Abstract
RNA sequencing has been extensively used to study specific gene expression patterns to discover potential key genes related to complex traits of interest in animals. Of note, a new regulatory mechanism builds a large-scale regulatory network among transcriptome, where lncRNAs act as competing endogenous RNAs (ceRNAs) to sponge miRNAs to regulate the expression of miRNA target genes post-transcriptionally. In this study, we sequenced the cDNA and sRNA libraries of nine liver samples from three Holstein cows during dry period, early lactation, and peak of lactation with HiSeq platform. As a result, we identified 665 genes, 57 miRNAs and 33 lncRNAs that displayed differential expression patterns across periods. Subsequently, a total of 41ceRNA pairs (lncRNA-mRNA) sharing 11 miRNAs were constructed including 30 differentially expressed genes. Importantly, 12 among them were presented in our large metabolic networks, and predicted to influence the lipid metabolism through insulin, PI3K-Akt, MAPK, AMPK, mTOR, and PPAR signaling pathways, thus, these genes were considered as the most promising candidates for milk fat formation. To our knowledge, this is first investigation to profile the ceRNA regulatory networks of liver transcriptome that could affect milk fat synthesis in bovine, providing a new view of the regulatory mechanism of RNAs.
Introduction
Milk and dairy products are an excellent source of rich nutrients for humans, providing protein, fatty acids, minerals, and vitamins1. Cow milk production has increased from 0.46 to 0.64 billion tonnes worldwide between 1961 and 2013 (FAOSTAT, Food and Agriculture Organization of the United Nations, 2016, http://faostat3.fao.org/browse/Q/QL/E). Milk production traits are the most important economic traits in the dairy industry. They include milk yield, fat yield, protein yield, fat percentage, and protein percentage2. Many researchers have attempted to identify the causal genes that have large effects on milk production traits with the aim of increasing production, including quantitative trait locus (QTL) mapping, candidate gene analysis, and genome-wide association studies (GWAS).
RNA sequencing (RNA-seq) has been used widely to study specific gene expression patterns in different tissues or at different developmental stages, to predict new transcripts3, to identify alternative splicing4, to detect single nucleotide polymorphisms (SNPs)5, and to discover insertion/deletions in transcripts6. Several RNA-seq studies of bovine transcriptomes in embryos, liver, mammary glands, and milk have been reported so far7–15. Different populations of RNAs have been subjected to RNA-seq, including total RNA16, long non-coding RNAs (lncRNAs)17, small non-coding RNAs such as microRNAs18, ribosomal RNAs19, and transfer RNAs20. Non-coding RNAs (ncRNAs) that cannot be translated to proteins but can act as functional RNAs have been recognized. The ncRNAs are divided into small RNAs (sRNAs) and lncRNAs depending on their length21. MiRNAs were found to be involved in regulating reproductive performances22, lactation characters23, 24, health25, and mammary gland development26 in Holstein cows. Regulatory functions of lncRNAs and identification of novel bovine lncRNAs have been reported in horn bud agenesis27, pigmented bovine skin28, and longissimus thoracic 29.
A new regulatory mechanism, which involves crosstalk between RNAs (both ncRNAs and coding RNAs) through miRNA response elements (MREs), builds a large-scale regulatory network among transcriptome sequences. LncRNAs may act as competing endogenous RNAs (ceRNAs) to sponge miRNAs, thereby regulating the expression of miRNA target genes post-transcriptionally30. The initial experimental evidence for ceRNA crosstalk was the tumor suppressor gene PTEN (phosphatase and tensin homolog), which can be regulated by the 3′-UTR of the pseudogene PTENP1 (phosphatase and tensin homolog pseudogene 1)31. Further, PTEN ceRNAs have been identified and validated in glioblastoma32, melanoma33, and prostate cancer34. The lncRNA ADNCR was shown to inhibit bovine adipocyte differentiation by functioning as a ceRNA for sponging miR-204, thereby increasing the expression of SIRT1 (sirtuin 1), which is the target of miR-204 and known to repress adipocyte differentiation and adipogenic gene expression35. A muscle-specific lncRNA, lncMD, functions as a ceRNA to sponge miR-125b, resulting in increasing expression of IGF2 (insulin like growth factor 2) to promote bovine muscle differentiation36.
Liver is a complex digestive gland in ruminant animals including dairy cattle, plays an important role in the metabolism of carbohydrates, fats, proteins, vitamins, hormones, and other substances. Nutrients absorbed from the digestive tract pass through the liver, enter the circulatory system, and finally arrive in the mammary glands of dairy cattle. During the lactation period, liver plays a critical role37–39. Dorland et al. reported the period of transition from late gestation to early lactation involved considerable metabolic adaptation in dairy cows and the liver, as a key role in adaptation, and supported pregnancy and lactation through coordination and interconversion of nutrients39. Smith et al. suggested that a part of the adaptation aimed to increase the hepatic pool of cholesterol, which was required for an increased formation of bile acids, and for synthesis and secretion of lipoproteins to provide the mammary gland with cholesterol and triglycerides for milk production40. In the present study, we isolated the total RNA and sRNAs from livers of Holstein cows 50 days before (dry period), and 10 days (early lactation) and 60 days (peak of lactation) after parturition for RNA-seq and sRNA-seq, respectively. Interactions among differentially expressed miRNAs, differentially expressed genes, and lncRNAs across the three lactation periods were identified. We built ceRNA regulatory networks via miRNAs to reveal the potential regulatory mechanisms controlling milk fat formation in liver of dairy cattle.
Results
Sequencing and mapping of the bovine liver transcriptome
We sequenced the cDNA and sRNA libraries of nine liver samples from three Holstein cows (A, B, and C) during three periods (dry period, −50 days; early lactation, +10 days; peak of lactation, +60 days). We acquired 80,470,996- 99,986,546 paired-end reads 100 bp in length for the mRNAs and lncRNAs, and 10,277,358- 15,107,016 single-end reads 50 bp in length for the miRNAs in the nine samples. After removing reads containing adapters, reads containing poly-N/A, and low quality reads from raw data, we obtained 97.56 gigabases (Gb) and 5.27 Gb high-quality clean data for the mRNAs/lncRNAs and miRNAs, respectively (see Supplementary Table S1). All the clean reads were aligned to the bovine reference genome assembly (UMD3.1.80) for further analysis.
We calculated the correlation coefficient (r 2) of the sequencing data among the three individual cows in each period based on the FPKM and TPM values, and found that the r 2 values were 0.860–0.995, 0.973–0.998, and 0.883–0.983 for the mRNAs, miRNAs, and lncRNAs, respectively, indicating that the similarity of the three biological replicates was sufficiently high (see Supplementary Table S2). We also found the average r 2 between the randomly selected 10 mRNA, 10 miRNA, and nine lncRNA expression levels from the qRT-PCR and the corresponding sequencing data were 0.72, 0.66, and 0.71 respectively, confirming the high reproducibility of the sequencing data in this study (see Supplementary Fig. S1).
Identification and function analysis of differentially expressed genes, miRNAs, and lncRNAs
We identified a total of 20,831 mRNAs corresponding to 16,857 known and 1,665 putative novel genes, 1,046 miRNAs (446 known and 600 novel with miRDeep2 scores ≥1.0), and 1,704 lncRNAs. Of these, totally 665 genes, 57 miRNAs, and 33 lncRNAs were found to be differentially expressed between at least two of the periods using Cuffdiff (v2.1.1) (Table 1; see Supplementary Table S3).
Table 1.
Differentially expressed RNAs during dry period, early lactation, and peak of lactation.
| RNA name | Known/novel | Early lactation vs. Dry period | Peak of lactation vs. Dry period | Peak of lactation vs. Early lactation | Total | |||
|---|---|---|---|---|---|---|---|---|
| Up | Down | Up | Down | Up | Down | |||
| mRNA | Known | 236 | 173 | 189 | 106 | 102 | 62 | 598 |
| Potential | 19 | 34 | 19 | 11 | 16 | 5 | 67 | |
| miRNA | Known | 8 | 0 | 5 | 1 | 1 | 2 | 12 |
| potential | 9 | 4 | 22 | 1 | 29 | 3 | 45 | |
| lncRNA | Known | 0 | 1 | 0 | 0 | 0 | 0 | 1 |
| potential | 6 | 13 | 5 | 9 | 5 | 1 | 32 | |
Up means that the expression of mRNAs, miRNAs, or lncRNAs were up-regulated, and down means the expression of mRNAs, miRNAs, or lncRNAs were down-regulated.
By scanning for conserved miRNA target sites with miRanda and RNAhybrid, we predicted 10,749 target genes for the differentially expressed miRNAs in common. In addition, we searched coding genes within the 100-kb upstream and downstream regions of each differentially expressed lncRNA and found 76 cis-acting genes. We also calculated the expression correlation coefficients between the differentially expressed lncRNAs and all identified mRNAs, and detected 31 lncRNAs associated with 852 potential trans-regulated genes (see Supplementary Table S4).
Further, we performed functional enrichment analysis on these differentially expressed genes (DEGs) and target genes of differentially expressed miRNAs and lncRNAs. As a result, GO categories and pathways for DEGs such as fatty acid, amino acid, carbohydrate and energy metabolism, immune response, PPAR, MAPK, AMPK, and PI3K-Akt signaling pathways were clustered in functional enrichment analysis (see Supplementary Tables S5 and 6).
Construction of ceRNA regulatory networks for milk composition
Among the predicted targets for 57 differentially expressed miRNAs, 251 DEGs were included targeted by 47 miRNAs. Subsequently, we scanned potential MREs in the 33 differentially expressed lncRNAs specifically targeted by the differentially expressed miRNA using miRanda v3.3a, and found 47 miRNA–lncRNA interaction pairs. Further, we constructed ceRNA regulatory networks that indicated the cross-regulations among lncRNA–miRNA–mRNA via shared miRNAs by calculating the likelihood of each potential ceRNA pair (lncRNA-mRNA) based on a hypergeometric test (P < 0.05). Consequently, we obtained 80 ceRNA pairs, including 16 between the dry period and early lactation, 59 between the dry period and peak of lactation, and five between the early and peak of lactation periods (Fig. 1; see Supplementary Table S7), in which lncRNAs competitively bound miRNAs thereby up-regulated the targeting mRNA expression, reversely, lncRNAs decreased the mRNA expression by releasing MREs. Further, we profiled the dynamic expression trends of all DEGs, miRNAs and lncRNAs in the identified ceRNA networks among three periods based on their FPKM and TPM values, and finally determined 41 ceRNA pairs that shared 11 miRNAs (Fig. 2). Two miRNAs (novel_chr13_7054 and novel_chr11_4054) and their targeting lncRNAs and genes which showed almost same expression trends were ruled out (Fig. 2). In total, 30 DEGs, 11 miRNAs and 9 lncRNAs were included in the ceRNA networks.
Figure 1.
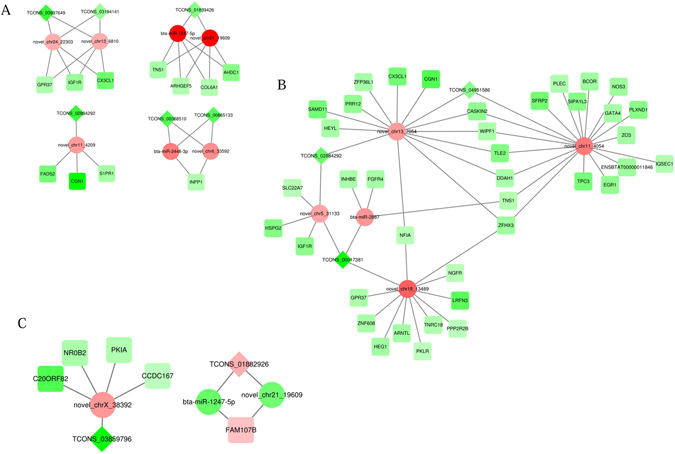
ceRNA regulatory networks among dry period, early lactation, and peak of lactation. (A) Early lactation vs. dry period; (B) peak of lactation vs. dry period; (C) peak of lactation vs. early lactation; red circles, squares and diamonds represent up-regulated miRNAs, mRNAs and lncRNAs, respectively; and the green circles, squares and diamonds represent down-regulated miRNAs, mRNAs and lncRNAs, respectively.
Figure 2.
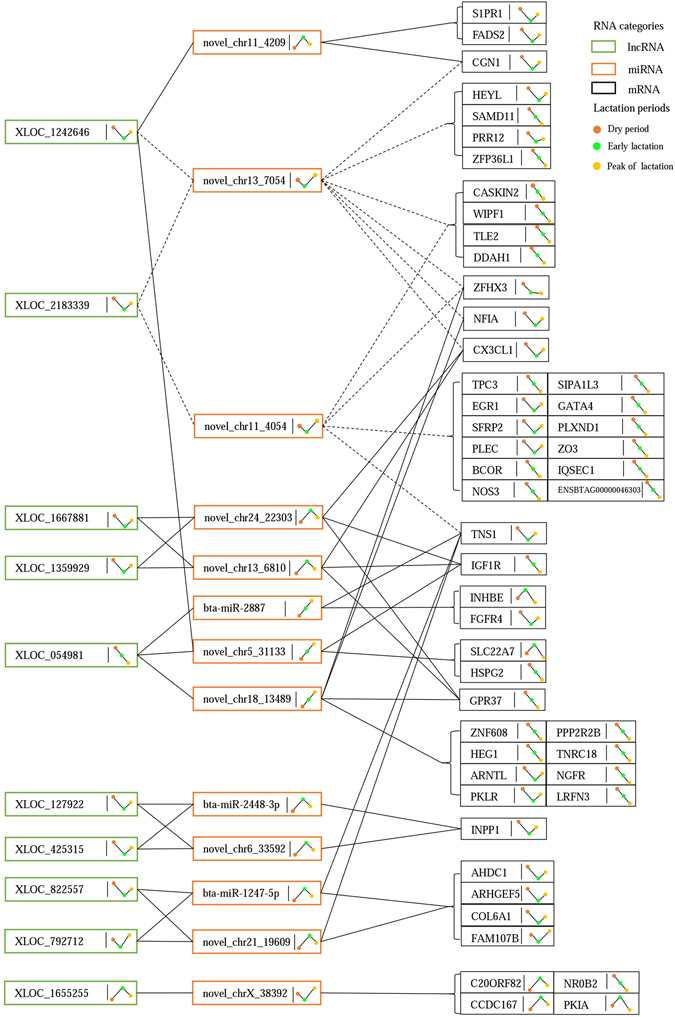
Definite expression of RNAs in ceRNA networks profiling their dynamic expression trends. The definite expression of lncRNAs, miRNAs, and mRNAs are shown based on FPKM, TPM, and FPKM values, respectively; green, orange, and black rectangles represent lncRNAs, miRNAs, and mRNAs, respectively; red, green, and yellow dots represent dry period, early lactation, and peak of lactation, respectively; the lines linked by these color dots show the dynamic expression trends of RNAs during periods; the full lines mean that interactions between RNAs follow the ceRNA regulatory mechanism; and the dash lines mean that the interactions between RNAs do not obey the ceRNA regulatory mechanism.
Construction of lipid metabolic network regulated by ceRNAs
We built a metabolic network comprising 12 out of 30 DEGs in ceRNA network that were involved in insulin, PI3K-Akt, AMPK, MAPK, mTOR, FoxO, and PPAR signaling pathways (Fig. 3; see Supplementary Table S8). As well-known, sterol regulatory element-binding proteins (SREBPs) and PPARα occupy the critical position in the lipid metabolism41, 42. Membranous receptors can influence SREBP-1c or PPARα through insulin, PI3K-Akt, AMPK, and PPAR pathways thereby modulating expression of lipid metabolic genes. As shown in Fig. 3, four membranous receptor genes (IGF1R, FGFR4, NGFR, and S1PR1) and COL6A1 in our ceRNA networks were down-regulated by six lncRNAs (Fig. 2), and induced the decreasing expression of PIK3R1, a critical node in PI3K-Akt pathway, so that SREBP-1c expression was correspondingly decreased through metabolic cascades. In addition, PPP2R2B, which was down-regulated by lnc-XLOC_054981, reduced SREBP-1c expression as well by increasing AMPK expression. Besides, the elevating AMPK up regulated the expression of CPT1 leading to the fatty acid oxidation through the differentially expressed MCD, and the decreasing PI3K modulated the expression of S1PR1 to affect the immune-regulation through PI3K-Akt and FoxO signaling pathways. Moreover, IGF1R, FGFR4, NGFR, and SIP1 protein reduced the expression of MNK1/2 and c-Myc via insulin, PI3K-Akt, and MAPK signaling pathways to modulate the proliferation and differentiation. The differentially expressed activin affected gonadal growth, embryo differentiation, and placenta formation through TGF-β signaling pathway. Bile acids entered into the cytoplasm through the receptor OATs that expressed differentially, and interacted with FXR to regulate the bile acid synthesis, transport, and detoxification, meanwhile, the nuclear receptor FXR also led to the differentially expression of SHP. Fatty acid and very low-density lipoprotein were transported into cytoplasm through CD36, and the unsaturated or saturated fatty acids and eicosanoid activated the expression of PPARα, which combined with RXR to modulate the expression of the lipid metabolic genes. As shown in Fig. 4, the downstream genes were up-/down-regulated by SREBP-1c and PPARα, among these, APOA1 (apolipoprotein A1), ACSL1 (acyl-CoA synthetase long-chain family member 1), CD36 (CD36 molecule), CYP7A1 (cytochrome P450, family 7, subfamily A, polypeptide 1), CPT1B (carnitine palmitoyltransferase 1B), SCD (stearoyl-CoA desaturase) and FADS2 (fatty acid desaturase 2) were evidenced to be differentially expressed, and eventually influenced the lipid and fatty acid transport, cholesterol metabolism, fatty acid oxidation, and lipogenesis, which belong to lipid metabolism in liver (see Supplementary Table S9).
Figure 3.
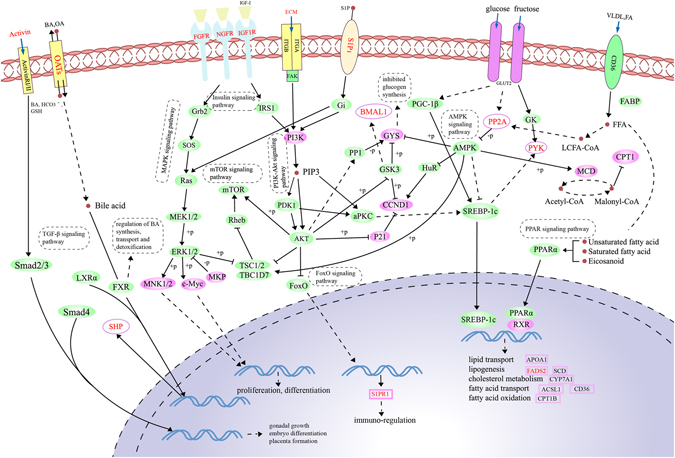
Metabolic networks involving in insulin, PI3K-Akt, AMPK, MAPK, mTOR, FoxO, and PPAR signaling pathways. Green ovals represent proteins involved in pathways; pink ovals (rectangles) represent the differentially expressed proteins (genes) identified in this study; the 12 genes in ceRNA networks are marked in red font, of these, BMAL1, ECM, IGF1R, NGFR, Activin, FGFR, PYK, SHP, S1P1, OATs, and PP2A protein in the networks respectively encoded by ARNTL, COL6A1, IGF1R, NGFR, INHBE, FGFR4, PKLR, NR0B2, S1PR1, SLC22A7 and PPP2R2B genes, and the remaining FADS2 gene is in the pink rectangles.
Figure 4.
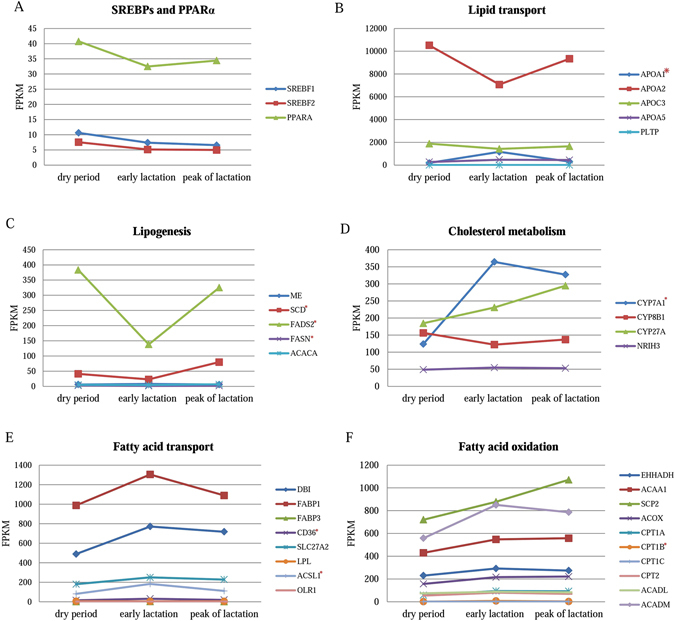
Expression profiles of the lipid metabolic genes during dry period, early lactation, and peak of lactation. The expression levels of lipid metabolic genes are shown based on FPKM values; the genes are divided into SREBPs and PPARα (A), lipid transport (B), lipogenesis (C), cholesterol metabolism (D), fatty acid transport (E), and fatty acid oxidation (F) categories; and the differentially expressed genes are labeled with an asterisk.
Based on biological functions, 12 among the 30 genes in the ceRNA networks were considered as keys participating in important lipid metabolic processes in liver, that were ARNTL (aryl hydrocarbon receptor nuclear translocator-like), COL6A1 (collagen, type VI, alpha 1), IGF1R (insulin like growth factor 1 receptor), NGFR (nerve growth factor receptor), INHBE (inhibin, beta E), FGFR4 (fibroblast growth factor receptor 4), PKLR (pyruvate kinase, liver and RBC), FADS2 (fatty acid desaturase 2), NR0B2 (nuclear receptor subfamily 0 group B member2), S1PR1(sphingosine-1-phosphate receptor 1), SLC22A7 (solute carrier family 22 member 7) and PPP2R2B (protein phosphatase 2 regulatory subunit Bbeta).
Discussion
We investigated the complexity of the liver transcriptome in Holstein cows at three levels, mRNA, miRNA, and lncRNA, using high-throughput RNA sequencing. We detected 665 genes, 57 miRNAs, and 33 lncRNAs that had different expression patterns among the dry period (−50 day), early lactation (+10 day), and peak of lactation (+60 days). Based on these results, we constructed 41 ceRNA pairs containing 30 DEGs, among which 12 genes were considered as the most important ones participating in lipid metabolism in liver of dairy cattle.
In this study, besides 665 DEGs, we identified a total of 10,749 target genes for 57 differentially expressed miRNAs, and 76 cis- and 852 trans-targets for 33 differentially expressed lncRNAs. By performing functional enrichment analysis for these genes, it was found that GO categories and pathways for DEGs such as fatty acid, amino acid, carbohydrate and energy metabolism, immune response, PPAR, MAPK, AMPK, and PI3K-Akt signaling pathways were enriched among three periods. Furthermore, genes including DEGs, and target genes of differentially expressed miRNAs between the dry period and early and/or peak of lactation were enriched mainly in metabolism-related functions, especially in processes involving carbohydrates, lipids and protein processes, suggesting that the genes regulating substance synthesis remained active to support the synthesis of a large number of milk compositions in both early and peak of lactation. Here, we found several pathways related to carbohydrate metabolism between dry period and early and/or peak of lactation including pyruvate metabolic process, glucocorticoid metabolic process, and regulation of glycogen catabolic process, and a study showed that gluconeogenesis in liver is vital to meet glucose requirements for dairy cows in perinatal period43. Additionally, fatty acids can be re-esterified to triglycerides in the liver of dairy cows44. The contents of triacylglycerol45 and blood nonesterified fatty acids (NEFA)46 were increased greatly during the perinatal period, and Pullen et al. reported that the free fatty acid absorption in liver was directly in proportion to NEFA47. Kristensen et al. suggested that milk fat proportion depended on the precursors freed by liver, including propionic acid, branched chain volatile fatty acids and mid/long chain fatty acids48. Our study also found that some genes were enriched into lipid metabolism in early and peak of lactation compared with dry period, such as bile secretion, biosynthesis of unsaturated fatty acids, fatty acid synthase activity, lipid homeostasis, carboxylic acid metabolic process, fatty acid metabolism, PPAR, and AMPK signaling pathways.
The recognition of ceRNAs has opened a new way of analyzing the role of RNAs in gene regulation. Here, we first constructed ceRNA networks across three periods in liver of dairy cows based on the differentially expressed mRNAs, miRNAs, and lncRNAs. In ceRNA networks, miRNAs targets RNA transcripts through MREs, which are approximately 22 nt in length and can be partially complementarity for target RNAs. If an RNA transcript has numerous MREs, miRNAs can function in a combinatorial manner30. As our ceRNA results showed, some lncRNA-mRNAs interacted with each other via several different miRNAs, and the expression trends of these pairs were the same, while opposite to the expression tendency of miRNAs. On the other hand, a single miRNA can also simultaneously target multiple lncRNAs or mRNAs. Such phenomenon indicated that a miRNA occupied the central position by supplying multiple intermediate bridges for lncRNAs and mRNAs. Li et al. first predicted genome-wide miRNA-mRNA and miRNA-lncRNA interactions in chicken, pigs and cows using the sequencing data from public databases, and found many SNPs located in miRNAs that may affect the interactions between miRNAs and mRNAs49. Li et al. and Sun et al. reported that lncRNAs act as ceRNAs to module mRNA expression by sponging miRNA in bovine adipocyte differentiation35 and muscle differentiation36, respectively. In liver cancer, Wang et al. demonstrated that lnc-HULC up-regulated cAMP responsive element binding (CREB) expression by interaction with miR-37250, and Li et al. found lnc-HULC functioned as a ceRNA to up-regulate zinc finger E-box binding homeobox 1(ZEB1)51. Our findings evidenced that 9 lncRNAs acted as ceRNAs to affect the expression of lipid metabolic genes by sponging miRNAs in bovine liver. Interestingly, we found that some lncRNAs might play predominant roles on affecting gene expression in the complicated ceRNA regulatory networks. As an example, IGF1R was a common target of three miRNAs which could combine with four lncRNAs as well; however, the lnc-XLOC_054981 mainly influenced IGF1R expression by sponging the miRNA (novel_chr5_31133).
Regarding the constructed ceRNA networks, we preferentially desired to explore their roles in milk composition formation. Based on biological functions of the 30 genes in ceRNAs, we constructed a large complicated metabolic network, and found that 12 out of 30 genes played critical roles in metabolism through several pathways, in which SREBP-1cand PPARα were the two keys in liver for lipid metabolism. Previous studies reported that SREBPs integrated signals from multiple pathways to control fatty acids, triglycerides, and cholesterol synthesis52, 53. PPARα, enriched in liver, was involved in peroxisomal and mitochondrial β-oxidation, fatty acid transport and catabolism, hepatic glucose production, lipogenesis and ketone body synthesis42. Our results also showed that the lipid metabolic genes involved in lipid transport, fatty acid oxidation and transport, and cholesterol metabolism were up-regulated since lactating, while the genes participating in lipogenesis were down-regulated, indicating that more fatty acids were transported to mammary gland for milk fat production since lactating, but the lipid synthesis in liver dropped off in order to meet homeostasis. Therefore, the 12 genes were inferred as the most promising candidate genes affecting milk fat formation, which were further described as follows.
IGF1R combines with insulin-like growth factor (IGF1). It was shown to be associated with milk production traits in dairy cattle54–56. In our previous GWAS, IGF1R was also included as one of promising genes for fatty acid traits in Chinese Holstein57. The protein encoded by FGFR4 is a member of the fibroblast growth factor receptor (FGFR) family. Previous studies found that integrated FGF15/FGFR4 and SHP signaling pathways regulated bile acid synthesis in the gut-liver axis58, and FGFR4 exerted a negative control on cholesterol metabolism and bile acid synthesis in liver59. NR0B2, also known as small heterodimer partner (SHP), is a member of the nuclear hormone receptor family and is involved in lipid homeostasis by regulating the expression levels of several genes in the lipid pathway60, 61. SNPs in NR0B2 was found to be associated with carcass- and fat-related traits in beef cattle62. As noted above, the integrated FGF15/FGFR4 and SHP signaling pathways show there is a relationship between FGFR4 and SHP (NR0B2). The protein encoded by PKLR is a pyruvate kinase that participates in the glycolysis and gluconeogenesis metabolic pathways (http://www.wikipathways.org/index.php/Pathway:WP534). A report showed that the high long-chain n-3 fatty acid in rat milk associated with low hepatic PKLR expression was relevant to hepatic metabolic regulation in a milk-fed neonate63. FADS2, belonging to the fatty acid desaturase (FADS) family, was involved in fatty acid synthesis and desaturation64–68. S1PR1, also known as endothelial differentiation gene 1 (EDG1), encodes a protein that is structurally similar to G protein-coupled receptors and is highly expressed in endothelial cells69. SNPs in the untranslated regions of EDG1 were related to marbling in Japanese Black beef cattle70. ARNTL (BMAL1) has been identified as a candidate gene for susceptibility to hypertension, diabetes, and obesity, and mutations in BMAL1 have been linked to infertility, gluconeogenesis and lipogenesis problems, and altered sleep patterns71, 72. Another study shows that the CLOCK/BMAL1 complex up regulates human LDLR promoter activity, suggesting the ARNTL gene also plays a role in cholesterol homeostasis73. Collagen VI is a major structural component of microfibrils, which is a heterotrimer of COL6A1, COL6A2, and COL6A3 protein. It was reported that the collagens, a superfamily of proteins, played a role in maintaining the integrity of various tissues and collagen IV is an important component of the extracellular matrix in the mammary glands74. NGFR plays a role in the regulation of the translocation of GLUT4 to the cell surface in adipocytes in response to insulin, and thereby contributes to the regulation of insulin-dependent glucose uptake75. INHBE may be up-regulated under conditions of endoplasmic reticulum stress76, and this protein may inhibit cellular proliferation77. SLC22A7 belongs to the solute carrier (SLC) family and is a liver-specific expressed gene78, 79. The protein encoded by PPP2R2B belongs to the phosphatase 2 regulatory subunit B family. Moreover, we identified that the 12 genes were markedly enriched into PPAR, MAPK, mTOR, insulin, AMPK, PI3K-Akt, and FoxO signaling pathways. Evidently, these 12 genes regulated by ceRNAs mechanism might be the crucial regulatory factors for milk fat formation in liver of dairy cattle.
Conclusions
In this study, we built a total of 41 ceRNA pairs with integrated analysis of the differentially expressed mRNAs, miRNAs, and lncRNAs across the three periods, and 12 functional genes in these ceRNA networks enriched in lipid metabolism were selected as the most promising candidates for milk fat formation. To our knowledge, this is first investigation to uncover the ceRNA regulatory networks of liver transcriptome that could affect milk fat synthesis in bovine.
Material and Methods
Ethics statement
All experiments, including all protocols for collection of the liver tissues of experimental individuals and phenotypic observations, were performed in accordance with relevant guidelines and regulations of the Institutional Animal Care and Use Committee (IACUC) at China Agricultural University (Permit Number: DK996) who have reviewed and approved the experiments. Samples were collected specifically for this study following standard procedures with the full agreement of the Hongda Dairy Farm in Mancheng of Hebei Province who owned the animals.
Samples collection
We selected three Chinese Holstein cows in their 2nd lactation from 1,300 Holstein cows maintained in the Hongda Dairy Farm in Mancheng, Hebei Province, China. To keep minimum variation between cows, the selection of similar milk production was based on their 305-day milk yield, milk protein percentage and milk fat percentage, which was calculated based on 10 test-day records in one lactation period using a multiple trait random regression test-day model by the Dairy Data Center of China (http://www.holstein.org.cn/). The 305-day milk yield, and milk fat and protein percentages in the milk of the three cows were 10,254–11,954 kg, 3.1–3.7%, and 2.7–2.9%, respectively.
We collected liver tissues using puncture biopsy in three periods: dry period (−50 days), early lactation (+10 days), and peak of lactation (+60 days) from each of the three cows. A 1.5–2.0 cm incision using a sterile scalpel blade was made between the 11th and 12th rib on the right side of each cow after injecting with 30 mL of 2% procaine on the sampling position as local anesthesia (China Agricultural University Veterinary Teaching Hospital). Following the skin incision, pressure was applied to the wound with sterile gauze until visual signs of the bleeding were gone. The liver biopsy was performed using a 4-mm Tru-Cut biopsy needle (Wuhan Anscitech Farming Technology Co. Ltd. Wuhan, China). The skin incision was closed using four or five Michel clips and erythromycin ointment was applied to the incision site. The liver tissue samples (about 0.5–1.0 g) per cow in every period were collected carefully for RNA isolation, placed into clean RNAase-free Eppendorf tubes, and stored in liquid nitrogen.
RNA isolation and quality assessment
We used TRIzol reagent (Life Technologies, CA, USA) to extract total RNA from the nine liver samples. RNase-free DNase (TIANGEN, Beijing, China) was used to remove DNA contamination from the extracted RNA. RNA degradation and contamination was monitored on 1% agarose gels and RNA purity was checked using a NanoPhotometer® spectrophotometer (IMPLEN, CA, USA). A Qubit® RNA Assay Kit in Qubit® 2.0 Fluorometer (Life Technologies) was used to measure the RNA concentration and a RNA Nano 6000 Assay Kit on a Bioanalyzer 2100 system (Agilent Technologies, CA, USA) was used to assess the RNA integrity.
RNA sequencing
mRNA and lncRNA library construction
A total of 3 μg RNA per sample was used for RNA sample preparation. The RNA integrity numbers of the nine samples from three cows in the three periods ranged from 7.3 to 7.8. Firstly, ribosomal RNA (rRNA) was removed using an EpicentreRibo-Zero™ rRNA Removal Kit (Epicentre, WI, USA) and the rRNA-free residue was obtained by ethanol precipitation. Subsequently, the sequencing libraries were generated using the rRNA-free RNA with a NEBNext® Ultra™ Directional RNA Library Prep Kit for Illumina® (New England Biolabs (NEB), MA, USA). The library fragments were purified with an AMPure XP system (Beckman Coulter, Beverly, USA) and cDNA fragments, preferentially 150–200 bp in length, were selected. Then, 3 μL USER Enzyme (NEB) was used with the selected adaptor-ligated cDNA at 37 °C for 15 min followed by 5 min at 95 °C before PCR. The PCRs were performed with Phusion High-Fidelity DNA polymerase, Universal PCR primers, and Index (X) Primer. The PCR products were purified (AMPure XP system) and library quality was assessed on an Agilent Bioanalyzer 2100 system.
miRNA library construction
From the same total RNA samples, a total of 10 μg RNA per sample was used for sRNA sample preparation. Firstly, sRNA was isolated from the total RNA with PEG8000 (Berry Genomics, Beijing, China). Then, the sequencing libraries were generated using a TruSeq Small RNA Sample Prep kit (Illumina, San Diego, CA, USA). A 3′ linker was ligated to the sequences and fractions of sRNAs 36–44 nt in length were extracted from the samples on a 15% denaturing polyacrylamide gel. The short RNAs were converted to DNA by reverse transcription-polymerase chain reaction (RT-PCR) after ligating a 5′ adaptor. The library fragments were purified on 3.5% agarose gels and cDNA fragments, preferentially 140–160 bp in length, were selected. The quality of the library products was assessed on a StepOnePlus PCR system (Applied Biosystems Inc., CA, USA).
Clustering and sequencing
For the lncRNAs and mRNAs, a TruSeq PE Cluster Kit v3-cBot-HS (Illumina) was used to cluster the index-coded samples. After cluster generation, the libraries were sequenced on a HiSeq. 2000 platform (Illumina) and 100-bp paired-end reads were obtained.
For the miRNAs, the index-coded samples were clustered using a HiSeq Rapid SBS Kit v2 (Illumina). Then, the libraries were sequenced on a HiSeq. 2500 platform (Illumina) and 50-bp single-end reads were obtained.
lncRNA and mRNA data analysis
Quality control and transcriptome assembly
Firstly, the raw reads in FASTQ format were processed with in-house Perl scripts. In this step, clean reads were obtained by removing reads containing adapters and poly-N, and low quality reads from the raw reads. The Q20 (proportion of bases with a Phred base quality score >20), Q30 (proportion of bases with a Phred base quality score >30), and GC content of the clean data were calculated. All subsequent analyses were based on the high-quality clean data. The bovine reference genome (UMD3.1.80) and gene model annotation files were downloaded from the Ensembl genome website (ftp://ftp.ensembl.org/pub/release-80). An index of the reference genome was built using Bowtie v2.0.680 and paired-end clean reads were aligned to the reference genome using TopHat v2.0.981. The mapped reads of each sample were assembled by both Scripture (beta2)82 and Cufflinks (v2.1.1)6 using a reference-based approach.
Coding potential analysis and target gene prediction for lncRNAs
The following four steps were used to select candidate lncRNAs for classification: (1) Transcript length ≥200 and exon number ≥2; (2) Minimal reads coverage ≥3 in at least one sample; (3) Filter known non-lncRNA annotations; (4) Classify the selected candidate lncRNAs, using four tools to predict their coding potential: CNCI (Coding-Non-Coding-Index) (v2) with default parameters83, CPC (Coding Potential Calculator) (0.9-r2) with the e-value set as ‘1e-10’84, Pfam Scan (v1.3) with default parameters of -E 0.001 (release 27; both Pfam A and Pfam B were used)85, and PhyloCSF (phylogenetic codon substitution frequency) (v20121028) with default parameters86. Transcripts predicted to have coding potential by all of the four prediction tools were filtered out, and transcripts with no coding potential were selected as the candidate set of lncRNAs.
To identify cis-acting lncRNAs (lncRNAs that act on neighboring genes), we searched for coding genes within the 100-kb upstream and downstream regions of the selected lncRNAs and then analyzed their functions. Additionally, we calculated the Pearson correlation between the differentially expressed lncRNAs and identified coding genes, and the potential trans-acting genes were selected based on the absolute value of correlation was higher than 0.95.
miRNA data analysis
Quality control and miRNA identification
We used the FASTX software (Fastx_toolkit-0.0.13.2) to pre-process the raw data as follows: remove low quality reads, remove 5′ joint pollution reads, remove 3′ splice reads and no insert fragment reads, remove reads <18-nt long, and remove reads that contain poly-A. The resultant clean reads (sRNAs) were compared against the known ncRNAs (i.e., rRNA, tRNA, snRNA, and snoRNA) in the Rfam database (v10.1) using BLAST (v2.2.26)87. The sRNAs that matched any of the sequences in Rfam database were removed. Then, the proportion of the four bases (A/T/G/C) in each position on the clean reads was calculated.
Mapping to the reference genome and target gene prediction
The remaining sRNAs that aligned to the bovine reference genome (UMD3.1.80) using TopHat v2.0.981 were searched against the known mature bovine miRNA sequences downloaded from miRBase (http://www.mirbase.org/) to identify known miRNAs. Novel miRNAs were predicted using mirDeep2 (v2.0.0.5) software88. To identify candidate target genes, we scanned for conserved miRNA target sites on the mRNAs identified in this study using miRanda (v3.3a)89 and RNAhybrid90.
Identification of differentially expressed lncRNAs, mRNAs, and miRNAs
We used Cuffdiff (v2.1.1) to calculate fragments per kilo-base of exon per million mapped fragments (FPKM) values for the lncRNAs and coding mRNAs (genes), and TPM (transcripts per million) scores for the miRNAs in each library6, 91. Gene FPKMs were computed by summing the FPKMs of transcripts in each lncRNA or mRNA group. The differential expression of the miRNAs was analyzed using the EdgeR software package92. We used the three cows as biological replicates and found differentially expressed lncRNAs, mRNAs, and miRNAs by comparing one period with another. MiRNAs with P-values < 0.05, absolute fold-change values >2.0, and miRDeep2 score ≥1, lncRNAs and mRNAs with corrected P-values < 0.05 in any of the pairwise comparisons were considered as significantly differentially expressed.
Quantitative real-time PCR (qRT-PCR)
To validate the repeatability and reproducibility of the RNA sequencing data, qRT-PCR was carried out on 10 DEGs, nine lncRNAs, and 10 miRNAs that were selected randomly using the total RNA that was used for sequencing. Primer3 (http://fokker.wi.mit.edu/primer3/input.htm) and Oligo 6.0 were both used to design specific primers (see Supplementary Table S9). The expression levels of the selected mRNAs and lncRNAs were normalized against the housekeeping gene, GAPDH, UXT and RPS9 and the expression levels of the miRNAs were normalized against U6 snoRNA. The qRT-PCRs were carried out in triplicate with the DyNAmo SYBR Green PCR kit (Applied Biosystems Inc.) on a LightCycler480 (Roche Applied Science, Penzberg, Germany) using the following program: 95 °C for 10 min; 45 cycles of 95 °C for 10 s, 60 °C for 10 s, and 72 °C for 10 s; 72 °C for 6 min.
Function enrichment analysis
We used the KOBAS software93 and SetRank94 for the Gene Ontology (GO) enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses of the differentially expressed genes (DEGs) and predicted target genes of the differentially expressed lncRNAs and miRNAs. Both GO terms and KEGG pathways with corrected P-values < 0.05 were considered to be significantly enriched.
Construction of ceRNA networks
Differentially expressed lncRNAs were scanned to find conserved MREs using miRanda v3.3a, as was done to predict the miRNA–mRNA target site. Based on the predicted miRNA–mRNA and miRNA–lncRNA regulatory pairs that were differentially expressed among the three periods, ceRNA networks were predicted in which lncRNAs and mRNAs interacted via shared miRNAs. We measured the likelihood of the ceRNAs using StarBase v2.095–97. Using the hypergeometric test32, we calculated the P-value for each potential ceRNA pair (lncRNA-mRNA) considering the number of shared miRNAs targeting individual components of the ceRNA pair with the following formula97:
where, N is the total number of miRNAs in the bovine reference genome, n is the number of miRNAs interacting with mRNAs (protein-coding), K is the total number of miRNAs interacting with lncRNAs, and c is the number of miRNAs shared between a ceRNA pair. All P-values were subjected to the false discovery rate (FDR) correction98.
Electronic supplementary material
Acknowledgements
This work was financially supported by the National Science and Technology Programs of China (2013AA102504, 2011BAD28B02, 2014ZX08009-053B), National Natural Science Foundation (31072016, 31472065), Beijing Natural Science Foundation (6152013), the Beijing Dairy Industry Innovation Team (BAIC06-2016), China Postdoctoral Science Foundation (2017M611057), and the Program for Changjiang Scholar and Innovation Research Team in University (IRT1191).
Author Contributions
D.S. and J.L. conceived and designed this experiment, Q.L. and Y.Y. collected the liver samples from cows, R.L. and Y.Y. isolated the RNA samples, R.L. and B.H. prepared the cDNA libraries for sequencing, R.L. performed qRT-PCR experiments, bioinformatics and statistical analysis with the help of B.H., the manuscript was prepared by R.L., B.H., and D.S. All authors read and approved the final manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Ruobing Liang and Bo Han contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-06634-w
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Tunick MH, Van Hekken DL. Dairy Products and Health: Recent Insights. J Agric Food Chem. 2015;63:9381–9388. doi: 10.1021/jf5042454. [DOI] [PubMed] [Google Scholar]
- 2.Spelman RJ, Coppieters W, Karim L, van Arendonk JA, Bovenhuis H. Quantitative trait loci analysis for five milk production traits on chromosome six in the Dutch Holstein-Friesian population. Genetics. 1996;144:1799–1808. doi: 10.1093/genetics/144.4.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Denoeud F, et al. Annotating genomes with massive-scale RNA sequencing. Genome Biol. 2008;9:R175. doi: 10.1186/gb-2008-9-12-r175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin, L. et al. Transcriptome sequencing reveals aberrant alternative splicing in Huntington’s disease. Hum Mol Genet, doi:10.1093/hmg/ddw187 (2016). [DOI] [PMC free article] [PubMed]
- 5.Trick M, Long Y, Meng J, Bancroft I. Single nucleotide polymorphism (SNP) discovery in the polyploid Brassica napus using Solexa transcriptome sequencing. Plant Biotechnol J. 2009;7:334–346. doi: 10.1111/j.1467-7652.2008.00396.x. [DOI] [PubMed] [Google Scholar]
- 6.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature biotechnology. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Canovas A, Rincon G, Islas-Trejo A, Wickramasinghe S, Medrano JF. SNP discovery in the bovine milk transcriptome using RNA-Seq technology. Mammalian Genome. 2010;21:592–598. doi: 10.1007/s00335-010-9297-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang, W. & Khatib, H. Comparison of transcriptomic landscapes of bovine embryos using RNA-Seq. Bmc Genomics11, doi:10.1186/1471-2164-11-711 (2010). [DOI] [PMC free article] [PubMed]
- 9.Robert C, et al. Combining Resources to Obtain a Comprehensive Survey of the Bovine Embryo Transcriptome Through Deep Sequencing and Microarrays. Mol Reprod Dev. 2011;78:651–664. doi: 10.1002/mrd.21364. [DOI] [PubMed] [Google Scholar]
- 10.Wickramasinghe, S. et al. Transcriptome Profiling of Bovine Milk Oligosaccharide Metabolism Genes Using RNA-Sequencing. Plos One6, doi:10.1371/journal.pone.0018895 (2011). [DOI] [PMC free article] [PubMed]
- 11.Wickramasinghe, S., Rincon, G., Islas-Trejo, A. & Medrano, J. F. Transcriptional profiling of bovine milk using RNA sequencing. Bmc Genomics13, doi:10.1186/1471-2164-13-45 (2012). [DOI] [PMC free article] [PubMed]
- 12.Canovas, A. et al. Comparison of five different RNA sources to examine the lactating bovine mammary gland transcriptome using RNA-Sequencing. Sci Rep-Uk4, doi:10.1038/srep05297 (2014). [DOI] [PMC free article] [PubMed]
- 13.Cui, X. G. et al. Transcriptional profiling of mammary gland in Holstein cows with extremely different milk protein and fat percentage using RNA sequencing. Bmc Genomics15, doi:10.1186/1471-2164-15-226 (2014). [DOI] [PMC free article] [PubMed]
- 14.Tizioto, P. C. et al. Global liver gene expression differences in Nelore steers with divergent residual feed intake phenotypes. Bmc Genomics16, doi:10.1186/s12864-015-1464-x (2015). [DOI] [PMC free article] [PubMed]
- 15.McCabe M, et al. RNA-seq analysis of differential gene expression in liver from lactating dairy cows divergent in negative energy balance. BMC Genomics. 2012;13:193. doi: 10.1186/1471-2164-13-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nagalakshmi U, et al. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320:1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He Y, et al. The conservation and signatures of lincRNAs in Marek’s disease of chicken. Sci Rep. 2015;5:15184. doi: 10.1038/srep15184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ahmed F, et al. Comprehensive analysis of small RNA-seq data reveals that combination of miRNA with its isomiRs increase the accuracy of target prediction in Arabidopsis thaliana. RNA Biol. 2014;11:1414–1429. doi: 10.1080/15476286.2014.996474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ingolia NT, Brar GA, Rouskin S, McGeachy AM, Weissman JS. The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat Protoc. 2012;7:1534–1550. doi: 10.1038/nprot.2012.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Su AA, Tripp V, Randau L. RNA-Seq analyses reveal the order of tRNA processing events and the maturation of C/D box and CRISPR RNAs in the hyperthermophile Methanopyrus kandleri. Nucleic Acids Res. 2013;41:6250–6258. doi: 10.1093/nar/gkt317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brosnan CA, Voinnet O. The long and the short of noncoding RNAs. Curr Opin Cell Biol. 2009;21:416–425. doi: 10.1016/j.ceb.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 22.Huang J, et al. Solexa sequencing of novel and differentially expressed microRNAs in testicular and ovarian tissues in Holstein cattle. Int J Biol Sci. 2011;7:1016–1026. doi: 10.7150/ijbs.7.1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Z, Liu H, Jin X, Lo L, Liu J. Expression profiles of microRNAs from lactating and non-lactating bovine mammary glands and identification of miRNA related to lactation. BMC Genomics. 2012;13:731. doi: 10.1186/1471-2164-13-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang M, et al. MicroRNA expression patterns in the bovine mammary gland are affected by stage of lactation. J Dairy Sci. 2012;95:6529–6535. doi: 10.3168/jds.2012-5748. [DOI] [PubMed] [Google Scholar]
- 25.Li Z, et al. Identification and characterization of novel and differentially expressed microRNAs in peripheral blood from healthy and mastitis Holstein cattle by deep sequencing. Anim Genet. 2014;45:20–27. doi: 10.1111/age.12096. [DOI] [PubMed] [Google Scholar]
- 26.Silveri L, Tilly G, Vilotte JL, Le Provost F. MicroRNA involvement in mammary gland development and breast cancer. Reprod Nutr Dev. 2006;46:549–556. doi: 10.1051/rnd:2006026. [DOI] [PubMed] [Google Scholar]
- 27.Allais-Bonnet A, et al. Novel insights into the bovine polled phenotype and horn ontogenesis in Bovidae. PLoS One. 2013;8:e63512. doi: 10.1371/journal.pone.0063512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weikard R, Hadlich F, Kuehn C. Identification of novel transcripts and noncoding RNAs in bovine skin by deep next generation sequencing. BMC Genomics. 2013;14:789. doi: 10.1186/1471-2164-14-789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Billerey C, et al. Identification of large intergenic non-coding RNAs in bovine muscle using next-generation transcriptomic sequencing. BMC Genomics. 2014;15:499. doi: 10.1186/1471-2164-15-499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poliseno L, et al. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033–1038. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sumazin P, et al. An extensive microRNA-mediated network of RNA-RNA interactions regulates established oncogenic pathways in glioblastoma. Cell. 2011;147:370–381. doi: 10.1016/j.cell.2011.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karreth FA, et al. In vivo identification of tumor- suppressive PTEN ceRNAs in an oncogenic BRAF-induced mouse model of melanoma. Cell. 2011;147:382–395. doi: 10.1016/j.cell.2011.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tay Y, et al. Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell. 2011;147:344–357. doi: 10.1016/j.cell.2011.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li M, et al. Long non-coding RNA ADNCR suppresses adipogenic differentiation by targeting miR-204. Biochim Biophys Acta. 2016;1859:871–882. doi: 10.1016/j.bbagrm.2016.05.003. [DOI] [PubMed] [Google Scholar]
- 36.Sun X, et al. The developmental transcriptome sequencing of bovine skeletal muscle reveals a long noncoding RNA, lncMD, promotes muscle differentiation by sponging miR-125b. Biochim Biophys Acta. 2016;1863:2835–2845. doi: 10.1016/j.bbamcr.2016.08.014. [DOI] [PubMed] [Google Scholar]
- 37.Graber M, et al. A field study on characteristics and diversity of gene expression in the liver of dairy cows during the transition period. J Dairy Sci. 2010;93:5200–5215. doi: 10.3168/jds.2010-3265. [DOI] [PubMed] [Google Scholar]
- 38.Schlegel G, Ringseis R, Keller J, Schwarz FJ, Eder K. Changes in the expression of hepatic genes involved in cholesterol homeostasis in dairy cows in the transition period and at different stages of lactation. J Dairy Sci. 2012;95:3826–3836. doi: 10.3168/jds.2011-5221. [DOI] [PubMed] [Google Scholar]
- 39.van Dorland HA, et al. Variation in hepatic regulation of metabolism during the dry period and in early lactation in dairy cows. J Dairy Sci. 2009;92:1924–1940. doi: 10.3168/jds.2008-1454. [DOI] [PubMed] [Google Scholar]
- 40.Smith JL, et al. Effect of pregnancy and lactation on lipoprotein and cholesterol metabolism in the rat. J Lipid Res. 1998;39:2237–2249. [PubMed] [Google Scholar]
- 41.Shao W, Espenshade PJ. Expanding roles for SREBP in metabolism. Cell Metab. 2012;16:414–419. doi: 10.1016/j.cmet.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hashimoto T, et al. Defect in peroxisome proliferator-activated receptor alpha-inducible fatty acid oxidation determines the severity of hepatic steatosis in response to fasting. The Journal of biological chemistry. 2000;275:28918–28928. doi: 10.1074/jbc.M910350199. [DOI] [PubMed] [Google Scholar]
- 43.Herbein JH, Aiello RJ, Eckler LI, Pearson RE, Akers RM. Glucagon, insulin, growth hormone, and glucose concentrations in blood plasma of lactating dairy cows. Journal of dairy science. 1985;68:320–325. doi: 10.3168/jds.S0022-0302(85)80828-6. [DOI] [PubMed] [Google Scholar]
- 44.Loor JJ, Bionaz M, Drackley JK. Systems physiology in dairy cattle: nutritional genomics and beyond. Annual review of animal biosciences. 2013;1:365–392. doi: 10.1146/annurev-animal-031412-103728. [DOI] [PubMed] [Google Scholar]
- 45.Graugnard DE, et al. Liver lipid content and inflammometabolic indices in peripartal dairy cows are altered in response to prepartal energy intake and postpartal intramammary inflammatory challenge. Journal of dairy science. 2013;96:918–935. doi: 10.3168/jds.2012-5676. [DOI] [PubMed] [Google Scholar]
- 46.Selim S, et al. Prepartal dietary energy alters transcriptional adaptations of the liver and subcutaneous adipose tissue of dairy cows during the transition period. Physiological genomics. 2014;46:328–337. doi: 10.1152/physiolgenomics.00115.2013. [DOI] [PubMed] [Google Scholar]
- 47.Pullen DL, Palmquist DL, Emery RS. Effect on days of lactation and methionine hydroxy analog on incorporation of plasma fatty acids into plasma triglycerides. Journal of dairy science. 1989;72:49–58. doi: 10.3168/jds.S0022-0302(89)79079-2. [DOI] [PubMed] [Google Scholar]
- 48.Kristensen NB. Splanchnic metabolism of volatile fatty acids in the dairy cow. Anim Sci. 2005;80:3–10. [Google Scholar]
- 49.Li A, et al. Genome-scale identification of miRNA-mRNA and miRNA-lncRNA interactions in domestic animals. Anim Genet. 2015;46:716–719. doi: 10.1111/age.12329. [DOI] [PubMed] [Google Scholar]
- 50.Wang J, et al. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res. 2010;38:5366–5383. doi: 10.1093/nar/gkq285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li, S. P. et al. LncRNA HULC enhances epithelial-mesenchymal transition to promote tumorigenesis and metastasis of hepatocellular carcinoma via the miR-200a-3p/ZEB1 signaling pathway. Oncotarget, doi:10.18632/oncotarget.9883 (2016). [DOI] [PMC free article] [PubMed]
- 52.Goldstein JL, DeBose-Boyd RA, Brown MS. Protein sensors for membrane sterols. Cell. 2006;124:35–46. doi: 10.1016/j.cell.2005.12.022. [DOI] [PubMed] [Google Scholar]
- 53.Shao W, Espenshade PJ. Expanding Roles for SREBP in Metabolism. Cell metabolism. 2012;16:414–419. doi: 10.1016/j.cmet.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Szewczuk M, Zych S, Czerniawska-Piatkowska E, Wojcik J. Association between IGF1R/i16/TaqI and IGF1/SnaBI polymorphisms and milk production traits in Polish Holstein-Friesian cows. Anim Sci Pap Rep. 2012;30:13–24. [Google Scholar]
- 55.Akis I, Oztabak K, Gonulalp I, Mengi A, Un C. IGF-1 and IGF-1r gene polymorphisms in East Anatolian Red and South Anatolian Red cattle breeds. Genetika. 2010;46:497–501. [PubMed] [Google Scholar]
- 56.Szewczuk, M. Polymorphism in exon 2 encoding the putative ligand binding pocket of the bovine insulin-like growth factor 1 receptor affects milk traits in four different cattle breeds. J Anim Breed Genet, doi:10.1111/jbg.12216 (2016). [DOI] [PubMed]
- 57.Li C, et al. Genome wide association study identifies 20 novel promising genes associated with milk fatty acid traits in Chinese Holstein. PLoS One. 2014;9:e96186. doi: 10.1371/journal.pone.0096186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Inagaki T, et al. Fibroblast growth factor 15 functions as an enterohepatic signal to regulate bile acid homeostasis. Cell Metab. 2005;2:217–225. doi: 10.1016/j.cmet.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 59.Yu C, et al. Elevated cholesterol metabolism and bile acid synthesis in mice lacking membrane tyrosine kinase receptor FGFR4. J Biol Chem. 2000;275:15482–15489. doi: 10.1074/jbc.275.20.15482. [DOI] [PubMed] [Google Scholar]
- 60.Chawla A, Repa JJ, Evans RM, Mangelsdorf DJ. Nuclear receptors and lipid physiology: opening the X-files. Science. 2001;294:1866–1870. doi: 10.1126/science.294.5548.1866. [DOI] [PubMed] [Google Scholar]
- 61.Brendel C, Schoonjans K, Botrugno OA, Treuter E, Auwerx J. The small heterodimer partner interacts with the liver X receptor alpha and represses its transcriptional activity. Mol Endocrinol. 2002;16:2065–2076. doi: 10.1210/me.2001-0194. [DOI] [PubMed] [Google Scholar]
- 62.Gill JL, Bishop SC, McCorquodale C, Williams JL, Wiener P. Identification of polymorphisms in the malic enzyme 1, NADP(+)-dependent, cytosolic and nuclear receptor subfamily 0, group B, member 2 genes and their associations with meat and carcass quality traits in commercial Angus cattle. Anim Genet. 2012;43:88–92. doi: 10.1111/j.1365-2052.2011.02216.x. [DOI] [PubMed] [Google Scholar]
- 63.Novak EM, Innis SM. Impact of maternal dietary n-3 and n-6 fatty acids on milk medium-chain fatty acids and the implications for neonatal liver metabolism. Am J Physiol Endocrinol Metab. 2011;301:E807–817. doi: 10.1152/ajpendo.00225.2011. [DOI] [PubMed] [Google Scholar]
- 64.Glaser C, Heinrich J, Koletzko B. Role of FADS1 and FADS2 polymorphisms in polyunsaturated fatty acid metabolism. Metabolism. 2010;59:993–999. doi: 10.1016/j.metabol.2009.10.022. [DOI] [PubMed] [Google Scholar]
- 65.Lattka E, Illig T, Koletzko B, Heinrich J. Genetic variants of the FADS1 FADS2 gene cluster as related to essential fatty acid metabolism. Curr Opin Lipidol. 2010;21:64–69. doi: 10.1097/MOL.0b013e3283327ca8. [DOI] [PubMed] [Google Scholar]
- 66.Gillingham LG, et al. Dietary oils and FADS1-FADS2 genetic variants modulate [13C]alpha-linolenic acid metabolism and plasma fatty acid composition. Am J Clin Nutr. 2013;97:195–207. doi: 10.3945/ajcn.112.043117. [DOI] [PubMed] [Google Scholar]
- 67.Ibeagha-Awemu, E. M., Akwanji, K. A., Beaudoin, F. & Zhao, X. Associations between variants of FADS genes and omega-3 and omega-6 milk fatty acids of Canadian Holstein cows. Bmc Genet15, doi:10.1186/1471-2156-15-25 (2014). [DOI] [PMC free article] [PubMed]
- 68.Bionaz M, Loor JJ. Gene networks driving bovine milk fat synthesis during the lactation cycle. BMC Genomics. 2008;9:366. doi: 10.1186/1471-2164-9-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Allende ML, Yamashita T, Proia RL. G-protein-coupled receptor S1P(1) acts within endothelial cells to regulate vascular maturation. Blood. 2003;102:3665–3667. doi: 10.1182/blood-2003-02-0460. [DOI] [PubMed] [Google Scholar]
- 70.Yamada T, et al. Novel SNP in 5′ flanking region of EDG1 associated with marbling in Japanese Black beef cattle. Anim Sci J. 2009;80:486–489. doi: 10.1111/j.1740-0929.2009.00665.x. [DOI] [PubMed] [Google Scholar]
- 71.Pappa KI, et al. The major circadian pacemaker ARNT-like protein-1 (BMAL1) is associated with susceptibility to gestational diabetes mellitus. Diabetes Res Clin Pract. 2013;99:151–157. doi: 10.1016/j.diabres.2012.10.015. [DOI] [PubMed] [Google Scholar]
- 72.Richards J, Diaz AN, Gumz ML. Clock genes in hypertension: novel insights from rodent models. Blood Press Monit. 2014;19:249–254. doi: 10.1097/MBP.0000000000000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lee YJ, Han DH, Pak YK, Cho SH. Circadian regulation of low density lipoprotein receptor promoter activity by CLOCK/BMAL1, Hes1 and Hes6. Exp Mol Med. 2012;44:642–652. doi: 10.3858/emm.2012.44.11.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Siu MK, Cheng CY. Extracellular matrix and its role in spermatogenesis. Adv Exp Med Biol. 2008;636:74–91. doi: 10.1007/978-0-387-09597-4_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Baeza-Raja B, Akassoglou K. Glucose homeostasis and p75NTR: the sweet side of neurotrophin receptor signaling. Cell Cycle. 2012;11:3151–3152. doi: 10.4161/cc.21590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dombroski BA, et al. Gene expression and genetic variation in response to endoplasmic reticulum stress in human cells. Am J Hum Genet. 2010;86:719–729. doi: 10.1016/j.ajhg.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Juckstock J, Kimmich T, Mylonas I, Friese K, Dian D. The inhibin-beta C subunit is down-regulated, while inhibin-beta E is up-regulated by interferon-beta 1a in Ishikawa carcinoma cell line. Arch Gynecol Obstet. 2013;288:883–888. doi: 10.1007/s00404-013-2848-2. [DOI] [PubMed] [Google Scholar]
- 78.Sekine T, et al. Identification of multispecific organic anion transporter 2 expressed predominantly in the liver. FEBS Lett. 1998;429:179–182. doi: 10.1016/S0014-5793(98)00585-7. [DOI] [PubMed] [Google Scholar]
- 79.Kok, L. D. et al. Assignment of liver-specific organic anion transporter (SLC22A7) to human chromosome 6 bands p21.2– > p21.1 using radiation hybrids. Cytogenet Cell Genet88, 76–77, 15489 (2000). [DOI] [PubMed]
- 80.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nature Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Guttman M, et al. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nature biotechnology. 2010;28:503–510. doi: 10.1038/nbt.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sun L, et al. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic acids research. 2013;41:e166–e166. doi: 10.1093/nar/gkt646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kong L, et al. CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic acids research. 2007;35:W345–349. doi: 10.1093/nar/gkm391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Punta M, et al. The Pfam protein families database. Nucleic acids research. 2012;40:D290–301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lin MF, Jungreis I, Kellis M. PhyloCSF: a comparative genomics method to distinguish protein coding and non-coding regions. Bioinformatics. 2011;27:i275–282. doi: 10.1093/bioinformatics/btr209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Altschul, S. F., Alejandro, T. L. M., Schäffer1, A. Jinghui Zhang, Zheng Zhang2, Webb Miller2 and David J. Lipman. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic acids research25 (1997). [DOI] [PMC free article] [PubMed]
- 88.Li Y, et al. Performance comparison and evaluation of software tools for microRNA deep-sequencing data analysis. Nucleic acids research. 2012;40:4298–4305. doi: 10.1093/nar/gks043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Anton Enright, J. et al. MicroRNA targets in Drosophila. Genome Biology5 (2003). [DOI] [PMC free article] [PubMed]
- 90.Kruger J, Rehmsmeier M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic acids research. 2006;34:W451–454. doi: 10.1093/nar/gkl243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Trapnell C, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nature protocols. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Robinson MDDJM, Gordon KS. EdgeR: a Bioconductor package for differential expression analysis of digital gene expression data. bioinformatics. 2010;26:2. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mao X, Cai T, Olyarchuk JG, Wei L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics. 2005;21:3787–3793. doi: 10.1093/bioinformatics/bti430. [DOI] [PubMed] [Google Scholar]
- 94.Simillion C, Liechti R, Lischer HE, Ioannidis V, Bruggmann R. Avoiding the pitfalls of gene set enrichment analysis with SetRank. BMC bioinformatics. 2017;18:151. doi: 10.1186/s12859-017-1571-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Roll JS. A user centered database for astronomy. Astr Soc P. 1996;101:536–539. [Google Scholar]
- 96.Yang JH, et al. starBase: a database for exploring microRNA-mRNA interaction maps from Argonaute CLIP-Seq and Degradome-Seq data. Nucleic Acids Res. 2011;39:D202–D209. doi: 10.1093/nar/gkq1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Li JH, Liu S, Zhou H, Qu LH, Yang J. H. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic acids research. 2014;42:D92–97. doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hochberg, Y. B. Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B57 (1995).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


