Abstract
Clustered class-I small heat-shock protein (sHSP) chaperone genes, SlHSP17.6, SlHSP20.0 and SlHSP20.1, in tomato are demonstrated to be transcriptionally regulated by ethylene during mature green (MG) fruit transition into ripening. These genes are constitutively expressed at MG fruit stage in two different tomato genotypes as well as in their ripening mutants, including rin, nor and Nr, and an ethylene-deficient transgenic line, ACS2-antisense. Notably, ethylene treatment of the MG fruit led to significant sHSP gene suppression in both wild-types, ACS2-antisense, nor/nor and Nr/Nr, but not the rin/rin mutant. Inability of ethylene to suppress sHSP genes in rin/rin mutant, which harbors MADS-RIN gene mutation, suggests that MADS-RIN transcription factor regulates the expression of these genes. Treatment of the wild type and ACS2-antisense fruit with the ethylene-signaling inhibitor, 1-methylcyclopropane (1-MCP), reversed the sHSP gene suppression. Transcripts of representative ethylene-responsive and ripening-modulated genes confirmed and validated sHSP transcript profile patterns. In silico analysis in conjunction with chromatin immunoprecipitation demonstrated MADS-RIN protein binding to specific CArG motifs present in the promoters of these chaperone genes. The results establish MADS-RIN protein as a transcriptional regulator of these chaperone genes in an ethylene-dependent manner, and that MADS-RIN protein-regulation of sHSPs is integral to tomato fruit ripening.
Introduction
Small heat shock proteins (sHSPs) are ubiquitous ancient proteins with conserved structural features, which evolved before the divergence of Archaea, Bacteria, and Eukarya1–7. Research has established that sHSPs function as molecular chaperones, assisting protein folding and preventing aggregation of their target proteins7–10. Although sHSPs were initially discovered among up-regulated proteins during heat stress, it is now known that they are also constitutively expressed10. Human sHSPs – HSP20, HSP22 and HSP27 - have biological roles varying from muscle contraction and metabolism (HSP20), regulation of apoptosis and carcinogenesis (HSP22) to oxidative stress protection and cytoskeleton regulation (HSP27)11. In plants, diverse sHSPs are found with distinct subfamilies, and their up-regulation by heat has presented an avenue to study their role in thermotolerance7. They are also induced by other stresses12.
Among eukaryotes, sHSPs are more abundant in land plants, ranging in size from 12–42 kD13. Our interest in exploring regulation of sHSPs in plant biology, particularly during ripening of fruits, emanated from previous studies that identified VISCOSITY 1 (VIS1), a tomato sHSP gene, as a regulator of pectin depolymerization effecting juice viscosity of the fruit14 and tomato sHSP21 as a stabilizer of photosystem II against oxidative stress and color change during tomato fruit ripening15. Moreover, a unique intron-less cluster of three sHSP chaperone genes, SlHSP17.6, SlHSP20.0 and SlHSP20.1, is resident on the short arm of chromosome 6 in tomato and is differentially expressed during tomato fruit ripening16. Interestingly, the 5′ end of each of these sHSP genes is enriched in sequence motifs responsive to hormones, i.e., ABA, ethylene, GA, and methyl jasmonate16. The presence of ethylene-responsive elements in these sHSP genes suggests that ethylene may regulate them during fruit ripening. Another sHSP protein, sHSP21, was shown to regulate pigment development in tomato fruit15. Ethylene directly or indirectly promotes transcription/translation of numerous ripening-related genes, including those associated with cell wall breakdown, carotenoid biosynthesis, aroma development, pigment accumulation, fruit softening, and flavor17, 18.
Tomato is an excellent system to dissect ethylene-mediated regulation of genes during fruit ripening19–24. More so, since several ripening tomato mutants, namely, rin, nor, Nr, alc, cnr, frm and DFD, are a good resource for characterization of fruit ripening19. Subsequent to the discovery that the rin mutation encodes a MADS-box transcription factor that regulates ripening25, the RIN protein was found to regulate a number of other genes involved in plant development25–28. Never-ripe 2 (Nr-2) mutation is a semi-dominant mutation in a tomato ethylene receptor orthologous to Arabidopsis AtETR1 ethylene receptor29, while the non-ripening (Nor) mutation is linked to a change in the expression of a NAC transcription factor, both affect ethylene and ripening-dependent gene expression30.
A plethora of genes are known to regulate fruit ripening. To the best of our knowledge, little is known about how sHSPs are transcriptionally regulated during tomato fruit ripening. To gain an insight into possible ethylene-mediated regulation of three clustered tomato sHSP genes, we studied their gene expression in wild-type Ailsa Craig variety and its isogenic ripening mutants – rin/rin, nor/nor and Nr/Nr, and in the wild-type Ohio8245 processing tomato line and its transgenic line harboring an anti-sense ACC synthase 2 gene (2AS-AS). Here, we present results demonstrating that the class 1 sHSP gene cluster on Chr. 6 is negatively regulated by ethylene prior to fruit ripening. Further, we demonstrate that all the three tomato sHSP genes (17.6, 20.0 and 20.1) harbor functional and interactive RIN-binding, CArG motifs in their promoters. The relevance of these findings to the control of ripening is discussed.
Materials and Methods
Plant Materials
Mutant tomato (Solanum lycopersicum cv Alisa Craig) lines of ripening-inhibitor (rin), non-ripening (nor) and Never-ripe (Nr) were repeatedly backcrossed into the cultivar Ailsa Craig to obtain near isogenic lines for these mutants. Mutant and wild type (WT) Ailsa Craig and a previously characterized ACC-synthase 2 (ACS2) silenced transgenic line (2AS-AS) in cultivar Ohio8245 and its azygous WT control31 were grown in a temperature-controlled greenhouse under natural light conditions. Tomato fruits at 5 days before breaker (−)5BR – which is equivalent to mature green (MG) stage, breaker (BR) and red ripe (7 days after breaker [BR + 7]) were analyzed by RNA gel-blot analysis. Fruits from the Ohio8245 WT and its 2AS-AS line were harvested at (−)5BR (MG stage), BR and BR + 8 and then treated and analyzed as described below.
Ethylene and 1-MCP treatments
Mature Green [MG, (−)5BR] tomato fruits were used for ethylene and 1-methylcyclopropene (1-MCP) treatment in three independent biological replicates. Briefly, MG tomatoes were harvested from each line, wiped with 70% ethanol and then rinsed with double distilled water. For ethylene treatment, fruits were placed in a known volume of air-tight jars and sealed with rubber septum glued to the lid. Ethylene was injected into the jars with a syringe needle to a final concentration of 25 ppm. A separate batch of MG fruit harvested at the same time was treated with 2 ppm 1-MCP to inhibit the ethylene response - SmartFresh (AgroFresh, Collegeville, PA, USA) is a powder that includes 1-MCP at 3.3%. The powder releases MCP as a gas when added to water. SmartFresh, 20 mg, was mixed vigorously with 380 mg of sucrose (1:20 w/w). The sucrose diluted SmartFresh (2.5 mg L−1) was put into a 1.5 mL tube that was hung inside the closed container and 0.5 mL of water injected into the 1.5 mL tube through the septum in the lid of the container. Corresponding control fruits (air) were enclosed in a similar container without ethylene or 1-MCP treatment. Fruits were collected at 0, 12 and 24 hours of treatment, peeled and the pericarp immediately frozen in liquid nitrogen. For ethylene treatment of leaves, 8–10 fully expanded leaves harvested from mature Ailsa Craig plants were treated with 25 ppm ethylene for 0, 24, 48, 72 and 96 hours in the dark at 25 °C. At the indicated times, leaf samples were removed, immediately frozen in liquid nitrogen and then stored at −70 °C.
Total RNA extraction, RNA gel blotting, cDNA synthesis and quantitative RT-PCR
Frozen tomato fruit and leaf samples were ground with liquid nitrogen to a fine powder. Total RNA was extracted from 100 mg tissue using Plant RNeasy kit according to manufacturer’s instructions (Qiagen), and treated with RNase-Free DNase (QIAGEN). Northern blotting was done as described previously25. A total of 2 µg RNA was used for cDNA synthesis using the iScript Advanced cDNA synthesis kit (Bio-Rad). cDNA was diluted 10-fold for further use. Quantitative Real-time PCR was performed using Sso Advanced Universal SYBR Green Supermix (Bio-Rad) in a Bio-Rad cycler (CFX96 Bio-Rad Real Time PCR machine). PCR conditions applied were: 95 °C for 10 min; 95 °C for 15 sec; and 60 °C for 60 sec (40 cycles), followed by melt curve analysis32. For relative quantification, Ct values of gene expression were quantified according to ∆∆CT method33. Ct/Cq quantification cycle was calculated by following the Bio-Rad CFX Manager 3.1 based on the MIQE (Minimum Information for publication of Quantitative real-time PCR Experiments) guidelines34. Relative fold changes were calculated as previously described35. Tomato actin (SlACT2) was used as a constitutively expressed gene to normalize the expression of the target genes32. Accession numbers and primer sequences of genes analyzed are listed in Supplementary Table S1. Data represent the average ± standard error from a minimum of three independent biological replicates for each gene transcript profiled.
In silico analysis of class I sHSP (17.6, 20.0 and 20.1) promoters for RIN protein binding (CArG motifs) sites
Promoters (≈3 kb of the 5′ upstream sequence from the start codon, ATG) of all the three class I tomato sHSPs, 17.6, 20.0 and 20.1, were identified based on information from the International Tomato Genome Sequencing Consortium (SGN; solgenomics.net) database (version ITAG 2.4). Two databases, Plant CARE relational database36 and PLACE (the plant-cis-acting regulatory DNA elements) database37 were used to identify putative regulatory motifs in the sHSP promoters, including the potential RIN binding, CArG-box motif sequences [{C(C/T) (A/T)6(A/G)G}, {C(A/T)8G} and {C(C/T)(A/T)G(A/T)4 (A/G)G}]38, 39. The consensus sequence for the CArG boxes was analyzed using the WebLogo 3 program (http://weblogo.threeplusone.com/)40, 41. The CArG motifs found in the sHSP gene promoters are listed in Supplementary Table S2.
Chromatin immunoprecipitation (ChIP) and quantification of enrichment via ChIP-qPCR
For chromatin cross-linking, breaker (BR) tomato fruit pericarp tissue was diced (1–2 cm2) and placed in a 50 mL falcon tube filled with 20 ml of 1% formaldehyde (Sigma). Vacuum infiltration was done at 62 cm of Hg for 10–15 min until air bubbles appeared in the formaldehyde solution. Cross-linking was stopped by adding 0.125 M glycine and vacuum infiltration continued for an additional 5 min. Cross-linked tissue was then rinsed thrice with water, frozen in liquid nitrogen and stored at −80 °C. For each ChIP reaction, 1 g of the cross-linked tissue was analyzed and 3 µL of RIN antibody was used42. A plant ChIP Kit (Epigentek) was used for ChIP assays according to manufacturer’s instructions with a few modifications43. Briefly, chromatin was isolated from 1 g of crosslinked tissue and then sheared by a 30 sec pulse at 30 second intervals for 10 cycles in a Branson 2200 water bath sonication machine at default settings (50/60HZ Power). The antibody coating step was performed at 4 °C for 8–10 h with anti-RIN antibody25. Rabbit IgG was used as negative control. Chromatin binding was carried out at 4 °C on an orbital shaker at 80–100 rpm for 8 h. Primers spanning each CArG motif present in each sHSP promoter (17.6, 20.0 and 21.0) are listed in Supplementary Table S3. Promoter primer specificity was checked with blast program [International Tomato Genome Sequencing Consortium (SGN; solgenomics.net) database, version ITAG 2.4] and their specificity tested using genomic DNA as template with PCR. PCR products were checked on agarose gels, and single amplicons representing the desired size were obtained. These primers were used to assay for the enrichment of CArG motifs with quantitative real-time PCR following the PCR conditions described above. Percent input method [100*2Cq (adjusted input) - Cq (IP)] was followed to calculate enrichment of fragments44 where n = 3 (three BR fruits from three different plants were used for chromatin immunoprecipitation to calculate the average and standard deviation in ChIP- q-PCR reaction).
Data analysis and statistics
Statistical analysis was carried out using two-tailed student’s t-test with P values < 0.05 treated as statistically significant.
Results
Upregulation of Class-I SlHSP17.6, SlHSP20.0 and SlHSP20.1 gene transcripts - transcriptomic and qRT-PCR analyses
To gain insight into the expression of the three clustered sHSP’ genes (17.6, 20.0, 20.1) during tomato fruit ripening, available transcriptome data (Solanum lycopersicum cv. Heinz) was retrieved from International Tomato Genome Sequencing Consortium (SGN; solgenomics.net) database (version ITAG 2.4). RPKM (reads per kilobase of transcript per million mapped reads) data obtained for the three transcripts during plant development and fruit ripening are presented as heat map in Fig. 1a. Transcripts of all the three SlHSP genes are highly up-regulated during ripening, abundance of each increasing from MG to BR stage [7.28-fold for SlHSP17.6, 5.35-fold for SlHSP20.0 and 17.40-fold for SlHSP20.1], with SlHSP20.1 being more abundant at the BR + 8 stage (Fig. 1a). Based on these transcriptomic data, it is noted that these sHSP genes are less abundant in other plant parts and during tomato development relative to that in the fruit and during ripening. Quantitative real time PCR (Q-PCR) analysis of fruit RNA from two tomato varieties, namely, S. lycopersicum cv. Ohio8245 and Alisa Craig during ripening stages was performed for the three SlHSP genes for further validation. Results in Fig. 1b confirm ripening-regulated expression of the three SlHSP (17.6, 20.0, 20.1) genes in the processing tomato Ohio8245 variety as well as in Alisa Craig.
Figure 1.
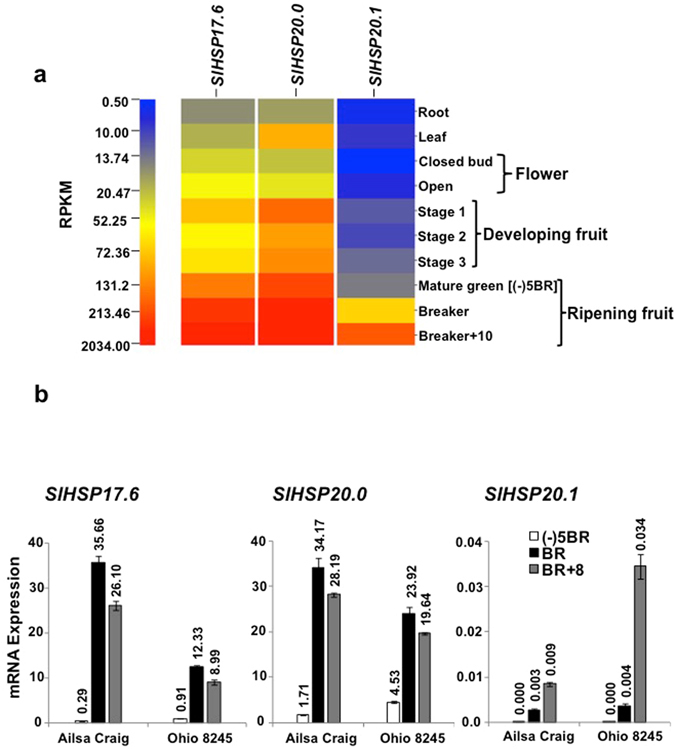
Transcriptome analysis and Q-PCR of class-I SlHSP genes (17.6, 20.0, 20.1) in wild type tomato during ripening. (a) RPKM values for the indicated SlHSP gene transcripts in tomato during plant growth and development and fruit ripening [root, leaf, bud, flower, 3 fruit developmental stages, mature green, breaker and red ripe (breaker + 10)] were derived from RNA-seq data in the SGN database (Solanum lycopersicum cv. Heinz). (b) Q-PCR analysis of SlHSP (17.6,20.0 and 20.1) genes was performed using total RNA isolated from Solanum lycopersicum cv. Ohio8245 and Ailsa Craig wild type fruits at the indicated ripening stages. A 10-fold diluted cDNA was used for transcript quantification using gene specific primers.
The Class-I SlHSP17.6, SlHSP20.0 and SlHSP20.1 gene transcripts were all more abundant in wild-type Ailsa Craig fruit than in the ripening mutants
RNA gel-blot analysis of HSP17.6, HSP20.0 and HSP20.1 genes in Ailsa Craig wild type (WT) and its near isogenic mutant lines, ripening-inhibitor (rin/rin), non-ripening (nor/nor) and Never-ripe (Nr/Nr), indicated differential expression and abundance during ripening (Fig. 2a,b). Abundance of SlHSP17.6 and SlHSP20.0 transcripts in the WT increased at the BR stage as compared to the MG fruit [(−)5BR], and remained more or less the same after 7 days (BR + 7; red ripe fruit) (Fig. 2a,b; WT, SlHSP17.6, SlHSP20.0). In contrast, SlHSP20.1 transcripts were undetectable at (−)5BR stage, increased at BR and showed reasonable abundance at BR + 7 stage, but lower than that found for SlHSP17.6 and SlHSP20.0 transcripts (Fig. 2a,b; WT, compare SlHSP20.1 with SlHSP17.6 and SlHSP20).
Figure 2.
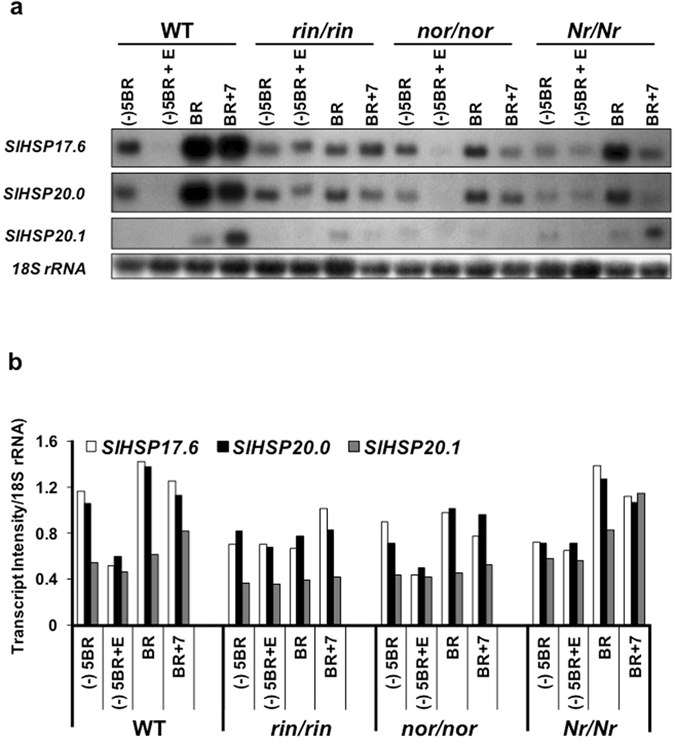
RNA blot-analysis of class-I SlHSP (17.6, 20.0 and 20.1) genes in wild type (Solanum lycopersicum cv. Ailsa Craig) and its ripening mutants. (a) Fruits from wild type tomato var. Ailsa Craig along with near isogenic lines of ripening-inhibitor (rin/rin), non-ripening (nor/nor) and Never-ripe (Nr/Nr) mutant fruits were harvested at mature green [5 days before breaker (−)5BR)], breaker (BR) and red ripe (7 days after breaker, BR + 7) stages as described25. RNA was isolated, separated on gels, blotted and northern blot analysis carried out25, using gene specific probes as previously described46. Fruits at mature green stage (−)5BR from the indicated lines were given ethylene treatment and designated as (−)5BR + E. (b) Quantification of northern blot analysis using Image J program (https://imagej.nih.gov/ij/). The band intensity was calibrated with the band intensity of 18 S rRNA internal control used in northern blotting.
In the rin/rin mutant, the abundance of SlHSP17.6 and SlHSP20.0 transcripts was apparent at (−)5BR but lower than the WT control, and remained more or less the same at BR and BR + 7 stages (Fig. 2a,b: rin/rin, SlHSP17.6 and SlHSP20.0). In contrast, SlHSP20.1 transcript abundance was drastically lower, with a minor but significant detection at BR stage (rin/rin, SlHSP20.1). In both nor/nor and Nr/Nr mutant fruit, abundance of SlHSP17.6 and SlHSP20.0 transcripts at the (−)5BR stage was lower than the WT control line, peaked at the BR stage and decreased thereafter; the abundance of SlHSP20.1 transcripts in these two mutants was barely detectable and no different than that in the rin/rin mutant (Fig. 2a,b; nor/nor, Nr/Nr). Since these mutants are deficient in ethylene production, it was surmised that the candidate class-I SlHSP gene transcript expression is regulated by ethylene.
Exogenous ethylene application suppresses SlHSP gene transcripts in MG fruit of WT, nor/nor and Nr/Nr but not in rin/rin mutant line
To delineate which genetic locus (considering the three mutants) regulates the expression of class-I sHSP genes, the (−)5BR (MG fruit) of WT and mutant lines were held in air-tight desiccators with 25 ppm ethylene (abbreviated as E) for 15 h, and the abundance of SlHSP17.6, SlHSP20.0 and SlHSP20.1 transcripts was analyzed by RNA gel-blot analysis. Surprisingly, transcripts of all the three SlHSP genes in Ailsa Craig WT as well as in nor/nor mutant fruit were found suppressed by ethylene treatment (Fig. 2a,b; WT and nor/nor, compare lane [−5BR + E with lane (−)5BR], and slightly in the Nr/Nr mutant [Nr/Nr, compare lane (−)5BR + E with lane (−)5BR]. However, ethylene treatment did not elicit inhibition of SlHSP17.6 and SlHSP20 transcripts in the rin mutant [Fig. 2; rin/rin, compare lane (−)5BR + E with lane (−)5BR]. These results indicated that RIN is likely associated with ethylene-induced suppression of class-I SlHSP17.6 and SlHSP20.0 chaperone gene transcripts.
Quantitative-Real Time PCR analysis confirms ethylene-mediated suppression of class-I SlHSP genes in wild type tomato and its anti-ACS2 homozygous transgenic line
For further studies on ethylene regulation of class I SlHSP17.6, SlHSP20.0 and SlHSP20.1 gene transcripts, we employed the Ohio8245 processing variety of tomato and its antisense-ACS2 transgenic line (designated as ACS2-AS, which is constitutively deficient in the in vivo ethylene production)31. MG fruit from Ohio8245 and the ACS2-AS homozygous line were incubated separately with ethylene or 1-MCP (inhibitor of ethylene signaling) or left in the air as described in the Materials and Methods section. At time 0, 12 and 24 h after treatment, fruit were sampled, their pericarp RNA isolated, and abundance of the SlHSP gene transcripts analyzed by Q-PCR. Expression of SlHSP17.6 was lower at 12 h of treatment with ethylene but higher in 1-MCP-treated Ohio8245 compared to samples in air, and by 24 h SlHSP17.6 expression was further suppressed in ethylene-treated samples (Fig. 3a, SlHSP17.6). This trend in SlHSP17.6 transcript levels was mimicked in the ACS2-AS, ethylene-deficient fruit, which registered much higher suppression in the ethylene-treated fruit (Fig. 3a, ACS2-AS). Interestingly, SlHSP17.6 and SlHSP20.0 transcript abundance was found decreased also in ethylene-treated tomato leaves for up to 96 h (Supplementary Fig. S1).
Figure 3.
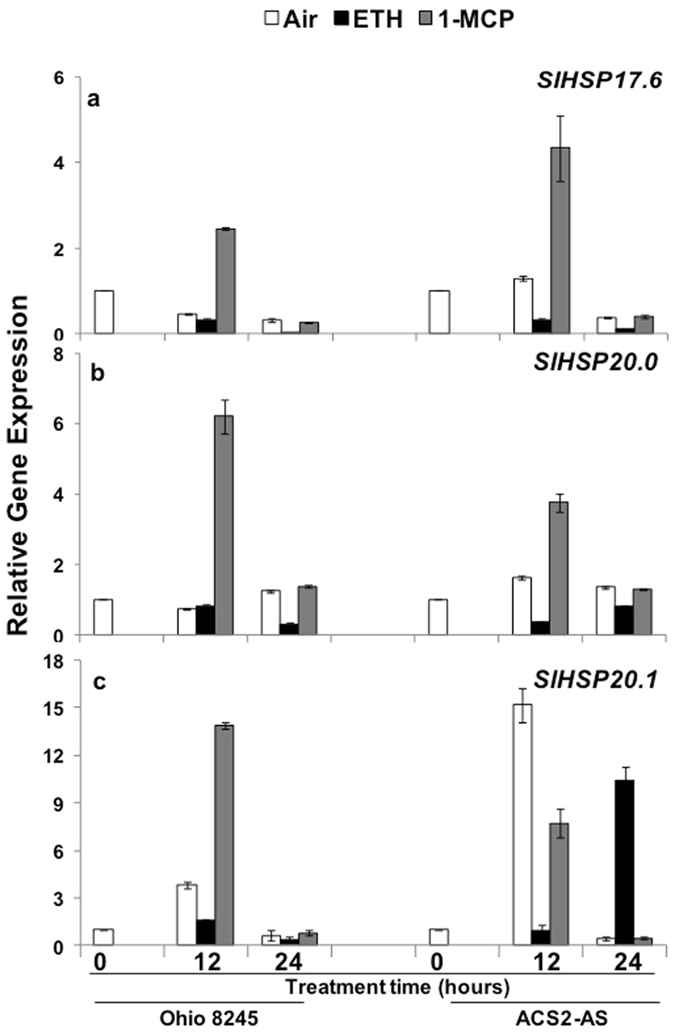
Quantitative RT-PCR expression analysis of class-I SlHSP (17.6, 20.0 and 20.1) genes in wild type tomato (Solanum lycopersicum cv. Ohio8245) and ethylene-deficient genotype (ACS2-AS). Fruits at mature green stage from two genotypes (WT - Ohio8245) and ethylene-deficient genotype (ACS2-AS) were treated with ethylene(ETH), 1-MCP (1-MCP), or left in air. Fruit RNA was isolated from three independent biological samples and Q-PCR analysis was carried out as described in the Materials and Methods section. Error bars indicate standard deviation of data from a minimum of three replicates.
In the case of SlHSP20.0 expression, suppression by ethylene was apparent at 24 h of ethylene treatment in both Ohio azygous line and ACS2-AS fruits, while 1-MCP-treated fruits had higher expression. Moreover, ethylene inhibition of these transcripts was clearly seen early at 12 h of treatment (Fig. 3b, SlHSP20.0). A similar trend of ethylene suppression was found for SlHSP20.1 at 12 h with elevated expression in the 1-MCP-treated fruit (Fig. 3c, SlHSP20.1). However, ethylene-mediated suppression of SlHSP20.1 transcripts in ACS2-AS line was reversed in 24 h-treated fruit, with 1-MCP treatment blocking this up-regulation. It appears that the suppression of SlHSP20.1 transcripts in ethylene-deficient ACS2-AS fruit on prolonged incubation (i.e., 24 h) may require a higher ethylene dose. However, this observation needs to be followed further in future experiments. These results confirm that ethylene suppresses these three class-I SlHSP genes.
Validation of ethylene and 1-MCP treatment effects by testing expression analysis of known ethylene-responsive genes
To insure reliable ethylene and 1-MCP treatment and thus accurate interpretation of our SlHSP gene expression results, we evaluated the expression of 7 additional genes previously shown to be regulated by ethylene (Fig. 3). ACS6 suppression by ethylene and up-regulation by 1-MCP treatment was previously shown45. Similar to these effects, ACS6 expression was found highly suppressed in ethylene-treated Ohio azygous fruit but was elevated in 1-MCP-treated fruit, particularly in the 24-h samples (Fig. 4a). TAG1 is also known to be slightly suppressed in response to exogenous ethylene46. This too was found to be the case in our experiments, with 1-MCP treatment giving an elevated TAG1 expression in 24-h samples (Fig. 4b). Data on TAGL11 expression indicated clear effects both with 12-h and 24-h treatments. Specifically, in the ethylene-treated fruit, TAGL11 was suppressed while in 1-MCP-treated fruit its levels were elevated, indicating that ethylene suppresses TAGL11 transcript (Fig. 4c). PG1, Polygalacturonase 1, is known to be induced by exogenous ethylene47, 48. This was evident in 24-h ethylene-treated samples and reflected by being inhibited in MCP-treated fruit (Fig. 4d). RIN was shown to be slightly up-regulated by exogenous ethylene25. Also, as expected, RIN transcripts were abundant within 12-h exposure of control fruit to exogenous ethylene and 1-MCP inhibited their expression (Fig. 4e). However, by 24 h of treatment, while air-exposed fruits had elevated RIN transcripts, ethylene-stimulation was already found reduced, which was in concurrence with stimulation in the presence of 1-MCP (Fig. 4e). NR gene transcripts are also induced by exogenous ethylene49, and likewise, NR transcripts accumulated in response to ethylene while 1-MCP suppressed its expression, indicating NR as an ethylene-inducible gene (Fig. 4f). SR3L is a calcium-signaling gene regulated by RIN and ethylene during ripening50. In our experiment, this gene was slightly up-regulated by ethylene treatment at 12 h but the up-regulation was not significantly different than in air at 12 h or 24 h (Fig. 4g). These data confirmed that our hormone and inhibitor treatment protocols delivered gene transcription data in line with those reported previously.
Figure 4.
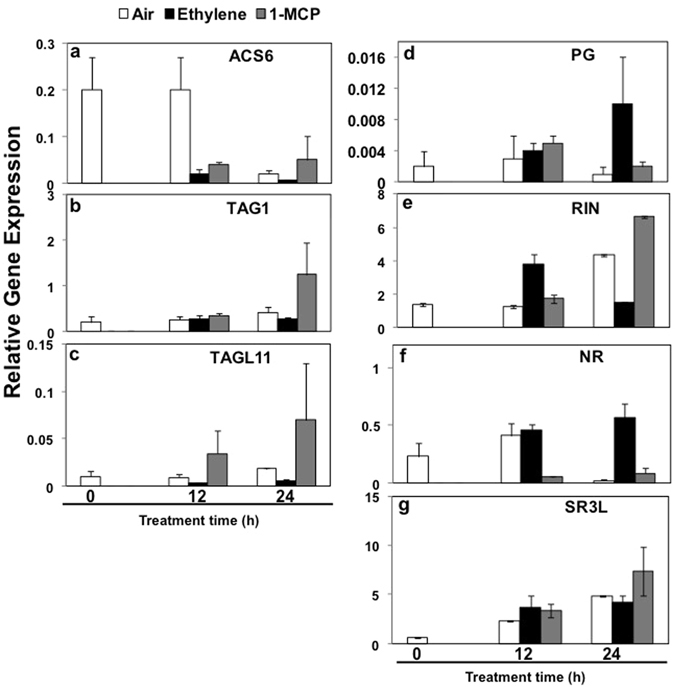
Quantitative RT-PCR analysis of ethylene responsive and nonresponsive genes in ethylene-treated wild type Ohio8245 tomato fruits. Fruits from wild type (Solanum lycopersicum cv. Ohio8245) control were exposed to air, ethylene or 1-MCP as described in the Materials and Methods section. RNA was isolated and transcripts for ACS6, TAG1, TAG11, PG, RIN, NR and SR3L genes were quantified by Q-PCR. Error bars indicate standard deviation from a minimum of three replicates.
In silico analysis of promoters of SlHSP17.6, SlHSP20.0 and SlHSP20.1 genes reveal putative RIN binding sites
Next, we analyzed the 3 Kb 5′ promoter regions of tomato SlHSP17.6, SlHSP20.0 and SlHSP20.1 genes for the presence of RIN binding ‘CArG’ motifs using PLACE37 and Plant Care databases36. All the three SlHSP genes were found decorated with the CArG RIN binding motif, with SlHSP17.6 and SlHSP20.0 each having 4 atypical motif types and SlHSP20.1 having 3 ‘atypical’ and 2 possible motif types (Supplementary Table S2). CArG cis element positions (denoted as ‘P’) for SlHSP17.6 were resident at −2221 (P1), −2306 (P2), −2345 (P3) and −2686 (P4) (Fig. 5a), for SlHSP20.0 at −303 (P1), −917 (P2), −1047 (P3) and −2014 (P4) (Fig. 5b), and for SlHSP20.1 at −901 (P1), −994(P2), −1323 (P3), −1341 (P4) and −1933 (P5) (Fig. 5c), respectively. Each of the four CArG cis-elements in the promoter of SlHSP17.6 and SlHSP20.0 genes fall in the category of ‘atypical’ [C(A/T)8G] cis elements, while in the promoter of SlHSP 20.1, three represent ‘atypical’ [C(A/T)8G] and the other two as ‘possible’ [C(C/T)(A/T)6(A/G)G] as previously noted28, 38, 39, 51.
Figure 5.
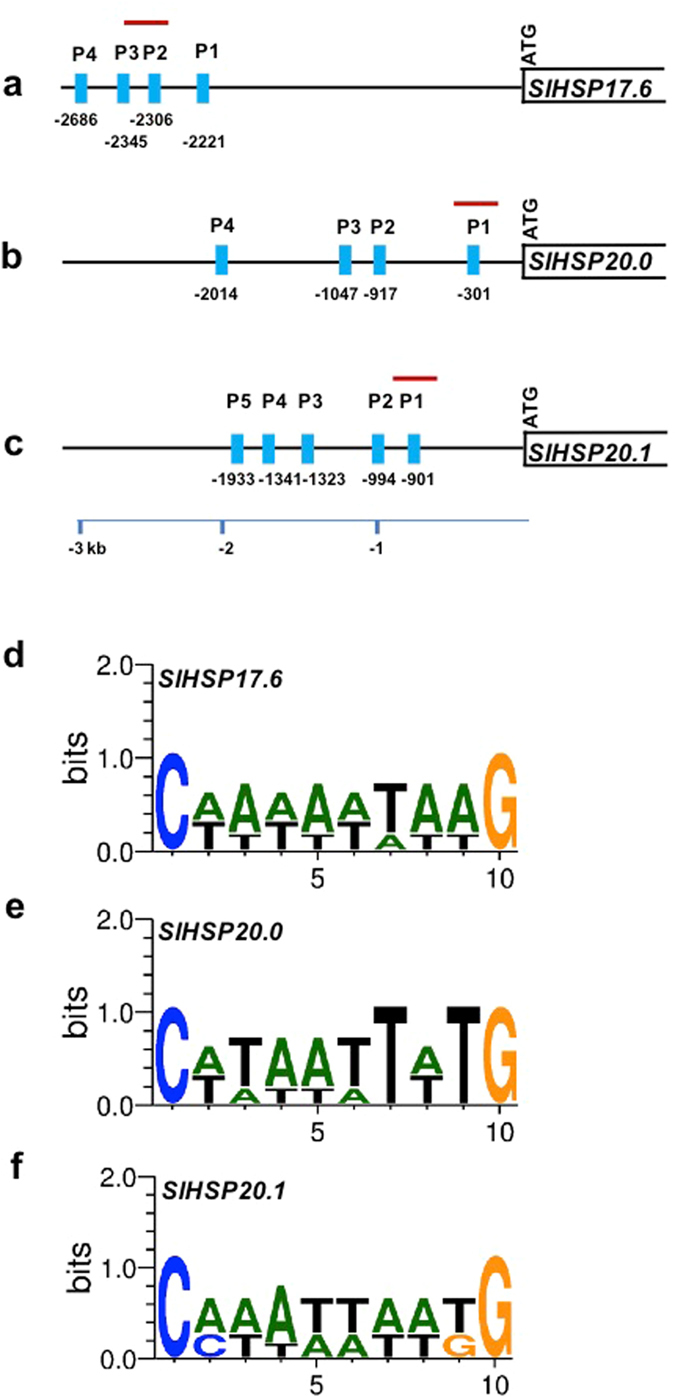
In silico analysis of class-I SlHSP (17.6, 20.0, 20.1) gene promoters. Promoter regions (≈3 kb of the 5′ upstream region of the start codon) of tomato SlHSP17.6, SlHSP20.0 and SlHSP20.1 genes were extracted using the International Tomato Genome Sequencing Consortium (SGN) database (version ITAG 2.4). Two databases, Plant CARE relational database35 and PLACE, the plant-cis-acting regulatory DNA elements database25 were used for plant cis-element search in the promoters of the described SlHSP genes. The possible RIN binding CArG-box motif sequences are [{C(C/T) (A/T)6(A/G)G}, {C(A/T)8G} and {C(C/T)(A/T)G(A/T)4 (A/G)G}]38, 39. All the CArG motifs found in SlHSP gene promoters are listed in Supplementary Table S2. The promoter position(s) significantly enriched in ChIP assay are highlighted with horizontal red line. (a) SlHSP17.6 gene promoter contains CArG cis element positions (here denoted as ‘P’), respectively at P1 (−2221), P2 (−2306), P3 (−2345) and P4 (−2686). (b) SlHSP20.0 gene promoter contains four CArG motifs positioned at P1 (−303), P2 (−917), P3 (−1047) and P4 (−2014), respectively. (c) SlHSP20.1 gene promoter harbors five CArG cis elements, positioned at P1 (−901), P2 (−994), P3 (−1323), P4 (−1341) and P5 (−1933). (d,e,f) show corresponding conserved locations and their distribution patterns in the 10 bp consensus sequence described in the text.
For prediction of the in vivo RIN binding CArG-box motif, we analyzed the collected CArG-box sequences from all the three class-I SlHSP gene promoters which yielded a 10-bp motif consensus sequence, C(T/A/C)(A/T)6(A/T/G)G (Fig. 5d,e,f). No conservation was observed in the flanking regions upstream and downstream of the 10-bp core. The presence of CArG cis elements in SlHSP genes suggested the possibility that they could be targets of MADS-box RIN transcription factor binding as previously characterized for other genes28, 42.
Chromatin immunoprecipitation (ChIP) identifies CArG motifs in the SlHSP17.6, SlHSP20.0 and SlHSP20.1 chaperone genes – confirmation that these are targets of SlMADS-RIN protein
Each CArG motif present in the promoter of the three SlHSP genes (Supplementary Table S2 and Fig. 5) was tested for binding to the RIN protein using a chromatin immunoprecipitation (ChIP) assay and an anti-RIN antibody42. The forward and reverse primer pairs spanning these cis motifs were designed (Supplementary Table S3) and tested with a nucleotide blast search in Solanaceae Genome Network (SGN) database for their unique and single hits. None of the primer pairs gave non-specific blast hits in SGN database. Further, single amplification product was obtained with the synthesized primers when tested with genomic DNA for their specificity. All CArG motifs were assayed for probable ChIP enrichment. Two positions from the SlHSP17.6 promoter, P2 (−2306) and P3 (−2345), were significantly enriched (P < 0.001) for the corresponding CArG cis element (Fig. 6a), whereas P1 position (−303) in SlHSP20.0 promoter was also significantly enriched (P < 0.03) (Fig. 6b). P1 (−901) position in SlHSP20.1 promoter was enriched with a significance P < 0.05 (Fig. 6c). The other predicted CArG cis element positions did not show significant enrichment in the ChIP assay. These data provide for the first time evidence that RIN protein interacts with the promoters of class-I SlHSP chaperone genes and mediates negative regulation of ethylene action.
Figure 6.
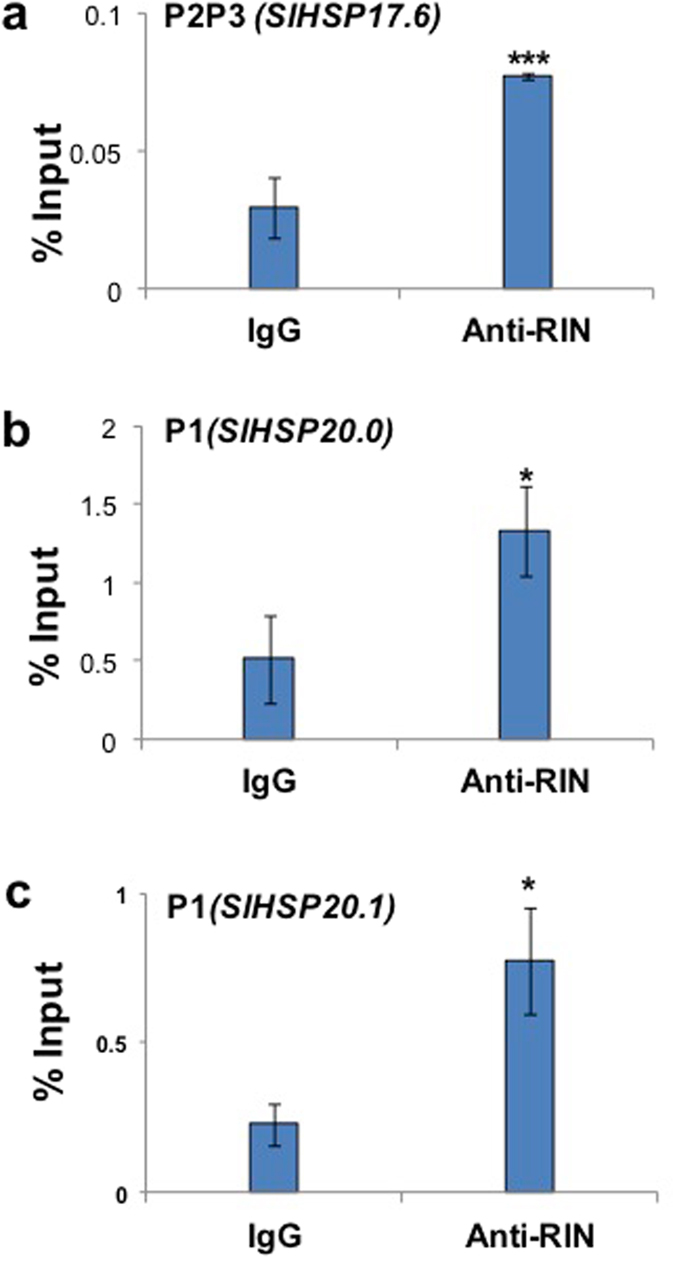
Chromatin immunoprecipitation (CHIP) and quantification of enrichment using CHIP-qPCR. Breaker stage Ailsa Craig tomato fruits were analyzed for chromatin cross linking. Plant CHIP Kit (Epigentek) was used for CHIP assays according to manufacturer’s instructions with few modifications43. Graph bars represent the relative DNA amounts of CArG-box sequences in the CHIP DNA recovered using either anti-RIN antibody or IgG antibody to those in the total input chromatin DNA. Data are the means of three independently prepared chromatin samples. Error bars indicate the standard deviation of each mean where n = 3. Rabbit IgG was used as a negative control. Asterisks indicate statistically significant differences, P < 0.05.
Discussion
We demonstrate here that ethylene and transcription of a subset of class-1 small heat-shock protein genes are interlinked, particularly at the initial phase of fruit ripening in tomato. Ethylene transiently suppressed SlHSP transcripts in two different tomato cultivars (Ailsa Craig and Ohio8245), A. Craig nor/nor, A. Craig Nr/Nr mutant and anti-ACS2 transgenic line in Ohio8245 background, but not in A. Craig rin/rin mutant. The insensitive nature of the rin ripening-deficient mutant to ethylene in modulating transcription of HSP genes suggests that they are associated with fruit ripening and regulated by ethylene albeit in a transient manner. The SlMADS-RIN transcription factor is known to regulate multiple processes involved in tomato fruit ripening42. Its mutation results in down regulation of many genes causing typical non-ripening phenotype beyond breaker stage28, 51. CArG cis elements are a common target of MADS box transcription factors (RIN protein) for regulating plant growth and development38, 39, 52. Interestingly, the promoter of each of the three SlHSP genes, SlHSP17.6, SlHSP20.0 and SlHSP20.1, are decorated with multiple CArG cis elements, and we demonstrated RIN-specific binding to a subset of these CArG motifs. Thus, RIN protein seems likely involved in the regulation of these SlHSP genes. SlHSP90 was previously reported to be a probable target of RIN protein28, and as shown here RIN protein has a larger regulatory network, which includes at least (but not limited to) three small heat shock protein genes. These class-I small heat shock proteins are encoded by nuclear genes and are resident in the chloroplast. It is noted here that ethylene-mediated and RIN-regulated transcriptional control for any chloroplast localized small heat shock protein has not been previously demonstrated. Additionally, this demonstrates a possible role of chloroplast localized proteins in tomato fruit ripening.
The class-1 sHSP genes studied here are basically intronless and uniquely clustered in a tandem repeat manner on the short arm of chromosome 6 in tomato16. Their differential expression during fruit ripening and the presence of many cis elements on their 5′ flanking region including motifs for ethylene perception are indicative of ethylene regulation1, 6. Previous studies with another class-I sHSP gene, HSP21, demonstrated its protein to function in plastid development (i.e., chloroplast to chromoplast transformation), protection of photosystem II under oxidative stress15 and heat stress53. In this regard, it is noted that Arabidopsis ethylene signaling mutants (ein2 and etr1) were found defective in basal thermotolerance but accumulated HSP101 and sHSP, suggesting that ethylene signaling may play a role in the thermal behavior of plants54. Although we have not pinned down the exact function of the intronless SlHSP genes, their expression patterns and ethylene regulation presented here implicate them in the ripening of tomato.
Small heat shock proteins have dynamic protein structure with diverse evolutionary origin and have been implicated not only in human disease but also stress acclimation in plants and other organisms7, 55. Therefore, ethylene-induced transient suppression of HSP genes mediated by RIN protein highlights their fundamental importance in plant biology, particularly fruit ripening. Our studies provide impetus to investigate yet unknown functional interactions of ethylene and HSP gene expression during fruit ripening. The transient nature of such a phenomenon could well indicate a role in the fruit’s transition from growth to ripening. Reports of transient suppression of a gene have appeared in the literature, each resulting in a specific function. For example, transiently regulated Rd22 gene expression in ABA-entrained plants indicated a probable memory in plants due to stress56. Similarly, cyanide produced concomitantly with ethylene biosynthesis results in transient transcriptional regulation of the cyanide-detoxifying gene, CYS-C1, in Arabidopsis, which then resulted in a cascade of downstream processes57. In addition, cell speciation in development is known to involve gene-regulatory responses to transient signals. Another example involves the transient accumulation of auxin which activates self-sustaining or hysteretic feedback system and results in unequivocal developmental responses58. Similarly, transient suppression of host gene expression due to viral infections in plants is a common phenomenon for viruses to establish the infection. PSbMV virus is known to transiently suppress the expression of host genes and inhibit host protein accumulation59.
Based on the above literature and data presented here, we hypothesize that ethylene-mediated transient down regulation of SlHSP genes at the onset of fruit ripening, when ethylene is just synthesized, is required for uninterrupted ripening. This contention is in tune with our demonstration that RIN regulates SlHSP gene transcription and the fact that RIN expression occurs at the onset of ripening of tomato fruit35. A model on the transcriptional regulation that interfaces ethylene, RIN and class I SlHSPs, including possible components involved, is presented in Fig. 7. This model builds on and shares some features with a previous fruit ripening model60. ACS2 regulates ethylene biosynthesis during fruit ripening and RIN is upstream of ethylene since RIN mutation abolishes ripening by inhibiting ethylene biosynthesis (pointed arrows) and ethylene inhibits SlHSP gene expression. A stronger inhibition of SlHSP gene expression in the ethylene-deficient ACS2-AS transgenic line adds support to the suggestion of an inhibitory role of ACS2 in regulating expression of the SlHSP genes (Fig. 7, blunt end arrows). Moreover, it is noted that the expression of SlHSP genes was not inhibited in the ethylene-treated RIN mutant, suggesting that RIN protein regulates the transcription of these genes. RIN mutant fruit does not ripen and thus RIN may primarily function at the transition stage at the onset of ripening. The data presented here implicate ethylene as a transcriptional regulator of SlHSPs genes in the fruit transition process from mature green to ripening. Further functional genomic studies are needed to characterize in planta the promoters of the sHSP genes studied here and use transgenic approaches to shed light on their specific role(s) in fruit physiology and ripening.
Figure 7.
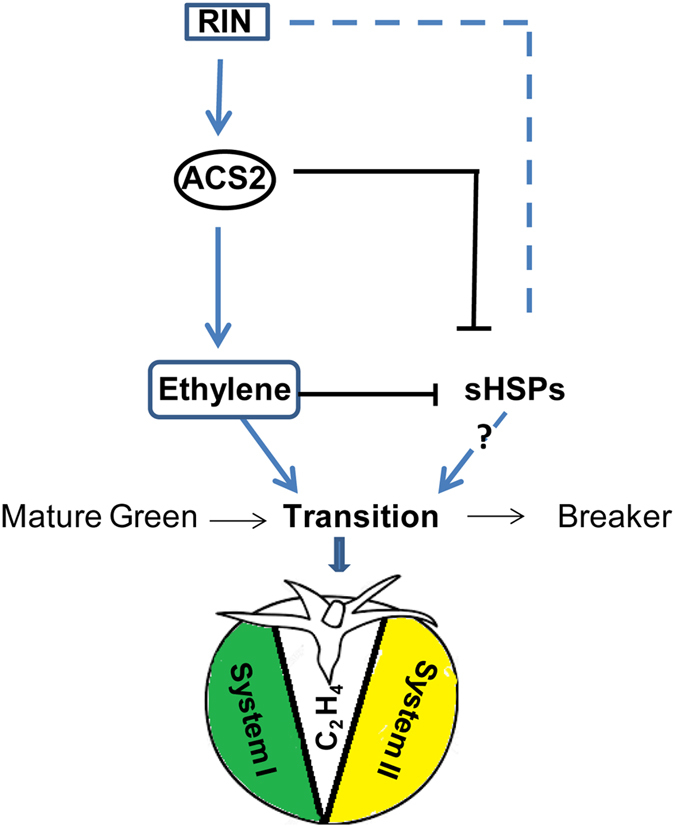
A schematic illustration showing additions to the previous fruit ripening model60 modified to include the SlHSP (17.6, 20.0, 20.1) genes with their proposed role in the transition from mature green to ripening phase of tomato fruit. RIN regulates ethylene biosynthesis in ripening tomato fruit involving ACS2 transcription (pointed blue arrows). During System1 ripening, low levels of auto-inhibitory ethylene are synthesized via SlACS1A,6 and SlACO1,3,4 60. At the transition stage, the ripening regulator RIN plays a critical role, where SlACS4 is induced to initiate a large increase in auto-catalytic ethylene that negatively feeds back on System I. SlACS2,4 and SlACO1,4 are involved in high ethylene production through System II60. SlHSP genes (sHPSs) are transiently suppressed by ethylene at the ripening-transition mediated by the RIN protein (blunt headed black arrows). Ethylene and ACS2 exert negative regulation of the three class-1 SlHSP gene transcripts via interactions involving RIN protein (blue dashes).
Electronic supplementary material
Acknowledgements
We thank Julia Vrebalov for her contribution to Fig. 2a. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture. USDA is an Equal Opportunity Provider and Employer.
Author Contributions
Conceived and designed the experiments: A.K.M., R.K.U., V.S. Performed the experiments: R.K.U., V.S., M.T. and J.J.G. Analyzed the data: R.K.U., V.S. and A.K.M. Facilitated the research: A.K.M. and S.V.R. Wrote the paper: A.K.M. and R.K.U. Revised and approved the final version of the paper: A.K.M., R.K.U., V.S., S.V.R., M.L.T. and J.J.G.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Vijaya Shukla and Rakesh K. Upadhyay contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-06622-0
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Plesofsky-Vig N, Vig J, Brambl R. Phylogeny of the alphacrystallin-related heat shock proteins. J. Mol. Evol. 1992;35:537–545. doi: 10.1007/BF00160214. [DOI] [PubMed] [Google Scholar]
- 2.Waters ER, Vierling E. Chloroplast small heat shock proteins: evidence for atypical evolution of an organelle-localized protein. Proc. Natl. Acad. Sci. USA. 1999;96:14394–14399. doi: 10.1073/pnas.96.25.14394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kappe G, Leunissen JA, de Jong WW. Evolution and diversity of prokaryotic small heat shock proteins. Prog. Mol. Subcell. Biol. 2002;28:1–17. doi: 10.1007/978-3-642-56348-5_1. [DOI] [PubMed] [Google Scholar]
- 4.Franck E, et al. Evolutionary diversity of vertebrate small heat shock proteins. J. Mol. Evol. 2004;59:792–805. doi: 10.1007/s00239-004-0013-z. [DOI] [PubMed] [Google Scholar]
- 5.Fu X, Jiao W, Chang Z. Phylogenetic and biochemical studies reveal a potential evolutionary origin of small heat shock proteins of animals from bacterial class A. J. Mol. Evol. 2006;62:257–266. doi: 10.1007/s00239-005-0076-5. [DOI] [PubMed] [Google Scholar]
- 6.Aevermann BD, Waters ER. A comparative genomic analysis of the small heat shock proteins in Caenorhabditis elegans and briggsae. Genetica. 2007;133:307–319. doi: 10.1007/s10709-007-9215-9. [DOI] [PubMed] [Google Scholar]
- 7.Waters ER. The evolution, function, structure, and expression of the plant sHSPs. J. Exp. Bot. 2013;64:391–403. doi: 10.1093/jxb/ers355. [DOI] [PubMed] [Google Scholar]
- 8.Becker J, Craig EA. Heat-shock proteins as molecular chaperones. Eur. J. Biochem. 1994;219:11–23. doi: 10.1111/j.1432-1033.1994.tb19910.x. [DOI] [PubMed] [Google Scholar]
- 9.Hartl FU. Molecular chaperones in cellular protein folding. Nature. 1996;381:571–580. doi: 10.1038/381571a0. [DOI] [PubMed] [Google Scholar]
- 10.Tyedmers J, Mogk A, Bukau B. Cellular strategies for controlling protein aggregation. Nat. Rev. Mol. Cell Biol. 2010;11:777–788. doi: 10.1038/nrm2993. [DOI] [PubMed] [Google Scholar]
- 11.Mymrikov EV, Seit-Nebi AS, Gusev NB. Large potentials of small heat shock proteins. Physiol. Rev. 2011;91:1123–1159. doi: 10.1152/physrev.00023.2010. [DOI] [PubMed] [Google Scholar]
- 12.Sorensen GJ, Kristensen NT, Loeschcke V. The evolutionary and ecological role of heat shock proteins. Ecol. Lett. 2003;6:1025–1037. doi: 10.1046/j.1461-0248.2003.00528.x. [DOI] [Google Scholar]
- 13.Waters ER, Rioflorido I. Evolutionary analysis of the small heat shock proteins in five complete algal genomes. J. Mol. Evol. 2007;65:162–174. doi: 10.1007/s00239-006-0223-7. [DOI] [PubMed] [Google Scholar]
- 14.Ramakrishna W, Deng Z, Ding C, Handa AK, Ozminkowski RHJ. A novel small heat shock protein gene, vis1, contributes to pectin depolymerization and juice viscosity in tomato fruit. Plant Physiol. 2003;131:725–735. doi: 10.1104/pp.012401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Neta-Sharir I, Isaacson T, Lurie S, Weiss D. Dual role for tomato heat shock protein 21: protecting photosystem II from oxidative stress and promoting color changes during fruit maturation. Plant Cell. 2005;17:1829–1838. doi: 10.1105/tpc.105.031914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goyal RK, et al. Features of a unique intron less cluster of class I small heat shock protein genes in tandem with box C/D snoRNA genes on chromosome 6 in tomato (Solanum lycopersicum) Planta. 2012;235:453–471. doi: 10.1007/s00425-011-1518-5. [DOI] [PubMed] [Google Scholar]
- 17.Gray JE, Picton S, Giovannoni JJ, Grierson D. The use of transgenic and naturally occurring mutants to understand and manipulate tomato fruit ripening. Plant Cell Environ. 1994;17:557–571. doi: 10.1111/j.1365-3040.1994.tb00149.x. [DOI] [Google Scholar]
- 18.Klee H, Giovannoni J. Genetics and control of tomato fruit ripening and quality attributes. Annu. Rev. Genet. 2011;45:41–59. doi: 10.1146/annurev-genet-110410-132507. [DOI] [PubMed] [Google Scholar]
- 19.Giovannoni JJ. Fruit ripening mutants yield insights into ripening control. Curr. Opin. Plant Biol. 2007;10:283–289. doi: 10.1016/j.pbi.2007.04.008. [DOI] [PubMed] [Google Scholar]
- 20.Alexander L, Grierson D. Ethylene biosynthesis and action in tomato: a model for climacteric fruit ripening. J. Exp. Bot. 2002;53:2039–2055. doi: 10.1093/jxb/erf072. [DOI] [PubMed] [Google Scholar]
- 21.Oeller PW, Lu MW, Taylor LP, Pike DA, Theologis A. Reversible inhibition of tomato fruit senescence by antisense RNA. Science. 1991;254:437–439. doi: 10.1126/science.1925603. [DOI] [PubMed] [Google Scholar]
- 22.Picton S, Barton SL, Bouzayen M, Hamilton AJ, Grierson D. Altered fruit ripening and leaf senescence in tomatoes expressing an antisense ethylene-forming enzyme transgene. Plant J. 1993;3:469–481. doi: 10.1111/j.1365-313X.1993.tb00167.x. [DOI] [Google Scholar]
- 23.Wilkinson JQ, Lanahan MB, Yen H, Giovannoni JJ, Klee HJ. An ethylene-inducible component of signal transduction encoded by Never-ripe. Science. 1995;270:14–16. doi: 10.1126/science.270.5243.1807. [DOI] [PubMed] [Google Scholar]
- 24.Razdan, M. K. & Mattoo, A. K. Genetic improvement of Solanaceous crops: Volume 2: Tomato. Science Publishers, Inc., Enfield, UK (2007).
- 25.Vrebalov JA, et al. MADS-Box gene necessary for fruit ripening at the tomato ripening-inhibitor (Rin) locus. Science. 2002;296:343–346. doi: 10.1126/science.1068181. [DOI] [PubMed] [Google Scholar]
- 26.Ng M, Yanofsky MF. Function and evolution of the plant MADS-box gene family. Nat. Rev. Genet. 2001;2:186–195. doi: 10.1038/35056041. [DOI] [PubMed] [Google Scholar]
- 27.Hileman LC, et al. Molecular and phylogenetic analyses of the MADS-box gene family in tomato. Mol. Biol. Evol. 2006;23:2245–2258. doi: 10.1093/molbev/msl095. [DOI] [PubMed] [Google Scholar]
- 28.Fujisawa M, Nakano T, Shima Y, Ito Y. A large-scale identification of direct targets of the tomato MADS box transcription factor RIPENING INHIBITOR reveals the regulation of fruit ripening. Plant Cell. 2013;25:371–386. doi: 10.1105/tpc.112.108118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hackett RM, et al. Antisense inhibition of the Nr gene restores normal ripening to the tomato Never-ripe mutant, consistent with the ethylene receptor-inhibition model. Plant Physiol. 2000;124:1079–1086. doi: 10.1104/pp.124.3.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moore S, Vrebalov J, Payton P, Giovannoni J. Use of genomics tools to isolate key ripening genes and analyse fruit maturation in tomato. J. Exp. Bot. 2002;53:2023–2030. doi: 10.1093/jxb/erf057. [DOI] [PubMed] [Google Scholar]
- 31.Sobolev AP, et al. Genetic introgression of ethylene-suppressed transgenic tomatoes with higher-polyamines trait overcomes many unintended effects due to reduced ethylene on the primary metabolome. Front. Plant Sci. 2014;5:632. doi: 10.3389/fpls.2014.00632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nambeesan S, et al. Overexpression of yeast spermidine synthase impacts ripening, senescence and decay symptoms in tomato. Plant J. 2010;63:836–47. doi: 10.1111/j.1365-313X.2010.04286.x. [DOI] [PubMed] [Google Scholar]
- 33.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 34.Bustin SA, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 35.Upadhyay RK, et al. SlERF36, an EAR-motif-containing ERF gene from tomato, alters stomatal density and modulates photosynthesis and growth. J. Exp. Bot. 2013;64:3237–3247. doi: 10.1093/jxb/ert162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lescot M, et al. Plant CARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30:325–327. doi: 10.1093/nar/30.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Higo K, Ugawa Y, Iwamoto M, Korenaga T. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 1999;27:297–300. doi: 10.1093/nar/27.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ito Y, et al. DNA-binding specificity, transcriptional activation potential, and the rin mutation effect for the tomato fruit-ripening regulator. RIN. Plant J. 2008;55:212–223. doi: 10.1111/j.1365-313X.2008.03491.x. [DOI] [PubMed] [Google Scholar]
- 39.Fujisawa M, Nakano T, Ito Y. Identification of potential target genes for the tomato fruit-ripening regulator RIN by chromatin immunoprecipitation. BMC Plant Biol. 2011;11:26. doi: 10.1186/1471-2229-11-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schneider TD, Stephens RM. Sequence logos: A new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martel C, Vrebalov J, Tafelmeyer P, Giovannoni JJ. The tomato MADS-Box transcription factor ripening inhibitor interacts with promoters involved in numerous ripening processes in a colorless nonripening-dependent manner. Plant Physiol. 2011;157:1568–1579. doi: 10.1104/pp.111.181107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yan D, et al. Curved chimeric PALEA 1 encoding an EMF1-like protein maintains epigenetic repression of OsMADS58 in rice palea development. Plant J. 2015;82:12–24. doi: 10.1111/tpj.12784. [DOI] [PubMed] [Google Scholar]
- 44.Petell CJ, et al. An epigenetic switch regulates de novo DNA methylation at a subset of pluripotency gene enhancers during embryonic stem cell differentiation. Nucleic Acids Res. 2016;44:7605–7617. doi: 10.1093/nar/gkw426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nakatsuka A, et al. Differential expression and internal feedback regulation of 1-aminocyclopropane-1-carboxylate synthase, 1-aminocyclopropane-1-carboxylate oxidase, and ethylene receptor genes in tomato fruit during development and ripening. Plant Physiol. 1998;118:1295–1305. doi: 10.1104/pp.118.4.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vrebalov J, et al. Fleshy fruit expansion and ripening are regulated by the tomato SHATTERPROOF gene TAGL1. Plant Cell. 2009;21:3041–62. doi: 10.1105/tpc.109.066936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dellapenna D, et al. Polygalacturonase isozymes and pectin depolymerization in transgenic rin tomato fruit. Plant Physiol. 1990;94:1882–1886. doi: 10.1104/pp.94.4.1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sitrit Y, Bennett AB. Regulation of tomato fruit polygalacturonase mRNA accumulation by ethylene: a re-examination. Plant Physiol. 1998;116:1145–1150. doi: 10.1104/pp.116.3.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kevany BM, Tieman DM, Taylor MG, Cin VD, Klee HJ. Ethylene receptor degradation controls the timing of ripening in tomato fruit. Plant J. 2007;51:458–467. doi: 10.1111/j.1365-313X.2007.03170.x. [DOI] [PubMed] [Google Scholar]
- 50.Yang T, Peng H, Whitaker BD, Conway WS. Characterization of a calcium/calmodulin-regulated SR/CAMTA gene family during tomato fruit development and ripening. BMC Plant Biol. 2012;12:19. doi: 10.1186/1471-2229-12-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fujisawa M, et al. Direct targets of the tomato-ripening regulator RIN identified by transcriptome and chromatin immunoprecipitation analyses. Planta. 2012;235:1107–1122. doi: 10.1007/s00425-011-1561-2. [DOI] [PubMed] [Google Scholar]
- 52.Zhong S, et al. Single-base resolution methylomes of tomato fruit development reveal epigenome modifications associated with ripening. Nature Biotech. 2013;31:154–159. doi: 10.1038/nbt.2462. [DOI] [PubMed] [Google Scholar]
- 53.Zhang J, et al. Constitutive expression of a tomato small heat shock protein gene LeHSP21 improves tolerance to high-temperature stress by enhancing antioxidation capacity in tobacco. Plant Mol. Biol. Report. 2015;34:399–409. doi: 10.1007/s11105-015-0925-3. [DOI] [Google Scholar]
- 54.Larkindale J, et al. Heat stress phenotypes of Arabidopsis mutants implicate multiple signaling pathways in the acquisition of thermotolerance. Plant Physiol. 2005;138:882–897. doi: 10.1104/pp.105.062257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Basha E, O’Neill H, Vierling E. Small heat shock proteins and alpha-crystallins: dynamic proteins with flexible functions. Trends Biochem. Sci. 2012;37:106–117. doi: 10.1016/j.tibs.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Goh CH, Gil NH, Shin PY. Stress memory in plants: A negative regulation of stomatal response and transient induction of rd22 gene to light in abscisic acid-entrained Arabidopsis plants. Plant J. 2003;36:240–255. doi: 10.1046/j.1365-313X.2003.01872.x. [DOI] [PubMed] [Google Scholar]
- 57.García I, Rosas T, Bejarano ER, Gotor C, Romero LC. Transient transcriptional regulation of the CYS-C1 gene and cyanide accumulation upon pathogen infection in the plant immune response. Plant Physiol. 2013;162:2015–2027. doi: 10.1104/pp.113.219436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lau S, Smet ID, Kolb M, Meinhardt H, Jürgens G. Auxin triggers a genetic switch. Nat. Cell Biol. 2011;13:611–615. doi: 10.1038/ncb2212. [DOI] [PubMed] [Google Scholar]
- 59.Wang D, Maule AJ. Inhibition of host gene expression associated with plant virus replication. Science. 1995;267:229–231. doi: 10.1126/science.267.5195.229. [DOI] [PubMed] [Google Scholar]
- 60.Cara B, Giovannoni JJ. Molecular biology of ethylene during tomato fruit development and maturation. Plant Sci. 2008;175:106–113. doi: 10.1016/j.plantsci.2008.03.021. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


