Summary
Mec1ATR mediates the DNA damage response (DDR), integrating chromosomal signals and mechanical stimuli. We show that the PP2A phosphatases, ceramide-activated enzymes, couple cell metabolism with the DDR. Using genomic screens, metabolic analysis, and genetic and pharmacological studies, we found that PP2A attenuates the DDR and that three metabolic circuits influence the DDR by modulating PP2A activity. Irc21, a putative cytochrome b5 reductase that promotes the condensation reaction generating dihydroceramides (DHCs), and Ppm1, a PP2A methyltransferase, counteract the DDR by activating PP2A; conversely, the nutrient-sensing TORC1-Tap42 axis sustains DDR activation by inhibiting PP2A. Loss-of-function mutations in IRC21, PPM1, and PP2A and hyperactive tap42 alleles rescue mec1 mutants. Ceramides synergize with rapamycin, a TORC1 inhibitor, in counteracting the DDR. Hence, PP2A integrates nutrient-sensing and metabolic pathways to attenuate the Mec1ATR response. Our observations imply that metabolic changes affect genome integrity and may help with exploiting therapeutic options and repositioning known drugs.
Keywords: DNA damage response, Mec1-ATR, protein phosphatase PP2A, TORC1, metabolism, genome stability, Rad53, Irc21
Graphical Abstract
Highlights
-
•
PP2A counteracts the DNA damage response
-
•
Cytochrome b5-like Irc21 attenuates the DDR through ceramide-induced PP2A activation
-
•
The TORC1-Tap42 axis contributes to DDR activation through PP2A inhibition
-
•
The SAM-Ppm1 pathway attenuates the DDR by activating PP2A
Ferrari et al. provide a model wherein cytoplasmic metabolic pathways (the TORC1-Tap42, Ceramide-Irc21, and SAM-Ppm1 axes) converge on PP2A and PP2A-like phosphatases to modulate the nuclear DNA damage response. This reveals a connection between cell metabolism and genome stability surveillance mechanisms.
Introduction
Tel1ATM and Mec1ATR mediate DNA damage checkpoints (Harrison and Haber, 2006) by activating the Chk1CHK1 and Rad53CHK2 kinases, which transduce the signal to downstream targets. Rad53 controls replication fork integrity and phosphorylates Dun1 to upregulate deoxy-nucleotide triphosphate (dNTP) levels (Bashkirov et al., 2003, Sogo et al., 2002). Checkpoint deactivation occurs during recovery or adaptation and is mediated by the PP1, PP2C, and PP4 phosphatases (Bazzi et al., 2010, Keogh et al., 2006, Leroy et al., 2003, O’Neill et al., 2007). Hydroxyurea (HU) inhibits replication by limiting dNTPs, causing replication stress and Mec1 activation (Slater, 1973, Sun et al., 1996).
PP2A and PP2A-like Ser/Thr phosphatases are ceramide-activated protein phosphatases (CAPPs) (Janssens and Goris, 2001, Jiang, 2006, Nickels and Broach, 1996). PPH21 and PPH22 encode PP2A catalytic subunits, whereas Tpd3 is a scaffolding subunit. The Cdc55 and Rts1 regulatory subunits are mutually exclusive and direct PP2As to distinct processes. The Ppm1 methyltransferase methylates and activates PP2A. Sit4 is a PP2A-like phosphatase that interacts with four activators known as Sit4-associating proteins (Saps). Pph21/Pph22 and Sit4 interact with Tap42, a target of TOR complex 1 (TORC1) (Di Como and Arndt, 1996). Rrd2 and Rrd1 are PP2A- and PP2A-like activators. Tip41 inhibits Tap42, and their interaction is influenced by Ptc1 phosphatase. The PP2A/PP2A-like signaling pathways are not fully characterized (Düvel et al., 2003). According to the model, TORC1 phosphorylates Tap42, which inhibits PP2As; PP2A dephosphorylates TORC1 effectors (Loewith and Hall, 2011). Gln3, Npr1, Nnk1, and Rtg3 are PP2A targets involved in nitrogen and amino acid metabolism (Hughes Hallett et al., 2014). PP2A influences the ataxia telangiectasia mutated (ATM)-dependent DNA damage response (DDR) (Freeman and Monteiro, 2010).
We show that Irc21, a cytochrome b5-like enzyme influencing genome stability (Alvaro et al., 2007, Gallego et al., 2010, Guénolé et al., 2013, Lee et al., 2005) activates PP2A by promoting the synthesis of dihydroceramide (DHC) and that PP2A is a central hub in a regulatory loop that couples three metabolic pathways dependent on Irc21, Ppm1 and TORC1, with the ataxia telangiectasia and Rad3-related (ATR)-mediated DDR.
Results
Irc21 Influences the Replication Stress Response
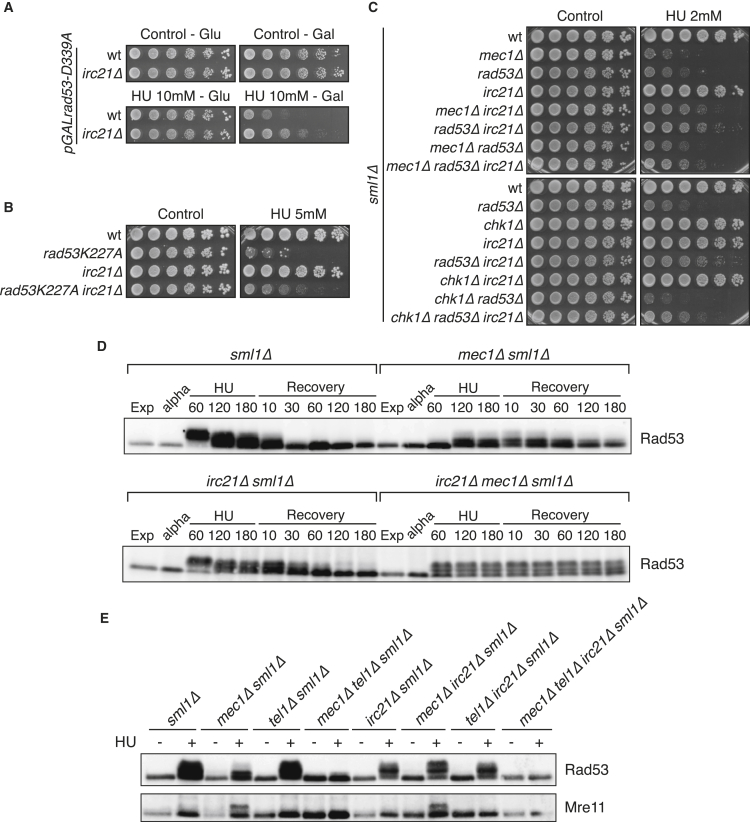
We identified irc21Δ as suppressor of the HU sensitivity of the dominant-negative rad53-D339A and the kinase-deficient rad53-K227A mutations (Bermejo et al., 2011, Fay et al., 1997). The suppression was validated in the W303 (Thomas and Rothstein, 1989) background (Figures 1A and 1B). irc21Δ also rescued the HU sensitivity of the mec1Δ sml1Δ, rad53Δ sml1Δ, mec1Δ rad53Δ sml1Δ, and chk1Δ rad53Δ sml1Δ strains (Figure 1C). Sml1 inhibits ribonucleotide reductase and its ablation bypasses the essential functions of MEC1 and RAD53 by increasing dNTP levels (Desany et al., 1998, Huang et al., 1998, Zhao et al., 1998). IRC21 and SML1 double ablation caused an additive suppression in a rad53-K227A background (Figure S1A), and irc21Δ mutants were hypersensitive to high HU doses (Figure S1B), suggesting that irc21Δ suppression does not depend on dNTP levels or intrinsic HU resistance.
Figure 1.
IRC21 Deletion Rescues Checkpoint Mutants
(A) Cells were grown on synthetic-defined (SD/-Ura) plates with glucose 2% or galactose 2% ± hydroxyurea (HU).
(B and C) Cells were grown on YPD plates with or without 5 mM HU (B) or 2 mM HU (C).
(D) sml1Δ, sml1Δ mec1Δ, sml1Δ irc21Δ, and sml1Δ mec1Δ irc21Δ cells were arrested in G1 with α-factor (alpha) and released into YPD containing 0.2 M HU. After 3 hr, cells were released into YPD. Samples were collected at the indicated times to detect Rad53 by western blot analysis.
(E) Cells were arrested with α-factor and released in YPD with or without 0.2 M HU. Cells were treated for 3 hr, and samples were collected to detect Rad53 and Mre11.
See also Figure S1.
We addressed whether IRC21 deletion influenced the Mec1-dependent Rad53 phosphorylation and dephosphorylation (Fiorani et al., 2008, Sanchez et al., 1996) during checkpoint activation and deactivation (Figure 1D). sml1Δ, mec1Δ sml1Δ, irc21Δ sml1Δ, and mec1Δ irc21Δ sml1Δ mutants were released from G1 into HU to activate Mec1 and released into medium without HU to recover (Pellicioli et al., 1999). In sml1Δ cells, Rad53 phosphorylation was obvious in HU and decreased during recovery. Rad53 phosphorylation was nearly abolished in mec1Δ sml1Δ mutants. IRC21 deletion restored Rad53 phosphorylation in mec1Δ sml1Δ cells and also delayed Rad53 dephosphorylation in sml1Δ mutants. The persistence of Rad53 phosphorylation in mec1Δ irc21Δ sml1Δ mutants recovering from HU correlated with the inability to efficiently complete S phase (Figure S1C). We addressed whether irc21Δ-mediated rescue of Rad53 phosphorylation in mec1Δ cells was dependent on Tel1, which phosphorylates Mre11 and shares overlapping functions with Mec1 (Usui et al., 2001). Rad53 was not phosphorylated in mec1Δ tel1Δ irc21Δ sml1Δ cells exposed to HU (Figure 1E), suggesting that the irc21Δ-mediated rescue of Rad53 phosphorylation in mec1Δ cells depends on Tel1. The irc21Δ suppression mechanism was not due to Tel1 hyperactivation because IRC21 deletion did not further elevate Tel1-dependent Mre11 phosphorylation (Ira et al., 2004; Figure 1E).
irc21Δ did not rescue the HU sensitivity of dun1Δ mutants (Figure S1D), further suggesting that Irc21 does not cause an increase in dNTP pools. Moreover, IRC21 deletion failed to fully phosphorylate Dun1 in mec1 or rad53 mutants (Figure S1E). Hence, the Tel1-mediated Rad53 phosphorylation in sml1Δ mec1Δ mutants is somewhat suboptimal because it prevents Rad53 from phosphorylating Dun1. This is consistent with the notion that certain phospho-isoforms of Rad53 are unable to phosphorylate Dun1 (Lee et al., 2003). irc21Δ mutants failed to efficiently recover from the HU treatment, as visualized by the fluorescence-activated cell sorting (FACS) profile and by the delayed dephosphorylation of Rad53 and Dun1 (Figure S1F).
Irc21 Affects Mitochondrial Functions and Lipid Biosynthesis
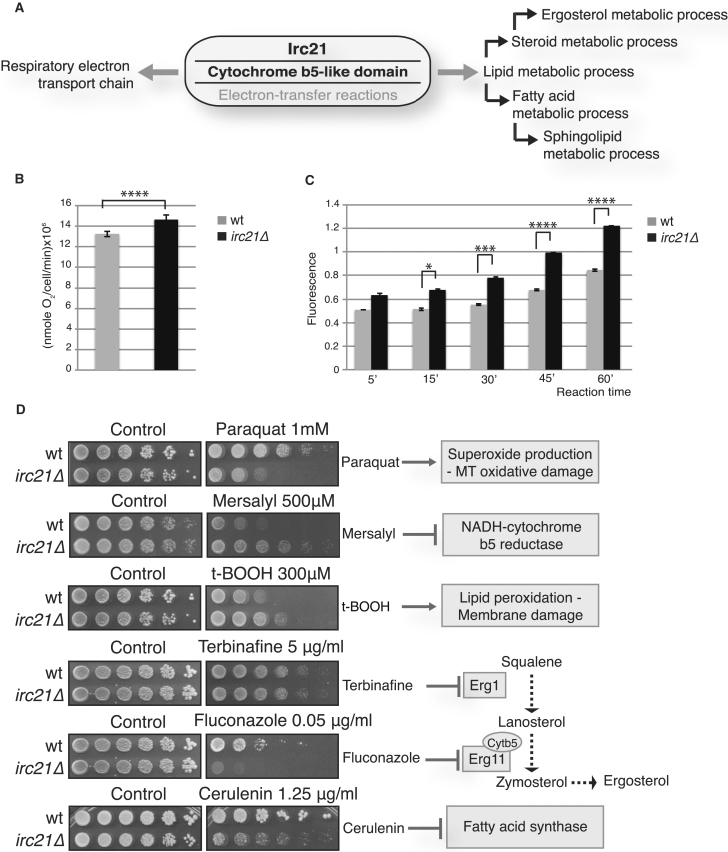
Irc21 contains an NADH-cytochrome b5 reductase domain (CBR); CBRs are involved in mitochondrial functions and lipid biosynthesis (Figure 2A). We measured the oxygen consumption rate of logarithmically growing cells and found that irc21Δ mutants showed a higher respiration rate compared with wild-type (WT) cells (Figure 2B). During respiration, mitochondria generate reactive oxygen species (ROS). Using the oxidant-sensing probe 2′,7′-dichlorodihydrofluorescein diacetate (DCFH-DA) to measure ROS levels (Davidson et al., 1996), we found that exponentially growing irc21Δ cells accumulated more ROS than WT cells (Figure 2C), as shown previously (Neklesa and Davis, 2008). Consistent with higher respiration activity, irc21Δ cells were hypersensitive to paraquat (1,1′-dimethyl-4,4’-bipyridinium dichloride), a redox cycler that stimulates respiration-dependent superoxide production (Cochemé and Murphy, 2009; Figure 2D). This did not reflect general ROS sensitivity because irc21Δ cells were resistant to tert-butyl hydroperoxide (t-BOOH), which produces ROS and damages a variety of cellular constituents, including lipids (Girotti, 1998; Figure 2D). In agreement with putative Irc21 CBR activity, irc21Δ cells were resistant to mersalyl, a mercurial diuretic that affects mitochondrial functions and inhibits the NADH-cytochrome b5 reductase (Bernardi and Azzone, 1981; Figure 2D). The cytochrome b5-dependent electron transport system is also involved in lipid metabolic processes such as cholesterol/ergosterol biosynthesis and desaturation and elongation of fatty acids (Vergéres and Waskell, 1995). In particular, heme is required for the enzymatic activities of Erg3p (sterol C5-6 desaturase), Erg5p (sterol C22–23 desaturase), and Erg11p (sterol 14α-demethylase). We analyzed the sensitivity of irc21Δ cells to inhibition of Cyb5-dependent Erg11 by fluconazole and Cyb5-independent Erg1 (squalene epoxidase) by terbinafine (Kontoyiannis, 2000, Lamb et al., 1999, Petranyi et al., 1984; Figure 2D). irc21Δ cells were specifically sensitive to fluconazole and not to terbinafine, suggesting that IRC21 ablation exacerbates the inhibitory effect of fluconazole on ergosterol synthesis. Fatty acid metabolism involves Cytb5-dependent reactions. We tested whether irc21Δ cells were sensitive to overall dampening of fatty acid production by cerulenin, an inhibitor of fatty acid synthase (FAS) that prevents the synthesis of medium- and long-chain fatty acids (MCFAs and LCFAs, respectively) and of very-LCFAs (VLCFAs) (Awaya et al., 1975). irc21 mutants were hypersensitive to cerulenin (Figure 2D), implying that Irc21 may affect fatty acid metabolism. Taken together, the sensitivity/resistance profile suggests that Irc21 influences CBR-dependent processes.
Figure 2.
IRC21 Deletion Impairs Cytb5-Dependent Processes
(A) Classification of putative Irc21 processes based on the gene ontology PANTHER (protein analysis through evolutionary relationships) classification system.
(B) Oxygen consumption rate of exponentially growing WT and irc21Δ cells. The results are shown as means ± SD of triplicates. ∗∗∗∗p < 0.0001.
(C) Determination of ROS levels using the DCFH-DA assay in WT and irc21Δ cells depending on the incubation period. The results are shown as means ± SD of triplicates. p values are indicated.
(D) Spot assay of WT and irc21Δ cells on YPD plates with or without paraquat, mersalyl, t-BOOH, terbinafine, fluconazole, or cerulenin at the indicated concentrations. Drug mechanisms of action are illustrated.
irc21Δ and PP2A Mutants Genetically Interact and Exhibit Similar Interactome Profiles
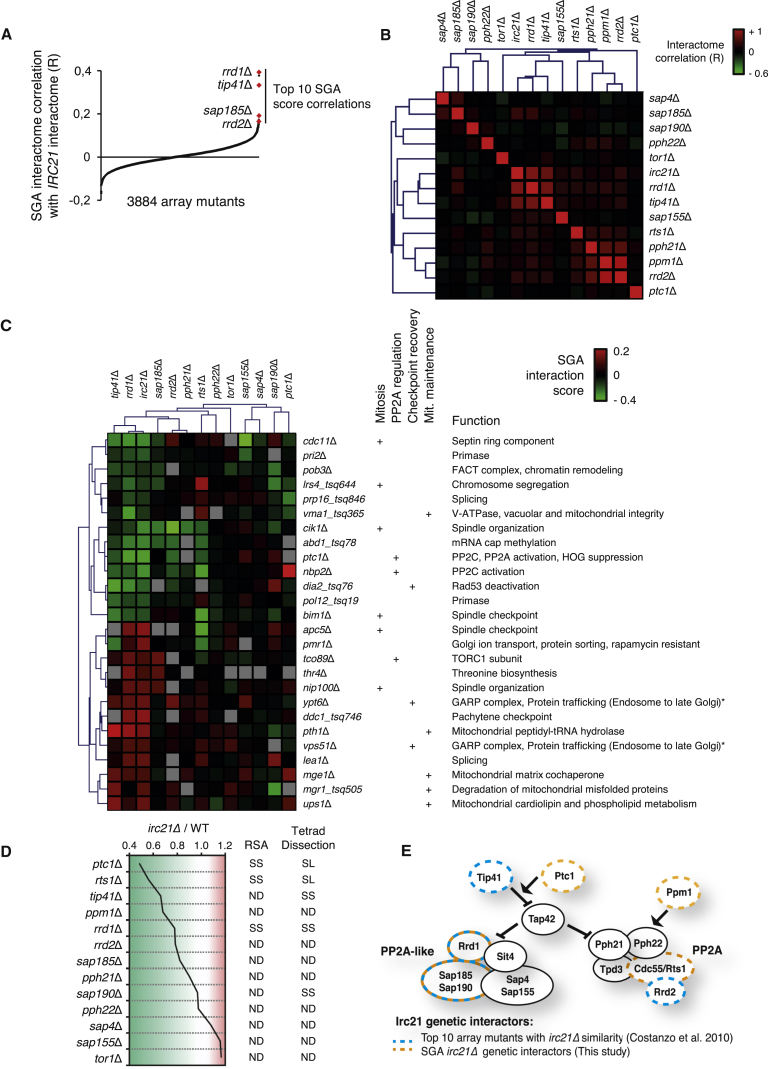
We compared the irc21Δ genetic interaction signatures with those of 3,884 other deletion strains (Costanzo et al., 2010) by calculating correlation scores (R) based on genetic interactions with 1,712 mutants. Among the top ten array strains with synthetic genetic array (SGA) interactomes most similar to irc21Δ mutants, we identified deletions in the RRD1 (R = 0.39), TIP41 (R = 0.33), SAP185 (R = 0.19), and RRD2 (R = 0.17) genes (Figures 3A and S2A), which encode PP2A and PP2A-like regulators (Luke et al., 1996, Van Hoof et al., 2005; Figure 3E). The interactome correlation between irc21Δ and rrd1Δ was quantitatively similar to the one between the two PP2A/PP2A-like activators rrd1Δ and tip41Δ (R = 0.37). The second hit with an irc21Δ interactome correlation similar to rrd1Δ was imp2′Δ (R = 0.38) (Figure S2A), in which the open reading frame (ORF) next to RRD1 is disrupted; likely, imp2′Δ impairs RRD1 expression and, thus, PP2A-like functionality. This is consistent with the high similarity of rrd1Δ and imp2′Δ interaction profiles (R = 0.69; data not shown). We also identified deletions in the genes encoding Bck2, involved in protein kinase C signaling (Lee et al., 1993); Rrm3, a replicative DNA helicase targeted by the Mec1-Rad53 pathway (Rossi et al., 2015, Torres et al., 2004); Rnh201, a ribonuclease H2 involved in Okazaki fragment processing (Qiu et al., 1999); and phosphatases Ptp2 and Pph3 (Guan et al., 1992, O’Neill et al., 2007). We compared the interactomes of irc21Δ and other PP2A and PP2A-like deletion mutants by calculating the pairwise interactome correlation scores and performed hierarchical clustering of R values (Figure 3B). The analysis revealed two clusters, the first containing pph21Δ, ppm1Δ, and rrd2Δ (loosely associated with rts1Δ) and the second containing irc21Δ, rrd1Δ, and tip41Δ (loosely associated with sap155Δ). These observations suggest that irc21Δ affects PP2A and PP2A-like phosphatase activities (Figure 3E). The signatures of genetic interactions shared between irc21Δ and either rrd1Δ or tip41Δ (Costanzo et al., 2010; Figure 3C) showed that common interactors were associated with PP2A/PP2A-like regulated processes, including mitosis, checkpoint recovery and adaptation, and mitochondrial maintenance. Other PP2A mutant strains, including sap185Δ, rrd2Δ, rts1Δ, pph21Δ, and pph22Δ, shared interactions with the irc21Δ/rrd1Δ/tip41Δ signatures to varying degrees (Figure 3C).
Figure 3.
Irc21 Interacts with PP2A and PP2A-like Phosphatases
(A) Comparison of the interactome of the irc21Δ array strain with the interactomes of 3,884 mutant array strains by calculating the correlation (R) value of their interaction scores with the 1,712 query mutants (datasets from Costanzo et al., 2010).
(B) Heatmap representing pairwise interactome correlation values of mutants with altered PP2A activity.
(C) Heatmap representing SGA interaction scores between query mutants of the irc21Δ and rrd1Δ and tip41Δ signature (rows) and PP2A-related array mutants (columns). Gray fields indicate that the interaction score has not been determined.
(D) Genetic interactions of IRC21 with PP2A components and regulators assessed by SGA screening. Left: quantitative effect of PP2A component deletions (rows) on the growth of irc21Δ mutants versus the WT. Right: summary of the confirmation of individual genetic interactions. SS, synthetic sick; SL, synthetic lethal. Interactions with ppm1 and tor1 were confirmed by gene targeting.
(E) Representation of PP2A and PP2A-like complex subunits and regulators (see text). The blue and orange dotted lines indicate irc21Δ mutant genetic interactors found in Costanzo et al. (2010) and in the present study, respectively.
See also Figures S2 and S3.
We conducted an independent SGA analysis mating the irc21Δ query strain to the haploid deletion library containing deletions of ∼4,700 non-essential genes (Figure 3D; Tong et al., 2001). We found 42 negative and 12 positive interactions, causing, respectively, synthetic growth defects or suppression of the mild slow growth phenotype of irc21Δ mutants (Figure S2B). In addition, we identified five high-confidence epistatic interactors of IRC21 by filtering potential epistatic interactors from our dataset, with the positive IRC21 interactors previously reported in a genome-wide, high-throughput screen (Costanzo et al., 2010; Figure S3A).
Several PP2A/PP2A-like components displayed negative interactions with irc21Δ (significance, ptc1Δ and rts1Δ; trend, tip41Δ, ppm1Δ, rrd1Δ, and rrd2Δ) (Figures 3D and 3E). We confirmed some of these interactions by random spore analysis and/or tetrad dissection (Figures 3D, S3B, and S3C). The largest categories of irc21Δ negative genetic interactors were metabolic (oxidative stress, TCA cycle, and lipids), and chromatin/checkpoint pathways, which were related to PP2A. In accordance with previous observations (Figure 2D; cerulenin sensitivity assay), irc21Δ displayed negative genetic interaction with FEN1 deletion (Figure S2B), encoding the fatty acid elongase required for ceramide biosynthesis (Oh et al., 1997); moreover, ceramide hydroxylase Scs7, which contains a cytochrome b5 domain like Irc21 (Mitchell and Martin, 1997), was identified among the top five high-confidence Irc21 epistatic interactors (Figure S3A). Rescuing interactors (Figure S2B) were involved in phospholipid (PGC1), sterol (NSG2), and respiratory (RGI2, TRX3) metabolism, mitochondrial localization/inheritance (JSN1), cell morphology (MGA1, DFG5), nuclear membrane (MLP2), and genome integrity (RAD51). Epistatic interactors (Figure S3A) were involved in spindle and organelle positioning (DYN3, NIP100), mitochondrial localization/inheritance (MMR1), and ion transport (PMR1).
Overall, the IRC21 interactome analysis supports a function for Irc21 in mitochondrial and lipid metabolism and in influencing PP2A activity, nuclear morphology, and genome integrity.
Irc21 Is Involved in the TORC1-PP2A Regulatory Axis
We next characterized the role of Irc21 in regulating PP2A/PP2A-like activities, which are negatively regulated by the TORC1 pathway (Di Como and Arndt, 1996). We found that tor1Δ and tco89Δ, defective in TORC1 components, were positive interactors of irc21Δ (Figures 3C and 3D). TORC1 controls Tap42 and Sch9 through phosphorylation (Urban et al., 2007; Figure 4A). Sch9 influences translation initiation and G0 events. Tap42 regulates PP2A and PP2A-like phosphatases, which control the phosphorylation state of Msn2/Msn4, involved in stress response; Rtg1/3, implicated in the retrograde pathway; and Npr1 and Gln3, connected with amino acid synthesis and nitrogen assimilation (Hughes Hallett et al., 2014).
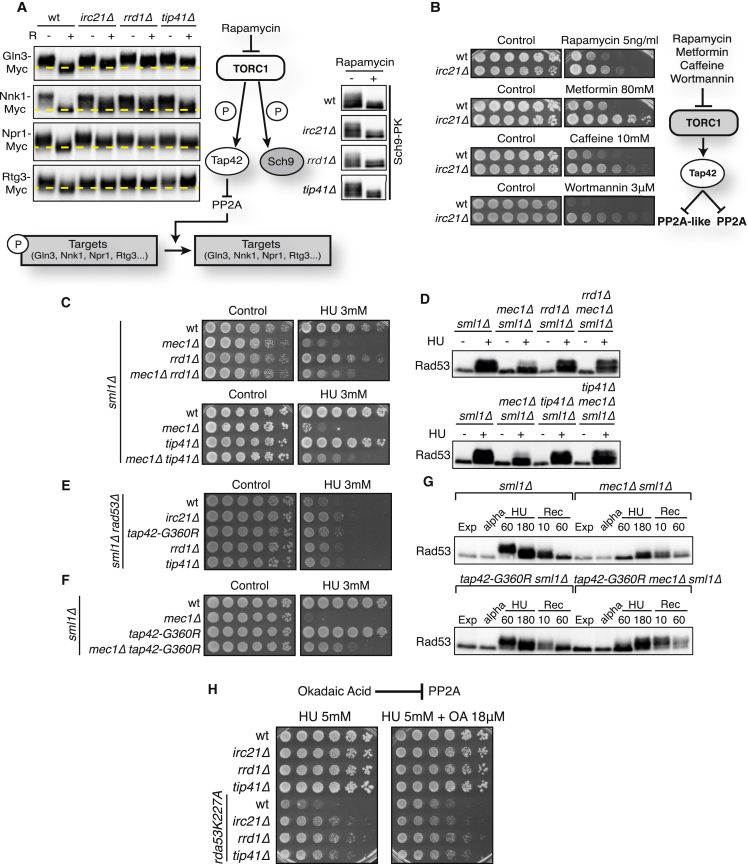
Figure 4.
Irc21 Inhibits DDR through PP2A Activation
(A) Cells were treated with 200 ng/mL rapamycin. Left: band-shift assays following phosphorylation of the PP2A branch proteins Gln3, Nnk1, Npr1, and Rtg3 after 30 min of rapamycin treatment. A horizontal line has been overlaid to assist in determining mobility shifts. Right: band-shift assays following the phosphorylation of Sch9 after 2 hr of rapamycin treatment.
(B) WT and irc21Δ cells were grown on YPD plates with or without rapamycin, metformin, caffeine, and wortmannin (left). All drugs are inhibitors of the Torc1-Tap42 pathway, represented at the right.
(C, E, and F) Cells were grown on YPD plates with or without 3 mM HU.
(D) Cells were arrested with α-factor and released in YPD with or without 0.2 M HU. Cells were treated for 3 hr and harvested to detect Rad53.
(G) Cells were arrested in G1 with α-factor and released in YPD with 0.2 M HU. After 3 hr, cells were released into YPD. Samples were collected at the indicated times to detect Rad53.
(H) Cells were grown on YPD + HU plates with or without okadaic acid (OA).
See also Figure S4.
To probe the Tap42/PP2A signaling branch in irc21Δ mutants, we selected four PP2A targets that exhibit clear modifications following TORC1 inhibition through rapamycin treatment: Gln3, Nnk1, Npr1, and Rtg3 (Figure 4A). As expected, rapamycin led to dephosphorylation of the four PP2A targets; in contrast, all four targets remained hyper-phosphorylated in irc21Δ, rrd1Δ, and tip41Δ cells even in the presence of rapamycin (Figure 4A, left). Thus, Irc21 participates in the activation of PP2A/PP2A-like pathways like Rrd1 and Tip41. To discern whether Irc21 activates PP2A directly or by inhibiting TORC1, we monitored the TORC1-Sch9 branch. Sch9 was normally dephosphorylated after rapamycin treatment in all mutant strains (Figure 4A, right). Hence, Irc21 is specifically involved in the activation of the PP2A/PP2A-like sub-pathways, which are also regulated by TORC1. Accordingly, irc21Δ mutants are resistant to treatment with a variety of TORC1 inhibitors, such as rapamycin, caffeine, metformin, and wortmannin (Figure 4B), in analogy to certain PP2A mutants (Rempola et al., 2000).
PP2A Influences the Checkpoint Response
The previous observations led us to hypothesize that, similar to Irc21, PP2A and PP2A-like control the Rad53-mediated response to replication stress. Ablation of the PP2A-positive regulators RRD1 and TIP41 mimicked irc21Δ in suppressing the HU sensitivity of rad53-D339A, rad53-K227A, rad53Δ sml1Δ, and mec1Δ sml1Δ mutant alleles (Figures S4A, S4B, 4C, and 4E). Moreover, deletion of either RRD1 or TIP41 in a mec1Δ sml1Δ background was able to rescue the crippled Rad53 phosphorylation (Figure 4D), similar to irc21Δ (Figure 1D). rrd1Δ and tip41Δ also exhibited delayed Rad53 deactivation following recovery from HU, similar to irc21Δ mutants (data not shown).
We performed a screen to find spontaneous extragenic suppressors of mec1-100 (Paciotti et al., 2001) lethality on HU and identified a mutation in TAP42. tap42-G360R rescued the HU sensitivity of mec1Δ and rad53Δ cells (Figures 4E and 4F) and abolished the defective Rad53 phosphorylation in HU-treated mec1Δ sml1Δ cells, mimicking the phenotype of irc21Δ, rrd1Δ, and tip41Δ (Figure 4G).
A gain-of-function mutation could account for the above results. To test whether the mutation caused a constitutive inhibition of PP2A, we analyzed the rapamycin and metformin sensitivity of tap42-G360R cells. As expected, and similar to irc21Δ, rrd1Δ, and tip41Δ, tap42-G360R mutants were partially resistant to both drugs (Figure S4C). Second, after rapamycin treatment, tap42-G360R cells showed defective Gln3, Nnk1, and Npr1 dephosphorylation (Figure S4D). Hence, the hyperactive Tap42 allele resembles the absence of PP2A activators.
Because PP2A appeared to target Rad53, we predicted that chemical compounds acting on PP2A activity should also affect rad53 HU sensitivity. Low doses of okadaic acid (OA) cause selective inhibition of PP2A (Zhang et al., 1994) and partially rescued the HU sensitivity of rad53-K227A mutants (Figure 4H).
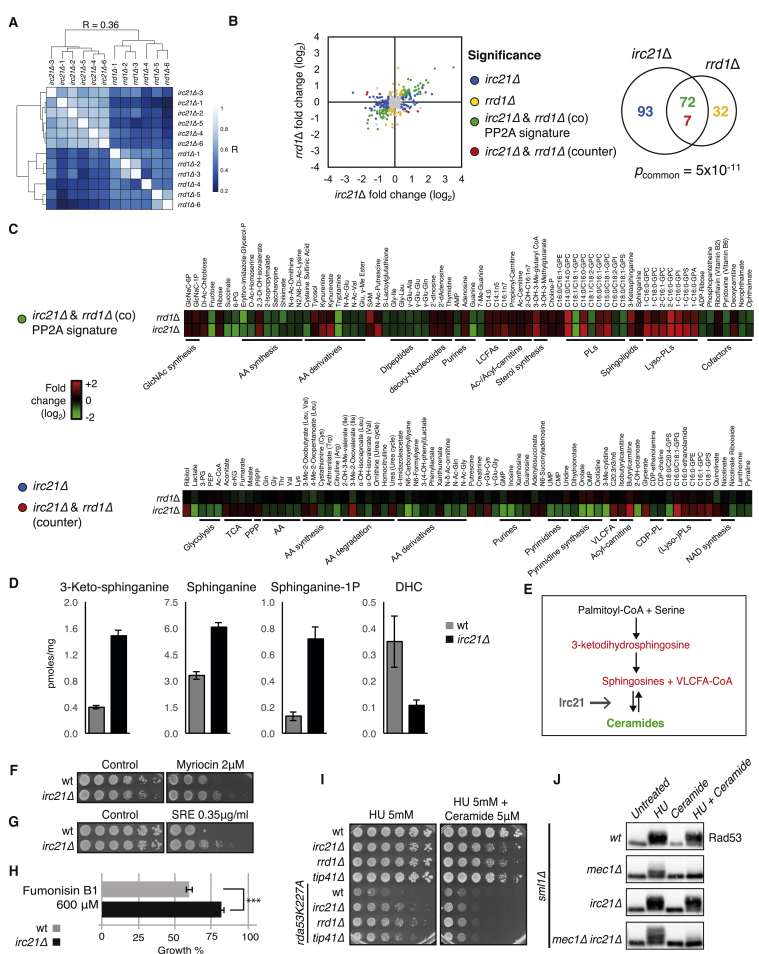
Irc21 and PP2A Metabolic Signatures
To characterize the relationship between PP2A and Irc21, we compared the global mass spectrometry metabolic profile of WT, irc21Δ, and rrd1Δ cells during logarithmic growth in rich medium. Unsupervised clustering by metabolite fold changes clearly grouped the replicates of irc21Δ and rrd1Δ by genotype but also revealed a degree of similarity between both mutants (Figure 5A). Of a total of 484 examined compounds, 172 and 111 were significantly changed in irc21Δ and rrd1Δ cells, respectively (linear model for microarray data [LIMMA], padj = 0.05) (Figure 5B). The two mutants shared a significant amount of metabolite alterations (79, p = 5 × 10−11). Importantly, nearly all of these were co-regulations (72 of 79), suggesting that these 72 alterations define a metabolic, shared Irc21-PP2A signature (Figure 5B). As expected for low PP2A activity (Staschke et al., 2010), the PP2A signature was characterized by a reduction of amino acid biosynthesis intermediates and dipeptides (Figure 5C, top). It also featured elevated levels of multiple lipids and lipid intermediates (long chain fatty acids, sterol biosynthesis intermediates, lyso-phospholipids, carnitine conjugates, sphingolipid precursors) and a shifted composition of phospholipids to a shorter fatty acid chain length (less than C18). Low PP2A activity correlated with high N-acetylglucosamine (GlcNAc) biosynthesis intermediates, high deoxy-nucleosides (but normal deoxy-nucleotides), and high levels of the methyl donor S-adenosyl-methionine (SAM).
Figure 5.
Irc21 Exerts PP2A-Dependent and PP2A-Activating Metabolic Regulations
(A) Unsupervised hierarchical clustering of irc21Δ and rrd1Δ mutants (six replicates each) based on metabolome alterations during logarithmic growth in rich medium.
(B) Summary of metabolome alterations of irc21Δ and rrd1Δ mutants during logarithmic growth in rich medium. Left: scatterplot of quantitative alterations of individual metabolites in irc21Δ and rrd1Δ mutants compared with a congenic WT identifies irc21Δ-specific (blue), rrd1Δ-specific (yellow), common (green, PP2A signature), and opposite (red) regulations. Right: Venn diagram representation of the intersection of metabolic alterations in irc21Δ and rrd1Δ mutants and intersection significance p value determined by chi-square test.
(C) Heatmap representation of altered metabolites by signature. Top: PP2A signature (common alterations in irc21Δ and rrd1Δ). Bottom: specific regulations in irc21Δ and opposite regulations in irc21Δ and rrd1Δ. As indicated, metabolites were grouped by class.
(D) 3-keto-sphinganine, sphinganine, sphinganine-1p, and dihydroceramide were quantified in WT and irc21Δ cells. Average values are shown, and error bars represent SEM.
(E) Simplified scheme representing ceramide biosynthesis in S. cerevisiae. Colored metabolites indicate an increase (red) or a decrease (green) of their amount in irc21Δ cells.
(F) WT and irc21Δ cells were grown on YPD plates with or without myriocin.
(G) WT and irc21Δ cells were grown on YPD plates with or without syringomycin E.
(H) WT and irc21Δ cells were grown in SD medium with or without Fumonisin B1.
(I) Cells were grown on YPD + HU with or without ceramide.
(J) Cells were arrested in G1 with α-factor and released in YPD with or without 0.2 M HU, 15 μM ceramide, or 0.2 M HU in combination with 15 μM ceramide for 3 hr.
See also Figure S5.
The metabolite alterations in irc21Δ that are not shared by rrd1Δ represent PP2A-independent functions of Irc21. We found that several metabolites related to CBR function were altered in irc21Δ cells. Accumulation of trichloroacetic acid (TCA) cycle intermediates (aconitate, α-ketoglutarate, fumarate, and malate) and a reduction in the late glycolysis intermediates and acetyl coenzyme A (Ac-CoA) were indicative of altered mitochondrial activity (Figures 2A and 5C, bottom). The levels of several amino acids derived from glycolysis (Gly and Val) and TCA (Gln, Thr, and Lys) and their derivatives were reduced, whereas urea cycle products accumulated (ornithine, urea). Purine and pyrimidine ribonucleosides and ribonucleotides as well as cytidine triphosphate (CTP)-dependent phospholipid precursors were also reduced, whereas the nucleotide synthesis substrate phosphoribosyl pyrophosphate (PRPP) increased. Cytb5 is also involved in fatty acid metabolism (Figure 2A), and we found that although fatty acid accumulation was common to irc21Δ and rrd1Δ, VLCFAs, which are used in the synthesis of ceramides, accumulated in irc21Δ cells. Accumulation of VLCFAs and the genetic interaction with the VLCFA synthesis enzyme FEN1 suggest that irc21Δ mutants inefficiently condense sphingolipids and VLCFAs into ceramides and, thus, fail to promote PP2A activation.
We performed a quantitative mass spectrometry analysis of ceramides and related lipid metabolites in WT and irc21Δ cells. DHC was more abundant than phytoceramides (PHCs) in WT cells (0.35 pmol/mg and 0.01 pmol/mg, respectively) (Figure S5A). Deletion of IRC21 reduced the level of DHC (Figures 5D and S5A–S5C), whereas the DHC precursors 3-ketodihydrosphingosine (3-keto-DHS), dihydrosphingosine (DHS), dihydrosphingosine-1-P (DHS-1-P), and VLCFA-CoA increased in irc21Δ cells (Figures 5C, 5D, and S5A–S5C). These observations confirmed that IRC21 deletion impairs ceramide biosynthesis and that the defective step corresponds to the DHS-DHC conversion (Figures 5E and S5C). Accordingly, irc21Δ mutants were resistant to myriocin (Figure 5F), an inhibitor of ceramide synthesis acting on serine palmitoyltransferase (SPT), the first enzyme in the sphingolipid biosynthesis pathway (Figure S5C; Huang et al., 2012). Moreover, irc21Δ cells were resistant to syringomycin E (SRE) (Figure 5G; Takemoto et al., 1993), supporting a defect in sphingolipid biosynthesis (Stock et al., 2000). Addition of exogenous DHC restored irc21Δ sensitivity to SRE (Figure S5D). Fumonisin B1, a ceramide synthase inhibitor, pheno-copies irc21Δ mutants by causing an increase in DHS and PHS levels and a concomitant decrease in ceramide and, therefore, DHC (Wu et al., 1995). irc21Δ mutants were resistant to fumonisin B1 (Figure 5H). This is consistent with the idea that the drug targets a pathway compromised in irc21Δ cells. We conclude that Irc21 promotes the condensation reaction leading to the formation of DHC.
Ceramides, SAM-Mediated Methylation, and TORC1 Inhibition Attenuate the Checkpoint Response by Promoting PP2A Activation
We investigated the possibility to abrogate irc21Δ HU resistance and PP2A inactivity phenotypes by exogenously providing ceramide. It is known that the cell-permeable ceramide analog C2-ceramide induces a dose-dependent activation of PP2A in yeast (Nickels and Broach, 1996). We first analyzed the effect of exogenous ceramide on the rescue of rad53-K227A HU sensitivity by irc21Δ, rrd1Δ, and tip41Δ. The sphingolipid suppressed the HU resistance of all mutants (Figure 5I). We then asked whether ceramide was also able to suppress the IRC21Δ-dependent rescue of Rad53 phosphorylation in mec1Δ cells. Ceramide-mediated PP2A activation abolished Rad53 phosphorylation, specifically in irc21Δ mec1Δ cells, and reduced it in WT and irc21Δ cells (Figure 5J). In addition, ceramide caused Rad53 dephosphorylation during recovery from HU treatment in irc21Δ mec1Δ mutants (Figure S5E).
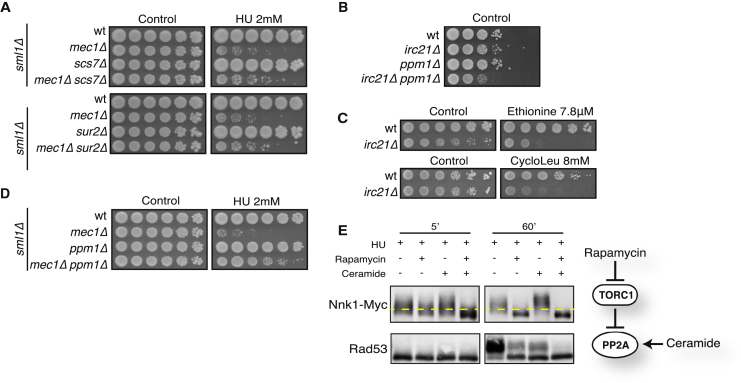
Sur2 and Scs7 are both required for ceramide hydroxylation and are members of the cytochrome b5-dependent enzyme family (Figure S5C; Haak et al., 1997). Scs7p contains a cytochrome b5-like domain, whereas cytochrome b5 may function to transfer electrons to Sur2. Both SCS7 and SUR2 deletions partially rescued mec1Δ sml1Δ HU sensitivities (Figures 6A). Thus, defective cytochrome b5-dependent enzymes, involved in ceramide biosynthesis, have beneficial consequences for checkpoint mutants exposed to replication stress.
Figure 6.
Ceramides, TORC1, Irc21, and Ppm1 Affect the HU-Induced DDR by Modulating PP2A Activity
(A and D) Cells were grown on YPD ± HU.
(B) Cells were grown on YPD. 1:10 dilutions were used to highlight growth rate differences.
(C) WT and irc21Δ cells were grown on YPD plates with or without ethionine or cycloleucine.
(E) Cells were arrested with α-factor and released into YPD containing 0.2 M HU alone or in combination with rapamycin (200 ng/mL), ceramide (15 μM), or rapamycin (200 ng/mL) + ceramide (15 μM). Cells were treated for 1 hr and harvested after 5 and 60 min to detect Nnk1 and Rad53.
See also Figure S6.
SAM levels are critical for methylation and activation of PP2A; in this process, Ppm1 methylates the C terminus of the PP2A catalytic subunit (Sutter et al., 2013). Accordingly, PPM1 ablation partially impaired dephosphorylation of PP2A targets (Figure S6A). We found a negative interaction between IRC21 and PPM1 (Figure 3D) and confirmed that irc21Δ and ppm1Δ are synthetic sick (Figure 6B). Moreover, irc21Δ and rrd1Δ mutants accumulate high levels of SAM (Figure 5C); accordingly, irc21Δ and rrd1Δ mutants were hypersensitive to SAM limitation caused by ethionine (a toxic analog of methionine) or cycloleucine (an inhibitor of methionine adenosyl transferase) (Figure 6C; data not shown). These results suggest that Irc21 and Ppm1 positively regulate PP2A through different mechanisms. Interestingly, like the deletion of IRC21, PPM1 ablation rescued the HU sensitivity of mec1Δ sml1Δ mutants (Figure 6D) and partially recovered the Rad53-defective phosphorylation in HU-treated mec1Δ cells (data not shown).
Both rapamycin and ceramide have been shown to promote PP2A activity (Loewith et al., 2002, Nickels and Broach, 1996). We tested whether rapamycin and ceramide treatments could modulate the HU-induced DDR response. Cells were released from G1 in the presence of HU alone or in combination with rapamycin and ceramide (Figure 6E). After 5 min, when Rad53 was still unphosphorylated, the concomitant presence of rapamycin and ceramide caused PP2A hyperactivation, as indicated by Nnk1 de-phosphorylation. At 60 min, Rad53 phosphorylation was evident in HU-treated cells, partial in HU + rapamycin and HU + ceramide, and abolished in HU + rapamycin and ceramide. We obtained analogous results by treating exponentially growing cells with HU in combination with rapamycin and/or ceramide (Figure S6B).
Altogether, these observations suggest that TORC1, Irc21, and Ppm1 influence the HU-induced DDR by regulating the activity of PP2A and that ceramide levels as well as SAM levels are crucial for Mec1 and Rad53 activation.
Discussion
Activation of Mec1ATR requires multiple post-translational modifications that integrate chromosomal signals and mechanical stimuli (Awasthi et al., 2016). Deactivation of Mec1ATR promotes cell-cycle recovery or adaptation (Bartek and Lukas, 2007). Fine-tuning of the Mec1ATR cascade prevents the deleterious consequences of unscheduled checkpoint activation (Harrison and Haber, 2006). Mec1 and ATR regulate nuclear and non-nuclear pathways (Hilton et al., 2015, Kumar et al., 2014, Matsuoka et al., 2007). In yeast, several phosphatases have been involved in DDR silencing, including PP2C (Ptc2/Ptc3) and PP4 (Pph3-Psy2), required for double-strand break (DSB) recovery, and PP1 (Glc7), which promotes HU recovery (Bazzi et al., 2010, Keogh et al., 2006, Leroy et al., 2003, O’Neill et al., 2007). PP2A has been genetically linked to the RAD53-MEC1 pathway but ruled out as one of the main phosphatases implicated in checkpoint control (Hustedt et al., 2015). PP2A shows activity toward γH2AX, ATM, p53, Chk1, and Chk2 (Chen et al., 2015, Dozier et al., 2004, Goodarzi et al., 2004). Here we demonstrate that PP2A inactivation is beneficial when the Mec1-Rad53 axis is defective. Moreover, PP2A/PP2A-like act in a network with Irc21 and TORC1 to integrate metabolic signals with phosphorylation and dephosphorylation events outside and inside the nucleus and to attenuate the Mec1ATR cascade in cells experiencing replication stress.
IRC21 ablation rescues mec1Δ, rad53Δ, and chk1Δ sensitivity to HU and promotes HU-induced Rad53 phosphorylation when Mec1 is absent through a process mediated by Tel1. Irc21 was previously connected to the DDR, but the mechanism remained unclear (Guénolé et al., 2013).
Irc21 is an uncharacterized protein that consists of a cytochrome b5 domain; in accordance, irc21Δ mutants influence the respiration rate and ROS levels, display resistance to mersalyl, and show metabolic alterations related to CBR functions. Interestingly, Irc21 localization is mainly cytoplasmic (Guénolé et al., 2013, Huh et al., 2003), although a fraction has been detected in the nucleus (Guénolé et al., 2013), in mitochondria and vacuoles (collection of yeast cells and localization patterns [CYCLoPs]; Koh et al., 2015). A key question is how Irc21 influences the checkpoint response.
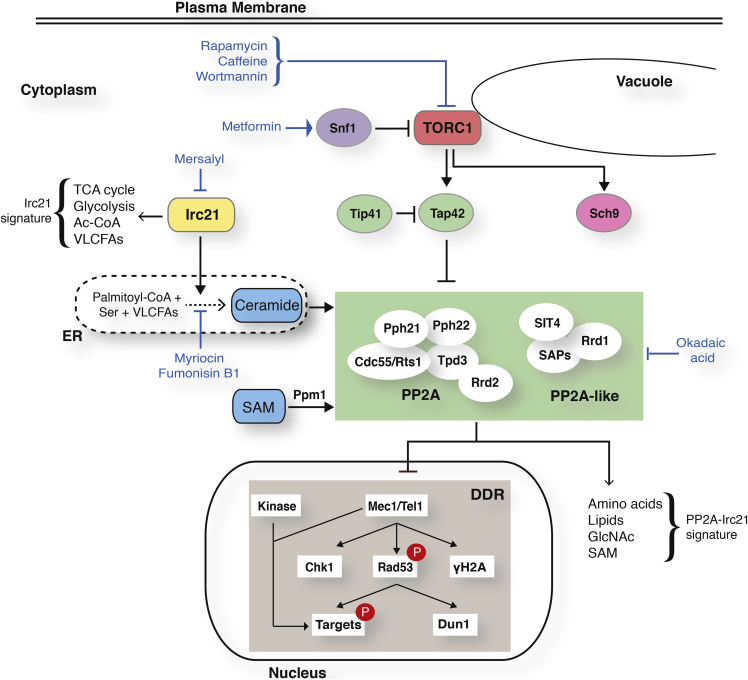
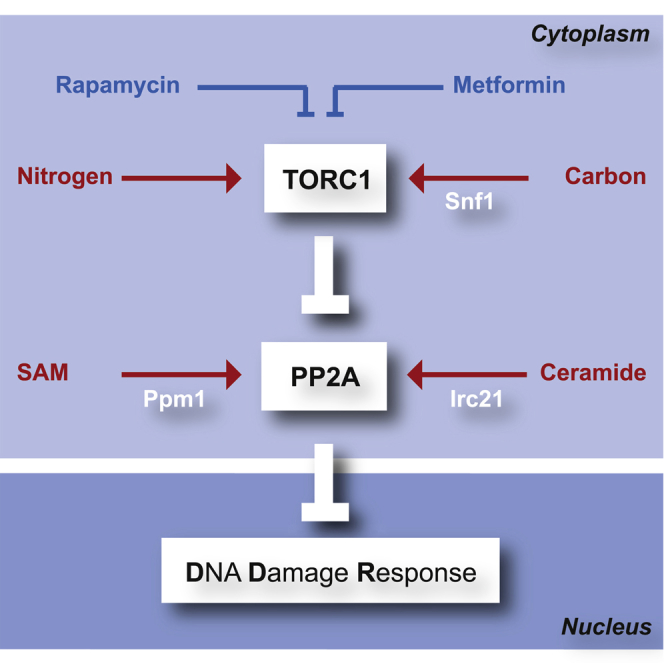
We show that Irc21 positively regulates PP2A/PP2A-like activities. IRC21 ablation causes resistance to TORC1 inhibitors but does not influence the TORC1-Sch9 axis, suggesting that Irc21 is not likely to act upstream of TORC1. The relationship between PP2A and TORC1 is complex and controversial. TORC1 negatively regulates PP2A/PP2A-like through Tap42 phosphorylation (Di Como and Arndt, 1996), whereas PP2A stimulates TORC1 through Npr2 phosphorylation (Laxman et al., 2014). irc21Δ mutants exhibit a negative genetic interaction with PP2A/PP2A-like activators (Rrd1, Rrd2, Tip41, Saps, and Ppm1) and positive genetic interactions with TORC1 components (Tco89 and Tor1) (Figure 7). We speculate that IRC21 ablation ameliorates TORC1-defective mutants by limiting PP2A/PP2A-like activities.
Figure 7.
Model: PP2A Links the DRR with Cell Metabolism
PP2A and PP2A-like phosphatases are regulated by TORC1 (nitrogen availability), Snf1AMPK (carbon availability), ceramide (sphingolipids and fatty acid availability), and SAM (methionine availability). The two PP2A complexes integrate the metabolic input with the control of DDR. Irc21 acts upstream of PP2A: it shares the PP2A signature but also displays specific metabolic functions (Irc21 signature).
We propose that Irc21 stimulates PP2A and, therefore, attenuates the DDR. Indeed, genetic and pharmacological inactivation of PP2A ameliorates the defective response to replication stress of checkpoint mutants. In addition, exogenous ceramide causes Rad53 dephosphorylation during recovery from HU treatment in mec1Δ irc21Δ sml1Δ mutants and abolishes irc21 rescue of Rad53 phosphorylation in mec1Δ irc21Δ sml1Δ cells during HU treatment.
The next key question is how Irc21 regulates PP2A activity. Among all metabolic alterations in irc21 mutants, elevated ROS are potential contributors to PP2A suppression; ROS accumulation causes PP2A inactivation (Nakahata and Morishita, 2014). We excluded this hypothesis because ROS scavengers did not affect the capability of irc21Δ to rescue the HU sensitivity of checkpoint mutants (data not shown).
IRC21 ablation alters sphingolipid metabolism and exhibits a reduction in DHC levels and an accumulation of DHC precursors (3-keto-DHS, DHS, DHS-1-P, and VLCFA-CoA). Hence, irc21Δ mutants are deficient in ceramide biosynthesis, and the defective step corresponds to the DHS-DHC conversion. DHC is produced by the condensation of DHS with VLCFAs, catalyzed by ceramide synthases (Lag1, Lac1, Lip1). The reverse reaction, hydrolysis of ceramides into sphingosine and fatty acid, is catalyzed by Ydc1 and Ypc1 ceramidases. A synthesis defect or elevated ceramidase activity would cause accumulation of DHS and a reduction in DHC, as we can observe in the absence of Irc21 (Mao et al., 2000b). Accordingly, irc21Δ mutants are resistant to fumonisin B1, a ceramide synthase inhibitor (Wu et al., 1995). Hence, we favor the hypothesis that Irc21 promotes the condensation reaction leading to the formation of DHC. One possibility is that Irc21 facilitates the activity of the ceramide synthase; intriguingly, LAC1 transcription is regulated by Rox1, a heme-dependent anaerobic repressor (Kolaczkowski et al., 2004). Another possibility is that Irc21 counteracts the activity of the Ydc1 and Ypc1 ceramidases that hydrolyze ceramides into sphingosine and fatty acid. Notably, Ydc1 or Ypc1 overexpression phenocopies the DHS accumulation and DHC reduction observed in irc21 mutants (Mao et al., 2000a, Mao et al., 2000b). Interestingly, Irc21 binds cardiolipin (CL), a mitochondrial phospholipid (Gallego et al., 2010) that is known to activate ceramidases (El Bawab et al., 2001).
Similar to irc21Δ, ablation of SUR2 and SCS7, two other members of the cytochrome b5-dependent enzyme family involved in the hydroxylation of sphingolipid long-chain bases and ceramides (Haak et al., 1997), ameliorates the replication stress sensitivity of checkpoint mutants. Intriguingly, we identified SCS7 as an epistatic interactor of IRC21. It is unlikely that this interaction reflects a role of Irc21 in ceramide hydroxylation because a hydroxylation defect would likely not lead to diminished DHC levels. Together, these observations suggest that both ceramide synthesis and hydroxylation are required for efficient PP2A-mediated checkpoint control.
Ceramides activate PP2A in yeast and mammals (Nickels and Broach, 1996). Hence, Irc21 might directly stimulate PP2A and attenuate the DDR by contributing to the production of ceramides. Interestingly, rapamycin and ceramide treatments cause a synergistic stimulation of PP2A activity, consistent with the view that TORC1 and Irc21 regulate PP2A activity in a negative and positive way, respectively (Figure 7).
PP2A activity is stimulated by the SAM-Ppm1 axis (Laxman et al., 2014). The synthetic sickness between ppm1 and irc21 mutants may therefore result from the simultaneous ablation of two independent positive regulatory pathways leading to PP2A activation (Figure 7). In this scenario, the low PP2A activity observed in ppm1 mutants would depend on the lack of PP2A methylation, whereas, in irc21 mutants, it may result from limiting ceramide levels. Intriguingly, we show that irc21 mutants display elevated SAM levels, raising the possibility that Ppm1-mediated PP2A methylation may facilitate basal PP2A activity in the absence of ceramide-dependent PP2A activation; consistently, irc21 mutants are particularly sensitive to treatments that limit SAM availability.
We propose that the DDR is attenuated by PP2A/PP2A-like, which are negatively regulated by the TORC1-Tap42 axis and positively regulated by the Irc21-ceramide and SAM-Ppm1 pathways (Figure 7).
Nutrients not only supply energy and building blocks for cellular growth but also exert crucial regulatory functions. PP2A controls the phosphorylation status of several targets involved both in cell metabolism and the DDR. It integrates signals from nitrogen and carbon metabolism as a central TORC1 effector (Hughes Hallett et al., 2014, Ramachandran and Herman, 2011), methionine metabolism by a methylation event that is sensitive to the levels of S-adenosylmethionine (Sutter et al., 2013), and sphingolipid metabolism as ceramide-activated phosphatase (Janssens and Goris, 2001, Nickels and Broach, 1996). Here we show that nutritional pathways impinge on the DDR by regulating PP2A, demonstrating a key role of PP2A in transducing metabolic signals to checkpoint kinases.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti Rad53 (clone EL7) (Dilution for western blot 1:5) | In house (Fiorani et al., 2008) | N/A |
| Rabbit polyclonal anti Mre11 (clone 263) (Dilution for western blot 1:15000) | In house (Ira et al., 2004) | N/A |
| Rabbit polyclonal anti Histone H2A phospho S129 (Dilution for western blot 1:500) | Abcam | Cat# ab15083; RRID: AB_301630 |
| Rabbit polyclonal anti Histone H2A (Dilution for western blot 1:3500) | Active Motif | Cat# 39235 |
| Mouse monoclonal anti c-MYC (clone 9E10) (Dilution for western blot 1:2000) | Santa Cruz Biotechnology | Cat# sc-40, RRID: AB_627268 |
| Mouse monoclonal anti-Viral V5-TAG (Clone SV5-Pk1) (Dilution for western blot 1:30000) | Bio-Rad | Cat# MCA1360G, RRID: AB_1172162 |
| Goat Anti-Mouse IgG (H + L)-HRP Conjugate (Dilution for western blot 1:15000) | Bio-Rad | Cat# 1706516 |
| Goat Anti-Rabbit IgG (H + L)-HRP Conjugate (Dilution for western blot 1:15000) | Bio-Rad | Cat# 1706515 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Hydroxyurea | Sigma | Cat# H8627 |
| Rapamycin | Sigma | Cat# R0395 |
| Metformin | Sigma | Cat# D150959 |
| Caffeine | Sigma | Cat# C8960 |
| Wortmannin | Sigma | Cat# W1628 |
| Okadayc acid | LC laboratories | Cat# O-2220 |
| C2 ceramide | Sigma | Cat# A7191 |
| Dihydroceramide C2 | Sigma | Cat# C7980 |
| Myriocin | Sigma | Cat# M1177 |
| Paraquat dichloride hydrate | Sigma | Cat# 36541 |
| Mersalyl | Sigma | Cat# M9784 |
| Tert-butyl hydroperoxide | Aldrich | Cat# 416665 |
| Terbinafine hydrochloride | Sigma | Cat# T8826 |
| Fluonazole | Sigma | Cat# F8929 |
| Cerulenin | Sigma | Cat# C2389 |
| Syringomycin E | Laboratory of Jon Takemoto | N/A |
| Fumonisin B1 | Enzo | Cat# BML-SL220 |
| L-Ethionine | Sigma | Cat# E1260 |
| Cycloleucine | Aldrich | Cat# A48105 |
| Deposited Data | ||
| Raw data: Metabolomics | This study; Mendeley data | http://dx.doi.org/10.17632/jfhwwhczx8.1 |
| Raw data: Synthetic Genetic Array (SGA) | This study; Mendeley data | http://dx.doi.org/10.17632/jfhwwhczx8.1 |
| Experimental Models: Organisms/Strains | ||
| All Saccharomyces cerevisiae yeast strains used in this study were W303 derivatives with the wild type RAD5 locus. They are listed in Table S1. | This study | N/A |
| MATa deletion mutant array (S288C) | OpenBiosystems | Cat#: YSC1053 |
| MATα query strain (S288C). See Table S1. | Laboratory of Charles Boone | N/A |
| Oligonucleotides | ||
| See Table S2 for a list of oligonucleotides used in this study. | This study | N/A |
| Software and Algorithms | ||
| MultiExperimentViewer 4.9.0 (MeV) | Saeed et al., 2003 | https://sourceforge.net/projects/mev-tm4/ |
| LIMMA (MeV module) | Smyth, 2004 | https://sourceforge.net/projects/mev-tm4/ |
| RStudio 1.0.136 | RStudio Team, 2016 | https://www.rstudio.com/products/rstudio/download2/ |
| pheatmap 1.0.8 (R package) | Kolde, 2015 | https://CRAN.R-project.org/package=pheatmap |
| HT Colony Grid Analyzer 1.1.7 | Collins et al., 2010 | https://sourceforge.net/projects/ht-col-measurer/ |
Contact for Reagents and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Marco Foiani (marco.foiani@ifom.eu).
Experimental Model and Subject Details
Yeast Strains
All yeast strains used in this study are listed in the Key Resources Table. All strains and genetic manipulations were verified by polymerase chain reaction (PCR) and phenotype. Gene deletions were carried out using either tetrad dissection or standard PCR-based strategies to amplify resistance cassettes with appropriate flanking sequences, and replacing the target gene by homologous recombination. Carboxy-terminal tags were similarly made with the PCR-based method to amplify resistance cassettes with flanking sequences. Primer sequences for gene deletions/carboxy-terminal tags and disruption/alteration controls are listed in the Key Resources Table or available upon request, respectively.
Method Details
Media and drug sensitivity assay
Unless otherwise stated, yeast strains were grown in yeast extract/peptone with 2% glucose (YPD). YPD agar plates were supplemented with adenine. Cells were synchronized in G1 with α-factor to a final concentration of 3 μg/ml. For drug sensitivity assay, cells were grown overnight. Serial 1:5 dilutions of stationary cultures were made and one drop of each dilution was pin-spotted onto agar plates, containing drugs. Plates were incubated for 2-3 days at 28°C.
For liquid drug sensitivity assay, yeast strains were grown in SD liquid medium at the initial concentration of 105 cell/ml in microtiter wells. Cultures were either left untreated (control-solvent) or were treated with the drug of interest. The absorbance (OD595) of untreated and treated cultures was measured after 12-18 hr. 3 independent repeats were performed. For ceramide experiments, we noticed a rapid response (few minutes for PP2A activity, within one hour for DDR activity).
Synthetic Genetic Array Screening
Synthetic genetic array (SGA) was carried out as described (Tong et al., 2001, Tong et al., 2004). Shortly, congenic irc21Δ (6 replicates) and wt (4 replicates) query strains were crossed with the haploid viable library (Tong et al., 2001). Colony sizes were quantified with the Colony Grid Analyzer (version 1.1.7) (Collins et al., 2010), and normalized to the intra-dish 80-percentile. A 1 x standard error separation of normalized wt and irc21Δ colony sizes was used to call candidate hits for negative interactions. A 10% increase of irc21Δ colony sizes in relation to the irc21Δ 80-percentile and the respective normalized wt colony size, and a 1 x standard error separation from the irc21Δ 80-percentile were used to call candidates for rescue interactions.
Tetrad dissection and random spore analysis
Standard procedures were used for tetrad dissection and random spore analysis (Abdullah and Borts, 2001, Tong and Boone, 2006). Mata and Matα cells were mixed on rich medium and allowed to mate at 28°C. Sporulation was induced at 23°C by transfer to VB sporulation medium. Ascal walls were removed by digestion with zymolase at 37°C. The four spores of each tetrad were separated from each other, germinated on YPD, and allowed to grow before scoring for genetic markers. For random spore analysis, a small amount of spores was inoculated in water and plated on marker selection plates.
Western Blotting
Protein extracts for western blotting were prepared following cell fixation using trichloroacetic acid and analyzed by SDS-polyacrylamide gel electrophoresis. Briefly, cells were quickly spun down and the pellet was resuspended in 20% TCA and lysed by bead beating. Lysate and precipitate/debris was mixed with 200 μL 5% TCA and pelleted. The pellet was resuspended in 100 μL Laemli buffer 1X (β-mercaptoethanol as reducing agent) and 100 μL Tris base 1M, boiled for 5 min. After centrifugation, the supernatant was transferred in new tubes.
Antibodies used for detection are listed in the Key Resources Table. Detection was done through electrogenerated chemiluminescence (ECL, GE- Healthcare).
Fluorescence-activated cell sorting (FACS) analysis
Cell cycle analysis was conducted as previously described (Pellicioli et al., 1999).
Cells were spun down and the pellet was resuspended in 250mM Tris-HCl pH 7.5, 70% ethanol and kept on ice for at least 30 min. The pellet was resuspended in 50mM Tris-HCl pH 7.5 and RNase A 1mg/ml and incubated at 37°C. The pellet was resuspended in 200mM TRIS-HCl pH 7.5, 200mM NaCl, 78mM MgCl2, Propidium Iodide. Before cytofluorimetric analysis, cells were diluted in Tris-HCl pH 7.5 in tubes and sonicated.
Budding index analysis
After sonication, cells were fixed by the addition of 3.7% formaldehyde and 0.9% NaCl. Cells were examined under a light microscope, by counting 200 cells per time point.
Measurement of intracellular ROS
ROS measurement was conducted as previously described (Rand and Grant, 2006). Shortly, exponentially growing cells were harvested and resuspended in PBS containing the oxidant-sensitive probe 2′,7′-dichlorofluorescin diacetate (DCFH-DA). Cells were broken with glass beads and fluorescence was measured with excitation and emission wavelengths of 485 and 528 nm, respectively.
Measurement of oxygen consumption
Respiration of log-phase S. cerevisiae cells was measured by polarographic analysis using a Clark’s type oxygen electrode (Hansatech Instrument Ltd, Pentney UK) according to standard procedures, upon addition of dinitrophenol as uncoupling agent of respiration/ATP synthesis.
Search for suppressors of mec1-100 sensitivity to HU
To search for suppressor mutations of the HU-sensitivity of mec1-100 mutant, 1x106 mec1-100 cells were plated on YEPD in the presence of 25mM HU. Survivors were crossed to wt cells to identify by tetrad analysis that the suppression events were due to single-gene mutations. Subsequent genetic analyses allowed grouping the single-gene suppression events in 4 classes. The class that showed the most efficient suppression was chosen and the mutations altering open reading frames within the reference S. cerevisiae genome were identified by next-generation Illumina sequencing (IGA technology services). To confirm that the tap42-G360R mutation was responsible for the suppression, a URA3 gene was integrated downstream of the tap42-G360R stop codon and the resulting strain was crossed to wt cells to verify by tetrad dissection that the suppression of the mec1-100 HU sensitivity co-segregated with the URA3 allele.
Metabolic analysis
Yeast cells were grown to logarithmic phase (1x107 cells/mL) in YPD medium in 6 replicates. 5x108 cells per replicate were harvested by centrifugation, washed in water, snap-frozen in liquid nitrogen and stored at −80°C. Metabolite extraction and Ultrahigh Performance Liquid Chromatography-Tandem Mass Spectroscopy analysis of 484 metabolites were performed by Metabolon (Durham, North Carolina) as previously described (Chaudhri et al., 2013). Missing metabolite raw intensity values were filled in with the lowest detectable intensity of the respective metabolite, and all raw intensities were normalized to the median intensity of the respective replicate. Fold changes and significant alterations were calculated with the LIMMA method implemented in MultiExperiment Viewer (MeV) software (version 4.9.0) (Saeed et al., 2003, Smyth, 2004) using an adjusted p value of 0.05 and a minimum fold-change of 1.3.
TrueMass Ceramide Panel
Metabolites were isolated from cell pellets by sequential chloroform/methanol extraction and aqueous potassium chloride liquid-liquid extraction. The chloroform/methanol solution contained internal standards (Cer12:0, Cer19:0, dhCer12:0, hexCer12:0, [Avanti Polar Lipids, Alabaster, AL]) The organic layer was evaporated in a stream of nitrogen, reconstituted and subjected to a solid phase extraction clean up step on silica [Si, 100 mg, Supelco, Bellefonte, PA]. The ceramide fraction was eluted, evaporated in a stream of nitrogen, reconstituted and an aliquot was injected onto an AB Sciex 4000 QTRAP (Sciex, Foster City, CA)/Acquity (Waters, Milford, MA) LC-MS/MS system equipped with a reversed phase UHPLC column [Zorbax Eclipse Plus C8, 2.1 × 150 mm, 1.8 μm, Agilent Technologies] using a gradient of 2mM ammonium formate/0.2% formic acid in water and 1 mM ammonium formate/0.2% formic acid in Acetonitrile:Isopropanol (60:40). The mass spectrometer was operated in MRM mode using positive electrospray ionization.
The peak areas of the analyte fragment ions were measured against the peak area of the respective fragment ions of the corresponding internal standards. For the purposes of this panel, the fragment ion m/z 264 was used for ceramides with a sphingosine backbone and the m/z 266 fragment was used for analytes with a sphinganine backbone. Quantitation was based on a series of five calibration standard samples that were included in each run. Calibration standards contained 26 reference compounds. For analytes for which calibration standards were not commercially available, a surrogate analyte from the same compound class was used for quantitation (e.g., quantitation of CER 22:1 is based on CER 20:0 calibration standards). A total of 56 analytes covering ceramides, dihydroceramides, hexosylceramides and lactosylceramides with different fatty acid composition (14:0, 16:0, 18:0, 18:1, 20:0, 20:1, 22:0, 22:1, 24:0, 24:1, 26:0, 26:1) were determined.
Quantification and Statistical Analysis
Interactome correlation analysis
Interactome correlation analysis of irc21Δ was performed with published genome-wide SGA scores (Costanzo et al., 2010). The genetic interaction scores between 3884 array and 1712 query strains were used to calculate correlation coefficients (R) of the query strain irc21Δ with all other query strains, using the CORREL function in Microsoft Excel 2013 (Figure 3A). The pairwise correlation coefficients (R) between PP2A subunits were calculated accordingly (Figure 3B). Genetic interactions underlying the interactome similarity between irc21Δ, rrd1Δ and tip41Δ were derived from the intermediate cutoff interactor table in (Costanzo et al., 2010; Figure 3C). Interactors were included if either rrd1Δ or tip41Δ showed the same interaction type as irc21Δ. Unsupervised hierarchical clustering and heatmap representation of correlation coefficients and genetic interaction scores were done in MeV.
Metabolome Analysis
Unsupervised hierarchical clustering of metabolome samples by Pearson correlation coefficient and heatmap representation were performed in R using the pheatmap library (version 1.0.8). Significance analysis was performed with LIMMA implemented in MeV as described above. Significances of the intersections of metabolite alterations were calculated by chi-square test. Heatmaps for visualization of altered metabolite classes were generated with MeV.
Statistical tests
The statistical significance was assessed using the indicated assays. Student’s t test was used for pairwise comparisons. ANOVA was used for comparisons of multiple groups. Number of replicates (n) and p values are specified in corresponding Figure legends.
Data and Software Availability
Raw data have been deposited to Mendeley Data and are available at http://dx.doi.org/10.17632/jfhwwhczx8.1.
Author Contributions
Conceptualization, E.F. and M.F.; Methodology, E.F. and M.F.; Formal Analysis, C.B. and W.V.C.; Investigation, E.F., M.P., C.C., S.M., and M.P.L.; Resources, E.F., C.B., M.E., G.S., C.L., R.B., and M.V.; Writing – Original Draft, E.F. and M.F.; Writing – Review & Editing, E.F., M.F., and C.B.; Visualization, E.F. and C.B.; Supervision, M.F.; Funding Acquisition, M.F.
Acknowledgments
We thank Jon Takemoto (Utah State University) for providing syringomycin; Marco Giorgio (IEO), Raluca Marcu (IEO), and Rachel Jossen (IFOM) for support during the experiments; and Sunil Laxman (INSTEM), Davide Cittaro (IFOM), Gianluca Deflorian (IFOM), and Lucilla Luzi (IEO) for helpful discussions. The work was supported by grants from the Associazione Italiana per la Ricerca sul Cancro (IG 16770 to M.F. and IG 15210 to M.P.L.), the European Union and Telethon-Italy (GGP12171 to M.F.), and Progetti di Ricerca di Interesse Nazionale (PRIN) 2015 (to M.F. and M.P.L). C.C. was supported by a fellowship from the Fondazione Italiana per la Ricerca sul Cancro (18125). C.B. was supported by a fellowship by Associazione Italiana per la Ricerca sul Cancro (AIRC) Fellowship i-Care (Marie Curie co-funded by the European Union)—16173.
Published: June 22, 2017
Footnotes
Supplemental Information includes six figures and two tables and can be found with this article online at http://dx.doi.org/10.1016/j.molcel.2017.05.027.
Supplemental Information
References
- Abdullah M.F., Borts R.H. Meiotic recombination frequencies are affected by nutritional states in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 2001;98:14524–14529. doi: 10.1073/pnas.201529598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvaro D., Lisby M., Rothstein R. Genome-wide analysis of Rad52 foci reveals diverse mechanisms impacting recombination. PLoS Genet. 2007;3:e228. doi: 10.1371/journal.pgen.0030228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awasthi P., Foiani M., Kumar A. ATM and ATR signaling at a glance. J. Cell Sci. 2016;129:1285. doi: 10.1242/jcs.188631. [DOI] [PubMed] [Google Scholar]
- Awaya J., Ohno T., Ohno H., Omura S. Substitution of cellular fatty acids in yeast cells by the antibiotic cerulenin and exogenous fatty acids. Biochim. Biophys. Acta. 1975;409:267–273. doi: 10.1016/0005-2760(75)90022-3. [DOI] [PubMed] [Google Scholar]
- Bartek J., Lukas J. DNA damage checkpoints: from initiation to recovery or adaptation. Curr. Opin. Cell Biol. 2007;19:238–245. doi: 10.1016/j.ceb.2007.02.009. [DOI] [PubMed] [Google Scholar]
- Bashkirov V.I., Bashkirova E.V., Haghnazari E., Heyer W.D. Direct kinase-to-kinase signaling mediated by the FHA phosphoprotein recognition domain of the Dun1 DNA damage checkpoint kinase. Mol. Cell. Biol. 2003;23:1441–1452. doi: 10.1128/MCB.23.4.1441-1452.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzi M., Mantiero D., Trovesi C., Lucchini G., Longhese M.P. Dephosphorylation of gamma H2A by Glc7/protein phosphatase 1 promotes recovery from inhibition of DNA replication. Mol. Cell. Biol. 2010;30:131–145. doi: 10.1128/MCB.01000-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermejo R., Capra T., Jossen R., Colosio A., Frattini C., Carotenuto W., Cocito A., Doksani Y., Klein H., Gómez-González B. The replication checkpoint protects fork stability by releasing transcribed genes from nuclear pores. Cell. 2011;146:233–246. doi: 10.1016/j.cell.2011.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardi P., Azzone G.F. Cytochrome c as an electron shuttle between the outer and inner mitochondrial membranes. J. Biol. Chem. 1981;256:7187–7192. [PubMed] [Google Scholar]
- Chaudhri V.K., Salzler G.G., Dick S.A., Buckman M.S., Sordella R., Karoly E.D., Mohney R., Stiles B.M., Elemento O., Altorki N.K., McGraw T.E. Metabolic alterations in lung cancer-associated fibroblasts correlated with increased glycolytic metabolism of the tumor. Mol. Cancer Res. 2013;11:579–592. doi: 10.1158/1541-7786.MCR-12-0437-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Lai Y., Zhu X., Ma L., Bai Q., Vazquez I., Xiao Y., Liu C., Li D., Gao C. Retraction. J. Cell Sci. 2015;128:421. [Google Scholar]
- Cochemé H.M., Murphy M.P. Chapter 22. The uptake and interactions of the redox cycler paraquat with mitochondria. Methods Enzymol. 2009;456:395–417. doi: 10.1016/S0076-6879(08)04422-4. [DOI] [PubMed] [Google Scholar]
- Collins S.R., Roguev A., Krogan N.J. Quantitative genetic interaction mapping using the E-MAP approach. Methods Enzymol. 2010;470:205–231. doi: 10.1016/S0076-6879(10)70009-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M., Baryshnikova A., Bellay J., Kim Y., Spear E.D., Sevier C.S., Ding H., Koh J.L., Toufighi K., Mostafavi S. The genetic landscape of a cell. Science. 2010;327:425–431. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson J.F., Whyte B., Bissinger P.H., Schiestl R.H. Oxidative stress is involved in heat-induced cell death in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 1996;93:5116–5121. doi: 10.1073/pnas.93.10.5116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desany B.A., Alcasabas A.A., Bachant J.B., Elledge S.J. Recovery from DNA replicational stress is the essential function of the S-phase checkpoint pathway. Genes Dev. 1998;12:2956–2970. doi: 10.1101/gad.12.18.2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Como C.J., Arndt K.T. Nutrients, via the Tor proteins, stimulate the association of Tap42 with type 2A phosphatases. Genes Dev. 1996;10:1904–1916. doi: 10.1101/gad.10.15.1904. [DOI] [PubMed] [Google Scholar]
- Dozier C., Bonyadi M., Baricault L., Tonasso L., Darbon J.M. Regulation of Chk2 phosphorylation by interaction with protein phosphatase 2A via its B′ regulatory subunit. Biol. Cell. 2004;96:509–517. doi: 10.1016/j.biolcel.2004.04.010. [DOI] [PubMed] [Google Scholar]
- Düvel K., Santhanam A., Garrett S., Schneper L., Broach J.R. Multiple roles of Tap42 in mediating rapamycin-induced transcriptional changes in yeast. Mol. Cell. 2003;11:1467–1478. doi: 10.1016/s1097-2765(03)00228-4. [DOI] [PubMed] [Google Scholar]
- El Bawab S., Birbes H., Roddy P., Szulc Z.M., Bielawska A., Hannun Y.A. Biochemical characterization of the reverse activity of rat brain ceramidase. A CoA-independent and fumonisin B1-insensitive ceramide synthase. J. Biol. Chem. 2001;276:16758–16766. doi: 10.1074/jbc.M009331200. [DOI] [PubMed] [Google Scholar]
- Fay D.S., Sun Z., Stern D.F. Mutations in SPK1/RAD53 that specifically abolish checkpoint but not growth-related functions. Curr. Genet. 1997;31:97–105. doi: 10.1007/s002940050181. [DOI] [PubMed] [Google Scholar]
- Fiorani S., Mimun G., Caleca L., Piccini D., Pellicioli A. Characterization of the activation domain of the Rad53 checkpoint kinase. Cell Cycle. 2008;7:493–499. doi: 10.4161/cc.7.4.5323. [DOI] [PubMed] [Google Scholar]
- Freeman A.K., Monteiro A.N. Phosphatases in the cellular response to DNA damage. Cell Commun. Signal. 2010;8:27. doi: 10.1186/1478-811X-8-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego O., Betts M.J., Gvozdenovic-Jeremic J., Maeda K., Matetzki C., Aguilar-Gurrieri C., Beltran-Alvarez P., Bonn S., Fernández-Tornero C., Jensen L.J. A systematic screen for protein-lipid interactions in Saccharomyces cerevisiae. Mol. Syst. Biol. 2010;6:430. doi: 10.1038/msb.2010.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girotti A.W. Lipid hydroperoxide generation, turnover, and effector action in biological systems. J. Lipid Res. 1998;39:1529–1542. [PubMed] [Google Scholar]
- Goodarzi A.A., Jonnalagadda J.C., Douglas P., Young D., Ye R., Moorhead G.B., Lees-Miller S.P., Khanna K.K. Autophosphorylation of ataxia-telangiectasia mutated is regulated by protein phosphatase 2A. EMBO J. 2004;23:4451–4461. doi: 10.1038/sj.emboj.7600455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan K., Deschenes R.J., Dixon J.E. Isolation and characterization of a second protein tyrosine phosphatase gene, PTP2, from Saccharomyces cerevisiae. J. Biol. Chem. 1992;267:10024–10030. [PubMed] [Google Scholar]
- Guénolé A., Srivas R., Vreeken K., Wang Z.Z., Wang S., Krogan N.J., Ideker T., van Attikum H. Dissection of DNA damage responses using multiconditional genetic interaction maps. Mol. Cell. 2013;49:346–358. doi: 10.1016/j.molcel.2012.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haak D., Gable K., Beeler T., Dunn T. Hydroxylation of Saccharomyces cerevisiae ceramides requires Sur2p and Scs7p. J. Biol. Chem. 1997;272:29704–29710. doi: 10.1074/jbc.272.47.29704. [DOI] [PubMed] [Google Scholar]
- Harrison J.C., Haber J.E. Surviving the breakup: the DNA damage checkpoint. Annu. Rev. Genet. 2006;40:209–235. doi: 10.1146/annurev.genet.40.051206.105231. [DOI] [PubMed] [Google Scholar]
- Hilton B.A., Li Z., Musich P.R., Wang H., Cartwright B.M., Serrano M., Zhou X.Z., Lu K.P., Zou Y. ATR plays a direct antiapoptotic role at mitochondria, which is regulated by prolyl isomerase Pin1. Mol. Cell. 2015;60:35–46. doi: 10.1016/j.molcel.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M., Zhou Z., Elledge S.J. The DNA replication and damage checkpoint pathways induce transcription by inhibition of the Crt1 repressor. Cell. 1998;94:595–605. doi: 10.1016/s0092-8674(00)81601-3. [DOI] [PubMed] [Google Scholar]
- Huang X., Liu J., Dickson R.C. Down-regulating sphingolipid synthesis increases yeast lifespan. PLoS Genet. 2012;8:e1002493. doi: 10.1371/journal.pgen.1002493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes Hallett J.E., Luo X., Capaldi A.P. State transitions in the TORC1 signaling pathway and information processing in Saccharomyces cerevisiae. Genetics. 2014;198:773–786. doi: 10.1534/genetics.114.168369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh W.K., Falvo J.V., Gerke L.C., Carroll A.S., Howson R.W., Weissman J.S., O’Shea E.K. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- Hustedt N., Seeber A., Sack R., Tsai-Pflugfelder M., Bhullar B., Vlaming H., van Leeuwen F., Guénolé A., van Attikum H., Srivas R. Yeast PP4 interacts with ATR homolog Ddc2-Mec1 and regulates checkpoint signaling. Mol. Cell. 2015;57:273–289. doi: 10.1016/j.molcel.2014.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ira G., Pellicioli A., Balijja A., Wang X., Fiorani S., Carotenuto W., Liberi G., Bressan D., Wan L., Hollingsworth N.M. DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature. 2004;431:1011–1017. doi: 10.1038/nature02964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens V., Goris J. Protein phosphatase 2A: a highly regulated family of serine/threonine phosphatases implicated in cell growth and signalling. Biochem. J. 2001;353:417–439. doi: 10.1042/0264-6021:3530417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y. Regulation of the cell cycle by protein phosphatase 2A in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 2006;70:440–449. doi: 10.1128/MMBR.00049-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keogh M.C., Kim J.A., Downey M., Fillingham J., Chowdhury D., Harrison J.C., Onishi M., Datta N., Galicia S., Emili A. A phosphatase complex that dephosphorylates gammaH2AX regulates DNA damage checkpoint recovery. Nature. 2006;439:497–501. doi: 10.1038/nature04384. [DOI] [PubMed] [Google Scholar]
- Koh J.L., Chong Y.T., Friesen H., Moses A., Boone C., Andrews B.J., Moffat J. CYCLoPs: a comprehensive database constructed from automated analysis of protein abundance and subcellular localization patterns in Saccharomyces cerevisiae. G3 (Bethesda) 2015;5:1223–1232. doi: 10.1534/g3.115.017830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolaczkowski M., Kolaczkowska A., Gaigg B., Schneiter R., Moye-Rowley W.S. Differential regulation of ceramide synthase components LAC1 and LAG1 in Saccharomyces cerevisiae. Eukaryot. Cell. 2004;3:880–892. doi: 10.1128/EC.3.4.880-892.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolde, R. (2015). pheatmap: Pretty Heatmaps. R Package Version 1.0.8. https://CRAN.R-project.org/package=pheatmap.
- Kontoyiannis D.P. Modulation of fluconazole sensitivity by the interaction of mitochondria and erg3p in Saccharomyces cerevisiae. J. Antimicrob. Chemother. 2000;46:191–197. doi: 10.1093/jac/46.2.191. [DOI] [PubMed] [Google Scholar]
- Kumar A., Mazzanti M., Mistrik M., Kosar M., Beznoussenko G.V., Mironov A.A., Garrè M., Parazzoli D., Shivashankar G.V., Scita G. ATR mediates a checkpoint at the nuclear envelope in response to mechanical stress. Cell. 2014;158:633–646. doi: 10.1016/j.cell.2014.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb D.C., Kelly D.E., Manning N.J., Kaderbhai M.A., Kelly S.L. Biodiversity of the P450 catalytic cycle: yeast cytochrome b5/NADH cytochrome b5 reductase complex efficiently drives the entire sterol 14-demethylation (CYP51) reaction. FEBS Lett. 1999;462:283–288. doi: 10.1016/s0014-5793(99)01548-3. [DOI] [PubMed] [Google Scholar]
- Laxman S., Sutter B.M., Shi L., Tu B.P. Npr2 inhibits TORC1 to prevent inappropriate utilization of glutamine for biosynthesis of nitrogen-containing metabolites. Sci. Signal. 2014;7:ra120. doi: 10.1126/scisignal.2005948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K.S., Hines L.K., Levin D.E. A pair of functionally redundant yeast genes (PPZ1 and PPZ2) encoding type 1-related protein phosphatases function within the PKC1-mediated pathway. Mol. Cell. Biol. 1993;13:5843–5853. doi: 10.1128/mcb.13.9.5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.J., Schwartz M.F., Duong J.K., Stern D.F. Rad53 phosphorylation site clusters are important for Rad53 regulation and signaling. Mol. Cell. Biol. 2003;23:6300–6314. doi: 10.1128/MCB.23.17.6300-6314.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W., St Onge R.P., Proctor M., Flaherty P., Jordan M.I., Arkin A.P., Davis R.W., Nislow C., Giaever G. Genome-wide requirements for resistance to functionally distinct DNA-damaging agents. PLoS Genet. 2005;1:e24. doi: 10.1371/journal.pgen.0010024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroy C., Lee S.E., Vaze M.B., Ochsenbein F., Guerois R., Haber J.E., Marsolier-Kergoat M.C. PP2C phosphatases Ptc2 and Ptc3 are required for DNA checkpoint inactivation after a double-strand break. Mol. Cell. 2003;11:827–835. doi: 10.1016/s1097-2765(03)00058-3. [DOI] [PubMed] [Google Scholar]
- Loewith R., Hall M.N. Target of rapamycin (TOR) in nutrient signaling and growth control. Genetics. 2011;189:1177–1201. doi: 10.1534/genetics.111.133363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewith R., Jacinto E., Wullschleger S., Lorberg A., Crespo J.L., Bonenfant D., Oppliger W., Jenoe P., Hall M.N. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol. Cell. 2002;10:457–468. doi: 10.1016/s1097-2765(02)00636-6. [DOI] [PubMed] [Google Scholar]
- Luke M.M., Della Seta F., Di Como C.J., Sugimoto H., Kobayashi R., Arndt K.T. The SAP, a new family of proteins, associate and function positively with the SIT4 phosphatase. Mol. Cell. Biol. 1996;16:2744–2755. doi: 10.1128/mcb.16.6.2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao C., Xu R., Bielawska A., Obeid L.M. Cloning of an alkaline ceramidase from Saccharomyces cerevisiae. An enzyme with reverse (CoA-independent) ceramide synthase activity. J. Biol. Chem. 2000;275:6876–6884. doi: 10.1074/jbc.275.10.6876. [DOI] [PubMed] [Google Scholar]
- Mao C., Xu R., Bielawska A., Szulc Z.M., Obeid L.M. Cloning and characterization of a Saccharomyces cerevisiae alkaline ceramidase with specificity for dihydroceramide. J. Biol. Chem. 2000;275:31369–31378. doi: 10.1074/jbc.M003683200. [DOI] [PubMed] [Google Scholar]
- Matsuoka S., Ballif B.A., Smogorzewska A., McDonald E.R., 3rd, Hurov K.E., Luo J., Bakalarski C.E., Zhao Z., Solimini N., Lerenthal Y. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- Mitchell A.G., Martin C.E. Fah1p, a Saccharomyces cerevisiae cytochrome b5 fusion protein, and its Arabidopsis thaliana homolog that lacks the cytochrome b5 domain both function in the alpha-hydroxylation of sphingolipid-associated very long chain fatty acids. J. Biol. Chem. 1997;272:28281–28288. doi: 10.1074/jbc.272.45.28281. [DOI] [PubMed] [Google Scholar]
- Nakahata S., Morishita K. PP2A inactivation by ROS accumulation. Blood. 2014;124:2163–2165. doi: 10.1182/blood-2014-08-594093. [DOI] [PubMed] [Google Scholar]
- Neklesa T.K., Davis R.W. Superoxide anions regulate TORC1 and its ability to bind Fpr1:rapamycin complex. Proc. Natl. Acad. Sci. USA. 2008;105:15166–15171. doi: 10.1073/pnas.0807712105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickels J.T., Broach J.R. A ceramide-activated protein phosphatase mediates ceramide-induced G1 arrest of Saccharomyces cerevisiae. Genes Dev. 1996;10:382–394. doi: 10.1101/gad.10.4.382. [DOI] [PubMed] [Google Scholar]
- O’Neill B.M., Szyjka S.J., Lis E.T., Bailey A.O., Yates J.R., 3rd, Aparicio O.M., Romesberg F.E. Pph3-Psy2 is a phosphatase complex required for Rad53 dephosphorylation and replication fork restart during recovery from DNA damage. Proc. Natl. Acad. Sci. USA. 2007;104:9290–9295. doi: 10.1073/pnas.0703252104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh C.S., Toke D.A., Mandala S., Martin C.E. ELO2 and ELO3, homologues of the Saccharomyces cerevisiae ELO1 gene, function in fatty acid elongation and are required for sphingolipid formation. J. Biol. Chem. 1997;272:17376–17384. doi: 10.1074/jbc.272.28.17376. [DOI] [PubMed] [Google Scholar]
- Paciotti V., Clerici M., Scotti M., Lucchini G., Longhese M.P. Characterization of mec1 kinase-deficient mutants and of new hypomorphic mec1 alleles impairing subsets of the DNA damage response pathway. Mol. Cell. Biol. 2001;21:3913–3925. doi: 10.1128/MCB.21.12.3913-3925.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellicioli A., Lucca C., Liberi G., Marini F., Lopes M., Plevani P., Romano A., Di Fiore P.P., Foiani M. Activation of Rad53 kinase in response to DNA damage and its effect in modulating phosphorylation of the lagging strand DNA polymerase. EMBO J. 1999;18:6561–6572. doi: 10.1093/emboj/18.22.6561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petranyi G., Ryder N.S., Stütz A. Allylamine derivatives: new class of synthetic antifungal agents inhibiting fungal squalene epoxidase. Science. 1984;224:1239–1241. doi: 10.1126/science.6547247. [DOI] [PubMed] [Google Scholar]
- Qiu J., Qian Y., Frank P., Wintersberger U., Shen B. Saccharomyces cerevisiae RNase H(35) functions in RNA primer removal during lagging-strand DNA synthesis, most efficiently in cooperation with Rad27 nuclease. Mol. Cell. Biol. 1999;19:8361–8371. doi: 10.1128/mcb.19.12.8361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran V., Herman P.K. Antagonistic interactions between the cAMP-dependent protein kinase and Tor signaling pathways modulate cell growth in Saccharomyces cerevisiae. Genetics. 2011;187:441–454. doi: 10.1534/genetics.110.123372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand J.D., Grant C.M. The thioredoxin system protects ribosomes against stress-induced aggregation. Mol. Biol. Cell. 2006;17:387–401. doi: 10.1091/mbc.E05-06-0520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rempola B., Kaniak A., Migdalski A., Rytka J., Slonimski P.P., di Rago J.P. Functional analysis of RRD1 (YIL153w) and RRD2 (YPL152w), which encode two putative activators of the phosphotyrosyl phosphatase activity of PP2A in Saccharomyces cerevisiae. Mol. Gen. Genet. 2000;262:1081–1092. doi: 10.1007/pl00008651. [DOI] [PubMed] [Google Scholar]
- Rossi S.E., Ajazi A., Carotenuto W., Foiani M., Giannattasio M. Rad53-mediated regulation of Rrm3 and Pif1 DNA helicases contributes to prevention of aberrant fork transitions under replication stress. Cell Rep. 2015;13:80–92. doi: 10.1016/j.celrep.2015.08.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RStudio Team. (2016). RStudio: Integrated Development for RStudio. http://www.rstudio.com/.
- Saeed A.I., Sharov V., White J., Li J., Liang W., Bhagabati N., Braisted J., Klapa M., Currier T., Thiagarajan M. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- Sanchez Y., Desany B.A., Jones W.J., Liu Q., Wang B., Elledge S.J. Regulation of RAD53 by the ATM-like kinases MEC1 and TEL1 in yeast cell cycle checkpoint pathways. Science. 1996;271:357–360. doi: 10.1126/science.271.5247.357. [DOI] [PubMed] [Google Scholar]
- Slater M.L. Effect of reversible inhibition of deoxyribonucleic acid synthesis on the yeast cell cycle. J. Bacteriol. 1973;113:263–270. doi: 10.1128/jb.113.1.263-270.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth G.K. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- Sogo J.M., Lopes M., Foiani M. Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science. 2002;297:599–602. doi: 10.1126/science.1074023. [DOI] [PubMed] [Google Scholar]
- Staschke K.A., Dey S., Zaborske J.M., Palam L.R., McClintick J.N., Pan T., Edenberg H.J., Wek R.C. Integration of general amino acid control and target of rapamycin (TOR) regulatory pathways in nitrogen assimilation in yeast. J. Biol. Chem. 2010;285:16893–16911. doi: 10.1074/jbc.M110.121947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock S.D., Hama H., Radding J.A., Young D.A., Takemoto J.Y. Syringomycin E inhibition of Saccharomyces cerevisiae: requirement for biosynthesis of sphingolipids with very-long-chain fatty acids and mannose- and phosphoinositol-containing head groups. Antimicrob. Agents Chemother. 2000;44:1174–1180. doi: 10.1128/aac.44.5.1174-1180.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z., Fay D.S., Marini F., Foiani M., Stern D.F. Spk1/Rad53 is regulated by Mec1-dependent protein phosphorylation in DNA replication and damage checkpoint pathways. Genes Dev. 1996;10:395–406. doi: 10.1101/gad.10.4.395. [DOI] [PubMed] [Google Scholar]
- Sutter B.M., Wu X., Laxman S., Tu B.P. Methionine inhibits autophagy and promotes growth by inducing the SAM-responsive methylation of PP2A. Cell. 2013;154:403–415. doi: 10.1016/j.cell.2013.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemoto J.Y., Yu Y., Stock S.D., Miyakawa T. Yeast genes involved in growth inhibition by Pseudomonas syringae pv. syringae syringomycin family lipodepsipeptides. FEMS Microbiol. Lett. 1993;114:339–342. doi: 10.1111/j.1574-6968.1993.tb06595.x. [DOI] [PubMed] [Google Scholar]
- Thomas B.J., Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989;56:619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- Tong A.H., Boone C. Synthetic genetic array analysis in Saccharomyces cerevisiae. Methods Mol. Biol. 2006;313:171–192. doi: 10.1385/1-59259-958-3:171. [DOI] [PubMed] [Google Scholar]
- Tong A.H., Evangelista M., Parsons A.B., Xu H., Bader G.D., Pagé N., Robinson M., Raghibizadeh S., Hogue C.W., Bussey H. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294:2364–2368. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- Tong A.H., Lesage G., Bader G.D., Ding H., Xu H., Xin X., Young J., Berriz G.F., Brost R.L., Chang M. Global mapping of the yeast genetic interaction network. Science. 2004;303:808–813. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- Torres J.Z., Schnakenberg S.L., Zakian V.A. Saccharomyces cerevisiae Rrm3p DNA helicase promotes genome integrity by preventing replication fork stalling: viability of rrm3 cells requires the intra-S-phase checkpoint and fork restart activities. Mol. Cell. Biol. 2004;24:3198–3212. doi: 10.1128/MCB.24.8.3198-3212.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban J., Soulard A., Huber A., Lippman S., Mukhopadhyay D., Deloche O., Wanke V., Anrather D., Ammerer G., Riezman H. Sch9 is a major target of TORC1 in Saccharomyces cerevisiae. Mol. Cell. 2007;26:663–674. doi: 10.1016/j.molcel.2007.04.020. [DOI] [PubMed] [Google Scholar]
- Usui T., Ogawa H., Petrini J.H. A DNA damage response pathway controlled by Tel1 and the Mre11 complex. Mol. Cell. 2001;7:1255–1266. doi: 10.1016/s1097-2765(01)00270-2. [DOI] [PubMed] [Google Scholar]
- Van Hoof C., Martens E., Longin S., Jordens J., Stevens I., Janssens V., Goris J. Specific interactions of PP2A and PP2A-like phosphatases with the yeast PTPA homologues, Ypa1 and Ypa2. Biochem. J. 2005;386:93–102. doi: 10.1042/BJ20040887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergéres G., Waskell L. Cytochrome b5, its functions, structure and membrane topology. Biochimie. 1995;77:604–620. doi: 10.1016/0300-9084(96)88176-4. [DOI] [PubMed] [Google Scholar]
- Wu W.I., McDonough V.M., Nickels J.T., Jr., Ko J., Fischl A.S., Vales T.R., Merrill A.H., Jr., Carman G.M. Regulation of lipid biosynthesis in Saccharomyces cerevisiae by fumonisin B1. J. Biol. Chem. 1995;270:13171–13178. doi: 10.1074/jbc.270.22.13171. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Zhao S., Long F., Zhang L., Bai G., Shima H., Nagao M., Lee E.Y. A mutant of protein phosphatase-1 that exhibits altered toxin sensitivity. J. Biol. Chem. 1994;269:16997–17000. [PubMed] [Google Scholar]
- Zhao X., Muller E.G., Rothstein R. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol. Cell. 1998;2:329–340. doi: 10.1016/s1097-2765(00)80277-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.