Abstract
The outgrowth of new dendritic spines is closely linked to the formation of new synapses, and is thought to be a vital component of the experience-dependent circuit plasticity that supports learning. Here, we examined the role of the RhoGEF Ephexin5 in driving activity-dependent spine outgrowth. We found that reducing Ephexin5 levels increased spine outgrowth, and increasing Ephexin5 levels decreased spine outgrowth in a GEF-dependent manner, suggesting that Ephexin5 acts as an inhibitor of spine outgrowth. Notably, we found that increased neural activity led to a proteasome-dependent reduction in the levels of Ephexin5 in neuronal dendrites, which could facilitate the enhanced spine outgrowth observed following increased neural activity. Surprisingly, we also found that Ephexin5-GFP levels were elevated on the dendrite at sites of future new spines, prior to new spine outgrowth. Moreover, lowering neuronal Ephexin5 levels inhibited new spine outgrowth in response to both global increases in neural activity and local glutamatergic stimulation of the dendrite, suggesting that Ephexin5 is necessary for activity-dependent spine outgrowth. Our data support a model in which Ephexin5 serves a dual role in spinogenesis, acting both as a brake on overall spine outgrowth and as a necessary component in the site-specific formation of new spines.
Keywords: dendritic spine, Ephexin5, activity-dependent, hippocampus, structural plasticity, proteasome
INTRODUCTION
Enhanced growth and stabilization of new spines has been closely linked with the acquisition of new skills (Xu et al., 2009; Yang et al., 2009; Roberts et al., 2010) and adaptation to changes in sensory input (Trachtenberg et al., 2002; Holtmaat et al., 2006; Hofer et al., 2009), supporting that new spine growth and stabilization is vital for experience-dependent plasticity. Indeed, nascent spines rapidly mature functionally (Zito et al., 2009; Kwon and Sabatini, 2011) and stabilize in response to defined patterns of neural activity (Hill and Zito, 2013), consistent with new spines serving as a major source of synaptic plasticity. Furthermore, targeted disruption of spines that grow during skill learning leads to loss of the newly acquired skill (Hayashi-Takagi et al., 2015), demonstrating that spine growth is necessary for learning. Thus, understanding the signaling mechanisms which induce the growth and stabilization of new dendritic spines is a vital step towards understanding how neural circuits are modified during experience-dependent plasticity.
Several studies have convincingly demonstrated that elevating synaptic or network activity leads to enhanced spine outgrowth (Engert and Bonhoeffer, 1999; Maletic-Savatic et al., 1999; Jourdain et al., 2003; Kato-Negishi et al., 2003; Hamilton et al., 2012). The signaling pathways linking enhanced neural activity to new spine growth are currently being elucidated; many converge upon Rho GTPases as critical regulators of spinogenesis (Tolias et al., 2011; Penzes and Cahill, 2012; Saneyoshi and Hayashi, 2012; Lai and Ip, 2013; Um et al., 2014; Kim et al., 2015; Nishiyama and Yasuda, 2015; Lee et al., 2016). In addition, we recently identified the proteasome as a key regulator of activity-dependent spine outgrowth (Hamilton et al., 2012). To define the signaling mechanisms that connect proteasome activation to new spine outgrowth, we searched for known targets of the proteasome that serve as negative regulators of spine density. Here, we focus on the guanine nucleotide exchange factor (GEF), Ephexin5. Like the other members of the Ephexin family, Ephexin5 functions as an activator of Rho family GTPases; specifically, Ephexin5 has been shown to activate RhoA in neurons (Margolis et al., 2010). Notably, Ephexin5 is ubiquitinated and degraded by the ubiquitin proteasome system (UPS) in response to stimulation of the EphB2 receptor tyrosine kinase (Margolis et al., 2010). Furthermore, Ephexin5 acts as a negative regulator of spine and synapse density in neuronal cultures (Margolis et al., 2010); however, whether Ephexin5 acts to inhibit spine outgrowth and synapse formation or to enhance spine and synapse loss remained unresolved. We hypothesized that Ephexin5 inhibits new spine outgrowth, and thus that proteasome-mediated degradation of Ephexin5 serves to connect activity-dependent proteasome activation with new spine outgrowth.
Using two-photon time-lapse imaging of neurons in cultured hippocampal slices combined with genetic manipulations of Ephexin5, we show that Ephexin5 inhibits spine outgrowth in a GEF-dependent manner. Furthermore, elevated neural activity leads to rapid degradation of dendritic Ephexin5-GFP. Surprisingly, we also found that dendritic Ephexin5-GFP levels are locally elevated at sites of spinogenesis, where it accumulates in the dendritic shaft prior to new spine outgrowth. Moreover, reducing Ephexin5 levels inhibits new spine outgrowth in response to both global enhancement of neural activity and local stimulation through glutamate uncaging. These data support a dual role for Ephexin5 in activity-dependent spinogenesis, where it operates both as a check on exuberant spinogenesis, and also as a necessary factor in driving new spine outgrowth.
RESULTS
Ephexin5 inhibits spine outgrowth
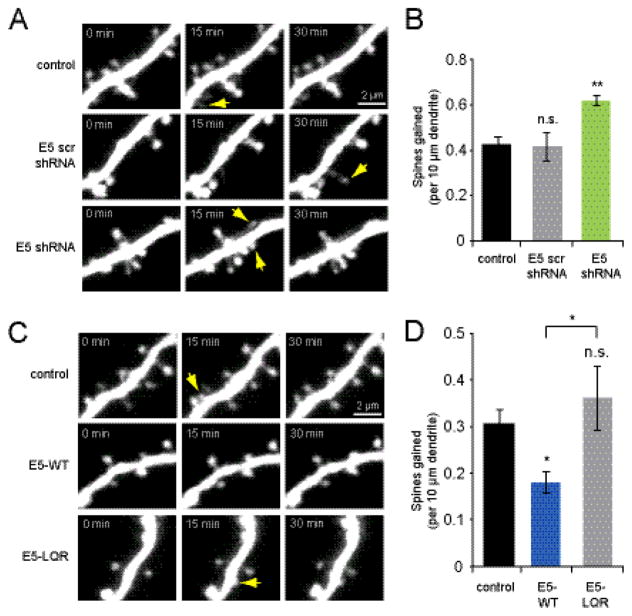
In order to determine whether Ephexin5 (E5) negatively regulates spinogenesis, we first tested the consequences of reducing E5 levels on the rate of new spine outgrowth. Hippocampal pyramidal neurons in slice culture (5-6 DIV) were co-transfected with EGFP and E5 shRNA or scrambled control shRNA, which we previously validated in dissociated hippocampal neuronal cultures (Margolis et al., 2010). Following 3–4 days of expression, we used two-photon time-lapse imaging at 15 min intervals to compare rates of new spine outgrowth in knockdown versus control (Fig. 1A). We found that shRNA-mediated knockdown of E5 led to increased spine outgrowth (0.62 ± 0.02 spines/10 μm/15 min) relative to EGFP alone (0.43 ± 0.04 spines/10 μm/15 min; p <0.001) or scrambled shRNA (0.42 ± 0.06 spines/10 μm/15 min; p < 0.05; Fig. 1B). Scrambled shRNA did not alter spine outgrowth relative to EGFP alone (p = 0.9). Thus, decreasing levels of E5 leads to enhanced spine outgrowth, suggesting that E5 acts to negatively regulate spine outgrowth.
Figure 1. Ephexin5 inhibits spine outgrowth in a GEF-dependent manner.
(A) Images of dendrites from hippocampal pyramidal neurons (DIV 9-10) transfected at DIV 5-6 with EGFP or co-transfected with EGFP + E5 shRNA scramble or E5 shRNA. Yellow arrows indicate sites of new spine outgrowth. (B) Transfection with E5 shRNA increased spine outgrowth (green bar; 7 cells, 101 spines) relative to EGFP controls (black bar; 9 cells, 88 spines), while E5 scrambled shRNA did not (gray bar; 9 cells, 86 spines; p = 0.9). (C) Images of dendrites from neurons (DIV 9-10) transfected at DIV 6-7 with EGFP or co-transfected with EGFP + E5-WT or E5-LQR. (D) Spine outgrowth was reduced by expression of E5-WT (blue bar; 11 cells, 48 spines) as compared to EGFP controls (black bar; 13 cells, 94 spines, p < 0.01) or E5-LQR (gray bar; 8 cells, 99 spines; p < 0.05). Overexpression of the GEF-inactive E5-LQR mutant did not alter spine outgrowth relative to EGFP controls (p = 0.5). *p<0.05, **p<0.001.
We next tested whether enhancing the levels of E5 would be sufficient to decrease the rate of new spine outgrowth. CA1 neurons were co-transfected with EGFP and myc-tagged variants of Ephexin5 and imaged 3–4 days later to monitor rates of spine outgrowth (Fig. 1C). We found that overexpression of the wild-type E5 reduced spine outgrowth (0.18 ± 0.02 spines/10 μm/15 min, p < 0.005) relative to cells transfected with EGFP alone (0.31 ± 0.03 spines/10 μm/15 min; Fig 1D). As E5 has been found to regulate spine density through activation of RhoA, we also examined the effect of the GEF-disabled Ephexin5-LQR (E5-LQR) mutant (Margolis et al., 2010) on spine outgrowth. We found that overexpression of E5-LQR had no significant effect on spine outgrowth (0.36 ± 0.07 spines/10 μm/15 min; p = 0.5) compared to EGFP-only controls (Fig. 1D), indicating that Ephexin5 inhibits spine outgrowth through its activity as a RhoGEF. As a control, we confirmed that the Ephexin5 constructs were expressing at comparable levels using immunostaining against the myc epitope (E5-WT: 84.7 ± 28.6 a.u., n = 6 cells; E5-LQR: 116.6 ± 20.9 a.u., n = 5 cells; p = 0.4). Thus, Ephexin5 acts to negatively regulate spine outgrowth through its RhoGEF activity.
Elevated neural activity rapidly induces proteasomal degradation of Ephexin5
E5 is ubiquitinated and degraded by the UPS in response to stimulation of the EphB2 receptor tyrosine kinase (Margolis et al., 2010). Because activity-dependent spine outgrowth is proteasome-dependent (Hamilton et al., 2012), we hypothesized that E5 might also be degraded in response to neural activity, thus releasing its brake on spinogenesis and leading to enhanced new spine outgrowth.
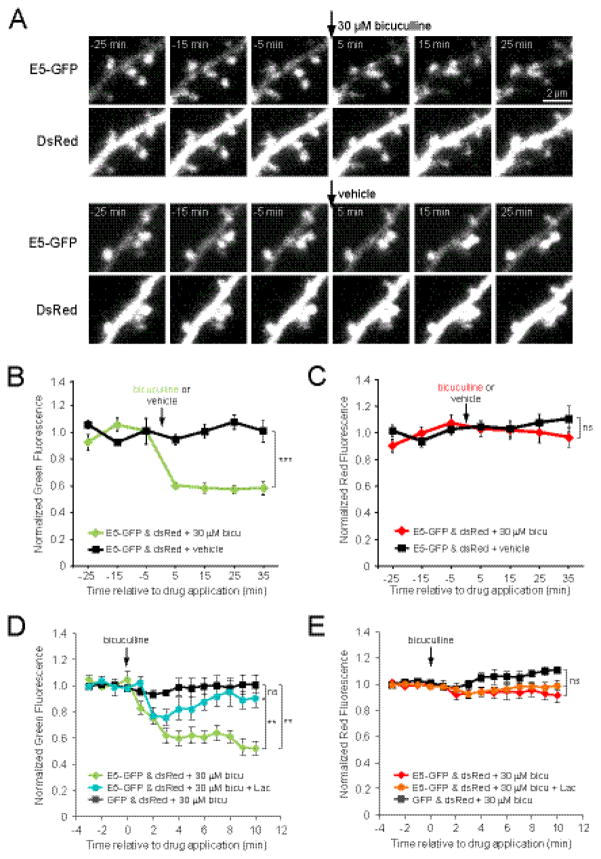
To address the regulation of E5 levels by neural activity, we co-transfected hippocampal slice cultures (DIV6-10) with Ephexin5-GFP (E5-GFP) and DsRedExpress (DsRed) as a cell fill. After 2–3 days of expression, we found that E5-GFP was present in the dendrite and concentrated in dendritic spines (Fig. 2A). To address whether enhanced neural activity would decrease E5 levels, we measured E5-GFP fluorescence in both the dendrite and in dendritic spines that were present across all images, before and after treatment with vehicle or the GABAA receptor blocker bicuculline (Fig. 2A), which rapidly increases neural activity in our cultured slices (Hamilton et al., 2012). While dendritic E5-GFP signal remained constant in vehicle-treated controls, treatment with 30 μM bicuculline resulted in a rapid, persistent decrease in dendritic green signal within 5 min of drug application (p < 0.001; heteroscedastic t-test of the average of all post drug time-points; Fig. 2B). In contrast, DsRedExpress signal did not change significantly between vehicle and bicuculline-treated conditions in the dendrite (p = 0.48; heteroscedastic t-test of the average of post-drug time-points; Fig. 2C), demonstrating that the drop in E5-GFP signal was not due to shrinkage of the dendrite. A reduction in green (but not red) signal was also observed in mature spine heads (p < 0.05, Fig S1), suggesting systemic degradation of E5 following enhanced neural activity.
Figure 2. Dendritic Ephexin5 levels are reduced in response to increased neural activity.
(A) Images of dendrites from neurons (DIV 9-10) co-transfected at DIV 7-8 with E5-GFP + DsRedExpress (DsRed) and treated with bicuculline or vehicle at t = 0 min. (B) Enhanced neural activity through application of 30 μM bicuculline (arrow) reduced E5-GFP (green fluorescence) levels in the dendrite (green line; 12 dendrites, 5 cells) relative to vehicle-treated controls (black line; 12 dendrites, 6 cells). (C) Application of 30 μM bicuculline (arrow) did not alter DsRed cell fill (red fluorescence) levels in the dendrite (red line) relative to vehicle-treated controls (black line; p > 0.1 for all post-treatment time-points). (D) Proteasome inhibition with lactacystin (10 μM) blocked the persistent decrease in E5-GFP fluorescence following bicuculline application (cyan line; 8 dendrites, 5 cells) compared to E5-GFP-expressing cells treated with bicuculline in the absence of lactacystin (green line; 8 dendrites, 7 cells; p < 0.01). Cells expressing GFP showed no significant decrease in green fluorescence in response to bicuculline (black line; 3 dendrites, 3 cells). (E) Application of bicuculline and lactacystin did not alter DsRed cell fill (red fluorescence) levels in the dendrites of cells expressing E5-GFP (red line, bicuculline alone; orange line, bicuculline + lactacystin) or GFP (black line, p = 0.18). Significance for panels B-E was determined using a one-way ANOVA of the average of the last 5 min for each cell followed by Bonferroni’s multiple comparison test, except for control cells expressing GFP, where a paired t-test was used to determine the significance of fluorescence changes at each time-point compared to baseline. *p<0.05, **p<0.001.
In order to establish whether this activity-induced reduction in E5-GFP signal was due to proteasomal degradation, we treated E5-GFP cells with 10 μM lactacystin to block proteasome activity (10 min preincubation and during the entire experiment), and found that this prevented the persistent reduction in E5-GFP signal observed with bicuculline alone (p < 0.05 for all points after 5 min post-bicuculline; Fig 2D). We also found that bicuculline had no effect on dendritic green signal in cells expressing GFP (p > 0.05 for all post-bicuculline time points; Fig 2D) nor on dendritic DsRedExpress signal (p > 0.05 for all post-treatment time points; Fig 2E). Taken together, these data demonstrate that elevated neural activity rapidly induces proteasomal degradation of Ephexin5, clearing the way for new spine outgrowth.
Ephexin5-GFP is enriched at sites of new spine formation
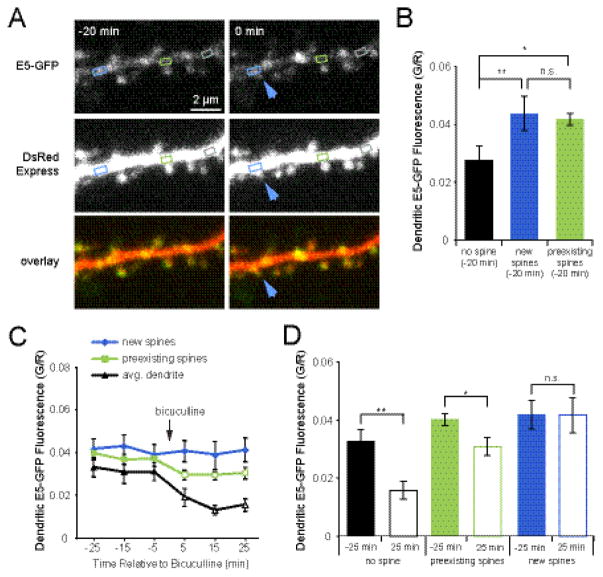
If dendritic E5 must be reduced to allow spinogenesis to occur, we hypothesized that E5 levels would be reduced locally at regions of the dendrite experiencing activity-dependent spine outgrowth. In order to examine local E5 content at sites of new spine outgrowth, we performed time-lapse imaging to identify new spines on pyramidal cells co-expressing E5-GFP and DsRedExpress (Fig. 3A). We then quantified the levels of E5-GFP on open regions of the dendrite and at the base of new and persistent spines. Green fluorescence intensity values were normalized to DsRedExpress to correct for fluctuations in laser intensity over time. Unexpectedly, we found that E5-GFP was enriched on the dendrite at the base of spines (G/R 0.042 ± 0.002) relative to open areas of the dendritic shaft (G/R 0.028 ± 0.005; p < 0.005; Fig. 3B). Remarkably, we also found that E5-GFP was enriched on the dendrite at sites of new spine outgrowth, even 20 min before the appearance of the new spine (G/R 0.044 ± 0.006; p < 0.001; Fig. 3A, B). The finding that new spine outgrowth occurs on the dendrite at sites with higher concentrations of Ephexin5 is especially surprising, considering that globally Ephexin5 acts to inhibit spinogenesis (Fig. 1).
Figure 3. Ephexin5-GFP is enriched at sites of new spine outgrowth.
(A) Images of dendrites from neurons (DIV 9-10) co-transfected at DIV 7-8 with E5-GFP + DsRedExpress (DsRed) and treated with bicuculline to enhance the rate of new spine outgrowth. Blue arrows indicate sites of new spine outgrowth. Time stamps are relative to time of new spine outgrowth. Boxes show representative regions of dendrite at the base of a new spine (blue), preexisting spine (green), or intervening dendrite (gray). (B) Twenty minutes prior to new spine outgrowth, E5-GFP is already concentrated in the dendritic shaft at sites of new spine growth (blue bar; 12 cells, 21 spines) above levels measured on dendritic regions between spines (black bar; 12 cells, 17 segments), but similar to levels observed at the base of preexisting spines (green bar; 12 cells, 75 spines; p = 0.1). (C) Within 5 min of treatment with 30 μM bicuculline, E5-GFP levels drop significantly (open symbols, p < 0.005) in the dendrite (black line) and at the base of preexisting spines (green line; p < 0.001), but not at sites of new spine outgrowth (blue line), compared to average pretreatment levels. (D) In response to bicuculline treatment, E5-GFP is reduced in the dendrite (open black bar; 12 cells, 17 segments; p < 0.001) and at the base of preexisting spines (open green bar; 12 cells, 75 spines; p < 0.001), but is not changed at the base of new spines (open blue bar; 12 cells, 21 spines; p = 0.8). *p<0.05, **p<0.001, Student’s paired t-test.
Considering that neural activity causes E5 degradation (Fig. 2), we next examined what happened at sites of new spine outgrowth following treatment with 30 μM bicuculline. As expected, we found that within 5 min of bicuculline application, E5 levels dropped rapidly in open dendritic regions and at the base of preexisting spines (−5 min vs. 5 min, p < 0.001; Fig. 3C, D). Unexpectedly, E5 levels at the base of new spines were unaffected by bicuculline (−25 min vs. 25 min, p = 0.8; Fig. 3C, D), suggesting an alternate state for Ephexin5 at these sites of new spine outgrowth, in which degradation or dispersal is limited.
Ephexin5 is necessary for activity-dependent spine outgrowth
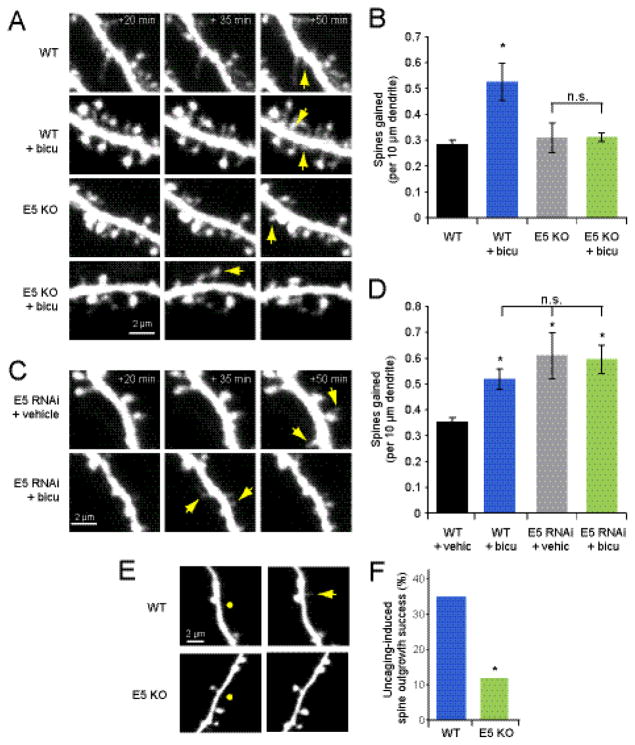
We have shown that E5 acts to inhibit spine outgrowth (Fig. 1); however, the finding that E5 is enriched at the base of new spines even prior to new spine outgrowth suggests that E5 may also play a positive role in spinogenesis. To test whether E5 is necessary for activity-dependent spine outgrowth, we treated hippocampal slices from WT and E5 KO (Margolis et al., 2010) mice with vehicle or 30 μM bicuculline. As expected, in WT neurons, spine outgrowth increased following bicuculline treatment (0.53 ± 0.07 spines/10 μm/15 min) relative to vehicle-treated controls (0.28 ± 0.02 spines/10 μm/15 min; p < 0.05; Fig. 4A, B). In contrast, there was no increase in spine outgrowth on E5 KO cells treated with bicuculline (0.31 ± 0.02 spines/10 μm/15 min) over that observed on their vehicle-treated counterparts (0.31 ± 0.06 spines/10 μm/15 min; p = 0.9; Fig. 4A, B). Thus, surprisingly, Ephexin5 both restricts spine outgrowth and is necessary for activity-dependent increases in spine outgrowth.
Figure 4. Ephexin5 is necessary for activity-induced spine outgrowth.
(A) Images of dendrites from neurons in slice cultures (DIV 9-10) from E5 KO mice or WT littermates transfected at DIV 6*7 with EGFP and treated with 30 μM bicuculline or vehicle at 0 min. Yellow arrows indicate sites of new spine outgrowth. (B) As expected, treatment with 30 μM bicuculline increased spine outgrowth on cells from WT littermates (blue bar; 4 cells, 68 spines) compared to vehicle-treated WT littermate controls (black bar; 4 cells, 39 spines). In contrast, no increase in spine outgrowth was observed following bicuculline treatment of E5 KO cells (green bar; 4 cells, 28 spines; p = 0.9) relative to vehicle-treated E5 KO controls (gray bar; 4 cells, 29 spines). (C) Images of dendrites from neurons in rat slice cultures (DIV 9-10) transfected at DIV 6-7 with EGFP or co-transfected with EGFP + E5 shRNA and treated with 30 μM bicuculline or vehicle at 0 min. Yellow arrows indicate sites of new spine outgrowth. (D) Bicuculline treatment increased spine outgrowth on EGFP-transfected cells (blue bar; 4 cells, 48 spines) relative to vehicle-treated EGFP controls (black bar; 7 cells, 59 spines; p < 0.05), but did not on E5 shRNA cells (green bar; 4 cells, 56 spines) relative to vehicle-treated E5 shRNA controls (gray bar; 6 cells, 83 spines; p = 0.9). (E) Images of dendrites of CA1 pyramidal cells in slices from WT and E5 KO mutant mice exposed to local uncaging (50 pulses of 4 ms at 5 Hz in 0 Mg2+) of MNI-glutamate (5 mM) at DIV 6-7. Yellow circles represent sites of localized uncaging stimulation. Yellow arrow indicates new spine. (F) Uncaging-induced spine formation success rate was lower on dendrites of E5 KO neurons (green bar; 10 cells, 4 of 34 segments) relative to that observed on dendrites from WT littermates (blue bar; 12 cells, 14 of 40 segments; p<0.05).
The lack of response to bicuculline in the E5 knockout mouse could be caused by altered connectivity, rather than a direct role for E5 in spine outgrowth. In order to determine whether E5 plays a cell autonomous role in activity-induced spine outgrowth, we examined the rate of bicuculline-induced spine outgrowth in neurons transfected with either EGFP alone or EGFP + E5 shRNA. We found that while 30 μM bicuculline increased spine outgrowth in EGFP neurons (0.52 ± 0.04 spines/10 μm/15 min) relative to vehicle-treated EGFP-only controls (0.35 ± 0.018 spines/10 μm/15 min; Fig. 4C, D), this stimulation had no significant effect on E5 shRNA cells (0.60 ± 0.06 spines/10 μm/15 min) relative to vehicle treated E5 shRNA controls (0.61 ± 0.09; p = 0.9, Fig 4C, D).
In addition to global stimulation of synaptic activity, we also examined the necessity for E5 in inducing spine outgrowth through targeted local dendritic stimulation by uncaging with MNI-glutamate. We found that while uncaging on the dendrites of EGFP-transfected CA1 neurons in slices from WT mice resulted in new spines at a frequency similar to previously published rates (Kwon and Sabatini, 2011; Hamilton et al., 2012), the same stimulation applied to E5 KO neurons resulted in dramatically lower rates of outgrowth (35% WT vs 11.8% E5 KO, p < 0.05, Fig. 4E, F). Thus, while Ephexin5 acts as a global brake on spine outgrowth, it also is required for spinogenesis in response to both circuit-wide and highly localized increases in neural activation.
Ephexin5 is not the sole target of the proteasome in regulating spine outgrowth
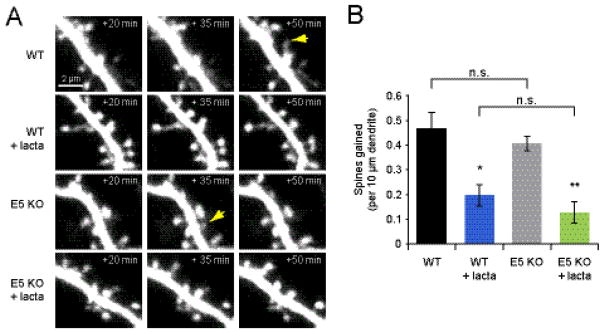
We previously proposed a model of activity-dependent spine outgrowth in which neural activity led to local proteasome activation that then caused the degradation of inhibitory factors blocking spine outgrowth (Hamilton et al., 2012). If E5 were the sole target of the proteasome in controlling spine outgrowth, the absence of E5 should render the UPS unnecessary for spine outgrowth. To test this hypothesis, we imaged EGFP-transfected hippocampal slices from E5 KO mice and WT littermates before and after the application of vehicle or the proteasome inhibitor lactacystin (Fig. 5A). Similar to previous results (Hamilton et al., 2012), spine outgrowth was decreased on WT neurons treated with 10 μM lactacystin (0.20 ± 0.04 spines/10 μm/15 min) relative to vehicle treated WT controls (0.47 ± 0.07 spines/10 μm/15 min; p <0.05; Fig. 5B). Similar to WT, E5-KO cells treated with lactacystin displayed a reduction in spine outgrowth (0.13 ± 0.04 spines/10 μm/15 min) relative to vehicle-treated E5 KO controls (0.41 ± 0.03 spines/10 μm/15 min; p < 0.001). Thus, while E5 may be one of the proteins degraded by the proteasome in order to allow for new spine outgrowth, it is not the only connection between the UPS and spinogenesis.
Figure 5. Ephexin5 is not the sole target of the proteasome during activity-dependent spine outgrowth.
(A) Images of dendrites from neurons in slice culture (DIV 9-10) from E5 KO or WT littermates transfected at DIV 6-7 with EGFP and treated with 10 μM lactacystin or vehicle at time 0 min. (B) Treatment of WT mice with 10 μM lactacystin reduced spine outgrowth (blue bar; 5 cells, 21 spines) relative to vehicle-treated WT controls (black bar; 5 cells, 56 spines; p < 0.05). A lactacystin-induced decrease in spine outgrowth was also observed in E5 KO cells (green bar; 5 cells, 15 spines) relative to vehicle-treated E5 KO cells (gray bar; 5 cells, 47 spines; p < 0.001). WT and KO vehicle treated (p=0.4) and lactacystin treated (p=0.3) groups were not significantly different from each other. *p<0.05, **p<0.001.
DISCUSSION
A complex role for Ephexin5 in regulation of activity-dependent spine outgrowth
Here, we demonstrate that reduced Ephexin5 levels increase spine outgrowth, and enhanced Ephexin5 levels decrease spine outgrowth, strongly supporting a role for Ephexin5 as a negative regulator of spinogenesis. We also show that elevated neural activity, which promotes spine outgrowth, is sufficient to rapidly induce Ephexin5 degradation by the ubiquitin proteasome system. However, the classification of Ephexin5 as a simple inhibitor of spinogenesis is not consistent with all of our findings. First, the concentration of GFP-tagged Ephexin5 is locally increased at sites of new spine outgrowth, contrary to our expectation that sites of new spine outgrowth would be associated with lower levels of Ephexin5. Second, while elevated neural activity results in reduced Ephexin5 content in open dendrite and at the base of preexisting spines, the Ephexin5 at the base of new spines is protected from degradation or dispersion, contradicting the hypothesis that Ephexin5 levels must be reduced to permit spine outgrowth. Third, reduced Ephexin5 levels inhibit activity-dependent spine outgrowth. These findings demonstrate that Ephexin5 is not a simple inhibitor that must be removed in order to permit spine outgrowth; instead, these data suggest that Ephexin5 acts both globally as an inhibitor of spinogenesis and locally as a necessary component in activity-dependent spinogenesis.
The precise mode of action of Ephexin5 in both restricting and permitting spine outgrowth remains unclear, although previous studies of the related GEF Ephexin1 provide an intriguing possible model. A closely related homolog of Ephexin5, Ephexin1 displays GEF specificity that is regulated by its phosphorylation state. Similarly to Ephexin5, Ephexin1 has been shown to target RhoA, but is also capable of activating Cdc42 and Rac1 (Shamah et al., 2001; Sahin et al., 2005), both of which enhance spine density (Tashiro et al., 2000; Wiens et al., 2005; Wegner et al., 2008; Kim et al., 2013; Ueda et al., 2013; Dhar et al., 2014; Raynaud et al., 2014; Evans et al., 2015; Jaudon et al., 2015; Kim et al., 2015; Valdez et al., 2016). Phosphorylation of Ephexin1 by Src kinase, however, causes a shift in Ephexin1 affinity away from Rac1 and Cdc42 towards RhoA, a switch which mediates EphA4-dependent growth cone collapse through RhoA activation (Shamah et al., 2001; Sahin et al., 2005). In a similar manner, spatially restricted phosphorylation of Ephexin5 may locally control a switch between enhancing and inhibiting spinogenesis. Indeed, while neuronal Ephexin5 has been shown to be specific to RhoA in vitro (Margolis et al., 2010), more recent studies have found that Ephexin5 activates Cdc42 in retinal endothelial cells (Kusuhara et al., 2012) and both Rac1 and Cdc42 in cancer cell lines (Fukushima et al., 2016). Perhaps neuronal Ephexin5 can also be induced to activate small GTPases other than RhoA, thus switching from inhibiting to activating spine outgrowth. As a reduction in Ephexin5 is not observed at the base of new spines following bicuculline stimulation, this local Ephexin5 population may also be protected from the forces driving Ephexin5 degradation in the surrounding dendrite. Further research into regulation of Ephexin5 activity and its binding partners will prove vital to understanding Ephexin5’s complex role in spine outgrowth.
A spinogenic complex on the dendrite prior to new spine outgrowth?
The accumulation of Ephexin5 on the dendrite prior to new spine outgrowth, and the necessity of Ephexin5 in activity-dependent spine outgrowth, suggest the presence of a nearby shaft synapse or of a spinogenic complex that assembles prior to new spine outgrowth. Which proteins might contribute to such a complex in addition to Ephexin5? The NMDA receptor (Engert and Bonhoeffer, 1999; Maletic-Savatic et al., 1999; Tolias et al., 2005; Kwon and Sabatini, 2011; Hamilton et al., 2012; Lin et al., 2013) and the EphB2 receptor tyrosine kinase (Penzes et al., 2003; Tolias et al., 2005; Tolias et al., 2007) both have demonstrated roles in facilitating spine outgrowth and have been shown to physically interact (Dalva et al., 2000; Tolias et al., 2005). Furthermore, Ephexin5 interacts physically with EphB2 (Margolis et al., 2010), and EphB2 stimulation has been shown to induce the recruitment of NMDA receptors and other synaptic proteins to EphB2 puncta (Dalva et al., 2000), suggesting that EphB2 could be a focal point for spinogenesis. Others have postulated such a complex for the control of spine and dendrite morphogenesis based upon the RhoGEFs kalirin-7 (Penzes et al., 2003) and Tiam1 (Tolias et al., 2005), which act as positive regulators of spinogenesis through activation of Rac1. Indeed, recent work has demonstrated that the Rac1 GEF Tiam1 and GAP Bcr form a complex with the EphB2 receptor, and that this complex regulates spine outgrowth in response to ephrinB1 stimulation (Um et al., 2014).
Molecular mechanisms of activity-dependent spine outgrowth
As an overall inhibitor of spine outgrowth which is degraded in response to increased neural activity, Ephexin5 is an excellent candidate for connecting activity-dependent control of proteolysis with the emergence of new dendritic spines (Hamilton et al., 2012). However, our observation that proteasome-dependent spine outgrowth occurs at similar levels in Ephexin5 knockout mice and their wild-type littermates strongly indicates that another target or targets must be degraded by the proteasome in order for activity-dependent spine outgrowth to occur. While a great many proteins involved in neuronal morphogenesis are known to be regulated by the ubiquitin-proteasome system (Ehlers, 2003; Hamilton and Zito, 2013), the small GTPases, regulators of the actin cytoskeleton, show exceptional promise as acute mediators of spine outgrowth (Tolias et al., 2011; Um et al., 2014).
Several small GTPases have been characterized as enhancers of spine density, especially Cdc42 (Tashiro et al., 2000; Wegner et al., 2008; Kim et al., 2013; Jaudon et al., 2015) and Rac1 (Tashiro et al., 2000; Penzes et al., 2001; Penzes et al., 2003; Tolias et al., 2007; Bacon et al., 2013; Ueda et al., 2013; Dhar et al., 2014; Raynaud et al., 2014; Evans et al., 2015; Jaudon et al., 2015; Kim et al., 2015; Valdez et al., 2016); others have been shown to inhibit spine density, in particular Ras (Yang et al., 2013; Lee et al., 2016) and RhoA (Tashiro et al., 2000; Margolis et al., 2010; Alder et al., 2013; Kim et al., 2015). Furthermore, modulators of activity for Ras (Pham and Rotin, 2001), Cdc42 (Hayakawa et al., 2008; Yamaguchi et al., 2008), and RhoA (Margolis et al., 2010; Lin et al., 2011; Papadimitriou et al., 2012) have been shown to be degraded by the proteasome, as well as Rac1 (Torrino et al., 2011; Zhao et al., 2013) and RhoA themselves (Wang et al., 2003; Cheng et al., 2011). Degradation of several of these proteins may be required in order to permit activity-dependent spine outgrowth, making small GTPases and their regulators a very promising avenue of future research into the molecular mechanisms of proteolysis- and activity-dependent spine outgrowth.
EXPERIMENTAL METHODS
Cultures and transfection
Organotypic hippocampal slice cultures were prepared as described (Stoppini et al., 1991) from postnatal day (P)6-7 Sprague-Dawley rats or wild-type and E5 KO C57BL/6 mice (Margolis et al., 2010) of both sexes. DNA constructs were delivered 3–4 days (EGFP and EGFP + E5 variants) or 2 days (DsRedExpress + E5-GFP) prior to imaging using biolistic gene transfer (130–180 PSI) as described (Woods and Zito, 2008). We coated 1.6 μm gold beads with 10–20 μg of EGFP (Clontech), 10 μg EGFP + 25 μg E5-WT, E5-LQR, E5 shRNA or E5 shRNA scramble (Margolis et al., 2010), or 10μg DsRedExpress (Clontech) + 5 μg E5-GFP.
Time-Lapse Imaging
Fluorescently-labeled pyramidal neurons (9–10 days in vitro [DIV]) were imaged at 15 min intervals for spine dynamics or 10 min or 1 min intervals for E5-GFP quantification, using a custom 2-photon microscope with a pulsed Ti::sapphire laser (Mai Tai: Spectra Physics) tuned to 930 nm. The microscope was controlled with ScanImage (Pologruto et al., 2003). Slices were imaged at 29 °C in artificial cerebrospinal fluid (ACSF) containing in mM: 127 NaCl, 25 NaHCO3, 25 D-glucose, 2.5 KCl, 1.25 NaH2PO4, 1 MgCl2, and 2 CaCl2, aerated with 95%O2/5%CO2. For each neuron, image stacks (512 × 512 pixels; 0.035 μm/pixel) with 1 μm steps were collected from 1–6 segments of secondary dendrites (apical and basal). All displayed images are maximum projections of three-dimensional (3D) image stacks.
Glutamate uncaging
Organotypic hippocampal slices prepared from P6-P8 wild-type or E5 KO littermates were transfected with 20 μg EGFP at DIV 3-4. CA1 pyramidal cells at 20–40 μm depth were imaged as above at DIV 6-7. Slices were bathed in Mg2+-free ACSF with 5 mM MNI-glutamate for at least 10 min before being exposed to uncaging stimulus, delivered by parking a 720 nm laser beam at a point 0.5 μm from an open segment of a secondary or tertiary dendrite. The uncaging stimulus consisted of 50 pulses of 4 ms at an interstimulus interval of 200 ms (5 Hz). A maximum of 4 dendrites (2 basal, 2 apical) per cell were stimulated in this manner. Spine outgrowth was scored blindly by 3 independent observers comparing pre- and post-uncaging 3-dimensional z stacks of the target dendrite. Rate of uncaging-induced spine outgrowth was compared between WT and E5 KO littermates.
Pharmacology
We prepared 1,000x stocks by dissolving bicuculline (Tocris) and lactacystin (EMD Biochemicals) in water. Vehicle controls were matched in identity and volume to the solution in which the inhibitors were dissolved.
Immunostaining
Immediately following imaging, slices were submerged in ice-cold 4% paraformaldehyde in 4% sucrose PBS and incubated at 4°C for 1 hr, washed and permeabilized in PBS containing 0.3% triton X-100 and 10% goat serum at 4°C overnight. Slices were blocked 4 hrs at RT in 10% goat serum PBS, and incubated with anti-myc (9E10 monoclonal; Covance; 30–50 μg/ml working concentration) overnight at 4°C, and then washed and incubated with goat anti-mouse IgG1-Alexa 594 for 3 to 4 hrs at RT, and finally washed and mounted in 100% glycerol. Neurons were imaged on a Zeiss LSM 500 META confocal microscope. All images were acquired using the same laser power, set to assure that pixel values were not saturated.
Image Analysis
Spine addition rates were blindly analyzed in 3D using custom software in MATLAB. For quantification of dendritic E5 levels in living neurons, integrated dendritic green (E5-GFP) and red (DsRedExpress) fluorescence intensities were measured from three boxed regions of interest (ROIs; ~ 1.0 – 0.5 μm2) on the dendritic segment in a single Z slice. Bleed-through corrected and background-subtracted (Woods et al., 2011) mean fluorescence intensities from the three ROIs for each dendritic segment were then normalized to the average of the three pre-treatment time-points. Preexisting spine head E5-GFP was quantified from all mature spines with good separation from the dendrite, and which were present for the entire duration of imaging. For quantification of myc-tagged E5 levels in fixed neurons, Alexa594 fluorescence intensity from anti-myc immunostaining was determined by measuring mean pixel intensity in a circular region entirely within the soma and background subtracted against the same circle dragged to a region devoid of target cell.
Statistics
Error bars represent standard error of the mean and significance was set at p = 0.05 (two-tailed, heteroscedastic t test, unless otherwise noted). All statistics were calculated across cells. *p < 0.05 and **p < 0.001. Uncaging-induced spine outgrowth was compared using a two-tailed Fisher’s exact test.
Supplementary Material
(A) In mature, preexisting spines, bicuculline treatment also reduced E5-GFP fluorescence (dark green line; 72 spines on 15 dendrites, 7 cells) compared to spines in vehicle-treated controls (black line; 94 spines on 18 dendrites, 5 cells) (B) Spine volume (red fluorescence) was not significantly altered by bicuculline treatment (dark red line; 72 spines on 15 dendrites, 7 cells) compared to vehicle-treated controls (black line; 94 spines of 18 dendrites, 5 cells). *p<0.05.
HIGHLIGHTS.
The RhoGEF, Ephexin5, is a negative regulator of dendritic spine outgrowth.
Neural activity drives a proteasome-dependent reduction in dendritic Ephexin5.
Ephexin5 accumulates on the dendrite prior to new spine outgrowth.
Ephexin5 is necessary for neural activity-dependent spine outgrowth.
Ephexin5 serves a dual role in spinogenesis.
Acknowledgments
This work was supported by the NIH (NS062736; T32GM007377). We thank J. Callis, A. Gomes, J. Trimmer, and members of the Zito lab for valuable discussion; J. Culp and L. Boudewyn for technical support; J. Culp and W.C. Oh for critical reading of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alder J, Kallman S, Palmieri A, Khadim F, Ayer JJ, Kumar S, Tsung K, Grinberg I, Thakker-Varia S. Neuropeptide orphanin FQ inhibits dendritic morphogenesis through activation of RhoA. Dev Neurobiol. 2013;73:769–784. doi: 10.1002/dneu.22101. [DOI] [PubMed] [Google Scholar]
- Bacon C, Endris V, Rappold GA. The cellular function of srGAP3 and its role in neuronal morphogenesis. Mechanisms of development. 2013;130:391–395. doi: 10.1016/j.mod.2012.10.005. [DOI] [PubMed] [Google Scholar]
- Cheng PL, Lu H, Shelly M, Gao H, Poo MM. Phosphorylation of E3 ligase Smurf1 switches its substrate preference in support of axon development. Neuron. 2011;69:231–243. doi: 10.1016/j.neuron.2010.12.021. [DOI] [PubMed] [Google Scholar]
- Dalva MB, Takasu MA, Lin MZ, Shamah SM, Hu L, Gale NW, Greenberg ME. EphB receptors interact with NMDA receptors and regulate excitatory synapse formation. Cell. 2000;103:945–956. doi: 10.1016/s0092-8674(00)00197-5. [DOI] [PubMed] [Google Scholar]
- Dhar M, Wayman GA, Zhu M, Lambert TJ, Davare MA, Appleyard SM. Leptin-induced spine formation requires TrpC channels and the CaM kinase cascade in the hippocampus. J Neurosci. 2014;34:10022–10033. doi: 10.1523/JNEUROSCI.2868-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers MD. Activity level controls postsynaptic composition and signaling via the ubiquitin-proteasome system. Nature neuroscience. 2003;6:231–242. doi: 10.1038/nn1013. [DOI] [PubMed] [Google Scholar]
- Engert F, Bonhoeffer T. Dendritic spine changes associated with hippocampal long-term synaptic plasticity. Nature. 1999;399:66–70. doi: 10.1038/19978. [DOI] [PubMed] [Google Scholar]
- Evans JC, Robinson CM, Shi M, Webb DJ. The guanine nucleotide exchange factor (GEF) Asef2 promotes dendritic spine formation via Rac activation and spinophilin-dependent targeting. The Journal of biological chemistry. 2015;290:10295–10308. doi: 10.1074/jbc.M114.605543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushima H, Yasumoto M, Ogasawara S, Akiba J, Kitasato Y, Nakayama M, Naito Y, Ishida Y, Okabe Y, Yasunaga M, Horiuchi H, Sakamoto E, Itadani H, Mizuarai S, Oie S, Yano H. ARHGEF15 overexpression worsens the prognosis in patients with pancreatic ductal adenocarcinoma through enhancing the motility and proliferative activity of the cancer cells. Molecular cancer. 2016;15:32. doi: 10.1186/s12943-016-0516-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton AM, Zito K. Breaking it down: the ubiquitin proteasome system in neuronal morphogenesis. Neural plasticity. 2013;2013:196848. doi: 10.1155/2013/196848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton AM, Oh WC, Vega-Ramirez H, Stein IS, Hell JW, Patrick GN, Zito K. Activity-dependent growth of new dendritic spines is regulated by the proteasome. Neuron. 2012;74:1023–1030. doi: 10.1016/j.neuron.2012.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayakawa M, Matsushima M, Hagiwara H, Oshima T, Fujino T, Ando K, Kikugawa K, Tanaka H, Miyazawa K, Kitagawa M. Novel insights into FGD3, a putative GEF for Cdc42, that undergoes SCF(FWD1/beta-TrCP)-mediated proteasomal degradation analogous to that of its homologue FGD1 but regulates cell morphology and motility differently from FGD1. Genes Cells. 2008;13:329–342. doi: 10.1111/j.1365-2443.2008.01168.x. [DOI] [PubMed] [Google Scholar]
- Hayashi-Takagi A, Yagishita S, Nakamura M, Shirai F, Wu YI, Loshbaugh AL, Kuhlman B, Hahn KM, Kasai H. Labelling and optical erasure of synaptic memory traces in the motor cortex. Nature. 2015;525:333–338. doi: 10.1038/nature15257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill TC, Zito K. LTP-induced long-term stabilization of individual nascent dendritic spines. J Neurosci. 2013;33:678–686. doi: 10.1523/JNEUROSCI.1404-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofer SB, Mrsic-Flogel TD, Bonhoeffer T, Hubener M. Experience leaves a lasting structural trace in cortical circuits. Nature. 2009;457:313–317. doi: 10.1038/nature07487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtmaat A, Wilbrecht L, Knott GW, Welker E, Svoboda K. Experience-dependent and cell-type-specific spine growth in the neocortex. Nature. 2006;441:979–983. doi: 10.1038/nature04783. [DOI] [PubMed] [Google Scholar]
- Jaudon F, Raynaud F, Wehrle R, Bellanger JM, Doulazmi M, Vodjdani G, Gasman S, Fagni L, Dusart I, Debant A, Schmidt S. The RhoGEF DOCK10 is essential for dendritic spine morphogenesis. Molecular biology of the cell. 2015;26:2112–2127. doi: 10.1091/mbc.E14-08-1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jourdain P, Fukunaga K, Muller D. Calcium/calmodulin-dependent protein kinase II contributes to activity-dependent filopodia growth and spine formation. J Neurosci. 2003;23:10645–10649. doi: 10.1523/JNEUROSCI.23-33-10645.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato-Negishi M, Muramoto K, Kawahara M, Hosoda R, Kuroda Y, Ichikawa M. Bicuculline induces synapse formation on primary cultured accessory olfactory bulb neurons. The European journal of neuroscience. 2003;18:1343–1352. doi: 10.1046/j.1460-9568.2003.02901.x. [DOI] [PubMed] [Google Scholar]
- Kim Y, Ha CM, Chang S. SNX26, a GTPase-activating protein for Cdc42, interacts with PSD-95 protein and is involved in activity-dependent dendritic spine formation in mature neurons. The Journal of biological chemistry. 2013;288:29453–29466. doi: 10.1074/jbc.M113.468801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Lee SE, Park J, Kim M, Lee B, Hwang D, Chang S. ADP-ribosylation factor 6 (ARF6) bidirectionally regulates dendritic spine formation depending on neuronal maturation and activity. The Journal of biological chemistry. 2015;290:7323–7335. doi: 10.1074/jbc.M114.634527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusuhara S, Fukushima Y, Fukuhara S, Jakt LM, Okada M, Shimizu Y, Hata M, Nishida K, Negi A, Hirashima M, Mochizuki N, Nishikawa S, Uemura A. Arhgef15 promotes retinal angiogenesis by mediating VEGF-induced Cdc42 activation and potentiating RhoJ inactivation in endothelial cells. PloS one. 2012;7:e45858. doi: 10.1371/journal.pone.0045858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon HB, Sabatini BL. Glutamate induces de novo growth of functional spines in developing cortex. Nature. 2011;474:100–104. doi: 10.1038/nature09986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai KO, Ip NY. Structural plasticity of dendritic spines: the underlying mechanisms and its dysregulation in brain disorders. Biochimica et biophysica acta. 2013;1832:2257–2263. doi: 10.1016/j.bbadis.2013.08.012. [DOI] [PubMed] [Google Scholar]
- Lee NJ, Song JM, Cho HJ, Sung YM, Lee T, Chung A, Hong SH, Cifelli JL, Rubinshtein M, Habib LK, Capule CC, Turner RS, Pak DT, Yang J, Hoe HS. Hexa (ethylene glycol) derivative of benzothiazole aniline promotes dendritic spine formation through the RasGRF1-Ras dependent pathway. Biochimica et biophysica acta. 2016;1862:284–295. doi: 10.1016/j.bbadis.2015.12.007. [DOI] [PubMed] [Google Scholar]
- Lin MY, Lin YM, Kao TC, Chuang HH, Chen RH. PDZ-RhoGEF ubiquitination by Cullin3-KLHL20 controls neurotrophin-induced neurite outgrowth. J Cell Biol. 2011;193:985–994. doi: 10.1083/jcb.201103015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YC, Yeckel MF, Koleske AJ. Abl2/Arg controls dendritic spine and dendrite arbor stability via distinct cytoskeletal control pathways. J Neurosci. 2013;33:1846–1857. doi: 10.1523/JNEUROSCI.4284-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maletic-Savatic M, Malinow R, Svoboda K. Rapid dendritic morphogenesis in CA1 hippocampal dendrites induced by synaptic activity. Science. 1999;283:1923–1927. doi: 10.1126/science.283.5409.1923. [DOI] [PubMed] [Google Scholar]
- Margolis SS, Salogiannis J, Lipton DM, Mandel-Brehm C, Wills ZP, Mardinly AR, Hu L, Greer PL, Bikoff JB, Ho HY, Soskis MJ, Sahin M, Greenberg ME. EphB-mediated degradation of the RhoA GEF Ephexin5 relieves a developmental brake on excitatory synapse formation. Cell. 2010;143:442–455. doi: 10.1016/j.cell.2010.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama J, Yasuda R. Biochemical Computation for Spine Structural Plasticity. Neuron. 2015;87:63–75. doi: 10.1016/j.neuron.2015.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadimitriou E, Vasilaki E, Vorvis C, Iliopoulos D, Moustakas A, Kardassis D, Stournaras C. Differential regulation of the two RhoA-specific GEF isoforms Net1/Net1A by TGF-beta and miR-24: role in epithelial-to-mesenchymal transition. Oncogene. 2012;31:2862–2875. doi: 10.1038/onc.2011.457. [DOI] [PubMed] [Google Scholar]
- Penzes P, Cahill ME. Deconstructing signal transduction pathways that regulate the actin cytoskeleton in dendritic spines. Cytoskeleton (Hoboken) 2012;69:426–441. doi: 10.1002/cm.21015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penzes P, Beeser A, Chernoff J, Schiller MR, Eipper BA, Mains RE, Huganir RL. Rapid induction of dendritic spine morphogenesis by trans-synaptic ephrinB-EphB receptor activation of the Rho-GEF kalirin. Neuron. 2003;37:263–274. doi: 10.1016/s0896-6273(02)01168-6. [DOI] [PubMed] [Google Scholar]
- Penzes P, Johnson RC, Sattler R, Zhang X, Huganir RL, Kambampati V, Mains RE, Eipper BA. The neuronal Rho-GEF Kalirin-7 interacts with PDZ domain-containing proteins and regulates dendritic morphogenesis. Neuron. 2001;29:229–242. doi: 10.1016/s0896-6273(01)00193-3. [DOI] [PubMed] [Google Scholar]
- Pham N, Rotin D. Nedd4 regulates ubiquitination and stability of the guanine-nucleotide exchange factor CNrasGEF. The Journal of biological chemistry. 2001;276:46995–47003. doi: 10.1074/jbc.M108373200. [DOI] [PubMed] [Google Scholar]
- Pologruto TA, Sabatini BL, Svoboda K. ScanImage: flexible software for operating laser scanning microscopes. Biomed Eng Online. 2003;2:13. doi: 10.1186/1475-925X-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raynaud F, Moutin E, Schmidt S, Dahl J, Bertaso F, Boeckers TM, Homburger V, Fagni L. Rho-GTPase-activating protein interacting with Cdc-42-interacting protein 4 homolog 2 (Rich2): a new Ras-related C3 botulinum toxin substrate 1 (Rac1) GTPase-activating protein that controls dendritic spine morphogenesis. The Journal of biological chemistry. 2014;289:2600–2609. doi: 10.1074/jbc.M113.534636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts TF, Tschida KA, Klein ME, Mooney R. Rapid spine stabilization and synaptic enhancement at the onset of behavioural learning. Nature. 2010;463:948–952. doi: 10.1038/nature08759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahin M, Greer PL, Lin MZ, Poucher H, Eberhart J, Schmidt S, Wright TM, Shamah SM, O’Connell S, Cowan CW, Hu L, Goldberg JL, Debant A, Corfas G, Krull CE, Greenberg ME. Eph-dependent tyrosine phosphorylation of ephexin1 modulates growth cone collapse. Neuron. 2005;46:191–204. doi: 10.1016/j.neuron.2005.01.030. [DOI] [PubMed] [Google Scholar]
- Saneyoshi T, Hayashi Y. The Ca2+ and Rho GTPase signaling pathways underlying activity-dependent actin remodeling at dendritic spines. Cytoskeleton (Hoboken) 2012;69:545–554. doi: 10.1002/cm.21037. [DOI] [PubMed] [Google Scholar]
- Shamah SM, Lin MZ, Goldberg JL, Estrach S, Sahin M, Hu L, Bazalakova M, Neve RL, Corfas G, Debant A, Greenberg ME. EphA receptors regulate growth cone dynamics through the novel guanine nucleotide exchange factor ephexin. Cell. 2001;105:233–244. doi: 10.1016/s0092-8674(01)00314-2. [DOI] [PubMed] [Google Scholar]
- Stoppini L, Buchs PA, Muller D. A simple method for organotypic cultures of nervous tissue. J Neurosci Methods. 1991;37:173–182. doi: 10.1016/0165-0270(91)90128-m. [DOI] [PubMed] [Google Scholar]
- Tashiro A, Minden A, Yuste R. Regulation of dendritic spine morphology by the rho family of small GTPases: antagonistic roles of Rac and Rho. Cerebral cortex. 2000;10:927–938. doi: 10.1093/cercor/10.10.927. [DOI] [PubMed] [Google Scholar]
- Tolias KF, Duman JG, Um K. Control of synapse development and plasticity by Rho GTPase regulatory proteins. Prog Neurobiol. 2011;94:133–148. doi: 10.1016/j.pneurobio.2011.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolias KF, Bikoff JB, Kane CG, Tolias CS, Hu L, Greenberg ME. The Rac1 guanine nucleotide exchange factor Tiam1 mediates EphB receptor-dependent dendritic spine development. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:7265–7270. doi: 10.1073/pnas.0702044104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolias KF, Bikoff JB, Burette A, Paradis S, Harrar D, Tavazoie S, Weinberg RJ, Greenberg ME. The Rac1-GEF Tiam1 couples the NMDA receptor to the activity-dependent development of dendritic arbors and spines. Neuron. 2005;45:525–538. doi: 10.1016/j.neuron.2005.01.024. [DOI] [PubMed] [Google Scholar]
- Torrino S, Visvikis O, Doye A, Boyer L, Stefani C, Munro P, Bertoglio J, Gacon G, Mettouchi A, Lemichez E. The E3 ubiquitin-ligase HACE1 catalyzes the ubiquitylation of active Rac1. Developmental cell. 2011;21:959–965. doi: 10.1016/j.devcel.2011.08.015. [DOI] [PubMed] [Google Scholar]
- Trachtenberg JT, Chen BE, Knott GW, Feng G, Sanes JR, Welker E, Svoboda K. Long-term in vivo imaging of experience-dependent synaptic plasticity in adult cortex. Nature. 2002;420:788–794. doi: 10.1038/nature01273. [DOI] [PubMed] [Google Scholar]
- Ueda S, Negishi M, Katoh H. Rac GEF Dock4 interacts with cortactin to regulate dendritic spine formation. Molecular biology of the cell. 2013;24:1602–1613. doi: 10.1091/mbc.E12-11-0782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Um K, Niu S, Duman JG, Cheng JX, Tu YK, Schwechter B, Liu F, Hiles L, Narayanan AS, Ash RT, Mulherkar S, Alpadi K, Smirnakis SM, Tolias KF. Dynamic control of excitatory synapse development by a Rac1 GEF/GAP regulatory complex. Developmental cell. 2014;29:701–715. doi: 10.1016/j.devcel.2014.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdez CM, Murphy GG, Beg AA. The Rac-GAP alpha2-chimaerin regulates hippocampal dendrite and spine morphogenesis. Molecular and cellular neurosciences. 2016;75:14–26. doi: 10.1016/j.mcn.2016.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HR, Zhang Y, Ozdamar B, Ogunjimi AA, Alexandrova E, Thomsen GH, Wrana JL. Regulation of cell polarity and protrusion formation by targeting RhoA for degradation. Science. 2003;302:1775–1779. doi: 10.1126/science.1090772. [DOI] [PubMed] [Google Scholar]
- Wegner AM, Nebhan CA, Hu L, Majumdar D, Meier KM, Weaver AM, Webb DJ. N-wasp and the arp2/3 complex are critical regulators of actin in the development of dendritic spines and synapses. The Journal of biological chemistry. 2008;283:15912–15920. doi: 10.1074/jbc.M801555200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiens KM, Lin H, Liao D. Rac1 induces the clustering of AMPA receptors during spinogenesis. J Neurosci. 2005;25:10627–10636. doi: 10.1523/JNEUROSCI.1947-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods G, Zito K. Preparation of gene gun bullets and biolistic transfection of neurons in slice culture. J Vis Exp. 2008 doi: 10.3791/675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods GF, Oh WC, Boudewyn LC, Mikula SK, Zito K. Loss of PSD-95 enrichment is not a prerequisite for spine retraction. J Neurosci. 2011;31:12129–12138. doi: 10.1523/JNEUROSCI.6662-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T, Yu X, Perlik AJ, Tobin WF, Zweig JA, Tennant K, Jones T, Zuo Y. Rapid formation and selective stabilization of synapses for enduring motor memories. Nature. 2009;462:915–919. doi: 10.1038/nature08389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi K, Ohara O, Ando A, Nagase T. Smurf1 directly targets hPEM-2, a GEF for Cdc42, via a novel combination of protein interaction modules in the ubiquitin-proteasome pathway. Biol Chem. 2008;389:405–413. doi: 10.1515/BC.2008.036. [DOI] [PubMed] [Google Scholar]
- Yang G, Pan F, Gan WB. Stably maintained dendritic spines are associated with lifelong memories. Nature. 2009;462:920–924. doi: 10.1038/nature08577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang K, Cao F, Sheikh AM, Malik M, Wen G, Wei H, Ted Brown W, Li X. Up-regulation of Ras/Raf/ERK1/2 signaling impairs cultured neuronal cell migration, neurogenesis, synapse formation, and dendritic spine development. Brain Struct Funct. 2013;218:669–682. doi: 10.1007/s00429-012-0420-7. [DOI] [PubMed] [Google Scholar]
- Zhao J, Mialki RK, Wei J, Coon TA, Zou C, Chen BB, Mallampalli RK, Zhao Y. SCF E3 ligase F-box protein complex SCF(FBXL19) regulates cell migration by mediating Rac1 ubiquitination and degradation. FASEB J. 2013;27:2611–2619. doi: 10.1096/fj.12-223099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zito K, Scheuss V, Knott G, Hill T, Svoboda K. Rapid functional maturation of nascent dendritic spines. Neuron. 2009;61:247–258. doi: 10.1016/j.neuron.2008.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) In mature, preexisting spines, bicuculline treatment also reduced E5-GFP fluorescence (dark green line; 72 spines on 15 dendrites, 7 cells) compared to spines in vehicle-treated controls (black line; 94 spines on 18 dendrites, 5 cells) (B) Spine volume (red fluorescence) was not significantly altered by bicuculline treatment (dark red line; 72 spines on 15 dendrites, 7 cells) compared to vehicle-treated controls (black line; 94 spines of 18 dendrites, 5 cells). *p<0.05.