Abstract
We participated in the Critical Assessment of Genome Interpretation eQTL challenge to further test computational models of regulatory variant impact and their association with human disease. Our prediction model is based on a discriminative gapped-kmer SVM (gkm-SVM) trained on genome-wide chromatin accessibility data in the cell type of interest. The comparisons with Massively Parallel Reporter Assays (MPRA) in lymphoblasts show that gkm-SVM is among the most accurate prediction models even though all other models used the MPRA data for model training, while gkm-SVM did not. In addition, we compare to other MPRA datasets and show that gkm-SVM is a reliable predictor of expression and that deltaSVM is a reliable predictor of variant impact in K562 cells and mouse retina. We further show that DHS (DNase-I Hypersensitive Sites) and ATAC-seq (Assay for Transposase-Accessible Chromatin using sequencing) data are equally predictive substrates for training gkm-SVM, and that DHS regions flanked by H3K27Ac and H3K4me1 marks are more predictive than DHS regions alone.
Keywords: enhancers, regulatory variation, gene regulation, machine learning, MPRA, eQTL analysis
Introduction
The contribution of regulatory variation to human disease is becoming an increasingly active area of research. This is motivated in part by the observation that the majority of variants associated with disease by Genome Wide Association Studies (GWAS) are located in intergenic and putative regulatory regions (Hindorff et al., 2009; Maurano et al., 2012; Gusev et al., 2014), and in part by a growing number of regulatory variants whose disease impact has been directly experimentally elucidated (Musunuru et al., 2010; Bauer et al., 2013; Huang et al., 2014; Canver et al., 2015; Soldner et al., 2016). To build a predictive model of how regulatory variants contribute to disease by modulating the activity of regulatory elements, my lab has developed a computational framework for systematically identifying the necessary set of transcription factor (TF) binding sites active in a given cell type, and quantifying the impact of modulation of these TF binding sites by genetic variants. These sequence changes can be naturally occurring SNPs, indels, or synthetic or CRISPR-induced sequence scrambling. Our discriminative gkm-SVM model is typically trained on a positive set of active regulatory regions in the cell type and a negative set of non-active regions. The gkm-SVM output is a score that can be summarized as the sum of weights for each k-mer occurring in the sequence to be evaluated. Sequence alterations change this set of k-mers, and change the score. We use deltaSVM to refer to the change of the gkm-SVM score induced by a sequence change, and deltaSVM is thus the gkm-SVM prediction of variant impact.
While we have previously shown that gkm-SVM can predict ChIP-seq binding for the complete set of ENCODE TFs (Ghandi et al., 2014), and that deltaSVM can predict variant impact (Lee et al., 2015) more accurately than alternative approaches (Kircher et al., 2014; Ritchie et al., 2014; Peterson et al., 2016), the Critical Assessment of Genome Interpretation eQTL challenge provided a rigorous test of this method in a blind control. Further, the eQTL challenge allowed us to assess gkm-SVM’s ability to predict expression levels directly, in addition to expression change, which we had not previously evaluated. Here we show on the eQTL challenge dataset and on previously published datasets that gkm-SVM is indeed a reliable predictor of expression levels, in addition to variant impact. As described in more detail in the eQTL challenge overview paper (Kreimer et al., 2016), the eQTL challenge dataset reports expression levels in Lymphoblast Cell Lines (LCLs) from a Massively Parallel Reporter Assay (MPRA) for both alleles of a set of 9116 150bp human DNA sequences encompassing variants which had been previously identified as eQTL loci in LCLs (Consortium, 2012; Lappalainen et al., 2013). Prediction groups were provided the expression levels of a subset of 3044 pairs of alleles as a training set to train parameters of the computational prediction models. In the first part of the challenge, an additional 3044 alleles were tested for expression, and groups were asked to submit predictions for which would be positive. In the second part of the challenge, 401 additional variants which were positive for expression were tested for allelic differences, and groups were asked to predict which among these pairs of alleles would be differentially expressed. Each group was allowed to submit predictions from several distinct models, putting forward one main model for primary scoring.
Our gkm-SVM method is unique among the submitted eQTL challenge prediction methods in that we did not use the MPRA training set to develop our main model, yet gkm-SVM and deltaSVM were among the most accurate predictors for parts one and two of the challenge. Gkm-SVM used chromatin accessibility data from DNase-seq for training, and we show below that ATAC-seq chromatin accessibility data produces equally accurate predictions in a MPRA in mouse retina. This has significant consequences for the utility of gkm-SVM in the design of future MPRA experiments to test disease associated variants in other cell types. The constructs targeted for MPRA in this eQTL challenge were designed based on the existence of previous experimental evidence that these loci were eQTLs in LCLs (Consortium, 2012; Lappalainen et al., 2013; Tewhey et al., 2016). For most other disease relevant cell types, such eQTL data does not exist, and may be quite difficult to acquire. The results in this paper suggest that gkm-SVM can be trained on more easily obtainable DHS or ATAC-seq data in the cell type of interest, and then gkm-SVM be used to both identify high confidence enhancers active in this cell type, and identify variants which will modulate cell-specific activity, greatly reducing the space of possible sequences required for testing in MPRA validation experiments.
Materials and Methods
gkm-SVM parameters and training sets
We ran the gkm-SVM R package (Ghandi et al., 2016) using default parameters (total word length l=10, k=6 informative columns, and d=3), and scored all possible 10-mers to generate 10-mer weights. We then calculated a gkm-SVM score for each construct tested using the full insert sequence. For the GM12878 cell line, we used a positive set of 22,384 300bp DHS regions defined by MACS (Zhang et al., 2008) and then optimized to maximize signal over the fixed 300bp interval, and used the average weights from training against each of five equal size GC and repeat matched (Fletez-Brant et al., 2013) negative genomic sequence sets, as described in (Lee et al., 2015). We choose the 300bp length to maximize cross-fold validation rates for DHS. To train on the MPRA data, we used the top expressing 1697 training set sequences as a positive set, and the bottom expressing 1851 training set sequences as a negative set, out of the total 6088 training sequences provided. For the K562 cell line, we used the top 10,000 300bp DHS (ENCODE Consortium, 2012) MACS peaks optimized in the same manner, but restricted to non-promoter sequences (>2kb from a TSS). For the Segway/ChromHMM training set we used all K562 Segway/ChromHMM regions which were between 300 and 500bp long (9320 sequences) to select a well-defined peak set of approximately equal size to the DHS training set, and we generated an equal size GC/repeat matched negative set. For K562 DHS regions flanked by H3K27Ac and H3K4me1, we selected the top 10000 300bp non-promoter DHS peaks with an average histone mark signal of least 4 and 3 reads in the 1000bp window centered on the DHS peak for H3K27Ac and H3K4me1, respectively. For the retinal expression comparison we used WT (wild-type) retina ATAC-seq (Mo et al., 2016), DNaseI hypersensitive sites (DHS) from 8 week old retina (Yue et al., 2014), and enhancer marks in three unrelated cell types as controls: P300-bound enhancers in melanocytes (Gorkin et al., 2012), GATA1-bound enhancers in megakaryocytes (Pimkin et al., 2014), and DHS regions in GM12878 described above (Lee et al., 2015). We trained the gkm-SVM classifier on the top 4000 distal (≥2 kb away from a TSS) WT retina ATAC-seq peaks, the top 10,000 distal retina DHSs, the top 2351 P300-bound melanocyte enhancers, the top 1230 megakaryocyte GATA1-bound enhancers, and the top 22384 GM12878 DHSs versus length, repeat, and GC-matched negative sets of 16000, 10,000, 9404, 4920, and 22384 sequences, respectively.
Results
eQTL Challenge: Predicting Expression and Variant Impact in GM12878
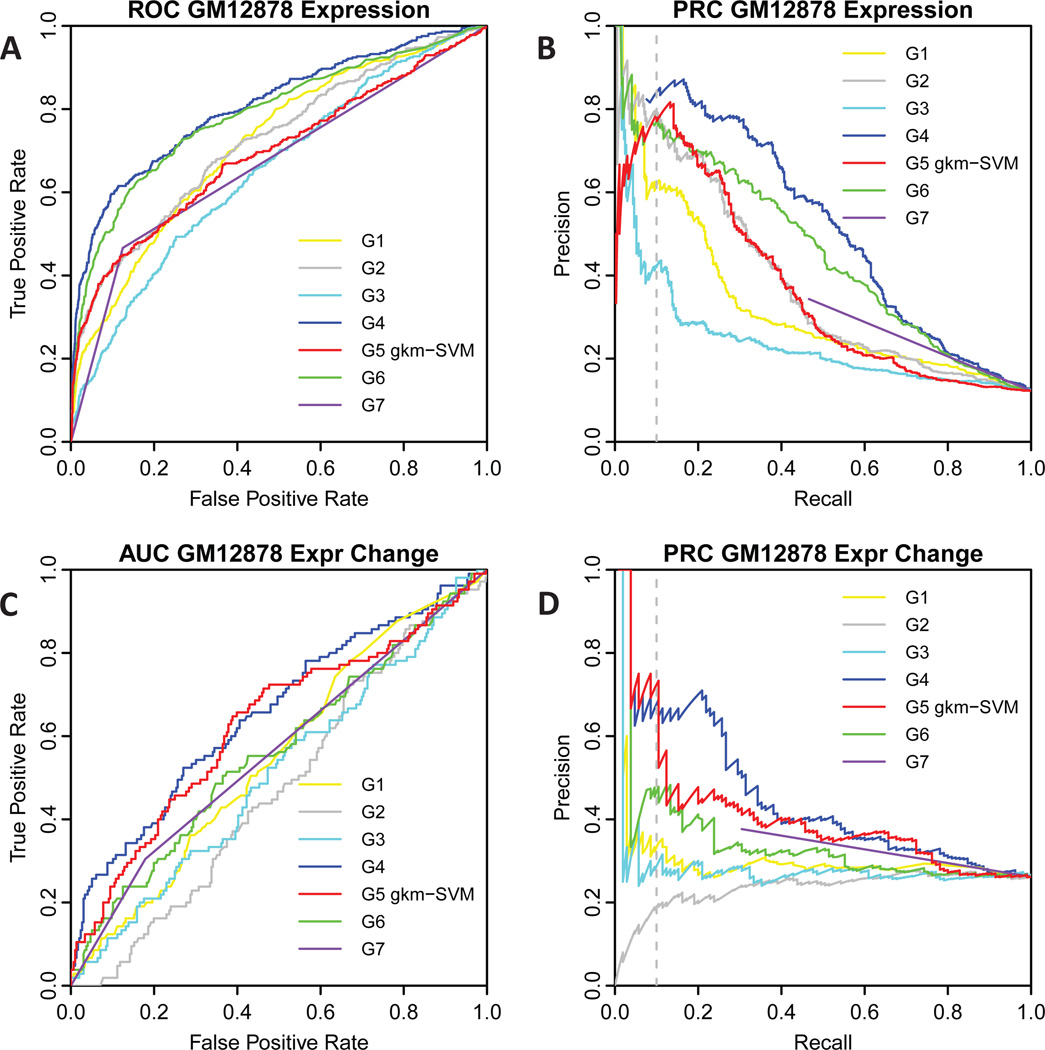
Our main model for the eQTL challenge (group 5, method 1) was a gkm-SVM trained on accessible chromatin regions as measured by UW ENCODE DHS in GM12878 LCLs (ENCODE Consortium, 2012), as described in (Lee et al., 2015) and in Materials and Methods. The regulatory vocabulary, or set of active TF binding sites, in LCLs are encapsulated in the set of weights for all 10-mers. The predicted expression score is then computed as the sum of all weights for all 10-mers in the 150bp tested expression construct. The Receiver Operating Characteristic (ROC) and Precision-Recall Curves (PRC) are shown in Figure 1. For evaluation, the positive set for part one (predicting expression, Figure 1AB) is defined to be those sequences where at least one allele drove sufficient expression (regulatory hits), and all other sequences are negatives. For part two (predicting regulatory impact, Figure 1CD) the positive set is defined to be sequences exhibiting statistically significant differences between the reference and alternate alleles (emVar hits, (Tewhey et al., 2016)), and all other sequence are negatives. As shown in Figure 1 and in Table 1, gkm-SVM (group 5, G5, red) was not the most accurate classifier, but was among the top three classifiers for both tasks: predicting expression and expression variation. For validation experiments, the precision of the predictions (how many predicted positives are actually positive) is usually the most useful measure of performance. These results are noteworthy because gkm-SVM was the only classifier that did not require the MPRA training dataset to achieve this level of performance.
Fig. 1. Comparison of ROC and PRC curves for gkm-SVM and other prediction methods on the two eQTL challenges.
A) ROC curve for eQTL challenge part one, predicting expression in GM12878. B) PRC curve for eQTL challenge part one, predicting expression in GM12878. C) ROC curve for eQTL challenge part two, predicting expression change in GM12878. D) PRC curve for eQTL challenge part two, predicting expression change in GM12878. Group numbers are labelled as in the eQTL challenge overview paper (Kreimer et al., 2016), gkm-SVM is group 5 (G5). The gkm-SVM predictions are among the most accurate for predicting both expression and variant impact, even though they do not use the MPRA training data.
Table I.
AUROC and AUPRC for all methods on both eQTL challenge tasks: predicting expression and predicting expression change.
| Predicting expression | Predicting expression change | |||
|---|---|---|---|---|
| Method | AUROC | AUPRC | AUROC | AUPRC |
| 1-1 | 0.716044 | 0.329400 | 0.550820 | 0.305258 |
| 2-1 | 0.723336 | 0.385369 | 0.477301 | 0.234723 |
| 3-1 | 0.652051 | 0.242830 | 0.511197 | 0.284360 |
| 4-1 | 0.807690 | 0.528288 | 0.655261 | 0.452561 |
| 5-1 gkm-SVM DHS | 0.693357 | 0.369462 | 0.626850 | 0.409730 |
| 5-2 gkm-SVM MPRA | 0.578095 | 0.189516 | 0.577220 | 0.369083 |
| 6-1,4 | 0.786722 | 0.461099 | 0.561953 | 0.345064 |
| 7-1 | 0.670681 | 0.437487 | 0.562854 | 0.431639 |
| gkm-SVM DHS+MPRA | 0.680054 | 0.377978 | 0.619772 | 0.458197 |
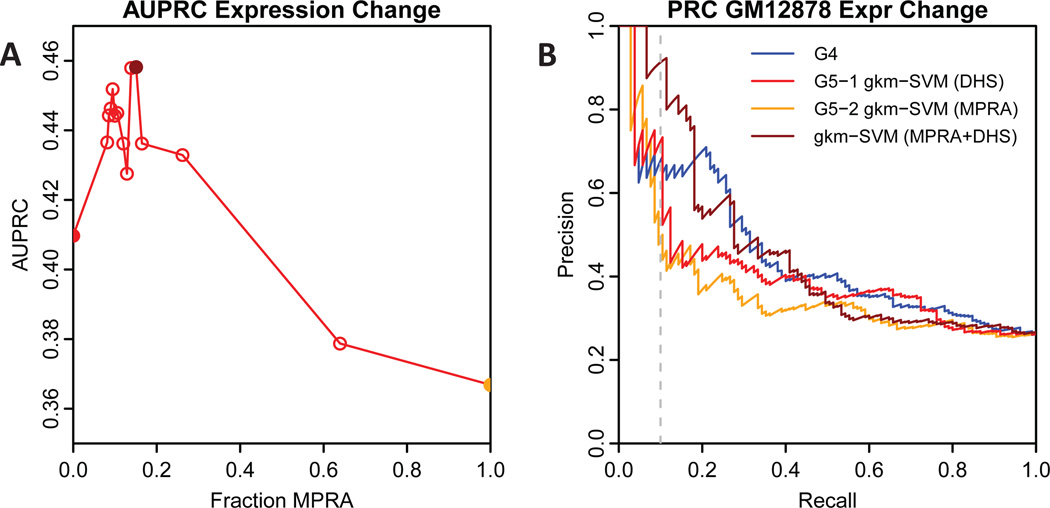
We also submitted a second method (group 5-method 2), which by contrast did train a gkm-SVM classifier on the MPRA training data by separating the constructs into a positive and negative set based on MPRA expression level. Because this yields a smaller training set of only a few thousand sequences compared to the tens of thousands of training sequences generated by DHS or ATAC-seq chromatin accessibility measurements, this classifier was slightly less accurate at predicting expression than our main method (group 5-method 1), as shown in Table 1, in the eQTL challenge overview manuscript (Kreimer et al., 2016), and in Figure 2 below, but was of comparable accuracy at predicting variant impact. The AUROC and AUPRC for all methods on both tasks are listed in Table 1.
Fig. 2. AUPRC for gkm-SVM trained on combined MPRA and DHS data.
A) AUPRC for predicting expression change as the fraction of MPRA training data is varied from zero (method 5-1, red filled circle) to one (method 5-2, orange filled circle). Maximum AUPRC is achieved near 15% MPRA (dark red filled circle). B) PRC curve comparison for the top submitted method (4-1, blue), the two gkm-SVM submitted methods (red and orange), and gkm-SVM trained on 15% MPRA+DHS (dark red).
Although not submitted as a prediction method for the original eQTL challenge, we subsequently trained a hybrid of the two methods described above, by training on the combination of the DHS data and the MPRA training data. We did this by training on the combination of all of the MPRA training sequences used for method 2 (3548 sequences, see Materials and Methods) supplemented by a variable amount of DHS regions (up to 22384 positive sequences, adding the strongest DHS signal sequences first, and an equal number of negative sequences). Figure 2A shows the AUPRC of gkm-SVM trained on the combined DHS+MPRA data as the fraction of MPRA data is varied. The extreme limits (0,1) of MPRA fraction match the previous two submitted methods (5-1 and 5-2, respectively), but using a mixture of DHS and MPRA data for training gkm-SVM outperforms both methods, and slightly outperforms all methods in the CAGI challenge (Table 1). The optimization curve in Figure 2A is slightly noisy as there are only 105 positive sequences in the validation set, and AUPRC can be sensitive to changes in just a few predictions. Figure 2B compares the precision-recall curves for the top submitted method (4-1) the two submitted gkm-SVM methods (5-1 and 5-2) and gkm-SVM trained on the combined DHS+MPRA data with 10,000 positive DHS regions (+10,000 negative) and 3548 MPRA sequences.
Predicting MPRA Expression in K562 cells
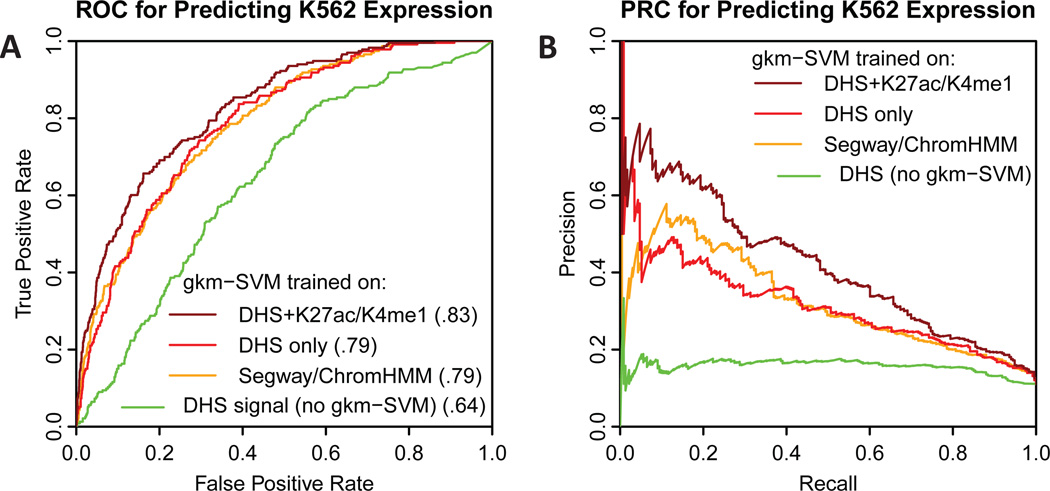
Encouraged by these results, we next sought to test whether gkm-SVM could predict expression levels in a previously published study of ENCODE enhancer predictions. In (Kwasnieski et al., 2014), ENCODE segmentation predictions were tested for enhancer activity by CRE-seq (Kwasnieski et al., 2012), a method very similar to the MPRA used in the eQTL challenge, in K562 cells. They selected 130bp regions labeled strong or weak enhancers from K562 Segway/ChromHMM (Ernst and Kellis, 2010; Hoffman et al., 2012) merged predictions, and also tested regions predicted to be H1-ESC enhancers, and matching scrambled sequence constructs as negative controls. All tested regions were predicted to be enhancers based on ENCODE segmentations, but only 233 of the 3236 constructs showed significant expression in this assay, or about 20% of the predicted enhancers (196/1200). We trained gkm-SVM on K562 DHS (see Materials and Methods) and scored each sequence tested, and found that gkm-SVM could predict the expressing positive set (with normalized expression > 1.9) with high accuracy based on the gkm-SVM score of the construct sequence (Figure 3A, red, AUROC=.79). Similarly, gkm-SVM trained on all 300–500bp size selected Segway/ChromHMM enhancers in K562 cells could also predict well (AUROC=.79), in spite of the fact that all tested constructs were within this set of Segway regions, yet validated at a low rate. As previously shown (Kwasnieski et al., 2014), the DHS signal within the tested regions is also a poor predictor of the positive expressing subset (Figure 3A, green, AUROC=.64). We are thus led to the surprising conclusion that our DNA sequence based model (gkm-SVM) predicts expression significantly more accurately than the raw data used to train the model (either DHS or Segway predictions). Several important factors contribute to this result. First, the gkm-SVM extracts and encapsulates the essential binding site vocabulary from all enhancers active in the cell type, while any specific individual region may have varying DHS signal because of experimental or biological noise. Second, after decoding the relevant TF vocabulary, the gkm-SVM can accurately determine whether or not the shorter tested 130bp DNA fragment within the broader DHS peak or Segway/ChromHMM prediction contains the combinations of features necessary to produce expression in the reporter assay, and does so with higher resolution than standard DHS (but comparable resolution to footprinting). An even better gkm-SVM prediction (AUROC=0.83) is obtained by training gkm-SVM on K562 DHS regions flanked by H3K27ac and H3K4me1 marks, well known to be associated with enhancer activity (Heintzman et al., 2007). Although the training set crossfold-validation for the histone flanked regions is slightly lower than training on DHS peaks alone, the accuracy predicting enhancer reporter expression is improved, demonstrating that training set accuracy does not necessarily translate to more accurate predictions of enhancer activity.
Fig. 3. Comparison of ROC and PRC curves for predicting MPRA expression in K562 cells.
A) ROC and B) PRC for predicting MPRA expression in K562 cells (Kwasnieski et al., 2014) using different methods. All tested regions were within Segway/ChromHMM enhancer predictions in K562, but only ~20% were positive. DHS in the tested regions is also weak predictor of expression (green). However a gkm-SVM trained on DHS regions or Segway/ChromHMM regions is reasonably accurate (red and orange). The most accurate predictor is a gkm-SVM trained on DHS regions flanked by H3K27Ac and H3K4me1 (dark red).
Predicting MPRA Expression in Mouse Retina
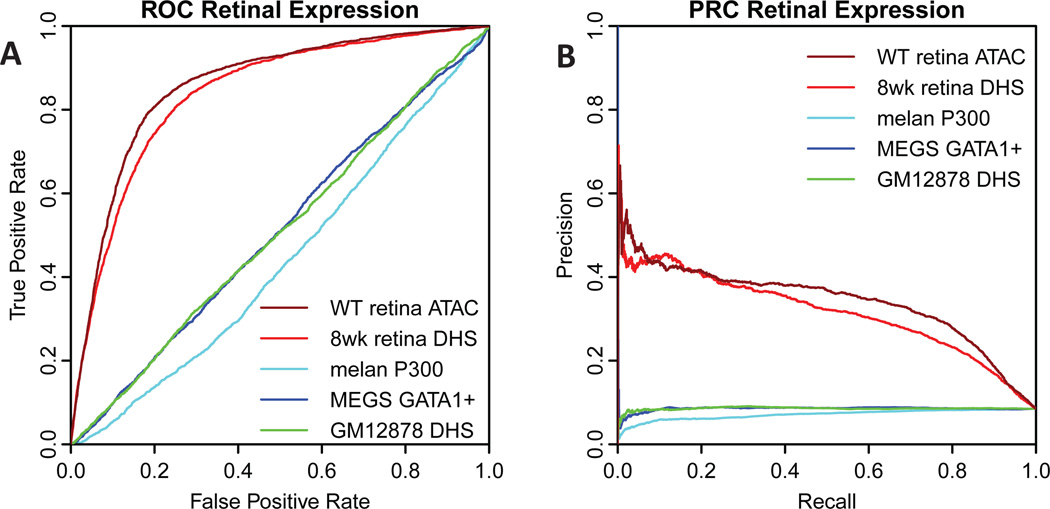
To demonstrate that gkm-SVM is also a robust predictor of expression when trained on other datatypes and whole tissue samples, we next compared to a recent MPRA dataset testing longer ~500bp fragments tiling across DHS peaks in mouse retina and cortex (Shen et al., 2016). Although in this experiment only 6% of the 36005 constructs tested produced detectable expression in each of three replicates, gkm-SVM was able to predict the consistently expressing constructs with high accuracy. We trained gkm-SVM on mouse retina DHS (Yue et al., 2014), retina ATAC-seq (Mo et al., 2016), and for comparison included three unrelated cell types which produced gkm-SVM classifiers which we have previously shown were predictive in their respective cell types: non-retinal melanocytes (Gorkin et al., 2012), megakaryocytes (Pimkin et al., 2014), and GM12878 lymphoblasts as described above. As shown in Figure 4, the reliably expressing constructs scored highly by a gkm-SVM trained on mouse retina accessible chromatin (DHS or ATAC-seq), but did not score highly when gkm-SVM was trained on unrelated cell types. For these ROC and PRC curves, the positive set was defined as those constructs which tested positive in all three replicates (2156 barcodes) and the negative set was defined as those constructs which were not detected in any replicate (23147 barcodes). The tested insert sequences are all DHS positive in retina or cortex, but are staggered across the DHS peaks. Gkm-SVM can reliably detect those sequences which contain the necessary TF binding sites for expression in vivo.
Fig. 4. Predicting MPRA expression in mouse retina.
A) ROC and B) PRC for predicting MPRA expression in mouse retina (Shen et al., 2016). Gkm-SVM trained on either retina ATAC-seq or DHS (dark red, red) predicts the expressing constructs with about 50% precision, but gkm-SVM trained on unrelated cell types does not (melanocytes, cyan; megakaryocytes, blue; lymphoblasts, green).
Discussion
The eQTL challenge comparisons of MPRA expression and gkm-SVM predictions, and comparisons to other datasets shown here, demonstrate that DNA sequence based models trained on the relevant cell type can predict expression and variant impact with precision around 50% in both cell lines and tissues. This level of agreement is quite encouraging and suggests that computational predictions used to select SNPs and target regions for MPRAs should greatly accelerate the discovery and validation of disease associated variants.
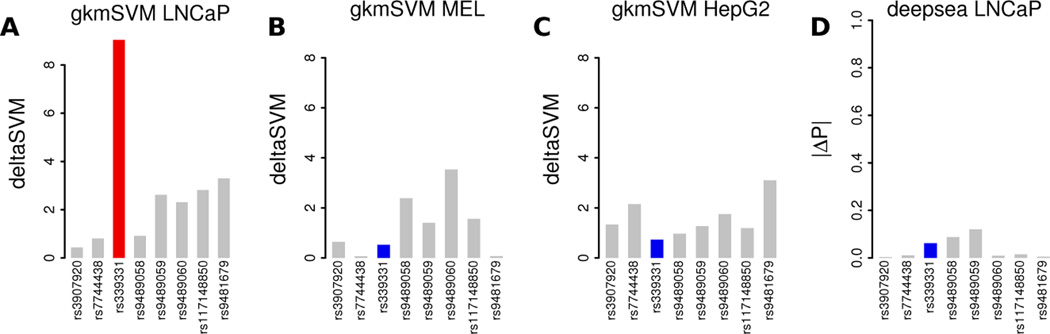
As described in (Ghandi et al., 2014), we proposed gkm-SVM as a DNA sequence based regulatory prediction method in order to evaluate the impact of disease associated human genetic variation, and showed that it outperformed existing methods. We then systematically compared the gkm-SVM predictions to previously published MPRA variant datasets (Patwardhan et al., 2012; Kheradpour et al., 2013), to our own direct validation experiments, and to validated GWAS associated loci in (Lee et al., 2015). One of the disease associated SNPs that gkm-SVM can explain is the common SNP rs339331, which was shown by GWAS to increase prostate cancer risk (Takata et al., 2010) (odds ratio=1.22, p=1.6×10−12). Further dissection of this locus (Huang et al., 2014) showed that the risk SNP allele TTTTATGAG is bound by HOXB13, which in combination with FOXA1 and AR activates RFX6 and promotes cell migration and metastatic disease, while the protective allele TTTCATGAG is not bound by HOXB13. As shown in (Lee et al., 2015) and in Figure 5A, deltaSVM is able to identify the validated SNP rs339331 (red), which has a very large deltaSVM score relative to flanking SNPs (grey) when gkm-SVM is trained on LNCaP DHS (a prostate adenocarcinoma cell line) because the weight for the risk allele TTTTATGAG is large and the weight for TTTCATGAG is small. In contrast, when gkm-SVM is trained on melanocytes (Gorkin et al., 2012) (Figure 5B) or the liver cell line HepG2 (Lee et al., 2015) (Figure 5C), deltaSVM for the causal SNP (blue) is small and comparable to flanking SNPs. After publication of deltaSVM (Lee et al., 2015), a similar DNA sequence based regulatory prediction method called Deepsea was reported (Zhou and Troyanskaya, 2015) which differs from gkm-SVM in two main respects: first, it uses a deep neural network (DNN) instead of an SVM, and second, it trains on all ENCODE cell types simultaneously, while gkm-SVM is trained separately for each cell type. In principle DNNs could produce more accurate classifiers than SVMs if trained on sufficient data. However, because certain classes of cell types are overrepresented in the ENCODE collection (e.g. blood and immune), it is possible that Deepsea training is biased toward (and quite accurate on) these overrepresented cell types, and might be less accurate on underrepresented cell types (such as LNCaP). While the Deepsea predictions have not been directly experimentally validated, Deepsea predictions can be generated from deepsea.princeton.edu, and Figure 5D shows that this is indeed the case for the RFX6 prostate cancer SNP, where Deepsea predicts that the validated variant has less predicted effect on LNCaP DHS than flanking non-causal SNPs, in spite of the fact that Deepsea reported high test-set cross-fold validation AUROC for LNCaP DHS.
Fig. 5. Predicting causal SNPs within the RFX6 prostate cancer locus.
deltaSVM using a gkm-SVM trained on a prostate cancer cell line LNCaP (A) can identify the causal SNP (red) from among flanking SNPs (grey), but a gkm-SVM trained on melanocytes (B) or HepG2 (C) cannot. Deepsea predictions include LNCaP cells in the training set but do not correctly identify the validated SNP (D).
While many validated GWAS SNPs are accurately predicted by deltaSVM (Lee et al., 2015), this requires training data for gkm-SVM in the cell type and time of development when the modulated enhancer is active. RET+3 (Emison et al., 2005) is not predicted well by gkm-SVM trained on fetal intestine or brain DHS (Roadmap Epigenomics Consortium et al., 2015), presumably because whole brain is not sufficiently representative of enteric neurons, which make up only a small fraction of intestinal tissue.
An advantage of gkm-SVM over neural network based approaches is the relative ease of interpreting biological mechanisms from the results. In our approach, each gapped k-mer can be assigned a weight directly from the support vectors, or more simply, each k-mer (we typically use length 10 for interpreting binding sites) can be assigned a weight based on its gkm-SVM score. The k-mers in the tails of this weight distribution typically fall into similar groups representing sets of binding sites for active TFs in the training cell-type. Large deltaSVM scores typically result from disrupting or creating one of these TF binding sites. In this regard, the improved prediction accuracy of gkm-SVM trained on combined DHS+MPRA compared to DHS or MPRA alone is interesting. With sufficient MPRA training data, a gkm-SVM trained on MPRA data should best predict the MPRA experiments. However, MPRA training data is limited to the alleles tested (existing or synthetic). DHS data it is easier to acquire (for this test we have about ten times more positive sequences from DHS compared to MPRA), and is more diverse (longer sequences of more varied composition). However, DHS data is less direct for the current prediction task, as it measures chromatin accessibility instead of reporter expression. While it remains to be shown whether or not reporter expression is more disease relevant than genomic chromatin accessibility, there may be features in the DHS data which contribute to genomic accessibility but not to reporter expression. For this experiment the optimal training set, with approximately 15% MPRA and 85% DHS sequences, appears to balance the assay specificity of the MPRA training set sequences with the increased diversity and size of the DHS training set sequences.
The successful comparisons with MPRA experiments are encouraging because our gkm-SVM model is relatively simple, is trained on easily obtainable chromatin accessibility data, and does not try to predict either the structure of the complexes of proteins interacting at the enhancer and promoter, specific binding site combinations, or spatial constraints between TF binding events. This experimental validation of the gkm-SVM model therefore supports the hypothesis that an accurate description of binding sites in the enhancers is sufficient to predict much of their activity. However, the precision of the gkm-SVM prediction of MPRA expression is still around 50%. Some of the errors may not in fact be problems with the model, but could be due to the difficulty of the expression measurements: either measurement noise, the episomal nature of the reporter assay, or other synthetic properties of the reporter constructs. Measurement noise from technical replicates could be used to estimate an upper bound on prediction accuracy. However, we also suspect that significant improvements to our DNA sequence based modeling (e.g. the combinatorial effects described above) could improve the overall prediction accuracy and resolve many of these differences.
Acknowledgments
This work was supported by US National Institutes of Health grant R01 HG007348 and U01 HG009380. The CAGI experiment coordination is supported by NIH U41 HG007446 and the CAGI conference by NIH R13 HG006650. MB acknowledges useful discussions with Dongwon Lee and an anonymous reviewer for suggesting training on the combined DHS and MPRA data.
References
- Bauer DE, Kamran SC, Lessard S, Xu J, Fujiwara Y, Lin C, Shao Z, Canver MC, Smith EC, Pinello L, Sabo PJ, Vierstra J, et al. An Erythroid Enhancer of BCL11A Subject to Genetic Variation Determines Fetal Hemoglobin Level. Science. 2013;342:253–257. doi: 10.1126/science.1242088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canver MC, Smith EC, Sher F, Pinello L, Sanjana NE, Shalem O, Chen DD, Schupp PG, Vinjamur DS, Garcia SP, Luc S, Kurita R, et al. BCL11A enhancer dissection by Cas9-mediated in situ saturating mutagenesis. Nature. 2015;527:192–197. doi: 10.1038/nature15521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium T 1000 GP. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emison ES, McCallion AS, Kashuk CS, Bush RT, Grice E, Lin S, Portnoy ME, Cutler DJ, Green ED, Chakravarti A. A common sex-dependent mutation in a RET enhancer underlies Hirschsprung disease risk. Nature. 2005;434:857–863. doi: 10.1038/nature03467. [DOI] [PubMed] [Google Scholar]
- ENCODE Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Kellis M. Discovery and characterization of chromatin states for systematic annotation of the human genome. Nat Biotechnol. 2010;28:817–825. doi: 10.1038/nbt.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletez-Brant C, Lee D, McCallion AS, Beer MA. kmer-SVM: a web server for identifying predictive regulatory sequence features in genomic data sets. Nucleic Acids Res. 2013;41:W544–W556. doi: 10.1093/nar/gkt519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghandi M, Lee D, Mohammad-Noori M, Beer MA. Enhanced Regulatory Sequence Prediction Using Gapped k-mer Features. PLoS Comput Biol. 2014;10:e1003711. doi: 10.1371/journal.pcbi.1003711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghandi M, Mohammad-Noori M, Ghareghani N, Lee D, Garraway L, Beer MA. gkmSVM: an R package for gapped-kmer SVM. Bioinformatics. 2016;32:2205–2207. doi: 10.1093/bioinformatics/btw203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorkin DU, Lee D, Reed X, Fletez-Brant C, Bessling SL, Loftus SK, Beer MA, Pavan WJ, McCallion AS. Integration of ChIP-seq and machine learning reveals enhancers and a predictive regulatory sequence vocabulary in melanocytes. Genome Res. 2012;22:2290–2301. doi: 10.1101/gr.139360.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusev A, Lee SH, Trynka G, Finucane H, Vilhjálmsson BJ, Xu H, Zang C, Ripke S, Bulik-Sullivan B, Stahl E Schizophrenia Working Group of the Psychiatric Genomics Consortium, SWE-SCZ Consortium, et al. Partitioning heritability of regulatory and cell-type-specific variants across 11 common diseases. Am J Hum Genet. 2014;95:535–552. doi: 10.1016/j.ajhg.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, Wang W, Weng Z, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, Manolio TA. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman MM, Buske OJ, Wang J, Weng Z, Bilmes JA, Noble WS. Unsupervised pattern discovery in human chromatin structure through genomic segmentation. Nat Methods. 2012;9:473–476. doi: 10.1038/nmeth.1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Q, Whitington T, Gao P, Lindberg JF, Yang Y, Sun J, Väisänen M-R, Szulkin R, Annala M, Yan J, Egevad LA, Zhang K, et al. A prostate cancer susceptibility allele at 6q22 increases RFX6 expression by modulating HOXB13 chromatin binding. Nat Genet. 2014;46:126–135. doi: 10.1038/ng.2862. [DOI] [PubMed] [Google Scholar]
- Kheradpour P, Ernst J, Melnikov A, Rogov P, Wang L, Zhang X, Alston J, Mikkelsen TS, Kellis M. Systematic dissection of regulatory motifs in 2000 predicted human enhancers using a massively parallel reporter assay. Genome Res. 2013;23:800–811. doi: 10.1101/gr.144899.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreimer A, Zeng H, Edwards MD, Guo Y, Tian K, Shin S. Predicting Gene Expression in Massively Parallel Reporter Assays: A Comparative Study. Hum Mutat. 2016 doi: 10.1002/humu.23197. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwasnieski JC, Fiore C, Chaudhari HG, Cohen BA. High-throughput functional testing of ENCODE segmentation predictions. Genome Res. 2014;24:1595–1602. doi: 10.1101/gr.173518.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwasnieski JC, Mogno I, Myers CA, Corbo JC, Cohen BA. Complex effects of nucleotide variants in a mammalian cis-regulatory element. Proc Natl Acad Sci. 2012;109:19498–19503. doi: 10.1073/pnas.1210678109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappalainen T, Sammeth M, Friedländer MR, AC’t Hoen P, Monlong J, Rivas MA, Gonzàlez-Porta M, Kurbatova N, Griebel T, Ferreira PG, et al. Transcriptome and genome sequencing uncovers functional variation in humans. Nature. 2013;501:506–511. doi: 10.1038/nature12531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D, Gorkin DU, Baker M, Strober BJ, Asoni AL, McCallion AS, Beer MA. A method to predict the impact of regulatory variants from DNA sequence. Nat Genet. 2015;47:955–961. doi: 10.1038/ng.3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurano MT, Humbert R, Rynes E, Thurman RE, Haugen E, Wang H, Reynolds AP, Sandstrom R, Qu H, Brody J, Shafer A, Neri F, et al. Systematic Localization of Common Disease-Associated Variation in Regulatory DNA. Science. 2012;337:1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo A, Luo C, Davis FP, Mukamel EA, Henry GL, Nery JR, Urich MA, Picard S, Lister R, Eddy SR, Beer MA, Ecker JR, et al. Epigenomic landscapes of retinal rods and cones. eLife. 2016;5:e11613. doi: 10.7554/eLife.11613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musunuru K, Strong A, Frank-Kamenetsky M, Lee NE, Ahfeldt T, Sachs KV, Li X, Li H, Kuperwasser N, Ruda VM, Pirruccello JP, Muchmore B, et al. From noncoding variant to phenotype via SORT1 at the 1p13 cholesterol locus. Nature. 2010;466:714–719. doi: 10.1038/nature09266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patwardhan RP, Hiatt JB, Witten DM, Kim MJ, Smith RP, May D, Lee C, Andrie JM, Lee S-I, Cooper GM, Ahituv N, Pennacchio LA, et al. Massively parallel functional dissection of mammalian enhancers in vivo. Nat Biotechnol. 2012;30:265–270. doi: 10.1038/nbt.2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson TA, Mort M, Cooper DN, Radivojac P, Kann MG, Mooney SD. Regulatory Single-Nucleotide Variant Predictor Increases Predictive Performance of Functional Regulatory Variants. Hum Mutat. 2016;37:1137–1143. doi: 10.1002/humu.23049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimkin M, Kossenkov AV, Mishra T, Morrissey CS, Wu W, Keller CA, Blobel GA, Lee D, Beer MA, Hardison RC, Weiss MJ. Divergent functions of hematopoietic transcription factors in lineage priming and differentiation during erythro-megakaryopoiesis. Genome Res. 2014;24:1932–1944. doi: 10.1101/gr.164178.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie GRS, Dunham I, Zeggini E, Flicek P. Functional annotation of noncoding sequence variants. Nat Methods. 2014;11:294–296. doi: 10.1038/nmeth.2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roadmap Epigenomics Consortium. Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, Ziller MJ, Amin V, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen SQ, Myers CA, Hughes AEO, Byrne LC, Flannery JG, Corbo JC. Massively parallel cis-regulatory analysis in the mammalian central nervous system. Genome Res. 2016;26:238–255. doi: 10.1101/gr.193789.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldner F, Stelzer Y, Shivalila CS, Abraham BJ, Latourelle JC, Barrasa MI, Goldmann J, Myers RH, Young RA, Jaenisch R. Parkinson-associated risk variant in distal enhancer of α-synuclein modulates target gene expression. Nature. 2016;533:95–99. doi: 10.1038/nature17939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata R, Akamatsu S, Kubo M, Takahashi A, Hosono N, Kawaguchi T, Tsunoda T, Inazawa J, Kamatani N, Ogawa O, Fujioka T, Nakamura Y, et al. Genome-wide association study identifies five new susceptibility loci for prostate cancer in the Japanese population. Nat Genet. 2010;42:751–754. doi: 10.1038/ng.635. [DOI] [PubMed] [Google Scholar]
- Tewhey R, Kotliar D, Park DS, Liu B, Winnicki S, Reilly SK, Andersen KG, Mikkelsen TS, Lander ES, Schaffner SF, Sabeti PC. Direct Identification of Hundreds of Expression-Modulating Variants using a Multiplexed Reporter Assay. Cell. 2016;165:1519–1529. doi: 10.1016/j.cell.2016.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue F, Cheng Y, Breschi A, Vierstra J, Wu W, Ryba T, Sandstrom R, Ma Z, Davis C, Pope BD, Shen Y, Pervouchine DD, et al. A comparative encyclopedia of DNA elements in the mouse genome. Nature. 2014;515:355–364. doi: 10.1038/nature13992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nussbaum C, Myers RM, Brown M, Li W, Liu XS. Model-based Analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Troyanskaya OG. Predicting effects of noncoding variants with deep learning-based sequence model. Nat Methods. 2015;12:931–934. doi: 10.1038/nmeth.3547. [DOI] [PMC free article] [PubMed] [Google Scholar]