Abstract
The objective of this study was to determine the effect of ionizing radiation (IR) exposure of parents on carcinogenesis of the next generation focusing on the epigenetic perspective to clarify the relationship between radiation dose and carcinogenesis in F1 generation SD rats. F1 generations from pregnant rats (F0) who were exposed to gamma rays were divided into three groups according to the dose of radiation: 10 rad, 30 rad, and untreated. They were intraperitoneally injected with 50 mg/kg of diethylnitrosamine (DEN). Carcinogenesis was analyzed by examining expression levels of tumor suppressor genes (TSG) and other related genes by methylation-specific polymerase chain reaction (MSP). DNA methylation in liver tissues was evaluated to discern epigenetic regulation of transgenerational carcinogenesis vulnerability following IR exposure. Numerous studies have proved that transcriptional inactivation due to hypermethylation of TSG preceded carcinogenesis. Results of this study revealed hypermethylation of tumor suppressor gene SOCS1 in group treated with 30 rad. In addition, genes related to DNA damage response pathway (GSTP1, ATM, DGKA, PARP1, and SIRT6) were epigenetically inactivated in all DEN treated groups. In the case of proto-oncogene c-Myc, DNA hypermethylation was identified in the group with low dose of IR (10 rad). Results of this study indicated that each TSG had different radiation threshold level (dose-independent way) and DEN treatment could affect DNA methylation profile irrelevant of ionizing radiation dose.
Keywords: Radiation, DNA methylation, carcinogenesis, tumor suppressor gene
It is generally known that radiation can destabilize the state of in vivo gene. In other words, irradiation can partly damage particular cells or give stress to various mammalian cells. It has been reported that such partial damage to genes caused by irradiation can increase the incidence of cancer when they are passed from generation to generation [1,2]. A lot of studies on carcinogenesis caused by radiation have provided new and important information about radiation-related carcinogenesis in the past decades [2,3]. However, experiments used to assess radiation effects on cancer, particularly in vivo experiments, still have many problems. For example, they cannot confirm the effect of low dose radiation on carcinogenesis [3,4]. Although numerous studies have reported cancer susceptibility of the F1 generation born after maternal radiation exposure, experimental evidence is still weak [4]. In addition, the dose-response relationship about cancer susceptibility for the offspring has not been established yet.
Eukaryotes use many ways to control gene expression. DNA methylation is one of these ways that regulates gene expression by epigenetical shut down. Methylation is a key element involved in a number of biological processes, including embryonic development, genomic imprinting, X-chromosome inactivation, and preservation of chromosome stability [5,6,7,8,9,10]. Cancer is characterized by general DNA hypomethylation and regional hypermethylation. DNA hypomethylation makes chromosomes unstable and activates proto-oncogenes which promote tumorigenesis in mouse models. CpG islands in the promoter region of genes are unmethylated in normal tissues. Methylation of these CpG islands causes tumors and deters the expression of tumor suppressor genes [11,12,13,14,15]. Therefore, cancer is considered to be caused by abnormal DNA which affects the function and expression of genes, especially cancer related genes.
Liver has been widely used to study cancer because carcinogenesis caused by chemical and radiation can be easily measured. In addition, recent study has shown that radiation not only can initiate liver cancer, but also can boost cancer [16]. Therefore, liver was selected for the present study to determine the effect of radiation on cancer.
The objective of this study was to determine the effect of ionizing radiation (IR) exposure of parents on the carcinogenesis of F1 generation focusing on the epigenetic perspective to clarify the relationship between radiation dose and carcinogenesis in F1 generation. Animal model enabling the evaluation of susceptibility to carcinogenesis in offsprings after maternal preconceptional exposure to gamma irradiation was applied in this study. Carcinogenesis was analyzed by examining expression levels of tumor suppressor genes (TSG) and other related genes by methylation-specific polymerase chain reaction (MSP).
Materials and Methods
Carcinogenesis
Sprague-Dawley rats were exposed to radiation on day 14 of pregnancy. After birth, F1 generation from irradiated rats was used in this study. Baby rats were separated and assigned into three groups: 1) group 1, pregnant rats were systemically exposed to 10 rad gamma-ray (60Co Teletherapy unit, Theratron-780) is designated to group 1; 2) group 2, pregnant rats were systemically exposed to 30 rad gamma-ray; 3) control group, pregnant rats were not exposed to any radiation. Diethylnitrosamine (DEN, Sigma Chemical Co. USA) (at 50 mg/kg) was injected into the peritoneal cavity of F1 rats at 4-5 weeks of age, including the control group. All F1 rats were sacrificed at 13 weeks and their livers were excised to prepare paraffin blocks. Animal experiments were approved by Kangwon National University Institutional Animal Care and Use Committee (KIACUC-130325-1).
DNA extraction & C-T conversion
Genomic DNA was extracted from paraffin-embedded liver tissue using G-SpinTM Total DNA extraction kit (iNtRON Biotechnology, Korea) according to the manufacturer's protocol. Briefly, after removing paraffin using xylene, 90% alcohol, and 70% alcohol, these paraffin-removed liver tissues were cut in diameter of 5 mm and crushed. After adding cell lysis buffer (200 µL) and proteinase K+RNase A (25 µL), the mixture was incubated at 57℃ for 30 min. Blood lysis buffer (200 µL) was the then added to the mixture, mixed well by inverting, and incubated at 70℃ for 15 min. After liver tissue was broken, spin-column was used to collect DNA. After going through centrifugation, washing, and dry processes, DNA was finally eluted with 100 µL of elution buffer.
To analyze only methylated genes, it is necessary to distinguish methylated genes from unmethylated genes. EZ DNA methylation-GoldTM kit (Zymo Research, CA, USA) was used for C-T conversion to differentiate methylated cytosine from unmethylated uracils. First, C-T conversion reagent was used for sulphonation of unmethylated DNA. For this reaction, the tube was incubated at 98℃ for 10 min and 64℃ for 2.5 hours. To collect and distinguished DNA sample with Zymo-spin column. M-binding buffer was used in the following procedure for hydrolytic deamination. Finally, M-desulphonation buffer was used. DNA samples (methylated and unmethylated) were then prepared separately following the manufacturer's instructions.
Methylation-specific polymerase chain reaction (MSP)
Using DNA sample that underwent C-T conversion, methylated DNA was differentially amplified using specifically designed primers (Table 1). They are specific to each desirable target gene. For MSP, 2X multiplex PCR premix (Bioassay Co., Korea), PCR oligo premix (Cosmogenetech, Korea), template DNA, and RNase-free HPLC water were used. PCR premix (8 µL) and oligo premix (4 µL) were added into 12×8 PCR tube/well (Denville Scientific, USA). Then 4 µL of DNA template was added to each well. PCR was performed with the following cycles: annealing at 95℃ for 15 min, 38 cycles of 95℃ for 20 s, 59℃ for 30 s, and 72℃ for 35 s for amplification, followed by a final extension step at 72℃ for 3 min. PCR products were subjected to 4% agarose gel electrophoresis and visualized under UV.
Table 1. Primer sequences used for MSP analysis.
| Genes | Allele | Sense Primers 5' to 3' | Antisense primers 5' to 3 |
|---|---|---|---|
| SOCS1 | Methylated | GAGAATTCGAAGGATTTGTCGG | ATTCCGAAACTACTATCGCGCA |
| Unmethylated | GAAATTTAAAGAGAATTTGAAGGATTTGTT | AAAATTCCAAAACTACTATCACACA | |
| c-Myc | Methylated | AGATTGGTAGAGATTCGATCGG | AAAAACGACTCCTTACCCCGTA |
| Unmethylated | AGATTGGTAGAGATTTGATTGG | TAAAAACAACTCCTTACCCCATA | |
| MGMT | Methylated | TTTTAGAGGGTTGATGAAGCGT | AAACCAAATTACAAAAAATTACGAA |
| Unmethylated | GTTTTAGAGGGTTGATGAAGTGT | AAACCAAATTACAAAAAATTACAAA | |
| COX2 | Methylated | CGAGTTGTGTTGTTTTGCGT | AATCCCTCCAAAAATACTTCGAA |
| Unmethylated | TTTGAGTTGTGTTGTTTTGTGT | AAAAAATCCCTCCAAAAATACTTCAAA | |
| SIRT6 | Methylated | TAAGGGTAAGTGCGGGTTGTTC | TCCCATTATCTAAACTCTATACTTCGTA |
| Unmethylated | AGGGTAAGTGTGGGTTGTTTGA | TCCCATTATCTAAACTCTATACTTCATA | |
| ATM | Methylated | TTTTTATTTTGGGAAGAGTCGG | TCAACAAAATAACTAAACGCGTC |
| Unmethylated | TTTTTTATTTTGGGAAGAGTTGG | ATTTCAACAAAATAACTAAACACATC | |
| PARP1 | Methylated | GCGCGTTTTTTGTAAGAAATGTAGC | TAACCCTCAAAAACCTCGACGAC |
| Unmethylated | GTGTGTTTTTTGTAAGAAATGTAGTGA | AACCCTCAAAAACCTCAACAAC | |
| GSTP1 | Methylated | TCGGTAGGAGTCGGGAGTTGTC | ATAAAAAACATTACGCCCCGCC |
| Unmethylated | TTTGGTAGGAGTTGGGAGTTGTTG | TTATAAAAAACATTACACCCCACC | |
| DGKA | Methylated | TATATGTTTTGGGTTAGTTCGG | AATACTTAAAATTTACCACATCGAA |
| Unmethylated | TTATATGTTTTGGGTTAGTTTGG | TTAATACTTAAAATTTACCACATCAAA |
Results
Results of MSP were analyzed by 4% agarose gel electrophoresis for various genes with different radiation doses. Some genes, especially those related to DNA damage showed methylation in all groups, including the control group.
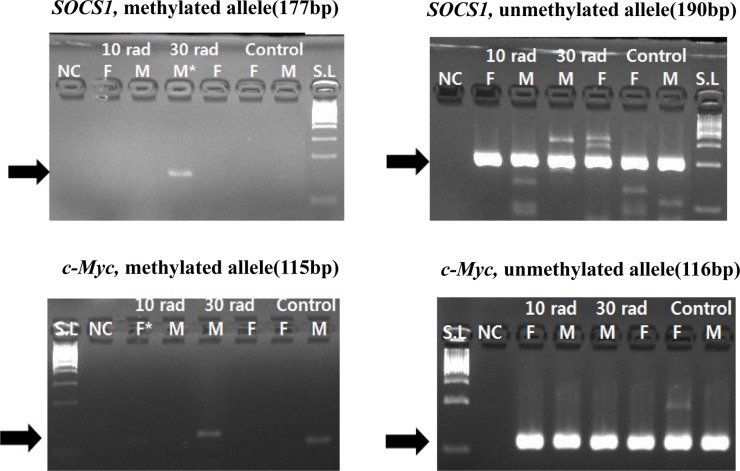
Suppressor of cytokine signaling 1 (SOCS1), a tumor suppressor gene, was methylated in the 30 rad group. Proto-oncogene c-Myc was also methylated in the 10 rad group (Figure 1). c-Myc involved in carcinogenesis has been reported to be hypomethylated in animal models [17]. However, in this study, c-Myc was hypermethylated in the 10 rad group.
Figure 1. Expressions of SOCS1 and c-Myc were detected by MSP. SOCS1 showed methylation from 30 rad (the second experimental group) and c-Myc showed methylation from 10 rad group. The sample having asterisk represents PCR positive. Abbreviations; S.L: 100bp size ladder, NC: negative control, F: female, M: male.
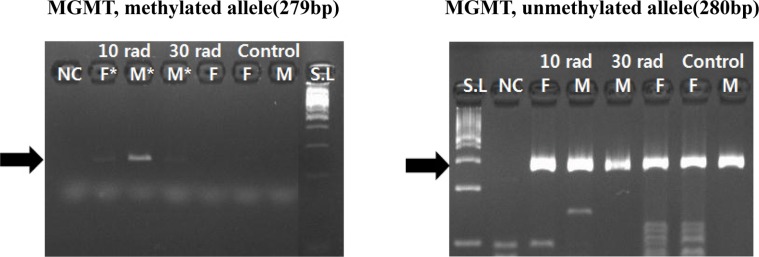
Regarding O6-methylguanine-DNA-methyltransferase (MGMT), its methylation was observed in all experimental groups. This suggests that methylation of MGMT is not dependent on IR concentration. It might be related to exposure to IR by itself. Methylation change of MGMT in the group with low dose of IR suggests that MGMT gene might be susceptible to IR (Figure 2).
Figure 2. Expression of MGMT was detected by MSP. Methylation change of MGMT was observed in all experimental groups. It suggests MGMT is not dependent on IR concentration, but related to exposure to IR by itself. Samples having asterisk represent PCR positives. Abbreviations; S.L: 100bp size ladder, NC: negative control, F: female, M: male.
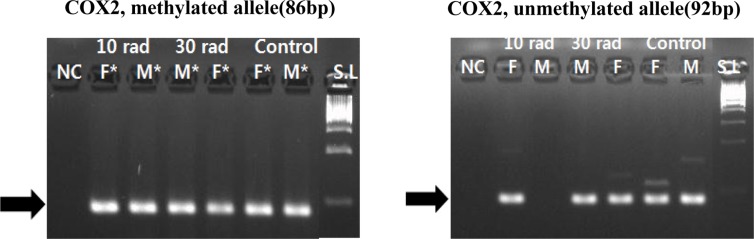
Cyclooxygenase 2 (COX2), a major gene related to inflammation response, showed methylation change in both IR experimental groups and the control group. This might be due to DEN injection (Figure 3).
Figure 3. Expression of COX2 was detected by MSP by MSP. COX2, a major gene related to inflammation response, showed methylation changes in all experimental and control groups. Samples having asterisk represent PCR positives. Abbreviations; S.L: 100bp size ladder, NC: negative control, F: female, M: male.
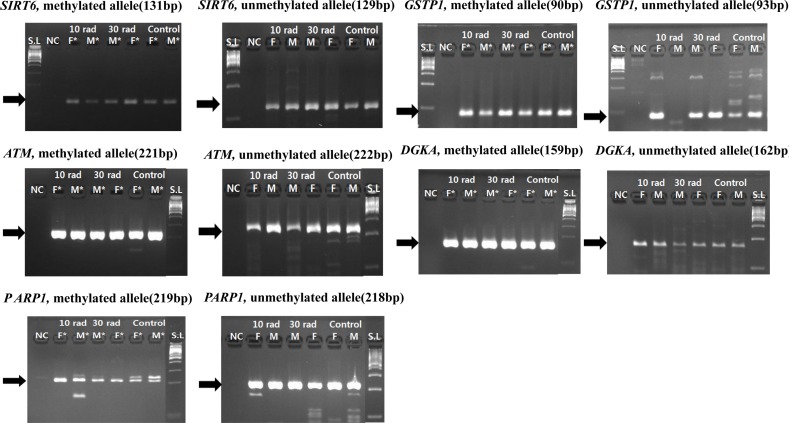
Genes (DGKA, SIRT6, ATM, PARP1, GSTP1) related to DNA damage also showed methylation in experimental groups and the control group (Figure 4). Since methylation was also observed in the control group without exposure to IR, DEN injection might have caused such results. Whether methylation of these genes in experimental groups was due to IR exposure or DEN injection was unclear. However, considering that methylation also appeared in the control group, methylation of these genes in IR experimental groups might also be due to DEN administration. In conclusion, genes that showed methylation pattern in IR exposure experimental groups and control groups were due to reaction to DEN administration, not IR exposure.
Figure 4. Expressions of genes (DGKA, SIRT6, ATM, PARP1, GSTP1) related to DNA damage were detected by MSP. Genes related to DNA damage also showed methylation in all experimental groups and controls. Methylation was also observed in the control group without exposure to IR, DEN injection affected the result. Samples having asterisk represent PCR positives. Abbreviations; S.L: 100bp size ladder, NC: negative control, F: female, M: male.
Discussion
Hypermethylation of tumor suppressor genes is a significant sign in organisms with cancer [18,19]. In this study, SOCS1, a tumor suppressor gene, was methylated in the group with irradiation of 30 rad. This result suggests that low dose of IR can influence carcinogenesis in the offspring. As a gene contributing to increased risk of cancer, c-Myc was also hypermethlylated in the group with irradiation of 10 rad. Hypermethylation of this gene indicates that oncogene c-Myc could not operate its functions well. However, a previous study has reported that c-Myc is hypomethylated in tumor [17], which is different from the result of c-Myc in the present study. Gene MGMT is a tumor suppressor gene related to gene repair. It is frequently methylated under tumor circumstances [20]. MGMT gene is also related to DNA damage. However, genes might have different methylation patterns according to their characteristics under the same condition. In this study, MGMT showed methylation pattern in the low dose IR group. This indicates that exposure to low dose of IR in maternal line might affect carcinogenesis and DNA damage in the offspring. However, it is currently unknown how susceptible MGMT is to IR exposure. This merits further studies.
In this study, SOCS1 showed methylation only in the 30 rad group while c-Myc only showed methylation in the 10 rad group. In the case of p16, although we performed MSP for p16, a tumor suppressor gene encoding a cyclin-dependent kinase (CDK) inhibitor [21], unfortunately we failed to observe methylation change in any experimental group for this gene. This suggests that different genes, particularly cancer-related genes, have various threshold levels against ionizing radiation. They might respond in a dose-independent way. For example, SOCS1 has 30 rad or lower reactive radiation threshold, which might have enabled us to observe its methylation change in this study. However, if reactive radiation threshold level for p16 gene is higher than 30 rad IR, p16 will not respond to IR dose at lower than 30 rad. Further studies are needed to improve our understanding about the relationship between radiation dose and methylation status of genes or carcinogenesis.
Acknowledgments
This study was supported by grants from Ministry of Science, ICT and Future Planning of Korea to the National Research Foundation of Korea (C1008955-01-03) and by Grant from Kangwon National University (520150281).
Footnotes
Conflict of interests: The authors declare that there is no financial conflict of interests to publish these results.
References
- 1.Little JB. Radiation carcinogenesis. Carcinogenesis. 2000;21(3):397–404. doi: 10.1093/carcin/21.3.397. [DOI] [PubMed] [Google Scholar]
- 2.Brenner DJ, Doll R, Goodhead DT, Hall EJ, Land CE, Little JB, Lubin JH, Preston DL, Preston RJ, Puskin JS, Ron E, Sachs RK, Samet JM, Setlow RB, Zaider M. Cancer risks attributable to low doses of ionizing radiation: assessing what we really know. Proc Natl Acad Sci U S A. 2003;100(24):13761–13766. doi: 10.1073/pnas.2235592100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Modan B. Low-dose radiation carcinogenesis. Eur J Cancer. 1992;28(6-7):1010–1012. doi: 10.1016/0959-8049(92)90442-5. [DOI] [PubMed] [Google Scholar]
- 4.Dasenbrock C, Tillmann T, Ernst H, Behnke W, Kellner R, Hagemann G, Kaever V, Kohler M, Rittinghausen S, Mohr U, Tomatis L. Maternal effects and cancer risk in the progeny of mice exposed to X-rays before conception. Exp Toxicol Pathol. 2005;56(6):351–360. doi: 10.1016/j.etp.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 5.Jones PA. DNA methylation errors and cancer. Cancer Res. 1996;56(11):2463–2467. [PubMed] [Google Scholar]
- 6.Gonzalo S. Epigenetic alterations in aging. J Appl Physiol. 2010;109(2):586–597. doi: 10.1152/japplphysiol.00238.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem Sci. 2006;31(2):89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 8.Counts JL, Goodman JI. Alterations in DNA methylation may play a variety of roles in carcinogenesis. Cell. 1995;83(1):13–15. doi: 10.1016/0092-8674(95)90228-7. [DOI] [PubMed] [Google Scholar]
- 9.Zingg JM, Jones PA. Genetic and epigenetic aspects of DNA methylation on genome expression, evolution, mutation and carcinogenesis. Carcinogenesis. 1997;18(5):869–882. doi: 10.1093/carcin/18.5.869. [DOI] [PubMed] [Google Scholar]
- 10.Lillycrop KA, Slater-Jefferies JL, Hanson MA, Godfrey KM, Jackson AA, Burdge GC. Induction of altered epigenetic regulation of the hepatic glucocorticoid receptor in the offspring of rats fed a protein-restricted diet during pregnancy suggests that reduced DNA methyltransferase-1 expression is involved in impaired DNA methylation and changes in histone modifications. Br J Nutr. 2007;97(6):1064–1073. doi: 10.1017/S000711450769196X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu WB, Liu JY, Ao L, Zhou ZY, Zhou YH, Cui ZH, Cao J. Epigenetic silencing of cell cycle regulatory genes during 3-methylcholanthrene and diethylnitrosamine-induced multistep rat lung cancer. Mol Carcinog. 2010;49(6):556–565. doi: 10.1002/mc.20621. [DOI] [PubMed] [Google Scholar]
- 12.Bird AP. DNA methylation and the frequency of CpG in animal DNA. Nucleic Acids Res. 1980;8(7):1499–1504. doi: 10.1093/nar/8.7.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu WB, Ao L, Zhou ZY, Cui ZH, Zhou YH, Yuan XY, Xiang YL, Cao J, Liu JY. CpG island hypermethylation of multiple tumor suppressor genes associated with loss of their protein expression during rat lung carcinogenesis induced by 3-methylcholanthrene and diethylnitrosamine. Biochem Biophys Res Commun. 2010;402(3):507–514. doi: 10.1016/j.bbrc.2010.10.061. [DOI] [PubMed] [Google Scholar]
- 14.Phillips T. The role of methylation in gene expression. Nat Educ. 2008;1(1):116. [Google Scholar]
- 15.Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358:1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 16.Saha A, Wittmeyer J, Cairns BR. Chromatin remodelling: the industrial revolution of DNA around histones. Nat Rev Mol Cell Biol. 2006;7(6):437–447. doi: 10.1038/nrm1945. [DOI] [PubMed] [Google Scholar]
- 17.Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, Schreiber SL, Mellor J, Kouzarides T. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419(6905):407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- 18.Liu WB, Liu JY, Ao L, Zhou ZY, Zhou YH, Cui ZH, Yang H, Cao J. Dynamic changes in DNA methylation during multistep rat lung carcinogenesis induced by 3-methylcholanthrene and diethylnitrosamine. Toxicol Lett. 2009;189(1):5–13. doi: 10.1016/j.toxlet.2009.04.022. [DOI] [PubMed] [Google Scholar]
- 19.Kim TY, Jong HS, Song SH, Dimtchev A, Jeong SJ, Lee JW, Kim TY, Kim NK, Jung M, Bang YJ. Transcriptional silencing of the DLC-1 tumor suppressor gene by epigenetic mechanism in gastric cancer cells. Oncogene. 2003;22(25):3943–3951. doi: 10.1038/sj.onc.1206573. [DOI] [PubMed] [Google Scholar]
- 20.Oue N, Sentani K, Yokozaki H, Kitadai Y, Ito R, Yasui W. Promoter methylation status of the DNA repair genes hMLH1 and MGMT in gastric carcinoma and metaplastic mucosa. Pathobiology. 2001;69(3):143–149. doi: 10.1159/000048769. [DOI] [PubMed] [Google Scholar]
- 21.Nuovo GJ, Plaia TW, Belinsky SA, Baylin SB, Herman JG. In situ detection of the hypermethylation-induced inactivation of the p16 gene as an early event in oncogenesis. Proc Natl Acad Sci U S A. 1999;96(22):12754–12759. doi: 10.1073/pnas.96.22.12754. [DOI] [PMC free article] [PubMed] [Google Scholar]