Abstract
In Streptococcus pneumoniae TIGR4, genes encoding a SecY2A2 accessory Sec system are present within a locus encoding a serine-rich repeat surface protein PsrP. Mutant strains deleted in secA2 or psrP were deficient in biofilm formation, while the ΔsecA2 mutant was reduced in binding to airway epithelial cells. Cell wall protein (CWP) fractions from the ΔsecA2 mutant, but not from the ΔpsrP mutant, were reduced in haemolytic (pneumolysin) activity. Contact-dependent pneumolysin (Ply) activity of wild type TIGR4 cells was ten-fold greater than that of ΔsecA2 mutant cells suggesting that Ply was not active at the ΔsecA2 cell surface. Ply protein was found to be present in the CWP fraction from the ΔsecA2 mutant, but showed aberrant electrophoretic migration indicative of protein modification. Proteomic analyses led to the discovery that the ΔsecA2 mutant CWP fraction was deficient in two glycosidases as well as other enzymes involved in carbohydrate metabolism. Taken collectively the results suggest that positioning of Ply into the cell wall compartment in active form, together with glycosyl hydrolases and adhesins, requires a functional accessory Sec system.
Keywords: SecA2, Streptococcus pneumoniae, Pneumolysin, Export, O-glycosidase, Biofilms
1. Introduction
Streptococcus pneumoniae, also known as the pneumococcus, colonizes the human respiratory tract. This can be asymptomatic, but in more susceptible individuals e.g. infants, the elderly, immunocompromised, the bacteria can move to other body sites and cause sinusitis, otitis media, pneumonia or meningitis. Pneumococci exhibit many of the properties of oral viridans streptococci, such as α-haemolysis on blood agar caused by hydrogen peroxide production, adherence to glycoproteins [1], biofilm formation [2] and natural competence for DNA-mediated transformation [3]. Components known to be essential for full virulence are the capsular polysaccharides, of which there are more than 90 serotypes [4], choline-binding proteins CbpA, LytA, PcpA, PspA and PspC [5], pore-forming toxin pneumolysin (Ply), and various cell-surface-associated proteins Eno, NanA, PavA, PavB, PsaA and PsrP [6], [7].
Ply is a highly conserved 53-kDa pore-forming toxin that is a member of a protein family known as the cholesterol-dependent cytolysins (CDCs). Members of this toxin family are expressed in Streptococcus, Clostridium and Listeria, and include streptolysin O, perfringolysin O and listeriolysin O. The CDC toxins bind to cholesterol in target membranes, and once inserted into the membrane they oligomerize to form pores (350–450 Å in diameter for Ply) [8], resulting in host cell lysis and tissue damage [9]. Ply is produced by virtually all clinical isolates of S. pneumoniae and is multifunctional. In addition to cytolytic activity, Ply modulates the immune system by activating the classical complement pathway [10] and neutrophil extracellular trap formation [11]. Ply also affects lysosomal integrity in epithelial cells [12], induces DNA damage and cell cycle arrest [13], and is reported to impact on biofilm formation [14]. However, a confounding issue about the production of Ply is that, unlike other CDC family members, Ply lacks an N-terminal leader peptide to direct secretion through the canonical Sec pathway. Consequently, it has been considered an intracellular protein, released only following cell lysis in vitro or in vivo [15]. More recently, evidence has emerged for autolysis-independent release of Ply [16], and for exported Ply to be localized mainly to the bacterial cell wall [17]. The current notion is that Ply is exported into the cell wall peptidoglycan matrix, within which branch-stem peptides act as a barrier to Ply release [18]. However, the precise mechanism for Ply export across the cell membrane has not yet been established.
Fusion of a canonical Sec signal sequence to Ply did not allow Sec-dependent Ply secretion in S. pneumoniae, although secretion occurred when the same construct was expressed in surrogate host Bacillus subtilis [19]. More importantly, B. subtilis secreted Ply with no leader peptide added, suggesting the existence of a conserved protein export system that is coupled to cell wall localization [19]. In L. monocytogenes, listeriolysin O is found extracellularly in complex with a putative chaperone protein (FbpA) and internalin B (InlB) [20]. FbpA protein is one of a number of leader-less proteins in L. monocytogenes that are secreted via an alternative pathway known as the accessory Sec system [21]. We hypothesized that the accessory Sec system in S. pneumoniae facilitates the export of Ply across the cytoplasmic membrane into the cell wall environment.
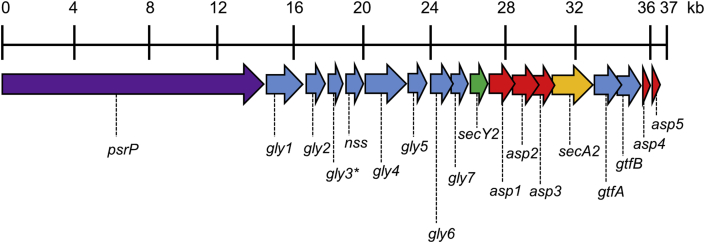
In S. pneumoniae TIGR4 (serotype 4), secY2 (transmembrane protein) and secA2 (ATPase) genes are found within a 37-kb pathogenicity island encoding cell-surface pneumococcal serine-rich repeat protein (PsrP, 4776 aa residues) (Fig. 1). An additional 10 genes encode glycosylation enzymes, and five asp genes encode transport complex proteins [22], [23]. A similar locus is found in several but not all sequenced pneumococcal genomes. By analogy to genomic loci in Streptococcus gordonii and Streptococcus parasanguinis encoding serine-rich repeat (SRR) proteins GspB [24], Hsa [25], [26] and Fap1 [27], PsrP in S. pneumoniae is predicted to become post-translationally glycosylated concomitantly with secretion via the alternate SecY2A2/Asp1-5 translocon [28]. The C-terminus of PsrP glycoprotein is cell wall-anchored while the N-terminal region is projected away from the cell surface and binds keratin [29]. In addition, PsrP plays a role in biofilm formation in vitro [30] and in vivo [31] by mediating direct cell–cell accumulation, or indirectly through binding extracellular DNA [32].
Fig. 1.
Schematic representation of the ∼36 kb accessory secY2A2 locus derived from the genome sequence of S. pneumoniae TIGR4 (GenBank™ accession number AE005672.3). Genes encode the following: serine-rich repeat protein psrP (SP_1772); glycosyltransferases gly1 (SP_1771), gly2 (SP_1770), gly3* (SP_1769), nss (SP_1768), gly4 (SP_1767), gly5 (SP_1766), gly6 (SP_1765), gly7 (SP_1764), gtfA (SP_1758, SP_RS08705), gtfB (SP_1757, SP_RS08700); accessory secretion proteins asp1 (SP_1762), asp2 (SP_1761), asp3 (SP_1760), asp4 (SP_1756), asp5 (SP_1755); secY2 (SP_1763) and secA2 (SP_1759, SP_RS08710). *gene contains a frame shift mutation.
To determine if Ply export was mediated or modulated by the SecY2A2 system in S. pneumoniae, the translocon function was inactivated by deletion of the secA2 gene. The effect of this was to inhibit biofilm formation, association to lung epithelial cells, and export of Ply to the cell wall compartment. Our results suggest that the SecY2A2 translocon is required for efficient localization of Ply to the cell surface.
2. Materials and methods
2.1. Bacterial strains and growth conditions
Bacteria and plasmids used are shown in Table S1. Pneumococci were grown on THY-blood agar (3.6% Todd-Hewitt broth, 5% yeast extract, 12% agar) with 5% defibrinated horse blood, or in THY broth, at 37 °C in 5% CO2, supplemented as appropriate with 100 μg spectinomycin (Sp) ml−1, 2 μg erythromycin (Em) ml−1 or 2 μg chloramphenicol (Cm) ml−1. Escherichia coli was cultivated in LB medium, with 100 μg ampicillin (Ap) ml−1 or 300 μg Em ml−1 as required, and was manipulated by standard protocols. Overnight cultures (10 ml) of all bacteria were grown for approximately 16 h unless stated otherwise. For long term storage (glycerol stocks), overnight cultures were pelleted and resuspended in appropriate broth containing 15% glycerol. Strains were either placed into the culture collection or aliquoted when needed for frequent use and stored at −70 °C until used. Cultures were always inoculated with standard amounts of frozen glycerol stock suspensions.
2.2. Generation of mutants
To generate a secA2 deletion, chromosomal DNA from S. pneumoniae TIGR4 (wild type, WT) was PCR-amplified (Expand Long Template, Roche Diagnostics, Mannheim, Germany) with primer pairs MBF7/MBR7 and MBF8/MBR8 (Table S2), producing 552-bp and 579-bp secA2-flanking regions. These were ligated by PCR, generating a central BamHI site, and cloned into pGEM-T in E. coli JM109. Spectinomycin-resistance cassette aad9 (1015 bp) was PCR-amplified from pFW5 [33] with primer pair BamSpecF/BamSpecR (Table S2) and cloned into the unique BamHI site within the flanking regions construct. This was purified, the insert was excised with SacII and SpeI, gel-purified and transformed [34] into S. pneumoniae TIGR4 selecting for SpR. Fidelity of strain UB2570 ΔsecA2 was confirmed by PCR-amplification with MBF7/MBR8 primers and DNA sequencing. A similar method was employed to produce a Δply mutant. Flanking regions of ply (624 bp and 940 bp) were PCR-amplified with primer pairs Up.PlyF/Up.PlyR and Down.PlyF/Down.PlyR (Table S2), ligated by PCR and cloned into pGEM-T in E. coli DH5α. Chloramphenicol resistance cassette cat (937 bp) was amplified from pR326 [35] with primers BamCatF2/BamCatR, cloned into the unique BamHI site within the flanking regions plasmid construct, and then excised (2513 bp), purified and transformed into S. pneumoniae as described above, selecting for CmR. Strain UB2706 Δply was confirmed by PCR with Up.PlyF/Down.PlyR and DNA sequencing. To generate a double mutant, the 2513-bp Δply::cat fragment was transformed into UB2570 with selection for CmR and SpR. Strain UB2707 ΔsecA2Δply was confirmed by PCR and DNA sequencing as before.
Complementation of ply in strains UB2706 and UB2707 was achieved as follows. The ply gene (1416 bp) was PCR-amplified using primer pair Nco1PlyF/Xho1PlyR (Table S2). Expression plasmid pMSP7517 [36], with nisin-inducible promoter, together with ply fragment, were digested with NcoI and XhoI restriction enzymes, ligated and transformed into E. coli DH5α selecting EmR (300 μg ml−1). The resulting pMSPply (denoted pply) was then transformed into UB2706 or UB2707 selecting EmR, generating UB2717 Δply/pply and UB2719 ΔsecA2Δply/pply. Expression of ply was induced with 10 ng nisin ml−1.
For mutagenesis of Thr63 to Ala, pply (10.116 kb) was PCR-amplified (Phusion®, New England Biolabs, Ipswich, MA) with primer pair PlyMutA.F/PlyMutA.R (Table S2) such that GCC (Ala codon) on primer PlyMutA.F replaced ACA (Thr) present in ply. The re-ligated plasmid was transformed into E. coli DH5α, purified, then transformed into pneumococcal strains UB2706 and UB2707 to generate UB2768 Δply/pply63 and UB2769 ΔsecA2Δply/pply63. All strains were confirmed by appropriate PCR amplifications and DNA sequencing of PCR fragments.
2.3. Phenotypic assays
2.3.1. Growth curve
S. pneumoniae strains were grown for 9 h in THY medium at 37 °C. Overnight cultures (250 μl) were sub-cultured into pre-warmed fresh broth (10 ml). A600 of the bacterial suspensions were measured at 1 h intervals until cultures reached stationary phase (≤9 h).
2.3.2. Biofilms
Biofilm assays were performed as previously described [26]. Sterile coverslips (19 mm diameter) were placed in each well of a 12-well polystyrene tissue culture plate (Greiner Bio-One), and 0.5 ml of 10% saliva [37] was added. Plates were incubated at 4 °C overnight. Overnight cultures were equilibrated to A600 = 0.1 with fresh broth. Saliva was removed from coverslips, and portions (0.5 ml) of culture was added to wells containing saliva-coated coverslips in quadruplicate for each strain. Biofilms were grown for 6 h anaerobically at 37 °C. Media were removed, and coverslips were rinsed in PBS. Biofilms were stained with 0.5% crystal violet (1 ml) for 15 min, washed with distilled H2O until excess stain was removed, and air-dried. For visualization of biofilms, coverslips were inverted and mounted onto microscope slides and viewed on a light microscope (Leica, Milton Keynes, UK) with attached colour view camera and images captured using CellD imaging software (Olympus, Southend-on-Sea, UK). For biomass quantification of biofilms, crystal violet was dissolved in 10% (v/v) acetic acid for 15 min, and 100 μl portions transferred to a microtiter plate (MTP). Absorbance at 595 nm (A595) was then measured on an iMark™ MTP reader (Bio-Rad). All studies were performed in triplicate, and mean biomass calculated from three independent experiments.
2.3.3. A549 association assay
A549 cells (lung pneumocytes) were maintained in DMEM (Dulbecco's Modified Eagle's Medium-F-12) supplemented with 10% foetal calf serum, 2 mM l-glutamine, 100 I.U. penicillin ml−1 and 100 μg streptomycin ml−1. Cells were grown to ≥80% confluence in 75 cm2 flasks and collected twice-weekly by trypsin–EDTA treatment. Detached cells were seeded into fresh flasks (106 cells per flask) and incubated for ∼3 d at 37 °C in humidified air-5% CO2. Association of S. pneumoniae, which encompasses both adhesion and invasion, to epithelial cells was determined by viable count assay [38]. Briefly, confluent monolayers of A549 cells in 24-well tissue culture plates were incubated with 1 × 106 bacterial cells, in triplicate, in serum-free DMEM for 2 h at 37 °C in humidified air. Monolayers were washed three times to remove non-adhering bacteria, and epithelial cells were detached by adding ice-cold sterile dH2O (20 min). Numbers of bacteria associated with A549 cells were determined as colony forming units (CFU) from agar plate dilution counts. Monolayers were visualised on an inverted microscope (Olympus, Southend-on-Sea, UK). Light micrographs were taken at each stage of the assay to monitor the state of the epithelial monolayer to ensure that 2 h of bacterial incubation and washing procedures did not affect pneumocyte cell layer integrity (Fig. S1).
2.3.4. Haemolytic assays
Haemolytic activity was measured as described elsewhere [17] with some modifications. Pneumococci in mid-exponential growth phase were harvested by centrifugation (10,000 × g, 10 min) washed with phosphate-buffered saline (PBS; pH 7.2), and suspended (∼2 × 1010 cells ml−1) in 350 μl cell wall digestion buffer (30% sucrose in 10 mM Tris–HCl pH 7.0, containing Sigma protease inhibitor cocktail, 1 mg lysozyme ml−1 and 1333 U mutanolysin ml−1). Suspensions were shaken gently for 3 h at 37 °C. Spheroplasts were sedimented by centrifugation (17,000 × g, 10 min) and supernatants containing cell wall proteins (CWPs) were retained. Spheroplasts were then lysed in 50 mM Tris–HCl pH 7.5, centrifuged as before, and the supernatant designated cytoplasmic protein (CP) fraction. Protein fractions were serially diluted two-fold in PBS containing 0.1% BSA and 10 mM dithiothreitol in 96-well U bottom plates. Suspensions of washed 2% sheep red blood cells were added (50 μl per well) and after 1 h at 37 °C the plates were visually assessed for haemolysis, then centrifuged (233 × g for 10 min), and absorbance of supernatants at 490 nm (A490) were measured.
To assay for contact-dependent haemolysis, S. pneumoniae cell suspensions in PBS (∼2 × 109 cells ml−1) were mixed with equal volumes of 25% sheep red blood cells, centrifuged at 9600 × g for 5 min at 4 °C to form close contacts, and incubated for 3 h at 37 °C. Non-contact controls (no centrifugation) were included. To control for centrifugation-induced lysis of S. pneumoniae, bacterial suspensions alone were centrifuged as above, then mixed with erythrocytes and incubated for 3 h at 37 °C. PBS controls were also included, and all assays were run in triplicate. To determine haemolytic activities, suspensions were centrifuged (9600 × g, 1 min, 4 °C) and A490 of supernatants measured.
2.4. Protein analysis
2.4.1. Cell fractionation
Bacteria were collected from 16 h cultures by centrifugation (5000 × g, 7 min) and identical cell concentrations were used in all extractions when comparing wild type and mutants. Pellets were suspended in TE buffer and disrupted with glass beads in a Precellys homogenizer (2 cycles of 5000 × g for 30 s, repeated twice with cooling on ice). Cryovials containing homogenates were pulse-centrifuged to sediment beads and debris, and the cloudy supernatants were retained at −20 °C as whole cell lysates. For fractionation of CWPs and cytoplasmic proteins (CPs) for SDS-PAGE, bacterial cell pellets were suspended in 20 mM Tris–HCl pH 6.8, containing 26% raffinose, 10 mM MgCl2, 0.4 mM phenylmethylsulfonyl fluoride and mutanolysin (500 U ml−1) and incubated for 15 min at 37 °C, centrifuged (17,000 × g, 15 min), and the supernatant (CWP fraction) was retained. The pellets were then vortex-mixed with 0.1 mm diameter glass beads to disrupt the cells, centrifuged (17,000 × g, 5 min) and the supernatant (CP fraction) was collected.
To obtain culture fluid (CF) proteins, overnight cultures of S. pneumoniae were sub-cultured in minimal medium TYG (0.5% tryptone, 0.5% yeast extract, 0.5% glucose, 4 g l−1 K2HPO4, pH.7.5) to mid-exponential phase to the same A600, pelleted by centrifugation, and the supernatant was filtered (0.22 μm pore), mixed with trichloroacetic acid (10% final concentration) and incubated on ice for 16 h. Precipitated proteins were recovered by centrifugation (20,000 × g, 30 min, 4 °C), washed three times with acetone, and stored at −20 °C.
2.4.2. Electrophoresis and immunoblot analyses
Protein samples were heated at 80 °C for 10 min in 20 mM Tris–HCl pH 6.8 containing 1% sodium dodecyl sulphate (SDS) and 10 mM dithiothreitol, mixed with loading dye and separated by SDS-PAGE at 120 V for 1 h. Mr markers were PageRuler Plus Prestained protein ladder (New England Biolabs). Proteins were transferred onto nitrocellulose membrane (113 V, 1 h) and membranes were blocked with 5% milk in TBS (1 mM Tris–HCl pH 8.0, 0.15 M NaCl) containing 0.05% Tween 20 (TBS-T) overnight at 4 °C. Membranes were washed with TBS-T, incubated with 1:1000-diluted anti-Ply monoclonal antibody (Pierce Antibodies, Thermo Fisher Scientific, Waltham, MA), and bound antibody was detected with 1:2000-diluted anti-mouse IgG HRP conjugated secondary antibody (Dako, Agilent Technologies, Santa Clara, CA) followed by enhanced chemiluminescence (Biological Industries, Beit Haemek, Israel).
2.4.3. Immunoprecipitation
CWP fraction (600 μg protein) was mixed with 15 μg anti-Ply antibody and incubated for 2 h with mixing. Negative controls contained no antibody. Magnetic beads (Pierce Protein A/G, 400 μg) were added together with TBS-T buffer, mixed gently, and the magnetic beads were collected to the tube sides. The suspension was removed, beads washed with TBS-T and magnetically-collected as before, and repeated twice. The antigen–antibody mixture was added to the pre-washed magnetic beads and incubated for 1 h with mixing. Ply was eluted by mixing the beads with 0.1 M glycine pH 2.0 for 10 min. The beads were magnetically separated and the supernatant was neutralized using 1 M Tris. Protein was subjected to SDS-PAGE and immunoblot analysis with anti-Ply antibody (see above).
2.4.4. Mass spectrometry
Briefly, for identifying total proteins, samples were subjected to SDS-PAGE for 10 min, and the top 2 cm of the gel was excised and fixed in 7% acetic acid-50% methanol for 30 min. Gel slices as above, or of Coomassie blue-stained single bands, underwent automated in-gel tryptic digestion (ProGest, Genomic Solutions, Ann Arbor, MI). For total proteins, peptides were analysed by reverse phase nano-liquid chromatography MS/MS using a LTQ-Orbitrap Velos (Thermo Fisher) mass spectrometer. The mass spectral data for each experiment were combined prior to database searching using an in-house Mascot server to identify proteins present. Data from the Orbitrap were received in Excel and further analysis was performed locally. The identified peptide data included accession number, sequence length of the protein, molecular mass, calculated pI, a description, coverage (percentage of the protein sequence covered by identified peptides), score (total score of the protein being the sum of the scores of the individual peptides), peptide spectrum matches (total number of identified peptide sequences for the protein, including those redundantly identified), and peptides (number of unique peptide sequences). When comparing between each sample, proteins detected in the sample with a spectral count of at least three was considered a true hit.
2.5. Statistics
All data are reported as mean ± standard deviation (SD) of at least two independent experiments. Significance between samples was determined using the paired two-tailed Student's t-test, and P values of <0.1 and <0.05 were considered statistically significant.
3. Results
3.1. Phenotype of secA2 mutant
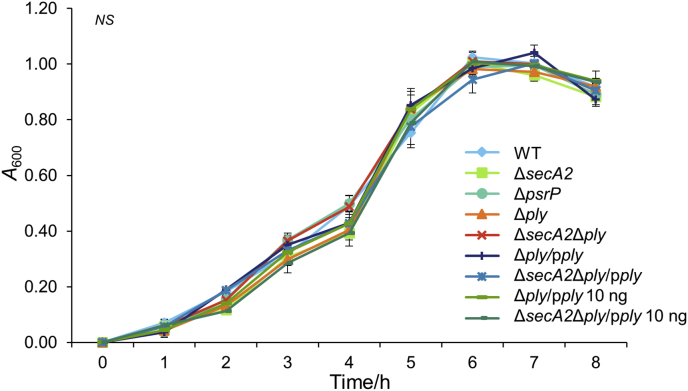
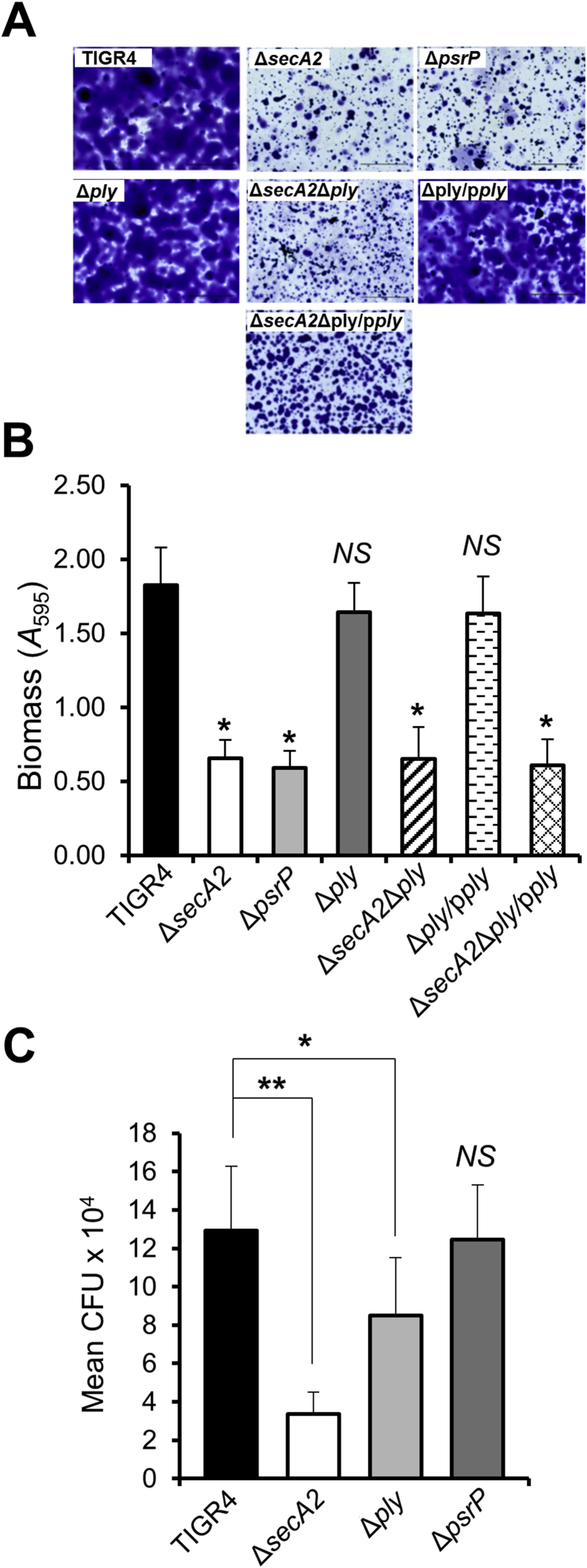
S. pneumoniae TIGR4 ΔsecA2 mutant was generated by allelic exchange of secA2 with aad9. Strain UB2570 ΔsecA2 cells showed no detectable differences in chain length, auto-aggregation or growth rate compared to wild type TIGR4 (Fig. 2). Biofilms formed by strains TIGR4, UB2570 ΔsecA2 and UB2312 ΔpsrP, the latter abrogated in SRR glycoprotein PsrP, were compared morphologically and quantitatively. There was approximately 60% reduced biomass for the ΔsecA2 and ΔpsrP mutant strains compared to WT (Fig. 3A and B). We also generated a Δply mutant for haemolytic assay controls (see below) and, by contrast, the Δply mutant and complemented mutant strain produced biofilms of similar morphology and biomass to WT (Fig. 3A and B). Biofilm formation was reduced by 60% in the ΔsecA2Δply mutant and ΔsecA2Δply/pply complemented strains (Fig. 3A and B). Therefore, under the conditions tested, expression of secA2 (or psrP), but not of ply, was necessary for normal biofilm formation.
Fig. 2.
Growth curves for S. pneumoniae WT, mutants and complemented strains. Growth rate analyses of S. pneumoniae wild type, ΔsecA2, ΔpsrP and Δply mutant strains were carried out to assess if inactivation of these genes affected bacterial growth that might influence subsequent assays. Complemented ply strains were induced with 0 and 10 ng ml−1 of nisin as mentioned in graph legend to show that addition of nisin had no effect on bacterial growth. Strains were grown in THY medium until cultures reached stationary phase. There are no significant differences (NS) between the WT, mutants, and nisin induced complemented strains in growth rates or growth yield. (n = 2).
Fig. 3.
Biofilm formation and association to lung cells by S. pneumoniae TIGR4, mutants and ply complemented strains. (A) Representative light micrographs of corresponding S. pneumoniae biofilms grown at 6 h and stained with crystal violet. Scale bar = 50 μm. (B) S. pneumoniae monospecies biofilms were grown on saliva coated cover slips for 6 h at 37 °C. Total biomass was quantified by crystal violet staining as described in Materials and Methods. Statistical significance compared to the wild type TIGR4 is indicated by an asterisk (*P < 0.05). No statistical significant difference from the wild type (NS) is also indicated. (n = 3). (C) S. pneumoniae association with A549 epithelial cells. A549 cells were grown to a confluence of 2 × 105 cells and infected with S. pneumoniae TIGR4 and mutant strains (1 × 106 cells ml−1 input) for 2 h at 37 °C. After removal of non-associated bacteria and washing of the epithelial cells, the numbers of bacterial cells associated with A549 cells were determined (CFU) by agar plate counts. P values: * <0.1, ** <0.05. No statistical significance to the wild type (NS) is also indicated. (n = 2).
Since PsrP binds to keratin 10 present on lung cells [29], [39] it was hypothesized that S. pneumoniae ΔsecA2 and ΔpsrP mutants would show decreased association to A549 type II pneumocyte monolayers. There was a statistically significant 70% decrease in association for ΔsecA2 mutant cells. However, the ΔpsrP mutant exhibited similar interaction levels to WT (Fig. 3C). Thus, PsrP is not required for association to pneumocytes. Deletion of ply resulted in a small reduction (35%) in association (Fig. 3C) and therefore ply may be required for optimal association of S. pneumoniae to pneumocytes.
3.2. secA2 expression and Ply export
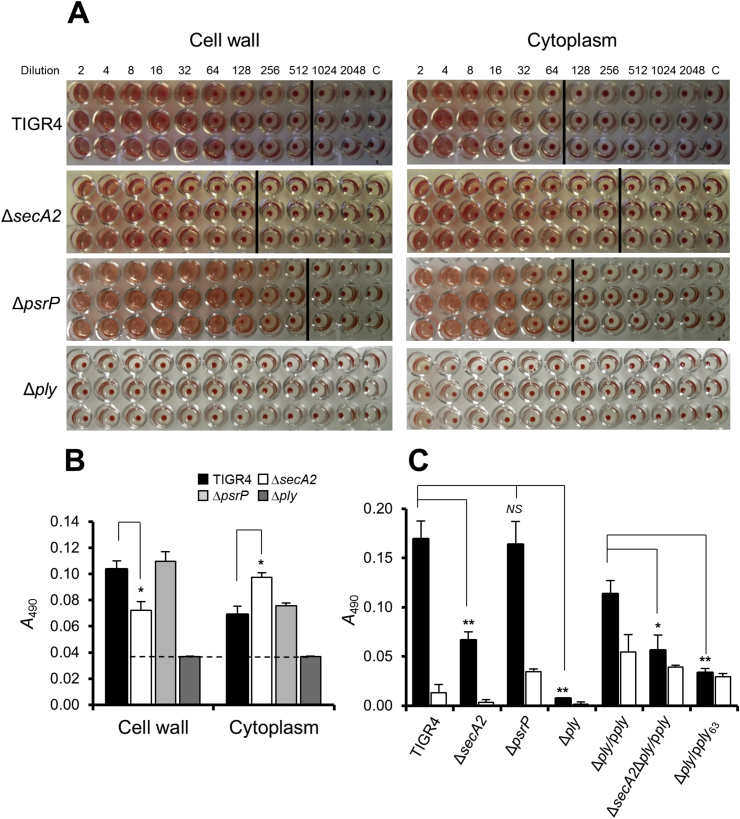
To determine if SecY2A2 was involved in Ply export we measured haemolytic activities present in CWP or CP fractions prepared from WT or ΔsecA2 mutant strains. Haemolytic activities were present in both fractions (Fig. 4A) but the haemolysis titre for the ΔsecA2 mutant CWP fraction was reproducibly two- to four-fold lower (Fig. 4A). Correspondingly, haemolytic activity within the CP fraction of the ΔsecA2 mutant was greater than WT (Fig. 4A), suggesting a decrease in export. The Δply mutant showed no haemolysis while the ΔpsrP mutant was similar to WT. Quantitative haemolytic assays demonstrated that Ply activity in the ΔsecA2 mutant CWP fraction was 50% lower than WT, taking into account the Δply negative control background and Ply activity in the ΔsecA2 mutant was 50% more in the CP fraction than the WT (Fig. 4B). Haemolytic activities for the ΔpsrP mutant fractions were similar to wild type (Fig. 4B). Therefore, cell wall-associated Ply (Plyw) activity appears to be associated with SecY2A2 function, but is not dependent upon secretion of PsrP.
Fig. 4.
Haemolytic activities of S. pneumoniae wild type and mutants strains. (A) Bacterial cells were fractionated into cell wall or cytoplasm fractions (as indicated) and two-fold serial dilutions were incubated with 2% sheep RBCs in replicates of three. The control was assay buffer only. No pellet was considered to be 100% lysis. Vertical lines show the detection endpoints of 50% lysis. (B) Quantitative haemolytic data of the absorbance (A490) values of the first 50% lysis endpoint and the corresponding WT or mutant absorbance value at the same dilution. Therefore, absorbance values for cell wall and cytoplasmic fractions were taken at dilution factors 128 and 64, respectively. The dotted line represents the limit of detection. *P < 0.05. (n = 3). (C) Haemolytic activities of S. pneumoniae TIGR4, mutants and complemented strains after contact (sedimentation with sheep RBCs; black bars) or non-contact (incubation with sheep RBCs without sedimentation; white bars). P values: * <0.1, ** <0.05. No statistical significance to the wild type (NS) is also indicated. (n = 3).
Plyw activity was also determined by contact-dependent haemolysis (CDH) assay. Brief centrifugation was employed to achieve close contact between erythrocytes (sheep RBCs) and bacterial cells, compared with simply gentle mixing (designated non-contact). Appropriate controls to correct for lysis of RBCs or bacteria were included (see Materials and Methods). CDH (Plyw) was ten-fold higher than the non-contact activity in TIGR4 wild type (Fig. 4C). There was 60% decrease in CDH by ΔsecA2 mutant Plyw compared to wild type (Fig. 4C), while the ΔpsrP mutant was unaffected, and the Δply mutant was negative. Complemented ply mutant strains Δply/pply and ΔsecA2Δply/pply were also included to validate the involvement of secA2 in Plyw. There was >90% reduction in Plyw activity in the ΔsecA2Δply/pply strain compared to the Δply/pply strain, following subtraction of the activity attributable to non-contact haemolysis (Fig. 4C). The complemented strain Δply/pply had lower Plyw activity than wild type, and higher non-contact activity, because the cells are weakened by nisin added to induce ply expression.
3.3. Post-translational modification of Ply
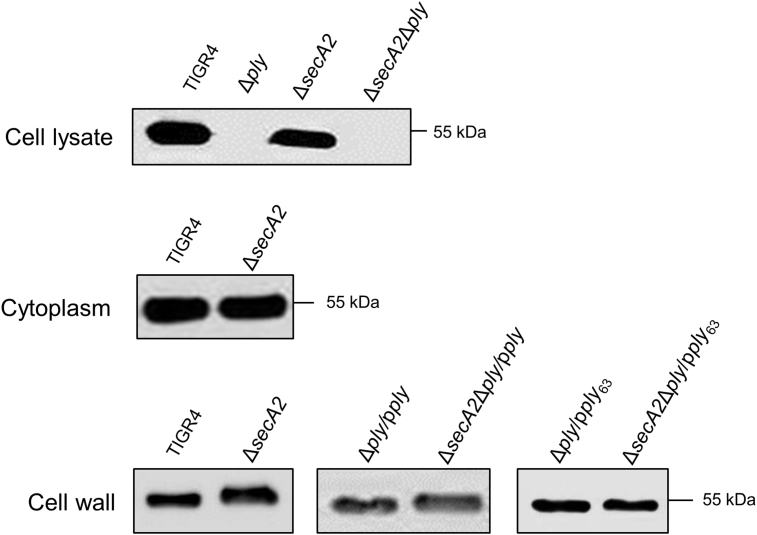
Since SecY2A2 is reported to secrete partially-glycosylated SRR proteins [24], [27], [28] we hypothesized that Ply might be post-translationally modified for export, thus explaining reduced Plyw activity in the ΔsecA2 mutant. Accordingly, whole cell lysates, CWP and CP fractions from WT and mutants were subjected to SDS-PAGE and immunoblot analysis with anti-Ply antibody. In Δply mutant whole cell lysates, 53-kDa Ply expression is ablated, thus validating the Ply-specific antibody (Fig. 5). Antibody-reactive bands were present in CP and in CWP fractions from WT and ΔsecA2 mutant (Fig. 5). No significant differences in relative quantities of Ply could be seen in these fractions between WT and ΔsecA2 mutant. However, Plyw in the ΔsecA2 mutant CWP fraction migrated slower by ∼2 kDa compared to WT (Fig. 5). Plyw also migrated more slowly in the CWP fraction of ΔsecA2Δply/pply complemented strain compared to the Δply/pply strain.
Fig. 5.
Western blot analysis of whole cell lysates and fractionated S. pneumoniae TIGR4, mutants and complement strains. Proteins were blotted onto nitrocellulose, reacted with anti-Ply antibody (1:1000 dilution) followed by anti-mouse HRP secondary antibody (1:2000). TIGR4 Ply is 53 kDa.
It was hypothesized that this difference might be associated with glycosylation of Ply. To investigate this, Ply was immunoprecipitated and purified from CWP fractions of TIGR4 and ΔsecA2 strains (see Materials and Methods) and subjected to SDS-PAGE and lectin-blot analysis. However, we could detect no reactivity with lectins PNA, RCA, UEA1, DBA, WGA, or SBA, variously specific to galactose, N-acetylgalactosamine (GalNAc), fucose, N-acetylglucosamine (GlcNAc) or mannose (data not shown). We then searched for potential O-, N- or C-linked glycosylation sites in Ply [40] and the most probable predicted O-linked glycosylation site was threonine residue 63 (Thr63) on the protein surface (Fig. S2). To determine if Thr63 was important for Plyw localization or activity, Thr was changed to Ala by site-directed mutagenesis of plasmid pMSPply. In CDH assays, there was 70% reduction in Plyw activity of S. pneumoniae Δply/pply63 compared with Δply/pply strain suggesting that Thr63 was important for Ply activity (Fig. 4C). However, the apparent Mr of Ply63 in CWP fractions from UB2769 ΔsecA2Δply/pply63 was similar to that in the ΔsecA2 mutant (Fig. 5). Plyw protein bands from WT and ΔsecA2 mutant were then subjected to in-gel digestion and LC-MS/MS as described previously [26]. There were no differences in distribution of peptides or sequences obtained from the protein samples (data not shown). Therefore, the difference in SDS-PAGE migration of Plyw in ΔsecA2 mutants remains unexplained.
3.4. Specificity of SecY2A2
In light of evidence that multiple secreted proteins are SecA2 pathway-dependent in L. monocytogenes [21], we undertook proteomic analysis of CWP fractions (see Fig. S3) and extracellular proteins from WT and ΔsecA2 mutant (see Tables S3 and S4). It was clear from these comparisons that the secA2 deletion had pleiotropic effects. While most proteins were present in similar quantities (numbers of unique peptide sequences) in WT and ΔsecA2 CWP fractions, there were several major differences. Two cell wall-anchored glycosidases (Q2MGH6 and Q97S90) were >90% reduced in the ΔsecA2 mutant, as well as a range of other proteins involved in sugar or cell wall metabolism e.g. galactokinase, M20 peptidase, glucuronyl hydrolase, as listed in Table 1. None of these were encoded by genes within the secY2A2 locus (Fig. 1). Other proteins of interest that could not be detected in the ΔsecA2 CWP extract were RrgC (ancillary pilus subunit) and Q97NT9, which has been implicated in transport of Ply from cytoplasm to cell wall compartment [41]. In the culture fluid (CF), a number of the proteins identified in both WT and mutant e.g. EF-Tu, GAPDH, PK are also found in extracellular vesicles [42]. However, the levels of ZmpC, PcpA, Eng and AgaS were >50% reduced in the ΔsecA2 mutant CF (Table S4).
Table 1.
Top 20 protein hits identified in S. pneumoniae TIGR4 CWP extracts that were <90% represented in corresponding ΔsecA2 mutant CWP extracts, compiled from data presented in Table S3. TIGR4 peptides are sorted highest to lowest number of unique peptide sequences.
| Acc.a (Gene name) |
pIb | Description | % Coveragec |
PSMd |
Peptidese |
|||
|---|---|---|---|---|---|---|---|---|
| WT | ΔsecA2 | WT | ΔsecA2 | WT | ΔsecA2 | |||
|
Q2MGH6 (SP_0368) |
6.07 | Endo-α-N-acetylgalactosaminidase (Eng) | 63.95 | 2.21 | 273 | 2 | 90 | 2 |
|
Q97S90 (SP_0498) |
5.31 | Putative endo-β-N-acetylglucosaminidase | 33.33 | 3.92 | 76 | 3 | 39 | 3 |
|
Q97NZ6 (SP_1853) |
5.08 | Galactokinase | 53.57 | 5.61 | 32 | 1 | 14 | 1 |
|
Q97NA0 (SP_2153) |
4.75 | Dipeptidase (M20/25/40 family) | 47.63 | 3.84 | 25 | 1 | 14 | 1 |
|
Q97NA2 (SP_2151) |
5.03 | Carbamate kinase (ArcC) | 61.90 | 4.76 | 41 | 1 | 13 | 1 |
|
Q97SL0 (SP_0322) |
5.43 | Glucuronyl hydrolase | 37.63 | 0 | 18 | 0 | 11 | 0 |
|
Q97SL2 (SP_0320) |
5.47 | Oxidoreductase, short chain dehydrogenase/reductase family | 54.61 | 0 | 25 | 0 | 10 | 0 |
|
P0CB58 (SP_0334) |
6.76 | Ribosomal RNA small subunit methyltransferase H | 25.00 | 0 | 6 | 0 | 5 | 0 |
|
Q97P42 (SP_1802) |
9.69 | Uncharacterized membrane protein (sugar-binding transcriptional regulator) | 23.68 | 0 | 6 | 0 | 5 | 0 |
|
Q97SC1 (SP_0464) |
6.05 | Cell surface anchor family protein RrgC (ancillary pilus subunit) | 13.23 | 0 | 9 | 0 | 5 | 0 |
|
P0A4M7 (SP_1890) |
9.10 | Oligopeptide transport system permease protein AmiC | 8.84 | 0 | 6 | 0 | 4 | 0 |
|
Q97NT9 (SP_1924) |
8.98 | Putative uncharacterized protein (ply locus) | 33.09 | 0 | 5 | 0 | 3 | 0 |
|
Q97QK2 (SP_1203) |
5.07 | Arginine repressor (ArgR2) | 29.37 | 0 | 3 | 0 | 3 | 0 |
|
Q97PP3 (SP_1560) |
8.02 | Putative uncharacterized lipoprotein (YbbR-like) | 23.17 | 0 | 3 | 0 | 3 | 0 |
|
Q97NS3 (SP_0775) |
4.82 | Ribosomal protein S16 | 19.73 | 0 | 5 | 0 | 3 | 0 |
|
Q97S82 (SP_0506) |
9.63 | Integrase/recombinase, phage integrase family | 13.21 | 0 | 3 | 0 | 3 | 0 |
|
Q97Q64 (SP_1366) |
6.11 | Glycosyl transferase, group 1 | 12.53 | 0 | 5 | 0 | 3 | 0 |
|
Q97T27 (SP_0129) |
4.93 | Probable tRNA threonylcarbamoyladenosine biosynthesis protein (Gcp) | 11.61 | 0 | 3 | 0 | 3 | 0 |
|
Q97NZ7 (SP_1852) |
5.36 | Galactose-1-phosphate uridylyltransferase 2 | 9.74 | 0 | 10 | 0 | 3 | 0 |
|
Q97SL4 (SP_0318) |
5.81 | Carbohydrate kinase (PfkB family) | 8.71 | 0 | 3 | 0 | 3 | 0 |
| Q97P71f (SP_1772) | 4.06 | Cell wall surface anchor family protein PsrP | 3.10 | 0 | 32 | 0 | 14 | 0 |
Protein accession number (gene locus).
Calculated isoelectric point.
Percent of the protein sequence covered by identified peptides.
Total number of identified peptide sequences (peptide spectrum matches) for the protein.
Number of unique peptide sequences.
Found in unfiltered data.
4. Discussion
The streptococcal accessory Sec systems function to secrete SRR glycoproteins. However, the mechanism by which SRR proteins are directed to the accessory Sec system is not fully understood. The SRR protein signal (leader) peptides do not contain a YSIRK-G/S motif, which is characteristic of cell wall-anchored proteins translocated via canonical Sec [43], and the leader peptides are unusually large e.g. 88–90 aa residues. These features may reduce the affinity of SRR proteins for canonical Sec. In the streptococcal accessory sec loci, gtfA and gtfB genes are located immediately downstream of the gene encoding the SecA2 protein (Fig. 1). The genes encode a GtfAB (glycosyltransferase) complex, which engages with and glycosylates the pre-protein [28]. In S. gordonii, GspB is glycosylated first with GlcNAc, then with other sugar residues, prior to or concomitant with SecY2A2-mediated transport across the cytoplasmic membrane [44]. Potentially therefore, proteins that are translocated via the accessory Sec need to be O-glycosylated. In the ΔsecA2 mutant generated here, aad9 insertion may have affected expression of downstream gtfA, gtfB, asp4 and asp5 genes since aad9 carried a potential transcriptional terminator stem loop.
The accessory Sec system appears also to be involved in the export and/or activation of Ply. The ΔsecA2 mutant cell surface is depleted of haemolytic activity (CDH) while the CWP fraction, obtained by enzymatic hydrolysis of the cell wall, is ∼50% reduced in haemolytic activity. The ΔsecA2 mutant CWP fraction contains Plyw protein but this displayed marginally slower electrophoretic mobility than WT Plyw. This suggests that in the ΔsecA2 mutant cell wall compartment, Plyw is present in a modified and less active form. Our data suggest the accessory Sec system is indirectly involved in Ply export because if it were directly involved we'd expect to see a 100% decrease in Ply export to the cell wall of the ΔsecA2 mutant.
Although we have been unable at this stage to demonstrate that Ply is glycosylated, the absence of two glycosidases in the ΔsecA2 mutant cell wall would be expected to impact on glycoprotein processing and on cell wall metabolism. Clearly Plyw is intimately associated with cell wall structure, since enzymatic digestion of peptidoglycan is necessary to liberate Ply from the cell wall [18].
The ΔsecA2 mutant cell walls were >90% deficient in two glycosidases, endo-α-N-acetylgalactosaminidase and endo-β-N-acetylglucosaminidase. The former, designated Eng [45], catalyses the release of Galβ1-3GalNAcα1 linked to serine or threonine residues found on O-linked glycoproteins. These enzymes are involved in hydrolysing oligo- or polysaccharides present in the environment, releasing sugars for bacterial cell growth, or exposing new oligosaccharide receptors for adhesion to host cells [45]. Eng has been shown to be necessary for pneumococcal binding to A549 cells, which may be a reason for why there was lower A549 association levels for the ΔsecA2 mutant. However, contrary to other work [39] we found that the ΔpsrP mutant was unaffected in adherence to A549 cells. The glycosidases both carry predicted leader peptides (37–38 aa residues) with YSIRK-G/S motifs, and cell wall-anchorage LPXTG motifs, suggesting export via canonical Sec [43]. Although there is evidence for functional cross-talk between canonical Sec and SecA2 [26], [46], it was nonetheless surprising to find amounts of these proteins >90% reduced in ΔsecA2 mutant cell walls. The mechanism for this is currently under investigation.
Other main components present in the WT CWP fraction and >90% reduced in the ΔsecA2 mutant were metabolic enzymes without predicted leader peptides. These were: galactokinase, converting galactose to galactose 1-phosphate in the Leloir pathway; dipeptidase M20/25/40, a potential virulence factor [47]; carbamate kinase involved in anaerobic arginine metabolism; glucuronyl hydrolase, a glycosidase acting on glycosaminoglycans; and oxidoreductase/short chain dehydrogenase, potentially membrane bound and involved in d-gluconate metabolism [48]. Additionally, a putative uncharacterized protein (Accession: Q97NT9), present in the WT only is a protein encoded by a gene transcriptionally linked to ply and has been suggested to assist Ply transport from cytoplasm to cell wall [41]. As mentioned previously, FbpA is a SecA2-dependent protein in L. monocytogenes that acts as a chaperone for listeriolysin O [20]. It is possible one or more of the genes in the ply locus (e.g. SP_1924 which encodes Q97NT9) encodes proteins that act as a chaperone for Ply export.
Proteins that were present in both WT and ΔsecA2 mutant included typical cell surface proteins such as PspA (Q97T39) and PcpA (Q97NB5) (Tables S3 and S4). We were not able to detect tryptic peptides of PsrP in the CWP fraction of wild type strain TIGR4 (Table S3), suggesting that PsrP is tightly bound within the cell wall. However, we found 14 unique PsrP peptides in the CWP fraction unfiltered data, i.e. identified at >5% false discovery rate, that were present in WT but not in the ΔsecA2 mutant (Table 1). Recently it has been shown that pyruvate oxidase (and H2O2 production) contributes to Ply release in a lysis-independent manner [49]. However, pyruvate oxidase was present in similar amounts in WT and ΔsecA2 mutant cell walls or culture fluid (data not shown).
In summary, we present data suggesting that the accessory Sec system is involved in the activation of Ply in the cell wall compartment of S. pneumoniae. One possibility is that Ply becomes transiently modified in transition from cytoplasmic to cell wall compartments. Alternatively, Ply becomes tightly associated with a peptidoglycan precursor [18] that requires SecY2A2 function for activation of Ply. The mechanisms of transcriptional regulation of the ply locus and Ply transport across the cytoplasmic membrane are thus still not understood. However, evidence that the accessory Sec system is involved in export of several potential virulence factors, including Ply, provides further motivation for identifying molecules that might selectively block the accessory Sec translocase.
Conflict of interest
The authors declare no conflicts of interest.
Acknowledgements
We would like to thank Lindsay Dutton and Kate Heesom (University of Bristol Proteomics Facility) for excellent technical assistance, and Gary Dunny, Jean-Pierre Claverys, Andreas Podbielski, and Tim Mitchell for kindly providing strains and plasmids. This work was supported by a Medical Research Council Doctoral Training Grant 2011-G1001606 to M.B.
Footnotes
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.micinf.2017.04.003.
Contributor Information
Ariel J. Blocker, Email: ariel.blocker@bristol.ac.uk.
Howard F. Jenkinson, Email: howard.jenkinson@bristol.ac.uk.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Thamadilok S., Roche-Håkansson H., Håkansson A.P., Ruhl S. Absence of capsule reveals glycan-mediated binding and recognition of salivary mucin MUC7 by Streptococcus pneumoniae. Mol Oral Microbiol. 2016;31:175–188. doi: 10.1111/omi.12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blanchette K.A., Shenoy A.T., Milner J., 2nd, Gilley R.P., McClure E., Hinojosa C.A. Neuraminidase A-exposed galactose promotes Streptococcus pneumoniae biofilm formation during colonization. Infect Immun. 2016;84:2922–2932. doi: 10.1128/IAI.00277-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Straume D., Stamsås G.A., Håvarstein L.S. Natural transformation and genome evolution in Streptococcus pneumoniae. Infect Genet Evol. 2015;33:371–380. doi: 10.1016/j.meegid.2014.10.020. [DOI] [PubMed] [Google Scholar]
- 4.Chaguza C., Andam C.P., Harris S.R., Cornick J.E., Yang M., Bricio-Moreno L. Recombination in Streptococcus pneumoniae lineages increase with carriage duration and size of polysaccharide capsule. mBio. 2016;7 doi: 10.1128/mBio.01053-16. e01053–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maestro B., Sanz J.M. Choline binding proteins from Streptococcus pneumoniae: a dual role as enzybiotics and targets for the design of new antimicrobials. Antibiotics. 2016;5:21. doi: 10.3390/antibiotics5020021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kadioglu A., Weiser J.N., Paton J.C., Andrew P.W. The role of Streptococcus pneumoniae virulence factors in host respiratory colonization and disease. Nat Rev Microbiol. 2008;6:288–301. doi: 10.1038/nrmicro1871. [DOI] [PubMed] [Google Scholar]
- 7.Feldman C., Anderson R. Epidemiology, virulence factors and management of the pneumococcus. F1000Res. 2016;5:2320. doi: 10.12688/f1000research.9283.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marshall J.E., Faraj B.H., Gingras A.R., Lonnen R., Sheikh M.A., El-Mezgueldi M. The crystal structure of pneumolysin at 2.0 Å resolution reveals the molecular packing of the pre-pore complex. Sci Rep. 2015;5:13293. doi: 10.1038/srep13293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Palmer M. The family of thiol-activated, cholesterol-binding cytolysins. Toxicon. 2001;39:1681–1689. doi: 10.1016/s0041-0101(01)00155-6. [DOI] [PubMed] [Google Scholar]
- 10.Mitchell T.J., Andrew P.W., Saunders F.K., Smith A.N., Boulnois G.J. Complement activation and antibody binding by pneumolysin via a region of the toxin homologous to a human acute-phase protein. Mol Microbiol. 1991;5:1883–1888. doi: 10.1111/j.1365-2958.1991.tb00812.x. [DOI] [PubMed] [Google Scholar]
- 11.Nel G., Jr., Theron A.J., Durandt C., Tintinger G.R., Pool R., Mitchell T.J. Pneumolysin activates neutrophil extracellular trap formation. Clin Exp Immunol. 2016;184:358–367. doi: 10.1111/cei.12766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Malet J.K., Cossart P., Ribet D. Alteration of epithelial cell lysosomal integrity induced by bacterial cholesterol-dependent cytolysins. Cell Microbiol. 2016;19:e12682. doi: 10.1111/cmi.12682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rai P., He F., Kwang J., Engelward B.P., Chow V.T. Pneumococcal pneumolysin induces DNA damage and cell cycle arrest. Sci Rep. 2016;6:22972. doi: 10.1038/srep22972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shak J.R., Ludewick H.P., Howery K.E., Sakai F., Yi H., Harvey R.M. Novel role for the Streptococcus pneumoniae toxin pneumolysin in the assembly of biofilms. mBio. 2013;4:e00655–13. doi: 10.1128/mBio.00655-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson M.K. Cellular location of pneumolysin. FEMS Microbiol Lett. 1997;2:243–245. [Google Scholar]
- 16.Balachandran P., Hollingshead S.K., James C., Briles D.E., Paton J.C. The autolytic enzyme LytA of Streptococcus pneumoniae is not responsible for releasing pneumolysin. J Bacteriol. 2001;183:3108–3116. doi: 10.1128/JB.183.10.3108-3116.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Price K.E., Camilli A. Pneumolysin localizes to the cell wall of Streptococcus pneumoniae. J Bacteriol. 2009;191:2163–2168. doi: 10.1128/JB.01489-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greene N.G., Narciso A.R., Filipe S.R., Camilli A. Peptidoglycan branch stem peptides contribute to Streptococcus pneumoniae virulence by inhibiting pneumolysin release. PLoS Pathog. 2015;11:e1004996. doi: 10.1371/journal.ppat.1004996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Price K.E., Greene N.G., Camilli A. Export requirements of pneumolysin in Streptococcus pneumoniae. J Bacteriol. 2012;194:3651–3660. doi: 10.1128/JB.00114-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dramsi S., Bourdichon F., Cabanes D., Lecuit M., Fsihi H., Cossart P. FbpA, a novel multifunctional Listeria monocytogenes virulence factor. Mol Microbiol. 2004;53:639–649. doi: 10.1111/j.1365-2958.2004.04138.x. [DOI] [PubMed] [Google Scholar]
- 21.Renier S., Chambon C., Viala D., Chagnot C., Hébraud M., Desvaux M. Exoproteomic analysis of the secA2-dependent secretion in Listeria monocytogenes EGD-e. J Proteomics. 2013;80:183–195. doi: 10.1016/j.jprot.2012.11.027. [DOI] [PubMed] [Google Scholar]
- 22.Tettelin H., Nelson K.E., Paulsen I.T., Eisen J.A., Read T.D., Peterson S. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science. 2001;293:498–506. doi: 10.1126/science.1061217. [DOI] [PubMed] [Google Scholar]
- 23.Obert C., Sublett J., Kaushal D., Hinojosa E., Barton T., Tuomanen E.I. Identification of a candidate Streptococcus pneumoniae core genome and regions of diversity correlated with invasive pneumococcal disease. Infect Immun. 2006;74:4766–4777. doi: 10.1128/IAI.00316-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yen Y.T., Cameron T.A., Bensing B.A., Seepersaud R., Zambryski P.C., Sullam P.M. Differential localization of the streptococcal accessory sec components and implications for substrate transport. J Bacteriol. 2013;195:682–695. doi: 10.1128/JB.01742-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takahashi Y., Konishi K., Cisar J.O., Yoshikawa M. Identification and characterization of hsa, the gene encoding the sialic acid-binding adhesin of Streptococcus gordonii DL1. Infect Immun. 2002;70:1209–1218. doi: 10.1128/IAI.70.3.1209-1218.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bandara M., Corey R.A., Martin R., Skehel J.M., Blocker A.J., Jenkinson H.F. Composition and activity of the non-canonical Gram-positive SecY2 complex. J Biol Chem. 2016;291:21474–21484. doi: 10.1074/jbc.M116.729806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou M., Zhu F., Dong S., Pritchard D.G., Wu H. A novel glucosyltransferase is required for glycosylation of a serine-rich adhesin and biofilm formation by Streptococcus parasanguinis. J Biol Chem. 2010;285:12140–12148. doi: 10.1074/jbc.M109.066928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shi W.W., Jiang Y.L., Zhu F., Yang Y.H., Shao Q.Y., Yang H.B. Structure of a novel O-linked N-acetyl-D-glucosamine (O-GlcNAc) transferase, GtfA, reveals insights into the glycosylation of pneumococcal serine-rich repeat adhesins. J Biol Chem. 2014;289:20898–20907. doi: 10.1074/jbc.M114.581934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shivshankar P., Sanchez C., Rose L.F., Orihuela C.J. The Streptococcus pneumoniae adhesin PsrP binds to Keratin 10 on lung cells. Mol Microbiol. 2009;73:663–679. doi: 10.1111/j.1365-2958.2009.06796.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sanchez C.J., Shivshankar P., Stol K., Trakhtenbroit S., Sullam P.M., Sauer K. The pneumococcal serine-rich repeat protein is an intra-species bacterial adhesin that promotes bacterial aggregation in vivo and in biofilms. PLoS Pathog. 2010;6:e1001044. doi: 10.1371/journal.ppat.1001044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blanchette-Cain K., Hinojosa C.A., Babu R.A.S., Lizcano A., Gonzalez-Juarbe N., Munoz-Almagro C. Streptococcus pneumoniae biofilm formation is strain dependent, multifactorial, and associated with reduced invasiveness and immunoreactivity during colonization. mBio. 2013;4 doi: 10.1128/mBio.00745-13. e00745–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schulte T., Mikaelsson C., Beaussart A., Kikhney A., Deshmukh M., Wolniak S. The BR domain of PsrP interacts with extracellular DNA to promote bacterial aggregation; structural insights into pneumococcal biofilm formation. Sci Rep. 2016;6:32371. doi: 10.1038/srep32371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Podbielski A., Spellerberg B., Woischnik M., Pohl B., Lütticken R. Novel series of plasmid vectors for gene inactivation and expression analysis in group A streptococci (GAS) Gene. 1996;177:137–147. doi: 10.1016/0378-1119(96)84178-3. [DOI] [PubMed] [Google Scholar]
- 34.Holmes A.R., McNab R., Millsap K.W., Rohde M., Hammerschmidt S., Mawdsley J.L. The pavA gene of Streptococcus pneumoniae encodes a fibronectin-binding protein that is essential for virulence. Mol Microbiol. 2001;41:1395–1408. doi: 10.1046/j.1365-2958.2001.02610.x. [DOI] [PubMed] [Google Scholar]
- 35.Claverys J.-P., Dintilhac A., Pestova E.V., Martin B., Morrison D.A. Construction and evaluation of new drug-resistance cassettes for gene disruption mutagenesis in Streptococcus pneumoniae, using an ami test platform. Gene. 1995;164:123–128. doi: 10.1016/0378-1119(95)00485-o. [DOI] [PubMed] [Google Scholar]
- 36.Hirt H., Erlandsen S.L., Dunny G.M. Heterologous inducible expression of Enterococcus faecalis pCF10 aggregation substance Asc10 in Lactococcus lactis and Streptococcus gordonii contributes to cell hydrophobicity and adhesion to fibrin. J Bacteriol. 2000;182:2299–2306. doi: 10.1128/jb.182.8.2299-2306.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jack A.A., Daniels D.E., Jepson M.A., Vickerman M.M., Lamont R.J., Jenkinson H.F. Streptococcus gordonii comCDE (competence) operon modulates biofilm formation with Candida albicans. Microbiology. 2015;161:411–421. doi: 10.1099/mic.0.000010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Molinari G., Talay S.R., Valentin-Weigand P., Rohde M., Chhatwal G.S. The fibronectin-binding protein of Streptococcus pyogenes, SfbI, is involved in the internalization of group A streptococci by epithelial cells. Infect Immun. 1997;65:1357–1363. doi: 10.1128/iai.65.4.1357-1363.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rose L., Shivshankar P., Hinojosa E., Rodriguez A., Sanchez C.J., Orihuela C.J. Antibodies against PsrP, a novel Streptococcus pneumoniae adhesin, block adhesion and protect mice against pneumococcal challenge. J Infect Dis. 2008;193:375–383. doi: 10.1086/589775. [DOI] [PubMed] [Google Scholar]
- 40.Caragea C., Sinapov J., Silvescu A., Dobbs D., Honavar V. Glycosylation site prediction using ensembles of support vector machine classifiers. BMC Bioinform. 2007;8:1–13. doi: 10.1186/1471-2105-8-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kimaro Mlacha S.Z., Romero-Steiner S., Hotopp J.C.D., Kumar N., Ishmael N., Riley D.R. Phenotypic, genomic, and transcriptional characterization of Streptococcus pneumoniae interacting with human pharyngeal cells. BMC Genomics. 2013;14:1–18. doi: 10.1186/1471-2164-14-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Olaya-Abril A., Prados-Rosales R., McConnell M.J., Martin-Peña R., Gonzáles-Reyes J.A., Jiménez-Munguia Characterization of protective extracellular membrane-derived vesicles produced by Streptococcus pneumoniae. J Proteomics. 2014;106:40–60. doi: 10.1016/j.jprot.2014.04.023. [DOI] [PubMed] [Google Scholar]
- 43.Bae T., Schneewind O. The YSIRK-G/S motif of staphylococcal protein A and its role in efficiency of signal peptide processing. J Bacteriol. 2003;185:2910–2919. doi: 10.1128/JB.185.9.2910-2919.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen Y., Seepersaud R., Bensing B.A., Sullam P.M., Rapoport T.A. Mechanism of a cytosolic O-glycosyltransferase essential for the synthesis of a bacterial adhesion protein. Proc Natl Acad Sci U S A. 2016;113:E1190–E1199. doi: 10.1073/pnas.1600494113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marion C., Limoli D.H., Bobulsky G.S., Abraham J.L., Burnaugh A.M., King S.J. Identification of pneumococcal glycoside that modifies O-linked glycans. Infect Immun. 2009;77:1389–1396. doi: 10.1128/IAI.01215-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou M., Zhang H., Zhu F., Wu H. Canonical SecA associates with an accessory secretion protein complex involved in biogenesis of a streptococcal serine-rich repeat glycoprotein. J Bacteriol. 2011;193:6560–6566. doi: 10.1128/JB.05668-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hava D.L., Camilli A. Large-scale identification of serotype 4 Streptococcus pneumoniae virulence factors. Mol Microbiol. 2002;45:1389–1406. [PMC free article] [PubMed] [Google Scholar]
- 48.Shi Z., Xuan C., Han H., Cheng X., Wang J., Feng Y. Gluconate-5-dehydrogenase (Ga5DH) participates in Streptococcus suis cell division. Protein Cell. 2014;5:761–769. doi: 10.1007/s13238-014-0074-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bryant J.C., Dabbs R.C., Oswalt K.L., Brown L.R., Rosch J.W., Seo K.S. Pyruvate oxidase of Streptococcus pneumoniae contributes to pneumolysin release. BMC Microbiol. 2016;16:271. doi: 10.1186/s12866-016-0881-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.