||||||
| never vs current | 6 | rs176095 | G | 1.12 [1.04, 1.21] | 3.3×10−3 | 0.33 | 0.83 [0.74, 0.94] | 4.3×10−3 | 0.94 | 7.1×10−5 |
| 14 | rs10400765 | G | 0.90 [0.83, 0.97] | 7.3×10−3 | 0.75 | 1.19 [1.06, 1.34] | 3.4×10−3 | 0.41 | 8.6×10−5 | |
| 16 | rs9940076 | A | 1.09 [1.02, 1.17] | 7.3×10−3 | 0.92 | 0.85 [0.77, 0.94] | 1.6×10−3 | 0.58 | 4.1×10−5 | |
|
| ||||||||||
| never vs former | 1 | rs6682359 | A | 1.12 [1.03, 1.22] | 5.8×10−3 | 0.94 | 0.91 [0.85, 0.98] | 9.0×10−3 | 0.56 | 1.5×10−4 |
Chr: chromosome; OR, p: genotypic odds ratio for exposure (see Methods) and p value from a fixed-effects inverse-variance meta-analysis, based upon center -specific Wald tests. Before meta-analysis, p values were individually adjusted for possible population stratification, following a genomic control approach. ph: heterogeneity (across study center) p value from a Cochrane Q test; p*: p value from a heterogeneity (Cochrane Q) test of OR differences in the CD and UC; 95% CI: 95% confidence interval.
By the time of our study, a total of 238 IBD-associated SNPs had been identified in GWAS.1,24 To assess their possible overlap with gene–smoking interactions, we quantified the level of linkage disequilibrium between the 64 unique SNPs identified in our gene–smoking interaction study with those 229 IBD-associated SNPs for which we had genotype data available. Some 29 interacting SNPs were found to be located within 1 Mb of a GWAS-identified SNP. However, only four pairs of SNPs (IBD-associated, smoking interacting) were found to be in moderate linkage disequilibrium (e.g. r2>0.05; Supplementary Table 9).
To further illustrate the validity of the case-only approach, we performed smoking-stratified case-control analyses, considering never vs current smoker contrast, for two selected SNPs from the NOD2 gene region (Supplementary Table 10). In the case of rs2270368, no association with CD was evident in current smokers (OR= 0.96, 95%CI= [0.78, 1.16]), but a protective effect of the minor allele emerged in never smokers (OR= 0.77, 95%CI= [0.69, 0.85]). Similarly, a smaller CD risk was found to be associated with rs17221417 in current smokers (OR= 1.28, 95%CI= [1.07, 1.52]) than in never smokers (OR= 1.74, 95%CI= [1.58, 1.91]).
HLA-Smoking Interaction
In our focused analyses of classical HLA-alleles, a gene–smoking interaction meeting our filtering criteria was observed for 4 alleles in CD, 1 allele in UC and 5 alleles in IBD (Table 4). Overall, unique alleles were identified with suggestive evidence of gene–smoking interaction with at least one smoking-status contrast. All of these alleles were found to comply with the G-E independence assumption in controls.
Table 4.
HLA alleles involved in gene–smoking interaction
| IBD type | HLA allele | never vs ever | never vs current | never vs former | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| OR [95% CI] | p | ph | OR [95% CI] | p | ph | OR [95% CI] | p | ph | ||
| CD | HLA-B*57 | 1.31 [1.14, 1.51] | 1.2×10−4 | 0.77 | 1.39 [1.19, 1.62] | 2.7×10−5 | 0.70 | 1.33 [1.09, 1.63] | 5.1×10−3 | 0.97 |
| HLA-B*57:01 | 1.32 [1.15, 1.52] | 1.1×10−4 | 0.70 | 1.40 [1.19, 1.63] | 2.9×10−5 | 0.62 | 1.34 [1.09, 1.65] | 4.7×10−3 | 0.97 | |
| HLA-DQA1*02:01 | 1.21 [1.12, 1.31] | 1.6×10−6 | 0.55 | 1.24 [1.14, 1.36] | 6.6×10−7 | 0.78 | 1.18 [1.05, 1.32] | 4.2×10−3 | 0.38 | |
| HLA-DRB1*07:01 | 1.20 [1.11, 1.30] | 2.5×10−6 | 0.52 | 1.24 [1.14, 1.35] | 1.3×10−6 | 0.74 | 1.18 [1.05, 1.32] | 4.0×10−3 | 0.34 | |
|
| ||||||||||
| UC | HLA-DRB3*91:01 | 0.81 [0.74, 0.89] | 1.8×10−5 | 0.50 | 0.77 [0.66, 0.90] | 7.9×10−4 | 0.84 | 0.84 [0.76, 0.94] | 1.7×10−3 | 0.45 |
|
| ||||||||||
| IBD | HLA-B*57 | 1.25 [1.12, 1.40] | 9.4×10−5 | 0.84 | 1.39 [1.22, 1.59] | 1.4×10−6 | 0.98 | 1.22 [1.06, 1.41] | 6.9×10−3 | 0.95 |
| HLA-B*57:01 | 1.25 [1.12, 1.40] | 1.0×10−4 | 0.79 | 1.40 [1.22, 1.61] | 1.1×10−6 | 0.97 | 1.22 [1.05, 1.41] | 8.8×10−3 | 0.94 | |
| HLA-DQA1*02:01 | 1.15 [1.08, 1.23] | 5.5×10−6 | 0.45 | 1.23 [1.14, 1.32] | 1.0×10−7 | 0.91 | 1.10 [1.02, 1.20] | 0.015 | 0.24 | |
| HLA-DQB1*02:02 | 1.14 [1.06, 1.22] | 3.7×10−4 | 0.37 | 1.21 [1.10, 1.31] | 2.6×10−5 | 0.64 | 1.09 [1.00, 1.20] | 0.058 | 0.45 | |
| HLA-DRB1*07:01 | 1.15 [1.08, 1.22] | 7.3×10−6 | 0.46 | 1.22 [1.13, 1.31] | 2.3×10−7 | 0.90 | 1.11 [1.02, 1.20] | 0.013 | 0.23 | |
For details, see legend to Table 2.
Functional Annotation of Interacting Variants
We functionally annotated the 64 SNPs identified as potentially interacting with smoking for at least one of three smoking contrasts (see Supplementary Methods). For 37 of the interaction signals (58%), the lead SNP mapped within the transcript of a known gene while 27 signals were located in intergenic regions (Supplementary Table 11). The interacting SNPs included one coding missense variant (rs41275313) in the SLU7 gene. However, all coding SNPs were predicted to be benign (Supplementary Table 12). Non-coding SNPs rs76903200 (ZNF804A; intronic), rs79716898 (intergenic between IRGM and ZNF300) and rs62407243 (intergenic between HLA-E and GNL1) were found to be potentially deleterious based upon the prediction of reduced organismal fitness (CADD scores; Supplementary Table 12). Among coding SNPs that were in strong linkage disequilibrium (r2>0.8 in the 1000 Genomes European samples; n=376 variants) with the 64 lead SNPs (see Methods), we identified another 5 synonymous SNPs at the ZNF300, IRGM, UHRF1BP1 and NOD2 (2x) gene loci and 1 non-synonymous SNP at the GRAMD2 gene (Supplementary Table 13).
We also examined which of the 64 lead SNPs and of the SNPs in strong linkage disequilibrium (r2>0.8) with the lead SNPs mapped to eQTLs from GTEx and Geuvadis eQTL studies,25 or to functional annotations of the noncoding genome in different cell types provided by the Roadmap Epigenomics26 and ENCODE27 projects, using web tool HaploReg28 (Supplementary Table 14; see Methods). The results are summarized in Supplementary Figure 13.
Pathway and Cell Type Enrichment Analyses
To ascertain whether genes at the putatively interacting loci were highly expressed in certain tissue/cell types, we conducted pathway and tissue/cell type enrichment analyses using DEPICT29 with 77,840 microarray gene expression profiles from human, rat and mouse and 209 tissue/cell type annotations30 (see Supplementary Methods). From DEPICT, we identified 24 gene sets (Supplementary Table 15) and 20 tissues (Supplementary Table 16) with significant enrichment of genes within the suggested interacting loci (false discovery rate <0.01). The results mainly point towards perturbation of immune response pathways in blood.
Loss of Nod2 and Il10 renders mice susceptible to intestinal inflammation in response to cigarette smoke exposure
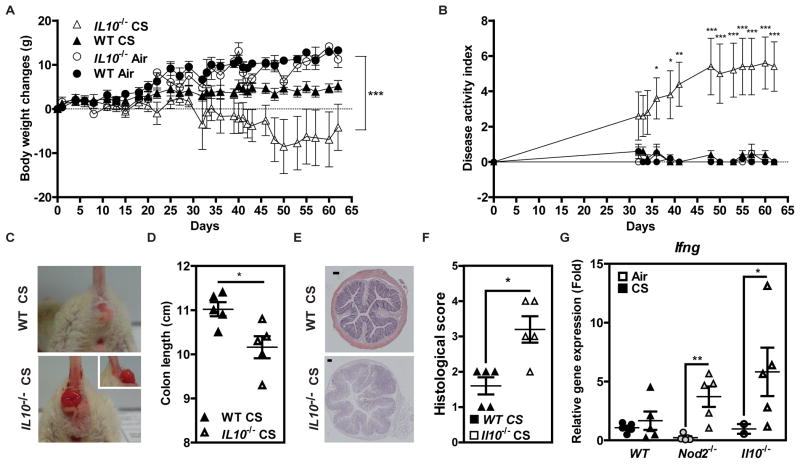
The possible impact of cigarette smoke on intestinal homeostasis was next evaluated in mice that were deficient for either Il10 or Nod2, two proteins encoded by genes that were identified as potential smoking-interacting genes in our human data (Table 2). While no signs of disease were apparent in wild-type (WT) mice after 8 weeks of cigarette smoke exposure, smoking Il10−/− mice experienced greater body-weight loss (Figure 3A), increased disease activity index (Figure 3B, significant gene-smoking interaction, p=0.01 at day 62), and an accelerated development of rectal prolapses (Figure 3C). Consequently, cigarette smoking markedly reduced the colon length of Il10−/− mice, compared to similarly treated WT animals (Figure 3D). While histological analysis failed to reveal any signs of inflammation in WT mice, a greater infiltration of immune cells within the colonic mucosa (Figures 3E and 3F) and an enhanced expression of Ifng transcipts in the ileum were observed in smoking Il10−/− mice (Figure 3G). Likewise, Nod2−/− mice also showed higher expression of Ifng in their ileum, but lacked signs of colitis and prolapse development (Figure 3G and Supplementary Figure 14). Taken together, our data suggest a strong protective role of both Il10 and Nod2 on intestinal homeostasis in response to tobacco smoking.
Figure 3. Impact of interleukin-10 and Nod2 deficiency on the risk of intestinal inflammation in cigarette smoke-exposed mice.
Wild-type (WT) and Il10−/− mice were exposed to cigarette smoke (CS), 5 days a week, over a period of 8 weeks. (A) Changes in body weight. (B) Disease Activity Index. (C) Prolapsus apparition under CS exposure in Il10−/− mice. (D) Colon length. (E) Representative H&E stainings. (F) Histological score. (G) Relative expression level of Ifng gene in the ileal tissue of WT, Nod2-deficient and Il10-deficent mice that were either exposed to CS or not. *p<0.05, ***p<0.001 (Kruskal-Wallis test). Data are representative of two independent experiments.
Discussion
We performed an extensive meta-analysis to investigate possible gene–smoking interactions in relation to the risk for CD, UC or IBD. To this end, we used the existing Immunochip-wide data collated by the IIBDGC. Our analysis identified 19 SNPs for CD, 25 SNPs for UC and 25 SNPs for IBD that potentially interact with regard to disease risk considering at least one of three smoking contrasts (never vs ever, never vs current, or never vs former). Interestingly, the largest number of interacting SNPs was identified with the never vs current contrast for UC, but with the never vs former contrast for CD.
Our findings are highly relevant to furthering the understanding of IBD etiology for various reasons. First, the discrepancies observed between the three smoking contrasts suggest that the precise mechanism by which the smoking-induced disease risk of an individual is modified by their genetic make-up differs between past and current smokers. This disparity has not been considered in previous epidemiologic studies.7,31–34 Second, we were able to show, for the first time, that some of the modification of smoking-induced IBD risk is brought about by more than one genetic factor located in the HLA region. Third, a clear-cut dependence upon smoking behavior became apparent in the HLA region for CD risk, but not UC risk. Such a differential role of G×E in the two IBD sub-entities may be a key to understanding why the smoking-induced risk for UC may still increase with time even decades after smoking cessation.6 Finally, the scope and nature of gene–smoking interaction in IBD may be exemplary for other diseases. For example, in line with our own results, BTNL2 and HLA-DRB5 were recently identified as candidate interaction signals as well in a rheumatoid arthritis SNP-smoking interaction study.35 Some 29 of the 64 unique interacting SNPs (45%) were found to lie in close vicinity (≤1Mb) of genes that were previously identified as being disease-associated in GWAS, including IL10 (Table 2).1,24 However, in view of the general lack of strong linkage disequilibrium between interacting and IBD-associated SNPs, we conclude that the respective association and interaction signals may highlight different genetic effects. Thus, even if two functionally relevant variants lie in the same gene or functional unit, the ensuing disease risk may still be modified by smoking for one variant, but not for the other. Along the same line, our focused analysis of the HLA region revealed that only a subset of the IBD-predisposing alleles16 was found to interact with smoking as well. We also identified seven SNPs that seemed to interact with smoking in opposite directions with regard to CD and UC. Since statistical interaction can be viewed from different angles, this difference may mean one of two things: Either a genetic mechanism predisposing to one of the two sub-entities is rendered protective against the other in the presence of smoking, or the effect of smoking in relation to one sub-entity is reversed in comparison to the other by that mechanism. Simply put, a certain genotype may simultaneously render smoking a risk factor for CD and a protective factor against UC.
We used web-based computational tools for evaluating the potential functional consequences of the interacting SNPs. Only few of the SNPs were found to have a known effect rendering a firm biological interpretation of the results difficult (Supplementary Tables 12–16). However, many of the interacting SNPs are located near or within genes that may be involved in, or interfere with, mucosal barrier function (e.g. NOD2, IRGM, CDH1 and GPSM336) or the adaptive immune response (e.g. IL2RA, CCL11, CCL8, MICB, IL10 and the HLA region). Several SNPs in the HLA region were also found to interact with smoking in relation to either CD, UC or IBD (Table 2), including one SNP (rs3129890) that had previously been found to be associated with a high risk of rheumatoid arthritis among smokers.35 Moreover, we identified six HLA alleles with suggestive gene–smoking interaction (namely HLA-DRB3*91:01, HLA-B*57, HLA-B*57:01, HLA-DQA1*02:01, HLA-DQB1*02:02 and HLA-DRB1*07:01). Of these six alleles, four were identified previously to have a main effect16 either on CD risk (HLA-B*57:01, HLA-DQA1*02:01 and HLA-DRB1*07:01) or on UC risk (HLA-DRB1*07:01, HLA-DQA1*02:01 and HLA-DQB1*02:02). The overlap between genetic interaction and main effect signals suggests that perturbation of the adaptive immune response may be one important mechanism by which smoking differentially confers risk to either CD or UC. Also of interest in this regard are the seven SNPs that interact with smoking in opposite directions in CD and UC (Table 3), which included one SNP (rs176095) that has been found to be associated with atopic dermatitis37 and asthma38 before. This SNP is located on chromosome 6 near the GPSM3 gene that regulates monocytes function and inhibits NLRP3-coupled inflammasome activation.36 NLRP3 is a member of the NOD-like receptor (NLR) family of intracellular sensors of danger signals, such as pathogen-associated molecular patterns (PAMPs), that controls IL-1α response to cigarette smoke exposure in mice.39
Our epidemiological results were exemplarily corroborated by the observation that Nod2- and Il10-deficient mice that were experimentally exposed to cigarette smoke had a greater risk of ileitis than similarly treated WT mice. One possible explanation for this difference could be that cigarette smoke exposure compromises the barrier function of the small intestine more effectively as a result of lower NOD2 gene expression and a consequent reduction of chemokine and antimicrobial peptides secretion.40 Likewise, long term exposure to cigarette smoke decreases the number of Foxp3+ cells and the expression of IL-10 which, in combination, represses IFN-γ production.41 Equally important is that a greater risk of colitis was observed in Il10-deficient mice that were exposed to cigarette smoke but not in wild-type and Nod2-deficient mice, suggesting a differential role of Il10 on disease location in response to cigarette smoke.
We employed the powerful albeit rarely used case-only design that relies upon two key assumptions, namely that (i) the disease is sufficiently rare (i.e. prevalence <5%) in the general population and that (ii) G and E are uncorrelated in the general population. Case-only studies offer a number of methodological advantages compared to traditional case-control studies, including higher per-sample power and better exposure data quality.18,42,43 The samples available to us provided 90% power for a small interaction effect (OR=1.15) in a case-only analysis, but only 3% power in a case-control analysis. However, since the validity of the results of case-only analyses depends upon the validity of the G-E independence assumption, the latter must be assessed empirically, for example, in control data from GWAS. This requirement cannot be obviated because many genetic variants are known to be associated with smoking behavior at the population level.44 Indeed, in our study, 15,196 SNPs violated the G-E independence assumption with at least one of the three smoking contrasts considered (never vs ever, never vs current, or never vs former). Only one of these, a synonymous SNP (rs1051730) in the nicotinic receptor gene CHRNA3 at 15q25, has been identified so far to be associated with smoking behavior.45 For all other G-E associations notified in the controls of our study, the underlying reason remains unclear because none of the respective SNPs coincides with any previously identified association with smoking behavior.44,45
One limitation of our study is that the smoking data were abstracted locally from existing clinical and/or research records which may have introduced some variability across centers in the way the data were initially recorded. To mitigate this, we focused on a clear-defined classification of smoking behavior as either (i) current (smoking within the past 3 months), (ii) ever (current or ex-smoker) or (iii) never as of the date of diagnosis (cases) or recruitment (controls). We also ascertained the year in which the subjects first started and finally stopped smoking, as applicable, and used this information in conjunction with the year of diagnosis to verify that each center was applying the smoking definition correctly. Since our study employed meta-analysis techniques to evaluate gene–smoking interactions at the center level, any remaining measurement error would have resulted only in a loss of power but not in an increased type 1 error rate. Moreover, the center-specific interaction estimates were not found to be related to the center-specific smoking rates for any of the interacting SNPs, an observation that reinforces our notion that differential assessment of smoking behavior was unlikely to affect the validity of our results. This notwithstanding, without accurate information on the actual number of cigarettes smoked (which would have been much more difficult to obtain), we were unable to account for potential dose-dependent effects of smoking.
In summary, our genome-wide study of G×E in IBD identified 64 SNPs with strong evidence for a complex modifying role in the smoking-related etiology of IBD. Functional studies in mice lend additional experimental support to these epidemiological findings by highlighting a direct effect of Il10 and Nod2 on disease risk in response to smoking. Our study thus sheds new light on the role of smoking as an important component of IBD pathogenesis interacting with the genetic background of at-risk individuals.
Supplementary Material
Acknowledgments
This project was supported by the Deutsche Forschungsgemeinschaft (DFG) through Excellence Cluster 306 ‘Inflammation at Interfaces’, Research and Training Group 1743 ‘Genes, Environment and Inflammation’, FOR 2107 and the PopGen Biobank. Additional support was provided by the German Federal Ministry of Education and Research (BMBF) within the framework of the e:Med research and funding concept (sysINFLAME grant 01ZX1306A). PS was supported by DCC grant (NIDDK grant 5U01DK062429) and VA by “Knud og Edith Eriksens Mindefond”. The mice studies were supported by the Fondation pour la Recherche Médicale (grant number DEQ20130326475) to MC and by the Conseil Régional Nord-Pas-de-Calais (Smo’X project, grant number 13001437) to PG. We thank all patients for their participation in the local studies. IIBDGC is gratefully acknowledged for providing access to the genotype and smoking data.
Footnotes
Author contributions
Pankaj Yadav, David Ellinghaus, Sandra Freitag-Wolf, Frauke Degenhardt, and Gabrielle Boucher performed statistical and computational analyses; Gaelle Remy, Anabelle Cesaro, Murielle Pichavant and Philippe Gosset undertook most of the animal experiments. Myriam Delacre performed qRTPCR analysis. Histological scoring was performed by Anabelle Cesaro and Mathias Chamaillard. Pankaj Yadav, David Ellinghaus, Astrid Dempfle, Vibeke Andersen, and Michael Krawczak wrote draft versions of the manuscript; Pankaj Yadav, David Ellinghaus, Sandra Freitag-Wolf, Frauke Degenhardt, Gabrielle Boucher, John D Rioux, Michael Krawczak, Philippe Gosset, Andre Franke, Mathias Chamaillard, Astrid Dempfle, and Vibeke Andersen conceived, designed and managed the study. The remaining authors contributed to the study conception, design, genotyping QC and/or writing of the manuscript. All authors saw, had the opportunity to comment on, and approved the final draft.
Conflicts of interest
Vibeke Andersen acknowledges receiving compensation from Merck (MSD) and Janssen as a consultant and advisory board member. The other authors declare that there is no conflict of interest.
Supplementary data include 16 tables (including five spreadsheets), 14 figures, a list of the IIBDGC members and their affiliations, methods, and references.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
Author names in bold designate shared co-first authorship.
- 1.Liu JZ, van Sommeren S, Huang H, et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet. 2015;47:979–86. doi: 10.1038/ng.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jostins L, Ripke S, Weersma RK, et al. Host–microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Franke A, McGovern DPB, Barrett JC, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat Genet. 2010;42:1118–25. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andersen V, Vogel U. Systematic review: interactions between aspirin, and other nonsteroidal anti-inflammatory drugs, and polymorphisms in relation to colorectal cancer. Aliment Pharmacol Ther. 2014;40:147–59. doi: 10.1111/apt.12807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andersen V, Holst R, Vogel U. Systematic review: diet-gene interactions and the risk of colorectal cancer. Aliment Pharmacol Ther. 2013;37:383–91. doi: 10.1111/apt.12180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Higuchi LM, Khalili H, Chan AT, et al. A prospective study of cigarette smoking and the risk of inflammatory bowel disease in women. Am J Gastroenterol. 2012;107:1399–406. doi: 10.1038/ajg.2012.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cosnes J. Tobacco and IBD: relevance in the understanding of disease mechanisms and clinical practice. Best Pract Res Clin Gastroenterol. 2004;18:481–496. doi: 10.1016/j.bpg.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 8.Parkes GC, Whelan K, Lindsay JO, et al. Smoking in inflammatory bowel disease: impact on disease course and insights into the aetiology of its effect. J Crohns Colitis. 2014;8:717–25. doi: 10.1016/j.crohns.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 9.Biedermann L, Brülisauer K, Zeitz J, et al. Smoking cessation alters intestinal microbiota: insights from quantitative investigations on human fecal samples using FISH. Inflamm Bowel Dis. 2014;20:1496–501. doi: 10.1097/MIB.0000000000000129. [DOI] [PubMed] [Google Scholar]
- 10.Benjamin JL, Hedin CRH, Koutsoumpas A, et al. Smokers with active Crohn’s disease have a clinically relevant dysbiosis of the gastrointestinal microbiota. Inflamm Bowel Dis. 2012;18:1092–100. doi: 10.1002/ibd.21864. [DOI] [PubMed] [Google Scholar]
- 11.Klareskog L, Stolt P, Lundberg K, et al. A new model for an etiology of rheumatoid arthritis: smoking may trigger HLA-DR (shared epitope)-restricted immune reactions to autoantigens modified by citrullination. Arthritis Rheum. 2006;54:38–46. doi: 10.1002/art.21575. [DOI] [PubMed] [Google Scholar]
- 12.Wagner CA, Sokolove J, Lahey LJ, et al. Identification of anticitrullinated protein antibody reactivities in a subset of anti-CCP-negative rheumatoid arthritis: association with cigarette smoking and HLA-DRB1 “shared epitope” alleles. Ann Rheum Dis. 2015;74:579–86. doi: 10.1136/annrheumdis-2013-203915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Helbig KL, Nothnagel M, Hampe J, et al. A case-only study of gene-environment interaction between genetic susceptibility variants in NOD2 and cigarette smoking in Crohn’s disease aetiology. BMC Med Genet. 2012;13:14. doi: 10.1186/1471-2350-13-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fowler EV, Doecke J, Simms LA, et al. ATG16L1 T300A shows strong associations with disease subgroups in a large Australian IBD population: further support for significant disease heterogeneity. Am J Gastroenterol. 2008;103:2519–26. doi: 10.1111/j.1572-0241.2008.02023.x. [DOI] [PubMed] [Google Scholar]
- 15.Doecke JD, Simms LA, Zhao ZZ, et al. Smoking behaviour modifies IL23r-associated disease risk in patients with Crohn’s disease. J Gastroenterol Hepatol. 2015;30:299–307. doi: 10.1111/jgh.12674. [DOI] [PubMed] [Google Scholar]
- 16.Goyette P, Boucher G, Mallon D, et al. High-density mapping of the MHC identifies a shared role for HLA-DRB1*01:03 in inflammatory bowel diseases and heterozygous advantage in ulcerative colitis. Nat Genet. 2015;47:172–9. doi: 10.1038/ng.3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Purcell S, Neale B, Todd-Brown K, et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Piegorsch WW, Weinberg CR, Taylor JA. Non-hierarchical logistic models and case-only designs for assessing susceptibility in population-based case-control studies. Stat Med. 1994;13:153–162. doi: 10.1002/sim.4780130206. [DOI] [PubMed] [Google Scholar]
- 19.Yadav P, Freitag-Wolf S, Lieb W, et al. Allowing for population stratification in case-only studies of gene–environment interaction, using genomic control. Hum Genet. 2015;134:1117–25. doi: 10.1007/s00439-015-1593-y. [DOI] [PubMed] [Google Scholar]
- 20.Benjamini Y, Hochberg Y, Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300. [Google Scholar]
- 21.Gauderman WJ. Sample size requirements for matched case-control studies of gene-environment interaction. Stat Med. 2002;21:35–50. doi: 10.1002/sim.973. [DOI] [PubMed] [Google Scholar]
- 22.Yang Q, Khoury MJ, Flanders WD. Sample size requirements in case-only designs to detect gene-environment interaction. Am J Epidemiol. 1997;146:713–720. doi: 10.1093/oxfordjournals.aje.a009346. [DOI] [PubMed] [Google Scholar]
- 23.Kraft P, Yen YC, Stram DO, et al. Exploiting gene-environment interaction to detect genetic associations. Hum Hered. 2007;63:111–119. doi: 10.1159/000099183. [DOI] [PubMed] [Google Scholar]
- 24.Ellinghaus D, Jostins L, Spain SL, et al. Analysis of five chronic inflammatory diseases identifies 27 new associations and highlights disease-specific patterns at shared loci. Nat Genet. 2016;48:510–518. doi: 10.1038/ng.3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.GTEx Consortium. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–60. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roadmap Epigenomics Consortium. Kundaje A, Meuleman W, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40:D930–4. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pers TH, Karjalainen JM, Chan Y, et al. Biological interpretation of genome-wide association studies using predicted gene functions. Nat Commun. 2015;6:5890. doi: 10.1038/ncomms6890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fehrmann RSN, Karjalainen JM, Krajewska M, et al. Gene expression analysis identifies global gene dosage sensitivity in cancer. Nat Genet. 2015;47:115–25. doi: 10.1038/ng.3173. [DOI] [PubMed] [Google Scholar]
- 31.Mahid SS, Minor KS, Soto RE, et al. Smoking and Inflammatory Bowel Disease: A Meta-analysis. Mayo Clin Proc. 2006;81:1462–1471. doi: 10.4065/81.11.1462. [DOI] [PubMed] [Google Scholar]
- 32.García Rodríguez LA, González-Pérez A, Johansson S, et al. Risk factors for inflammatory bowel disease in the general population. Aliment Pharmacol Ther. 2005;22:309–15. doi: 10.1111/j.1365-2036.2005.02564.x. [DOI] [PubMed] [Google Scholar]
- 33.Birrenbach T, Böcker U. Inflammatory bowel disease and smoking: a review of epidemiology, pathophysiology, and therapeutic implications. Inflamm Bowel Dis. 2004;10:848–59. doi: 10.1097/00054725-200411000-00019. [DOI] [PubMed] [Google Scholar]
- 34.Thomas GA, Rhodes J, Green JT, et al. Role of smoking in inflammatory bowel disease: implications for therapy. Postgrad Med J. 2000;76:273–9. doi: 10.1136/pmj.76.895.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang X, Källberg H, Chen Z, et al. An Immunochip-based interaction study of contrasting interaction effects with smoking in ACPA-positive versus ACPA-negative rheumatoid arthritis. Rheumatology (Oxford) 2016;55:149–55. doi: 10.1093/rheumatology/kev285. [DOI] [PubMed] [Google Scholar]
- 36.Giguère PM, Gall BJ, Ezekwe EAD, et al. G Protein signaling modulator-3 inhibits the inflammasome activity of NLRP3. J Biol Chem. 2014;289:33245–57. doi: 10.1074/jbc.M114.578393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weidinger S, Willis-Owen SAG, Kamatani Y, et al. A genome-wide association study of atopic dermatitis identifies loci with overlapping effects on asthma and psoriasis. Hum Mol Genet. 2013;22:4841–56. doi: 10.1093/hmg/ddt317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li L, Kabesch M, Bouzigon E, et al. Using eQTL weights to improve power for genome-wide association studies: a genetic study of childhood asthma. Front Genet. 2013;4:103. doi: 10.3389/fgene.2013.00103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pauwels NS, Bracke KR, Dupont LL, et al. Role of IL-1α and the Nlrp3/caspase-1/IL-1β axis in cigarette smoke-induced pulmonary inflammation and COPD. Eur Respir J. 2011;38:1019–28. doi: 10.1183/09031936.00158110. [DOI] [PubMed] [Google Scholar]
- 40.Aldhous MC, Soo K, Stark LA, et al. Cigarette Smoke Extract (CSE) Delays NOD2 Expression and Affects NOD2/RIPK2 Interactions in Intestinal Epithelial Cells Vij N, ed. PLoS One. 2011;6:e24715. doi: 10.1371/journal.pone.0024715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Duan M-C, Zhang J-Q, Liang Y, et al. Infiltration of IL-17-Producing T Cells and Treg Cells in a Mouse Model of Smoke-Induced Emphysema. Inflammation. 2016;39:1334–1344. doi: 10.1007/s10753-016-0365-8. [DOI] [PubMed] [Google Scholar]
- 42.Gatto NM, Campbell UB, Rundle AG, et al. Further development of the case-only design for assessing gene-environment interaction: evaluation of and adjustment for bias. Int J Epidemiol. 2004;33:1014–24. doi: 10.1093/ije/dyh306. [DOI] [PubMed] [Google Scholar]
- 43.Yadav P, Freitag-Wolf S, Lieb W, et al. The role of linkage disequilibrium in case-only studies of gene–environment interactions. Hum Genet. 2015;134:89–96. doi: 10.1007/s00439-014-1497-2. [DOI] [PubMed] [Google Scholar]
- 44.Wain LV, Shrine N, Miller S, et al. Novel insights into the genetics of smoking behaviour, lung function, and chronic obstructive pulmonary disease (UK BiLEVE): a genetic association study in UK Biobank. Lancet Respir Med. 2015;3:769–781. doi: 10.1016/S2213-2600(15)00283-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tobacco and Genetics Consortium. Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nat Genet. 2010;42:441–7. doi: 10.1038/ng.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.