Introduction
Despite evidence that genotype-guided therapy (GGTx) can improve outcomes, advances in genotyping technology and decreasing costs, integration into practice is limited. We describe a successful implementation framework using genotype-guided antiplatelet therapy as an example. We outline how we developed consensus across stakeholders, obtained institutional endorsement, built genotyping capability, and provide decision support to guide treatment selection; and demonstrate how to leverage such efforts to assess clinical effectiveness and cost-effectiveness and bolster research infrastructure.
Keywords: Precision medicine, personalized medicine, pharmacogenetics, implementation, clinical decision support, genotype-guided therapy, CYP2C19, antiplatelet therapy
Introduction
Precision medicine entails tailoring treatment based on patients’ unique characteristics. As drug therapy constitutes the cornerstone of treatment for most chronic diseases, pharmacogenomics (PGx), the study of genetic variation influencing individual response to drugs, is an important components of precision medicine. Over the past decade, investigations have identified genes and single nucleotide polymorphisms (SNPs) and quantified their effect on drug response. Parallel development of point-of-care (POC) genotyping platforms has enabled the interrogation of the genes/SNPs within a timeline conducive to the provision of care. Despite these advances, the pace of integration of genotype-guided drug therapy (GGTx) into practice has faced significant challenges. These include difficulty in identifying SNPs with sufficiently robust evidence to guide clinical decision making, lack of clinician training on how to order and use genotype data, lack of clinical decision support (CDS) to guide treatment, and limited reimbursement. The University of Alabama at Birmingham’s (UAB) efforts in precision medicine were initiated to address these challenges and improve the health of the racially diverse patients we treat.
As with any large initiative, we started by identifying the needs of the population we care for. Herein, we present our framework to assess our patient population and identify opportunities where GGTx can improve outcomes, develop consensus across stakeholders in the clinical enterprise and obtain institutional endorsement, build genotyping capability within our CLIA-certified molecular diagnostic laboratory, conduct testing within a timeline conducive to the provision of care, and provide CDS to guide treatment selection. We also present methodology to assess clinical and process-related outcomes to enable evaluation of clinical effectiveness and cost-effectiveness, and leverage the implementation efforts to fuel research and discovery.
Identifying an opportunity for improvement - the starting point
We started by understanding the distribution of disease in the patient population receiving care at the UAB Hospital (UABH) a 1,157-bed tertiary care hospital located in downtown Birmingham, Alabama. In 2015, 205,684 unique patients were cared for at UAB, contributing to over 50,000 hospital admissions and 1.4 million outpatient clinic visits. Within this population we identified medications for which there is consistent evidence that genotype influences drug response.1–5 We prioritized GGTx implementation based on frequency of use, available Food and Drug Administration (FDA) guidance and Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines, the availability of alternative drugs or dosing, and the feasibility of testing along a timeline conducive for clinical care.
Given the burden of coronary heart disease (CHD), the volume and racial diversity (30% African American) of patients presenting with acute coronary syndromes (ACS) undergoing percutaneous coronary intervention (PCI; ~1000 annually), the availability of CPIC guidelines,6 the potential to improve outcomes by guiding treatment selection, and availability of POC genotyping platforms, we chose to implement cytochrome P450 2C19 (CYP2C19) guided antiplatelet therapy for the patients presenting with ACS and those undergoing PCI.
It is standard-of-care for ACS and PCI patients to be prescribed dual-antiplatelet therapy (DAPT), most frequently aspirin and clopidogrel. Clopidogrel is a prodrug that requires bioactivation. CYP2C19 plays a key role in this process.7,8 The CYP2C19 harbors several variants that result in loss-of-function (LOF). The most common LOF allele is CYP2C19*2 (c.681G>A; rs4244285), a splice site variant which leads to the production of a truncated, non-functioning protein. The second most common LOF allele is CYP2C19*3 (c.636G>A; rs4986893), a variant that results in a premature stop codon. In contrast, CYP2C19*17 (c.-806C>T; rs12248560) is a common gain-of-function (GOF) variant.6 Table 1 presents the minor allele frequencies (MAF) for LOF and GOF alleles observed in our patient population and allows comparisons with MAFs reported in the literature.6
Table 1.
Expected and observed frequencies of CYP2C19 alleles by race
| Allele | Change (rs#) |
Effect on CYP2C19 |
Expected Allele Frequency1 |
Observed Allele Frequency in the UABH patients |
p-value for differences in observed vs. expected minor allele frequencies |
|||
|---|---|---|---|---|---|---|---|---|
| Europeans Americans |
African Americans |
Europeans Americans |
African Americans |
EA | AA | |||
| *1 | No change | No change |
63.6% | 68.5% | 63.6% | 56.5% | 0.99 | 0.25 |
| *2 | c.681G>A rs4244285 |
Loss-of- function |
15% | 15% | 14.8% | 17.7% | 0.95 | 0.58 |
| *3 | c.636G>A; rs4986893 |
Loss-of- function |
0.4% | 0.5% | 0% | 0% | 0.42 | 0.58 |
| *17 | c.-806C>T; rs12248560 |
Gain-of- function |
21% | 16% | 21.7% | 25.8% | 0.84 | 0.05 |
Scott SA, Sangkuhl K, Stein CM, et al. Clinical Pharmacogenetics Implementation Consortium guidelines for CYP2C19 genotype and clopidogrel therapy: 2013 update. Clin Pharmacol Ther 2013;94:317–23.
It is the combination of the two inherited alleles that determines an individual’s metabolizer status (Table 2). To inform DAPT selection, the CPIC guidelines categorizes individuals into four phenotypes based on CYP2C19 genotypes.6,9 Individuals with two copies of the wild-type allele (*1/*1) are extensive metabolizers (EM).6 Those with a single LOF allele (e.g., *1/*2, *1/*3) are considered intermediate metabolizers (IM), whereas those homozygous for LOF alleles (*2/*2, *3/*3) and compound heterozygous (*2/*3) are poor metabolizers (PM) with absent to minimal CYP2C19 enzyme activity.6 Patients with GOF allele (*1/*17, *17/*17) are considered ultra-rapid metabolizers (UM) and may be at an increased bleeding risk; however the clinical impact is equivocal.10–13 Patients possessing one GOF and one LOF allele (e.g.:*2/*17) are also considered IM as*17 is unable to completely compensate for the *2 allele.6,14
Table 2.
CPIC phenotype assignment based on CYP2C19 genotype and recommended antiplatelet therapy
| CYP2C19 genotype | Metabolizer Phenotype |
Treatment recommendation |
|---|---|---|
| *1/*1 | Extensive (Normal) | Usual dose of Clopidogrel |
| *1/*17, *17/*17 | Ultra-rapid | Usual dose of Clopidogrel |
| *1/*2, *2/*17 or *1/*3 | Intermediate | Alternative therapy (e.g., prasugrel, ticagrelor) |
| *2/*2, *2/*3 or *3/*3 | Poor | Alternative therapy (e.g., prasugrel, ticagrelor) |
Possession of one or two copies of CYP2C19 LOF alleles (intermediate, poor-metabolizer) confers an increased risk for Major Adverse Cardiovascular Events (MACE), defined as non-fatal stroke,15 non-fatal myocardial infarction (MI),16 and death secondary to any cardiovascular cause and stent thrombosis in clopidogrel treated patients.17–19 These findings prompted the FDA to issue a warning for clopidogrel in 2010,20 stating that patients possessing CYP2C19 LOF alleles may have suboptimal response to clopidogrel and should be considered for alternative platelet aggregation inhibitors, namely ticagrelor (Brillinta®) or prasugrel (Effient ®). In response, the American College of Cardiology Foundation (ACCF) and American Heart Association (AHA), expressed concerns about the label update citing lack of clinical evidence to recommend routine genetic in all patients receiving clopidogrel. Their guidelines state that genetic testing may be considered in patients at high risk for poor clinical outcomes with clopidogrel. In such patients alternate antiplatelet agents e.g. prasugrel or ticagrelor may be considered.21
Lessons learned: Evaluation of patient population served and catchment area can help identify clinical needs and guide initial implementation of precision interventions.
Developing consensus, engaging faculty and garnering institutional endorsement
At UAB, the standard-of-care is to initiate DAPT without CYP2C19 testing (non-GGAT) in ACS/PCI patients. Therefore, the initial steps of this implementation program focused on engaging faculty interventional cardiologists and intensivists.
This consensus building exercise spanned several months beginning with presenting the state of the evidence for CYP2C19 guided antiplatelet therapy with the cardiology faculty and pharmacists followed by more focused discussions with interventional cardiologists. Following the presentations, prescribing cardiologists were surveyed on their preferences with regard to using/not using genotype to guide antiplatelet selection. The consensus was not to use GGAT in all patients routinely. Clinicians opted to implement a selective approach and use genetic testing in high risk patients. High risk patients included patients with ACS and high risk PCI patients (e.g. bifurcation site PCI, multi-vessel PCI, history of adverse outcome) as reported in the ACCF/AHA recommendations.22 There are several prediction models 23–26 that identify patients at high risk for poor outcomes among ACS/PCI patients. Interventionists usually consider patients with multiple risk factors and those with bifurcation site PCI or multi-vessel PCI as high risk. Whether genotype-guided antiplatelet therapy improves outcomes; i.e. demonstrates benefit in high-risk patients is one of the aims for implementation efforts such as this one.
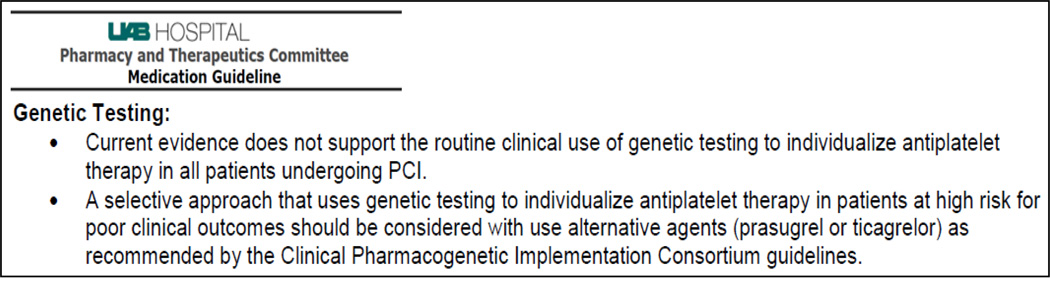
Finally, the current evidence for CYP2C19 guided antiplatelet therapy and the consensus opinion of UAB cardiologists was presented to the UAB Pharmacy and Therapeutics Committee. The proposed changes to the “Platelet Aggregation Inhibitors” medication guidelines were accepted and approved by the P&T committee. These guidelines support a selective approach to use CYP2C19 genetic testing in high risk patients (Figure 1) rather than as routinely ordered tests in all patients.
Figure 1.
Recommendations for genetic testing - Pharmacy and Therapeutics Committee guidelines for platelet aggregation inhibitors.
Non-physicians were involved in different steps in planning the implementation process. This included informal discussion with pharmacists and vascular lab nurses was conducted to understand patient flow and guide process of implementation. For example these discussions identified that patients are in the post-PCI observation unit for 45–60 minutes before being moved to the cardiac care unit or the interventional cardiac care unit. This provided a window wherein test sample can be collected at the same time as post-PCI labs such as fluid balance/troponin levels etc. Discussion with the health informatics and laboratory personnel informed the integration of results in our EMR. Discussions with cardiologists, laboratory and health informatics informed the design and verbiage of the alert so as to convey clinical decision information succinctly and clearly.
Lessons learned: We recommend engaging stakeholders across disciplines including clinicians, pharmacists, laboratory technicians and research coordinators early in the planning process. Departmental and institutional leadership within clinical and research arenas can facilitate both: clinical implementation and discovery efforts.
Building genotyping capability within our CLIA-certified molecular diagnostic laboratory
We had a choice between two strategies for implementing GGTx; preemptive GGTx and reactive GGTx. Preemptive GGTx involves genotyping patients so that information is available prior to the event that requires institution of treatment. Usually this involves genotyping samples in batches with interrogation of a multitude of SNPs including all relevant variants in actionable gene-drug pairs. Reactive GGTx involves genotyping patients at the time of the clinical event that requires institution of treatment and requires assessment of genotype quickly within a timeline that is conducive to the delivery of care. Given the availability of FDA-approved POC platforms, we chose to implement reactive GGTx for CYP2C19.
Evaluation of the Verigene and Spartan CYP2C19 assays
In parallel to the clinical consensus building process, we conducted validation genotyping using the Verigene® CYP2C19 Nucleic Acid Test (October 2013 to April 2014; Nanosphere Inc Northbrook, IL), and Spartan Rx (October to December 2014; Spartan Biosciences Inc, Ottawa ON). All testing was conducted in our CLIA certified Molecular Diagnostic Laboratory according to manufacturer’s procedures using peripheral blood was used for Verigene validation and buccal swab was used for Spartan validation. Only exception was Coriell genomic DNA controls for genotype *3, as we did not find a person with this rare genotype. Assay performance and genotypes results, reported as wild type (expressed as *1), heterozygous, or homozygous on *2, *3 and *17 alleles, were compared between the two platforms.
Three Coriell Institute quality control and 8 volunteer samples (Table 3) were evaluated by both assays. When genotype resulted from testing the concordance between the two platforms was 100%. The p-value not calculated as there is no difference in test results. However, samples tested on Verigene® CYP2C19 produced a 20% (8 of 40 tests) no call rate and one processing error. Spartan accurately identified the genotype 100% of the time on all 33 specimens (p = 0.0064) and was consistent among operators across three runs. Citations of the unacceptably high no-call rate resulted in a class 3 recall of the Verigene assay (August 2014) with subsequent product withdrawn from the US market. Based these developments, the Molecular Diagnostic Laboratory started offering reactive CYP2C19 test using Spartan RX in January 2015 to guide antiplatelet therapy selection in PCI patients.
Table 3.
Summary of Validation results from Verigene and Spartan CYP2C19 Tests
| Nanosphere’s Verigene CYP2C19 Nucleic Acid Test |
Spartan RX CYP2C19 Test | |||||||
|---|---|---|---|---|---|---|---|---|
| Sample | Number of tests performed |
Number of No Calls |
No Call Rate |
Number of tests performed |
Number of No Calls |
No Call Rate |
Result | Phenotype |
| Volunteer 1 | 11 | 3 | 27.3% | 3 | 0 | 0% | *1/*2 | Intermediate Metabolizer |
| Volunteer 2 | 5 | 2 | 40% | 3 | 0 | 0% | *1/*2 | Intermediate Metabolizer |
| Volunteer 3 | 2 | 0 | 0% | 3 | 0 | 0% | *1/*2 | Intermediate Metabolizer |
| Volunteer 4 | 1 | 0 | 0% | 3 | 0 | 0% | *1/*2 | Intermediate Metabolizer |
| Volunteer 5 | 3 | 0 | 0% | 3 | 0 | 0% | *2/*17 | Intermediate Metabolizer |
| Volunteer 6 | 4 | 1 | 25% | 3 | 0 | 0% | *17/*17 | Ultra-rapid Metabolizer |
| Volunteer 7 | 3 | 0 | 0% | 3 | 0 | 0% | *1/*17 | Ultra-rapid Metabolizer |
| Volunteer 8 | 8 | 1 | 12.5% | 3 | 0 | 0% | *1/*1 | Extensive Metabolizer |
| Coriell (gDNA) 1 |
1 | 0 | 0% | 3 | 0 | 0% | *1/*1 | Extensive Metabolizer |
| Coriell (gDNA) 2 |
1 | 0 | 0% | 3 | 0 | 0% | *2/*3 | Poor Metabolizer |
| Coriell (gDNA) 3 |
1 | 1 | 100% | 3 | 0 | 0% | *2/*3 | Poor Metabolizer |
Lessons learned: Before implementing genotype-guided drug therapy (GGTx), new technologies must be clinically validated in a stringent CLIA environment. We recommend a longer timeline should there be any unexpected problems and a need to evaluate multiple platforms. Allow for individual laboratories to gain valuable experience and facilitate informed decisions that work best with their laboratory and institutional workflow.
Implementation of workflow into electronic medical record and genotype guided clinical decision support
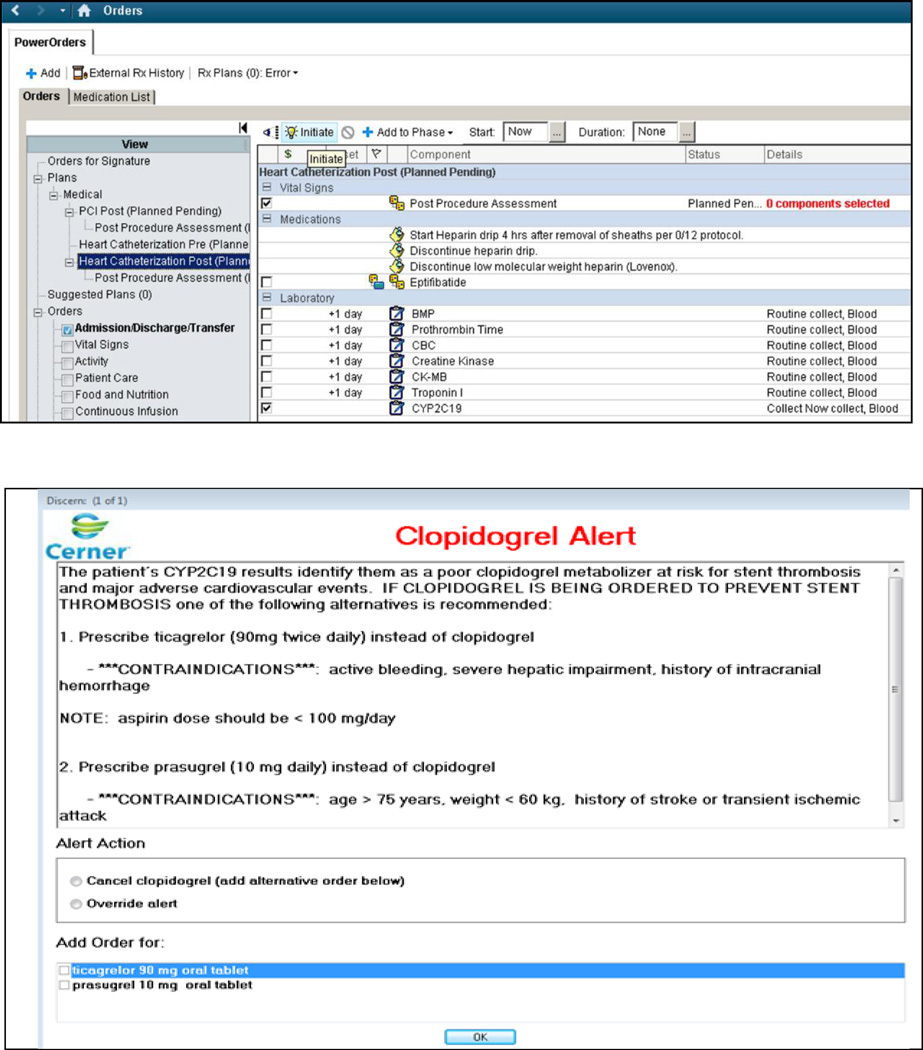
In parallel to the genotyping validation studies, the ability to order genotype test was built into the post-catheterization order sets (Figure 2A) and clinical decision support (CDS) developed (Figure 2B). The ability to order the test is currently active in the EHR, and the CDS will alert the ordering physician if two conditions are met; namely the prescription of clopidogrel in the setting of ACS/PCI, and the possession of a CYP2C19 LOF allele. Physicians are asked during electronic order entry for clopidogrel whether clopidogrel is being ordered in the setting of ACS/PCI. Once they confirm then the CDS is triggered based on the above conditions. This limits the number of alerts triggered while facilitating the selection of alternative agents or the option to override the recommendation.
Figure 2.
a) CYP2C19 test order built into the post-cardiac-catheterization order sets.
b) Clinical decision support guiding antiplatelet selection in post-PCI patients possessing CYP2C19 loss-of-function alleles*
It is important to emphasize that we have implemented reactive genotype guided therapy where interventionists order the CYP2C19 test only after PCI as they cannot predict whether or not a patient will undergo PCI prior to angiography. After completion of the intervention, along with the post-PCI orders (Figure 2a) the physician can order the CYP2C19 test. To ensure antiplatelet coverage, the current practice is to place the orders for DAPT before the CYP2C19 genotype is available. In our workflow, the laboratory technologist notifies the treating physician by pager if the patient possesses LOF alleles. Under our current clinical care timeline and reactive-genotyping, the genotype data is not available at the time DAPT orders are recorded in the EHR. In the future when genotype data are available prior to patient experiencing ACS / undergoing PCI, the CDS support functionality to alert clinicians to the existing genotypic data will be activated in the EHR.
Lessons learned: Genotyping strategy (reactive or preemptive) and the timeline of care provision will inform institutions on how to best deliver the intervention and clinical decision support for their local workflow.
Genotype results and acceptance of recommended treatment
From January 28, 2015 through July 22, 2016, a total of 231 clinical tests were performed using buccal swabs collected post-PCI. The test turnaround time was 70 min (range of 65–75 min) with the results presented under the laboratory menu in Cerner with notification by pager to the attending physician. Six patients underwent repeat testing (5 due to inconclusive results and 1 due to a failed positive control). The distribution of CYP2C19 genotype and treatment prescribed is presented in Table 4. Most patients genotyped were European Americans (71.9%) or African Americans (26.8%). Genotype frequencies did not differ by race (all p-values >0.18; data not shown). CYP2C19 *3 was not encountered among the 231 patients tested. Of the patients tested, 39.4% possessed the normal metabolizer phenotype (*1/*1), and 32.9% possessed the ultra-rapid metabolizer phenotype (*1/*17, *17/*17). Three percent of patients possessed the poor metabolizer (*2/*2) and 24.7% possessed intermediate metabolizer phenotype (*1/*2), bringing the total ACS/PCI patients with actionable genotypes to 27.7%.
Table 4.
Genotype results and treatment assignment among patients undergoing CYP2C19 testing.
| CYP2C19 Genotype* |
Total (%) | Race (number (%) | Aspirin + Clopidogrel |
Aspirin + Ticagrelor |
Aspirin + Prasugrel |
||
|---|---|---|---|---|---|---|---|
| African American |
European American |
Other (%) |
|||||
| *1/*1 | 91 (39.4) | 23 (37.1) | 67 (40.4) | 1 (33.3) | 63 | 28 | 0 |
| *1/*17 | 62 (26.8) | 14 (22.6) | 46 (27.7) | 2 (66.7) | 44 | 18 | 0 |
| *17/*17 | 14 (6.1) | 6 (9.7) | 8 (4.8) | 0 (0.0) | 8 | 6 | 0 |
| *1/*2 | 41 (17.7) | 10 (16.1) | 31 (18.7) | 0 (0.0) | 16 | 22 | 3 |
| *2/*17 | 16 (6.9) | 6 (9.7) | 10 (6.0) | 0 (0.0) | 8 | 7 | 1 |
| *2/*2 | 7 (3.0) | 3 (4.8) | 4 (2.4) | 0 (0.0) | 0 | 6 | 1 |
| TOTAL | 231 | 62 | 166 | 3 | 140 | 86 | 5 |
Genotype frequencies did not differ by race (all p-values>0.18)
All patients homozygous for LOF alleles were prescribed an alternative antiplatelet agent, most often ticagrelor, in combination with aspirin. Among patients heterozygous for the LOF allele (n=57), a majority (58%) were prescribed an alternative antiplatelet agent. Reasons for not following the recommendation for the remaining 24 patients heterozygous for the LOF allele included contraindications to alternative agents (n=6), cost related issues (n=9), or physician decision based on a patient’s bleeding tendency and co-therapy with warfarin (n=2). For seven patients, no reasons could be identified.
Lessons learned: Adoption and acceptance of genotype-based interventions requires buy-in from technicians and clinicians. Acceptance of genotype-guided therapy may not be universal at the beginning. Implementation initiatives should include ongoing educational efforts to inform clinicians on how to use this new information into treatment decisions.
Assessing impact on clinical outcomes
To evaluate whether genotype-guided therapy improves outcomes over a one-year follow-up, we also enrolled ACS/PCI patients receiving GGTx, and those not receiving GGTx in a prospective cohort study with IRB approval. Patients were consented for a one-year follow-up and a DNA and plasma sample was archived for future research. After discharge, patients are followed prospectively for up to one year to document the clinical course and changes in medication therapy. During follow-up each patient was tracked in the UAB-EMR for events reviewing all readmission, emergency room visits, and office visits. As discussed during the process of informed consent, all patients or family members are contacted at 6 month intervals to ascertain if encounters outside the UAB Health System occurred. Primary care physicians are contacted to document the clinical course for each patient under signed medical release authorization. This enabled verification of patient reported events and documentation of events not reported by patients.
Thromboembolic and hemorrhagic events encountered during the follow-up period are adjudicated by an interventional cardiologist blinded to patient genotype.15,27–29 Thromboembolic complications include stent thrombosis as defined by Academic Research Consortium15 and MACE: defined as non-fatal stroke,15 non-fatal myocardial infarction (MI),16 and death secondary to any cardiovascular cause.15 This includes deaths from MI, sudden cardiac death, heart failure, stroke, and other cardiovascular causes. For hemorrhage, 72 individual elements from commonly used bleeding scales were collected (e.g., ≥10% hematocrit and/or ≥2g/dL hemoglobin drop, transfusion, surgical intervention). These elements can then be used to derive bleeding as defined by commonly employed classification systems.30–32
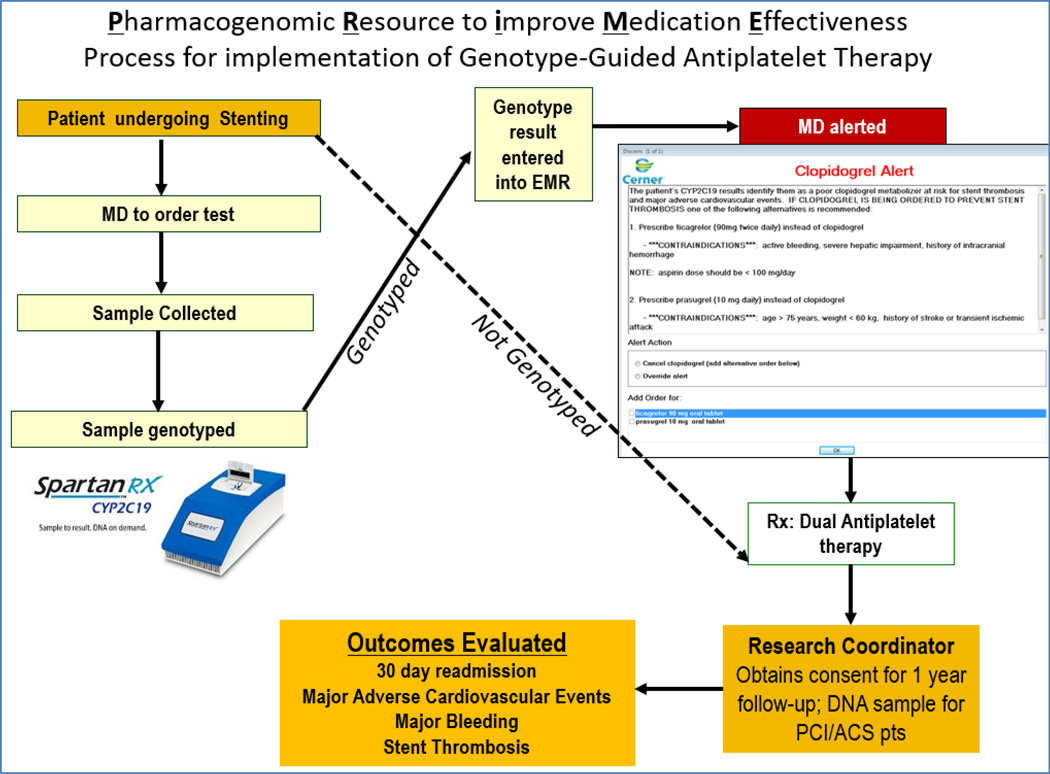
Implementation efforts can provide an opportunity to bolster institutional resources. Since the CYP2C19 test is not ordered in all patients as standard-of-care, we leveraged this implementation initiative to enhance our research infrastructure. As shown in Figure 3, along with patients receiving GGTx, those prescribed DAPT without genotyping are enrolled into a prospective cohort. A total of 548 patients (21.4% African American; 28.8% women) have been enrolled with a one-year prospective follow-up ongoing. Of these 231 received genotype-guided therapy. Plasma and DNA are archived within 24 hours of PCI, along with clinical course over the prospective one-year follow-up will provide a valuable resource for future research. Over this duration approximately 1440 patients underwent PCI. While it would be ideal to have all patients enrolled in the study, the limited research staff and the patient discharge within 24–36 hours post procedures did not allow time for all patients to be approached for enrollment. Similarly patients undergoing PCI on the weekends or holidays were not approached for enrollment.
Figure 3.
Clinical and research workflow for the implementation of genotype-guided antiplatelet therapy.
Lessons learned: Implementation efforts can provide a unique opportunity to bolster institutional resources. Engaging clinicians and researchers together in a robust assessment of outcomes will create resources which can be leveraged for future discovery.
Conclusion and future directions
We present our framework to demonstrate that implementation of GGTx, integrating test25 ordering and reporting can be built into the EHR, along with CDS. We describe how we offered genotype-guided antiplatelet therapy for patients undergoing PCI at UAB in a timeline conducive for optimizing clinical care. This process requires multidisciplinary collaboration across the clinical, administrative and research enterprise, including clinicians, pharmacists, pathologists, informaticians and researchers. In the specific example of CYP2C19 guided antiplatelet therapy, treatment recommendations were accepted in all poor clopidogrel metabolizers and a majority of intermediate metabolizers. Valid reasons for not following institutional guideline recommendations were identified in most instances.
Recognition of the barriers and challenges in integration of pharmacogenetics to guide treatment has fueled efforts on multiple fronts. For instance, the FDA has provided label updates for medications where genes/SNPs may help tailor dosing, improve efficacy and reduce adverse effects. To help clinicians understand and incorporate pharmacogenomics in making drug therapy decisions, CPIC synthesizes the evidence into guidelines for gene-drug pairs to enable the translation of test results into actionable decisions for specific drugs.3,33 To facilitate reporting and sharing pharmacogenetic test results across laboratories and EHRs, allele function and phenotypes are being standardized 2,34 and incorporated into well curated and machine-readable database of pharmacogenomic knowledge.35 The Implementing GeNomics In pracTicE (IGNITE; funded by NIH) network identifies and disseminates best practices for implementation and integration of pharmacogenomics to the broader community.36
Investigative teams have leveraged these resources to build consortia to address challenges in precision medicine and develop and disseminate best practices for implementation. Among the first such efforts, the IGNITE Pharmacogenetics team, is conducting a collaborative analysis using data from nine sites implementing genotype-guided antiplatelet therapy in patients undergoing PCI at their respective institutions. The data collection has been harmonized to capture the clinical course over a 1-year post-PCI period including MACE, bleeding, stent thrombosis. This analysis will shed light on wwhether genotype-guided antiplatelet therapy improves outcomes; i.e. demonstrate benefit in high-risk patients.
This provides a US based implementation cohort to allow robust assessment of clinical effectiveness of GGTx. This analysis will be followed by an economic analysis which will incorporate test costs (ranging from $100-250/test), reimbursement ($290/test by Medicare) along with cost of treatment for encountered MACE and bleeding events during follow-up. Such analysis will be vital in demonstrating evidence of clinical utility of GGTx and developing sustainable models for broader implementation.
Lessons learned: Implementation initiatives can provide a unique opportunity to conduct collaborative analysis. Pooling data across similar efforts provides a larger sample size and enables robust assessment of clinical effectiveness and cost-effectiveness of GGTx interventions.
Implementation efforts have moved beyond “one-gene-at-a-time” to combinatorial pharmacogenomics.37–40 A new challenge is to leverage data obtained through next-generation sequencing technologies to optimize oncology drug and dose selection based on germline pharmacogenetic variants.41 Pre-emptive genotyping efforts in adult and pediatric populations, including efforts in EHRs linked to biobanks where CDS is triggered are ongoing.42–47 Multidisciplinary teams must include pharmacists, clinicians, and pathologists to incorporate genomic information in drug therapy management 48–51 and work with dissemination scientists to study processes and understand factors influencing organizational adoption and implementation of clinical genetic services.52–54
Challenges faced and solutions developed by early-adopter institutions can provide valuable insight for institutions introducing similar programs. While much of the earlier work was initially done in isolation, structured collaborations such as the IGNITE pharmacogenetics consortium will be crucial in accessing clinical outcomes and informing reimbursement strategies. In order for precision medicine to materialize into reality, the utility of genomic information in clinical care must be demonstrated using robust implementation and real-world outcome studies.55,56 Pharmacogenetics is necessarily a pioneering field within a broader future of genomic guided medicine.
Acknowledgments
Funding/Support: This work was supported in part by UAB’s Health Service Foundations' General Endowment Fund and Hugh Kaul Personalized Medicine Institute and by grants from the National Heart Lung and Blood Institute (RO1HL092173; K24HL133373), and the National Institutes of Health (NIH) Clinical and Translational Science Award (CTSA) program (UL1TR000165).
References
- 1.Dotson WD, Douglas MP, Kolor K, et al. Prioritizing genomic applications for action by level of evidence: a horizon-scanning method. Clin Pharmacol Ther. 2014;95:394–402. doi: 10.1038/clpt.2013.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Caudle KE, Dunnenberger HM, Freimuth RR, et al. Standardizing terms for clinical pharmacogenetic test results: consensus terms from the Clinical Pharmacogenetics Implementation Consortium (CPIC) Genet Med. 2016 doi: 10.1038/gim.2016.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Relling MV, Klein TE. CPIC: Clinical Pharmacogenetics Implementation Consortium of the Pharmacogenomics Research Network. Clin Pharmacol Ther. 2011;89:464–467. doi: 10.1038/clpt.2010.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vladutiu GD. The FDA announces new drug labeling for pharmacogenetic testing: is personalized medicine becoming a reality? Mol Genet Metab. 2008;93:1–4. doi: 10.1016/j.ymgme.2007.10.133. [DOI] [PubMed] [Google Scholar]
- 5.Lesko LJ, Schmidt S. Clinical implementation of genetic testing in medicine: a US regulatory science perspective. Br J Clin Pharmacol. 2014;77:606–611. doi: 10.1111/bcp.12299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scott SA, Sangkuhl K, Stein CM, et al. Clinical Pharmacogenetics Implementation Consortium guidelines for CYP2C19 genotype and clopidogrel therapy: 2013 update. Clin Pharmacol Ther. 2013;94:317–323. doi: 10.1038/clpt.2013.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Close SL. Clopidogrel pharmacogenetics: metabolism and drug interactions. Drug Metabol Drug Interact. 2011;26:45–51. doi: 10.1515/DMDI.2011.002. [DOI] [PubMed] [Google Scholar]
- 8.Farid NA, Kurihara A, Wrighton SA. Metabolism and disposition of the thienopyridine antiplatelet drugs ticlopidine, clopidogrel, and prasugrel in humans. J Clin Pharmacol. 2010;50:126–142. doi: 10.1177/0091270009343005. [DOI] [PubMed] [Google Scholar]
- 9.Desta Z, Zhao X, Shin JG, Flockhart DA. Clinical significance of the cytochrome P450 2C19 genetic polymorphism. Clin Pharmacokinet. 2002;41:913–958. doi: 10.2165/00003088-200241120-00002. [DOI] [PubMed] [Google Scholar]
- 10.Sibbing D. Cytochrome 2C19*17 allelic variant, platelet aggregation, bleeding events, and stent thrombosis in clopidogrel-treated patients with coronary stent placement. Circulation. 2010;121:512–518. doi: 10.1161/CIRCULATIONAHA.109.885194. [DOI] [PubMed] [Google Scholar]
- 11.Cuisset T, Loosveld M, Morange PE, et al. CYP2C19*2 and *17 alleles have a significant impact on platelet response and bleeding risk in patients treated with prasugrel after acute coronary syndrome. JACC Cardiovasc Interv. 2012;5:1280–1287. doi: 10.1016/j.jcin.2012.07.015. [DOI] [PubMed] [Google Scholar]
- 12.Harmsze AM, van Werkum JW, Hackeng CM, et al. The influence of CYP2C19*2 and *17 on on-treatment platelet reactivity and bleeding events in patients undergoing elective coronary stenting. Pharmacogenet Genomics. 2012;22:169–175. doi: 10.1097/FPC.0b013e32834ff6e3. [DOI] [PubMed] [Google Scholar]
- 13.Li Y, Tang HL, Hu YF, Xie HG. The gain-of-function variant allele CYP2C19*17: a double-edged sword between thrombosis and bleeding in clopidogrel-treated patients. J Thromb Haemost. 2012;10:199–206. doi: 10.1111/j.1538-7836.2011.04570.x. [DOI] [PubMed] [Google Scholar]
- 14.Sibbing D, Gebhard D, Koch W, et al. Isolated and interactive impact of common CYP2C19 genetic variants on the antiplatelet effect of chronic clopidogrel therapy. J Thromb Haemost. 2010;8:1685–1693. doi: 10.1111/j.1538-7836.2010.03921.x. [DOI] [PubMed] [Google Scholar]
- 15.Cutlip DE, Windecker S, Mehran R, et al. Clinical end points in coronary stent trials: a case for standardized definitions. Circulation. 2007;115:2344–2351. doi: 10.1161/CIRCULATIONAHA.106.685313. [DOI] [PubMed] [Google Scholar]
- 16.Thygesen K, Alpert JS, Jaffe AS, et al. Third universal definition of myocardial infarction. Circulation. 2012;126:2020–2035. doi: 10.1161/CIR.0b013e31826e1058. [DOI] [PubMed] [Google Scholar]
- 17.Shuldiner AR, O'Connell JR, Bliden KP, et al. Association of cytochrome P450 2C19 genotype with the antiplatelet effect and clinical efficacy of clopidogrel therapy. Jama. 2009;302:849–857. doi: 10.1001/jama.2009.1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen BL, Zhang W, Li Q, et al. Inhibition of ADP-induced platelet aggregation by clopidogrel is related to CYP2C19 genetic polymorphisms. Clin Exp Pharmacol Physiol. 2008;35:904–908. doi: 10.1111/j.1440-1681.2008.04915.x. [DOI] [PubMed] [Google Scholar]
- 19.Mega JL, Simon T, Collet JP, et al. Reduced-function CYP2C19 genotype and risk of adverse clinical outcomes among patients treated with clopidogrel predominantly for PCI: a meta-analysis. Jama. 2010;304:1821–1830. doi: 10.1001/jama.2010.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perry E. Clopidogrel hyporesponsiveness and the FDA boxed warning: detection and management of patients with genetic polymorphisms. Am J Health Syst Pharm. 2011;68:529–532. doi: 10.2146/ajhp100422. [DOI] [PubMed] [Google Scholar]
- 21.Holmes DR, Jr, Dehmer GJ, Kaul S, Leifer D, O'Gara PT, Stein CM. ACCF/AHA Clopidogrel clinical alert: approaches to the FDA [ldquo]boxed warning[rdquo]: a report of the American College of Cardiology Foundation Task Force on Clinical Expert Consensus Documents and the American Heart Association. Circulation. 2010;122:537–557. doi: 10.1161/CIR.0b013e3181ee08ed. [DOI] [PubMed] [Google Scholar]
- 22.Levine GN, Bates ER, Blankenship JC, et al. 2011 ACCF/AHA/SCAI guideline for percutaneous coronary intervention: Executive Summary. Catheterization and Cardiovascular Interventions. 2012;79:453–495. doi: 10.1002/ccd.23438. [DOI] [PubMed] [Google Scholar]
- 23.Kovacic JC, Limaye AM, Sartori S, et al. Comparison of six risk scores in patients with triple vessel coronary artery disease undergoing PCI: competing factors influence mortality, myocardial infarction, and target lesion revascularization. Catheter Cardiovasc Interv. 2013;82:855–868. doi: 10.1002/ccd.25008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bawamia B, Mehran R, Qiu W, Kunadian V. Risk scores in acute coronary syndrome and percutaneous coronary intervention: a review. Am Heart J. 2013;165:441–450. doi: 10.1016/j.ahj.2012.12.020. [DOI] [PubMed] [Google Scholar]
- 25.McNeely C, Markwell S, Vassileva CM. Readmission after inpatient percutaneous coronary intervention in the Medicare population from 2000 to 2012. Am Heart J. 2016;179:195–203. doi: 10.1016/j.ahj.2016.07.002. [DOI] [PubMed] [Google Scholar]
- 26.Wasfy JH, Rosenfield K, Zelevinsky K, et al. A prediction model to identify patients at high risk for 30-day readmission after percutaneous coronary intervention. Circ Cardiovasc Qual Outcomes. 2013;6:429–435. doi: 10.1161/CIRCOUTCOMES.111.000093. [DOI] [PubMed] [Google Scholar]
- 27.Schulman S, Kearon C. Definition of major bleeding in clinical investigations of antihemostatic medicinal products in non-surgical patients. J Thromb Haemost. 2005;3:692–694. doi: 10.1111/j.1538-7836.2005.01204.x. [DOI] [PubMed] [Google Scholar]
- 28.Seto AC, Kenyon K, Wittkowsky AK. Discrepancies in identification of major bleeding events in patients taking warfarin. Pharmacotherapy. 2008;28:1098–1103. doi: 10.1592/phco.28.9.1098. [DOI] [PubMed] [Google Scholar]
- 29.Wiviott SD, Trenk D, Frelinger AL, et al. Prasugrel compared with high loading- and maintenance-dose clopidogrel in patients with planned percutaneous coronary intervention: the Prasugrel in Comparison to Clopidogrel for Inhibition of Platelet Activation and Aggregation-Thrombolysis in Myocardial Infarction 44 trial. Circulation. 2007;116:2923–2932. doi: 10.1161/CIRCULATIONAHA.107.740324. [DOI] [PubMed] [Google Scholar]
- 30.Hicks KA, Stockbridge NL, Targum SL, Temple RJ. Bleeding Academic Research Consortium consensus report: the Food and Drug Administration perspective. Circulation. 2011;123:2664–2665. doi: 10.1161/CIRCULATIONAHA.111.032433. [DOI] [PubMed] [Google Scholar]
- 31.An international randomized trial comparing four thrombolytic strategies for acute myocardial infarction. The GUSTO investigators. N Engl J Med. 1993;329:673–682. doi: 10.1056/NEJM199309023291001. [DOI] [PubMed] [Google Scholar]
- 32.James S, Akerblom A, Cannon CP, et al. Comparison of ticagrelor, the first reversible oral P2Y(12) receptor antagonist, with clopidogrel in patients with acute coronary syndromes: Rationale, design, and baseline characteristics of the PLATelet inhibition and patient Outcomes (PLATO) trial. Am Heart J. 2009;157:599–605. doi: 10.1016/j.ahj.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 33.Tutton R. Pharmacogenomic biomarkers in drug labels: what do they tell us? Pharmacogenomics. 2014;15:297–304. doi: 10.2217/pgs.13.198. [DOI] [PubMed] [Google Scholar]
- 34.Kalman LV, Agundez J, Appell ML, et al. Pharmacogenetic allele nomenclature: International workgroup recommendations for test result reporting. Clin Pharmacol Ther. 2016;99:172–185. doi: 10.1002/cpt.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hoffman JM, Dunnenberger HM, Kevin Hicks J, et al. Developing knowledge resources to support precision medicine: principles from the Clinical Pharmacogenetics Implementation Consortium (CPIC) Journal of the American Medical Informatics Association : JAMIA. 2016;23:796–801. doi: 10.1093/jamia/ocw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weitzel KW, Alexander M, Bernhardt BA, et al. The IGNITE network: a model for genomic medicine implementation and research. BMC Med Genomics. 2016;9:1. doi: 10.1186/s12920-015-0162-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Altar CA, Carhart JM, Allen JD, Hall-Flavin DK, Dechairo BM, Winner JG. Clinical validity: Combinatorial pharmacogenomics predicts antidepressant responses and healthcare utilizations better than single gene phenotypes. Pharmacogenomics J. 2015;15:443–451. doi: 10.1038/tpj.2014.85. [DOI] [PubMed] [Google Scholar]
- 38.Johnson JA, Klein TE, Relling MV. Clinical Implementation of Pharmacogenetics: More than One Gene at a Time. Clin Pharmacol Ther. 2013;93:384–385. doi: 10.1038/clpt.2013.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hicks JK, Bishop JR, Sangkuhl K, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for CYP2D6 and CYP2C19 Genotypes and Dosing of Selective Serotonin Reuptake Inhibitors. Clin Pharmacol Ther. 2015;98:127–134. doi: 10.1002/cpt.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muller DJ, Kekin I, Kao AC, Brandl EJ. Towards the implementation of CYP2D6 and CYP2C19 genotypes in clinical practice: update and report from a pharmacogenetic service clinic. International review of psychiatry (Abingdon, England) 2013;25:554–571. doi: 10.3109/09540261.2013.838944. [DOI] [PubMed] [Google Scholar]
- 41.Gillis NK, Patel JN, Innocenti F. Clinical implementation of germ line cancer pharmacogenetic variants during the next-generation sequencing era. Clin Pharmacol Ther. 2014;95:269–280. doi: 10.1038/clpt.2013.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peterson JF, Bowton E, Field JR, et al. Electronic health record design and implementation for pharmacogenomics: a local perspective. Genet Med. 2013;15:833–841. doi: 10.1038/gim.2013.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dunnenberger HM, Crews KR, Hoffman JM, et al. Preemptive Clinical Pharmacogenetics Implementation: Current Programs in Five United States Medical Centers. Annu Rev Pharmacol Toxicol. 2014 doi: 10.1146/annurev-pharmtox-010814-124835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.O'Donnell PH, Danahey K, Jacobs M, et al. Adoption of a clinical pharmacogenomics implementation program during outpatient care--initial results of the University of Chicago "1,200 Patients Project". American journal of medical genetics Part C, Seminars in medical genetics. 2014;166c:68–75. doi: 10.1002/ajmg.c.31385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ji Y, Skierka JM, Blommel JH, et al. Preemptive Pharmacogenomic Testing for Precision Medicine: A Comprehensive Analysis of Five Actionable Pharmacogenomic Genes Using Next-Generation DNA Sequencing and a Customized CYP2D6 Genotyping Cascade. J Mol Diagn. 2016;18:438–445. doi: 10.1016/j.jmoldx.2016.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rasmussen-Torvik LJ, Stallings SC, Gordon AS, et al. Design and anticipated outcomes of the eMERGE-PGx project: a multicenter pilot for preemptive pharmacogenomics in electronic health record systems. Clin Pharmacol Ther. 2014;96:482–489. doi: 10.1038/clpt.2014.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hoffman JM, Haidar CE, Wilkinson MR, et al. PG4KDS: a model for the clinical implementation of pre-emptive pharmacogenetics. American journal of medical genetics Part C, Seminars in medical genetics. 2014;166c:45–55. doi: 10.1002/ajmg.c.31391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Owusu-Obeng A, Weitzel KW, Hatton RC, et al. Emerging roles for pharmacists in clinical implementation of pharmacogenomics. Pharmacotherapy. 2014;34:1102–1112. doi: 10.1002/phar.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hicks JK, Stowe D, Willner MA, et al. Implementation of Clinical Pharmacogenomics within a Large Health System: From Electronic Health Record Decision Support to Consultation Services. Pharmacotherapy. 2016;36:940–948. doi: 10.1002/phar.1786. [DOI] [PubMed] [Google Scholar]
- 50.Johnson JA, Weitzel KW. Advancing Pharmacogenomics as a Component of Precision Medicine: How Where, and Who? Clin Pharmacol Ther. 2016;99:154–156. doi: 10.1002/cpt.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Eadon MT, Desta Z, Levy KD, et al. Implementation of a pharmacogenomics consult service to support the INGENIOUS trial. Clin Pharmacol Ther. 2016;100:63–66. doi: 10.1002/cpt.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Haga SB, LaPointe NM, Cho A, et al. Pilot study of pharmacist-assisted delivery of pharmacogenetic testing in a primary care setting. Pharmacogenomics. 2014;15:1677–1686. doi: 10.2217/pgs.14.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hamilton AB, Oishi S, Yano EM, Gammage CE, Marshall NJ, Scheuner MT. Factors influencing organizational adoption and implementation of clinical genetic services. Genet Med. 2014;16:238–245. doi: 10.1038/gim.2013.101. [DOI] [PubMed] [Google Scholar]
- 54.Kapoor R, Tan-Koi WC, Teo YY. Role of pharmacogenetics in public health and clinical health care: a SWOT analysis. Eur J Hum Genet. 2016 doi: 10.1038/ejhg.2016.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ioannidis JPA. This I believe in genetics: discovery can be a nuisance, replication is science, implementation matters. Frontiers in Genetics. 2013:4. doi: 10.3389/fgene.2013.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pinnock H, Epiphaniou E, Sheikh A, et al. Developing standards for reporting implementation studies of complex interventions (StaRI): a systematic review and e-Delphi. Implementation science : IS. 2015;10:42. doi: 10.1186/s13012-015-0235-z. [DOI] [PMC free article] [PubMed] [Google Scholar]