Abstract
H19 is involved in tumor metastasis and associated with tumor progression. Enhancer of zest homolog 2 (EZH2) is overexpressed in multiple cancer types and correlates with tumor proliferation, epithelial-mesenchymal transition, and poor prognosis. However, the interaction between H19 and EZH2 to promote tongue squamous cell carcinoma (TSCC) progression remains largely uncharacterized. Insitu hybridization and quantitative reverse-transcription PCR (qRT-PCR) were performed to measure H19 expression in primary TSCC and adjacent normal tissues and cell lines. EZH2 expression was determined by immunohistochemistry in matched primary TSCC and adjacent normal tissues. The correlation between H19 and EZH2 expression and clinicopathological characteristics were analyzed. The roles of H19 in cell proliferation, apoptosis, and invasion were analyzed using a H19-targeted lentivirus. Western blot and qRT-PCR were carried out to detect downstream signal pathway changes. Expression levels of downstream signaling proteins in primary TSCC tissues and adjacent normal tissues were analyzed by immunohistochemistry. H19 and EZH2 were upregulated in TSCC tissues compared to matched normal tissues, and significantly correlated with WHO grade, lymph node metastasis, and poor prognosis. H19 silencing attenuated cell proliferation, apoptosis, and invasion in vitro. H19 knockdown inhibited the activation of β-catenin/GSK-3β/cyclin D1/c-myc, upregulated E-cadherin and zonula occludens-1 (ZO-1), and inhibited N-cadherin, vimentin, Snail1, Twist1, and ZEB1. Silencing H19 expression also inhibited tumor progression and lung metastasis in an animal model. Our findings indicate that H19 promotes TSCC progression through association with EZH2, and affects downstream β-Catenin/GSK3β/EMT signaling, suggesting that H19 inhibition might be a potential target for the treatment of TSCC.
Keywords: H19, EZH2, TSCC, metastasis, EMT
Introduction
Tongue squamous cell carcinoma (TSCC) is one of the most common invasive and lethal malignant tumors, causing approximately 400,000 patient fatalities worldwide [1-3]. Despite tremendous advancements in therapeutic strategies for oral cancer (including surgery, chemotherapy and radiotherapy) over the past decades, the 5-year overall survival rate has not yet improved [4,5]. Cervical lymph node metastasis is the most prevalent factor affecting cancer recurrence and poor prognosis in patients [6,7]. Therefore, it is essential to reveal the underlying mechanisms and explore new biomarkers and therapeutic targets involved in tumor metastasis in TSCC.
There is increasing evidence that the abnormal expression of long non-coding RNAs (lncRNAs) play pivotal roles in carcinogenesis and cancer metastasis [8-11]. A better understanding of the aberrant expression of lncRNAs can provide molecular-level information that can inform diagnosis and therapy in TSCC. Several studies have shown that the aberrant expression of H19 is closely associated with tumor progression [12,13] in various cancer types, including bladder cancer [14,15], breast cancer [16], colorectal cancer [17,18], esophageal squamous cell carcinoma (ESCC) [19], gastric cancer [20], hepatocellular carcinoma (HCC) [21,22], ovarian carcinoma [23,24], pancreatic cancer [25] and prostate cancer [26].
EZH2 is a core component of the polycomb repressive complex 2 (PRC2), which can epigenetically regulate gene expression [27]. Previous studies have illustrated that aberrant EZH2 expression often correlates with advanced stages and poor prognosis in multiple cancer types [28-30]. In addition, the upregulation of EZH2 can enhance tumor proliferation, EMT, and metastasis, which results in poor prognosis [31-33].
β-catenin is a significant component of the Wnt/β-catenin signaling pathway, which has been demonstrated to regulate cell proliferation, differentiation and invasion [34,35]. Increasing evidence indicates that the abnormal activation of β-catenin is a common phenomenon in a range of cancers [36].
However, whether H19 is involved in the aberrant expression of EZH2, and the activation of β-catenin in TSCC, remains unknown. This study aimed to investigate the expression and function of H19 and EZH2 in TSCC. We also explored the relationship between H19, EZH2 and β-catenin in TSCC, seeking a potential therapeutic target for TSCC.
Materials and methods
Tissue specimens
123 TSCC tissues and 50 adjacent non-tumor tissues were respectively collected in this study. The patients were histopathologically diagnosed and verified at the Sun Yat-sen Memorial Hospital, Sun Yat-sen University from 2009 to 2015. Each TSCC tissue and adjacent non-cancerous tissue were obtained from resected tumors and adjacent non-cancerous tongue tissue, respectively. None of the patients had undergone chemotherapy or radiotherapy prior to surgery. This study was approved by the local ethics committee (Sun Yat-sen Memorial Hospital, Sun Yat-sen University); each patient signed a written informed consent.
Cell lines and cell cultures
Six human TSCC cell lines-Cal27, SCC9, SCC15, SCC25, Tca8113, and UM1-were used in this study. Tca8113 was purchased from Shanghai Jiaotong University and UM1 is available in our lab. The other five cells were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA) and human oral keratinocytes (ScienCell Research Laboratories, Carlsbad, CA, USA) were used as normal control. All cell lines were grown in Dulbecco’s modified Eagle medium (Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum. For all TSCC cell lines, 1% penicillin/streptomycin was added to the culture medium and all were cultured at 37°C in a humidified atmosphere containing 5% CO2.
RNA extraction, reverse transcription, and quantitative reverse-transcription polymerase chain reaction
Total RNA was extracted from fresh surgical tongue tissues and cells using the Trizol reagent (Invitrogen), and reverse transcription (RT) was performed using the Prime ScriptTM RT reagent kit according to the manufacturer’s instructions (Takara Biotechnology, Shiga, Japan). RT-PCR products were analyzed via 2.0% agarose gel electrophoresis and stained with ethidium bromide for visualization using ultraviolet light. qRT-PCR was performed using the Light Cycler Real Time PCR System (Roche Diagnostics, Basel, Switzerland).
Insitu hybridization (ISH)
The ISH assay was performed according to the manufacturer’s procedures (Exiqon, Vedbaek, Denmark). Briefly, after demasking, H19 was hybridized to 5’-DIG-labeled LNATM probes. Then, the digoxigenins were recognized by a specific anti-DIG antibody that is directly conjugated with alkaline phosphatase. The nuclei were counterstained with hematoxylin.
Lentivirus infection
Lentivirus against H19 and negative control were synthesized and purchased from GenePharma Company (GenePharma, Shanghai, China). Lentiviral production and transduction were performed according to the manufacturer’s instructions. Gene silencing efficiency was detected by qRT-PCR after infection for 48 h.
Cell proliferation and cell apoptosis assay
Cells were seeded in 96-well plates and then infected with lentivirus against H19 or NC. Cell proliferation assays were performed every 24 h using CCK-8 (Dojindo Molecular Technologies, Kumamoto, Japan) according to the manufacturer’s instructions. Cells were harvested and stained using an Annexin V-FITC apoptosis detection kit according to the manufacturer’s protocols. Stained cells were analyzed immediately by flow cytometry. Cells were analyzed by FACS Calibur and Flowjo software (BD Biosciences, Franklin Lakes, NJ, USA).
Transwell assay
The invasion Transwell assay was performed in 24-well plates containing 8-mm membrane filter inserts (Corning Inc., Corning, NY, USA) coated with Matrigel (BD Biosciences) according to the manufacturer’s instructions. A total of 1 × 105 cells was added to the upper chamber and 600 μl medium was added in the lower chamber. The plates were incubated for 18 h, and the cells that entered the bottom chamber were fixed with 4% paraformaldehyde, stained, and counted.
RNA immunoprecipitation
RNA immunoprecipitation (RIP) was performed to detect the interaction between H19 and EZH2. Briefly, the EZH2 antibody was used (Abcam, Cambridge, MA, USA) to precipitated EZH2, and co-precipitated RNAs were detected by RT-PCR. The isotype IgG was used as control. The primers for detecting H19 was: Forward: 5’-ACTGCACTACCTGACTCAGGAAT-3’; reverse: 5’-AAGAGACAGAAGGATGAAAAAGA-3’.
Western blotting
Cellular protein extracts were prepared using lysis buffer containing protease inhibitor cocktail and phenylmethylsulfonyl fluoride. Equal quantities of protein extracts were separated on 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). Proteins were transferred to polyvinylidene difluoride (PVDF) membranes (Amersham Pharmacia Biotech, Little Chalfont, UK), which were then probed with antibodies against E-cadherin, N-cadherin, ZO-1, vimentin, cyclin D1 and c-Myc (Santa Cruz); EZH2, p-β-catenin, p-GSK-3β (Cell Signaling, Danvers, MA, USA), and GAPDH (Proteintech, Wuhan, China), respectively. The membranes were then treated with the peroxidase-conjugated secondary antibody, (Proteintech) and the signals were visualized using an enhanced chemiluminescence kit (GE, Fairfield, CT, USA) according to the manufacturer’s instructions. GAPDH was used as a loading control.
Immunohistochemistry
Immunohistochemistry (IHC) was performed on paraffin-embedded normal human tongue tissues as well as on various differentiated stage cancer tissue sections to evaluate the expression of E-cadherin, vimentin, β-catenin, and EZH2. For negative controls, isotype-matched antibodies were applied. Diaminobenzidine tetrahydrochloride (DAB) was used as a chromogen. All slices were then counterstained with hematoxylin and observed under a Zeiss AX10-Imager A1 microscope (Carl Zeiss, Thornwood, NY), and all images were captured using AxioVision 4.7 microscopy software (Carl Zeiss, Thornwood, NY).
Tumor xenograft model
A total of 1 × 106 UM1 or UM1-shH19 cells were injected subcutaneously into the armpit of 5-week-old BALB/c-nu mice. After tumors were detected, the tumor size was measured and calculated using the following equation: volume (mm3) = length × width2 × 0.5. Tumor xenografts, as well as whole lung tissues, were then harvested and weighed. The lung tissues were snap-frozen in liquid nitrogen, and cryosections (5 μm) were stained with hematoxylin and eosin (H&E). These procedures were approved by the Sun Yat-sen University Animal Care and Use Committee.
Statistical analysis
All data were analyzed with one-way analysis of variance using GraphPad software (Prism 6.0). Each experiment was repeated three times, with all the data presented as the mean ± standard deviation. Comparisons were performed with student’s t-test and one-way ANOVA and correlation analysis was evaluated with Pearson correlation analysis. Survival curves were plotted by the Kaplan-Meier method and compared using the log-rank test. P<0.05 was considered statistically significant in all cases.
Results
Relationship between H19 expression and clinical pathological characteristics in TSCC patients
To investigate H19 expression status in tongue tissues, ISH was performed in TSCC tissues with and without metastasis, as well as adjacent normal tissue. The staining results showed that the expression level of H19 was much higher in TSCC tissue than in adjacent normal tongue tissue; in particular, TSCC with metastasis exhibited higher levels of H19 expression than that TSCC tissues without metastasis (Figure 1A). Furthermore, the expression of H19 by qRT-PCR was measured in 50 matched pairs of fresh adjacent non-cancerous and TSCC tissue samples. The expression level of H19 was notably higher in tumor tissues than in matched adjacent non-cancerous tissues (P<0.0001, Figure 1B). To explore the association between H19 expression and clinicopathological features, 123 patient TSCC tissues were determined. We observed that the H19 expression level was gradually increased in TSCC patients according with WHO stage (P=0.008, P=0.013, Figure 1C). H19 expression level was also upregulated in late-stage tumor tissues. In addition, the H19 expression level was positively associated with lymph node metastasis status (P=0.035, Figure 1D).
Figure 1.
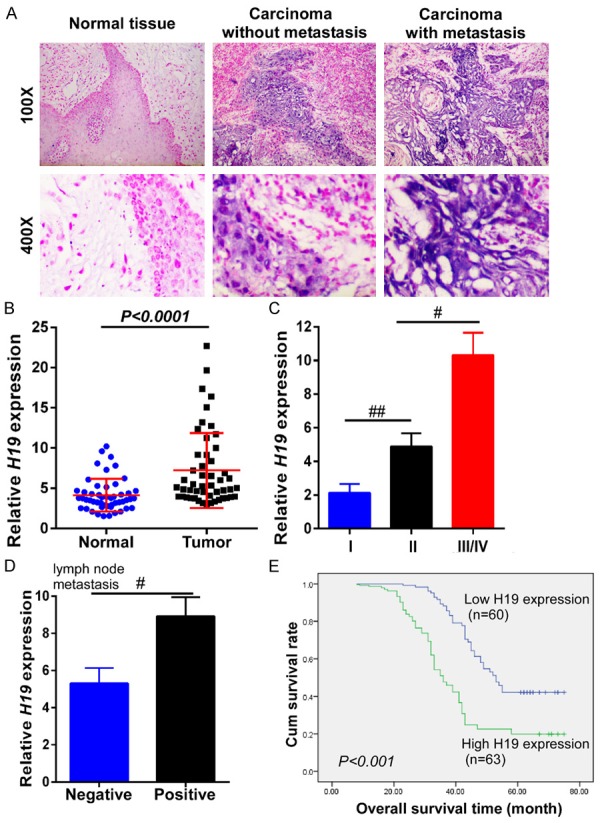
LncRNA H19 was overexpressed in TSCC tissues and significantly associated with poor prognosis. A. Insitu hybridization was performed to detect H19 expression in TSCC tissues with and without metastasis and in adjacent tongue tissues. The carcinoma with metastasis had a higher expression of H19 than that in the carcinoma without metastasis. B. The relative expression of H19 was examined by qRT-PCR in 50 TSCC tissues and matched adjacent non-cancerous tissues. The relative gene expression was normalized to that of GAPDH. C. The relative expression of H19 was much higher at advanced pathological stages in TSCC patients. D. The relative expression of H19 was analyzed in tissues from TSCC patients with or without lymph node metastasis. E. Kaplan-Meier overall survival curves were generated for patients with different H19 expression levels. The higher expression of H19 had a much shorter lifespan. Data represent the mean ± SD. #P<0.05, ##P<0.01, ###P<0.001.
To further explore the prognostic value of H19 in patient survival, 123 TSCC patients were divided into two groups according to H19 expression levels (high expression and low expression), and the correlation between H19 expression level and overall survival was evaluated through Kaplan-Meier analysis and log rank test. As shown in Figure 1E, the overall survival time varied significantly between patients with high and low H19 expression, with the low H19 expression group having longer overall survival times, compared with those with high expression levels of H19 (P<0.001).
Downregulation of H19 attenuated TSCC cancer cell proliferation, apoptosis, and invasion in vitro
To understand the effect of H19 in the progression of TSCC, the H19 expression profile was examined in six TSCC cell lines. Human oral keratinocytes were used as the normal control. The results of RT-PCR analysis revealed that the mRNA expression levels of H19 were increased in all six TSCC cell lines, compared with human oral keratinocyte cells, and that UM1 exhibited the highest expression of H19 (Figure 2A). This finding is consistent with results from our qRT-PCR analysis (Figure 2B).
Figure 2.
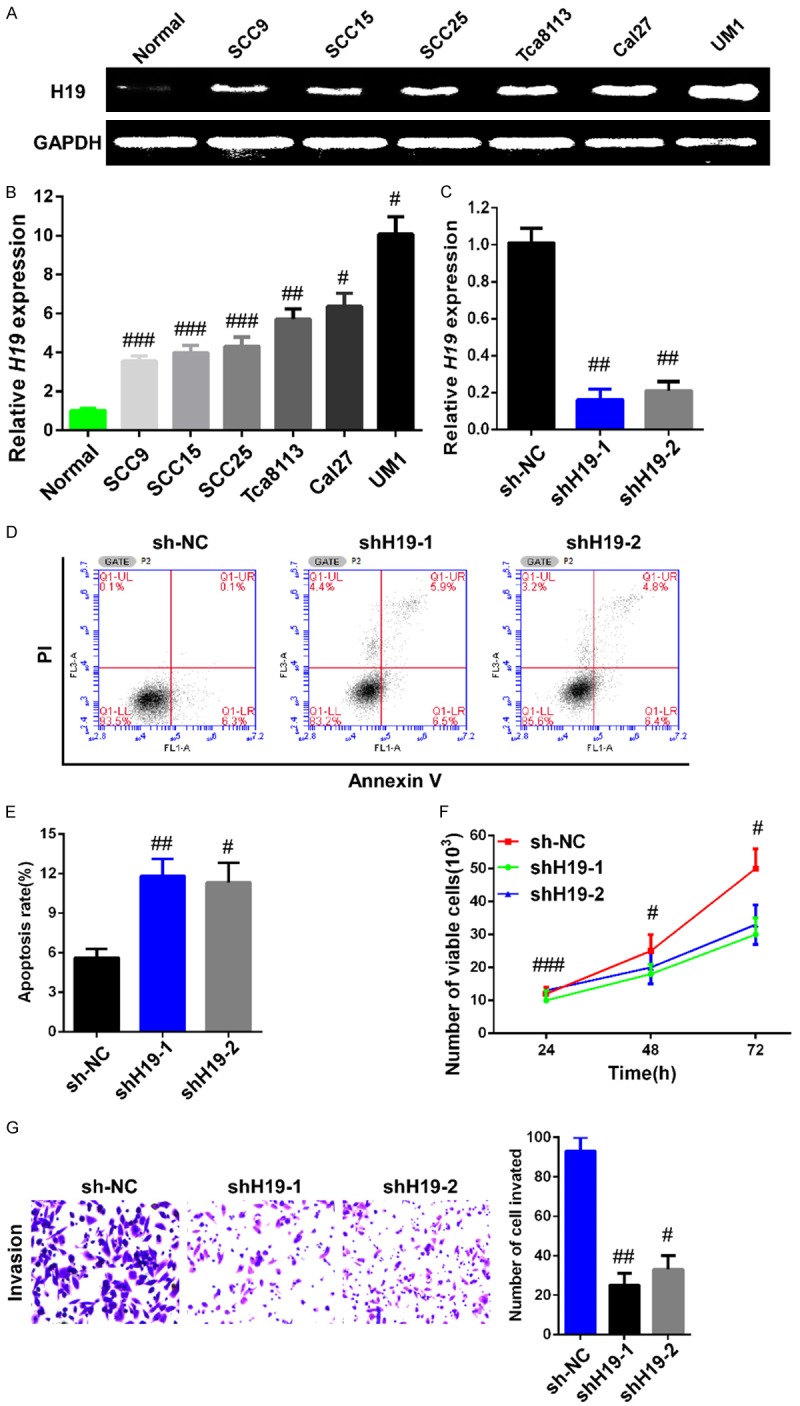
Downregulation of H19 inhibited malignant potential in UM1 cells. A. RT-PCR was performed to detect H19 mRNA expression in six TSCC cell lines and in human oral keratinocyte cells. The high invasive ability cancer cell line UM1 had a much higher expression profile of H19 in TSCC cell lines. B. H19 mRNA expression was examined by qRT-PCR in six TSCC cell lines and in human oral keratinocyte cells. C. UM1 cells were infected with lentivirus shH19 or sh-NC and the interference efficiency was measured by qRT-PCR. D. The percentage of apoptotic cells infected with lentivirus containing shRNA against H19 or NC were detected by flow cytometric analysis of annexin V/PI staining. E. Apoptosis rate was evaluated by counting the percentage of early apoptotic and late apoptotic cells. F. Silencing of H19 suppressed the proliferation of UM1 cells in vitro. The CCK-8 assay was carried out to determine cell growth. G. The Transwell invasion assay was performed to detect motility in UM1 cells. UM1 cells infected with sh-H19 exhibited lower invasive capacity compared with those infected with sh-NC. Data are representative of three independent experiments and represent the mean ± SD. #P<0.05, ##P<0.01, ###P<0.001.
To further investigate the function of H19 in TSCC progression, two lentiviruses against H19 (shH19-1 and shH19-2) and negative control (sh-NC) were designed, synthesized, and infected into UM1 cells, respectively. As shown in Figure 2C, cells infected with two different shH19s both showed a significant (>80%) decrease in the mRNA expression of H19 compared to the sh-NC group (P=0.0056). CCK-8 assays were performed to detect cell proliferation ability; the results revealed that cell growth was suppressed in UM1 cells infected with shH19 lentiviruses compared with the sh-NC group (Figure 2D). The apoptotic rate of cells infected with shH19 lentiviruses was notably elevated, compared with the sh-NC group (P=0.0061, Figure 2E). In addition, an invasion assay was carried out to analyze the role of H19 in cell invasion capability, and the results showed that UM1 cells infected with shH19 lentiviruses were significantly less migratory than cells infected with sh-NC (P=0.0008, Figure 2F).
Silencing H19 blocked β-catenin/GSK-3β activation, attenuated EZH2, cyclin D1, c-Myc, and mesenchymal markers, and enhanced epithelial marker expression in TSCC cells
It has been previously reported that H19 can interact with EZH2 and cause the upregulation of EZH2 in bladder cancer [14]. To determine the association between H19 and EZH2 in TSCC, RNA immunoprecipitation (RIP) was carried out in UM1 cells. Using an antibody specifically targeted to EZH2 to precipitate the complex, followed by qRT-PCR with specific primer, H19 was detected; as in addition, the total RNA input was also measured. However, no interaction was observed between the isotype antibody and H19 (Figure 3A). Meanwhile, it has been well established that EZH2 overexpression is associated with Wnt/β-catenin/GSK-3β phosphorylation, leading to tumor cell cycle dysfunction, oncogenic gene activation, and tumor invasion and metastasis [36,37]. Our results revealed that the overexpression of H19 significantly accelerated TSCC cell proliferation, apoptosis, and invasion ability. However, the underlying mechanism was unclear. To investigate the downstream signaling status, western blot was performed to detect the activation of β-catenin and the phosphorylation of GSK-3β, cyclin D1, and c-myc. The results showed that silencing H19 severely inhibited the activation of β-catenin and GSK-3β, as well as the cell cycle genecyclin D1 and the oncogenic gene c-Myc (Figure 3B). The EMT signal pathway expression status was also measured by western blot; the data indicate that when H19 expression is knocked down, epithelial markers (E-cadherin and ZO-1) are upregulated and mesenchymal markers (N-cadherin and vimentin) are downregulated (Figure 3C). qRT-PCR was used to evaluate the expression of transcriptional factors that positively promoted tumor cell EMT, and we found that Snail1, Twist1, and ZEB1 were downregulated when silencing H19 expression. In addition, knockdown H19 expression can also attenuate N-cadherin and vimentin expression and accelerate E-cadherin and ZO-1 expression at the mRNA level (Figure 3D).
Figure 3.
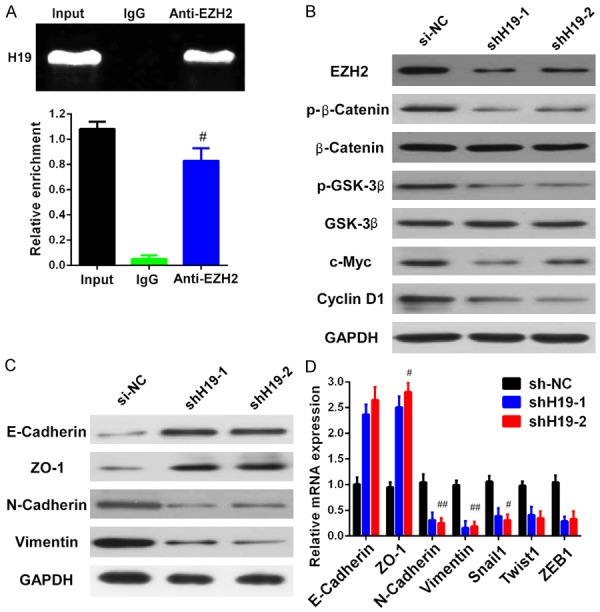
Silencing of H19 affected the expression levels of EZH2/β-catenin/GSK-3β/EMT. A. RNA immunoprecipitation was performed to analyze the interaction between H19 and EZH2. Total RNA input and isotype antibody were used as positive and negative control, respectively. After immunoprecipitation, the expression of H19 was detected by qRT-PCR in input, isotype and anti-EZH2 antibody groups. The antibody against EZH2 canimmunoprecipitate H19. B. The expression of EZH2, activated β-catenin, phosphorylated GSK-3β, cyclin D1, and c-myc were significantly decreased upon silencing H19 expression, as measured by western blot. C. Knockdown of H19 suppressed N-cadherin and vimentin expression, while upregulated E-cadherin and ZO-1 measured by western blot. D. The mRNA expression level of EMT markers (E-cadherin, ZO-1, N-cadherin and vimentin) and EMT promoting transcriptional factors (Snail1, Twist1 and ZEB1) were detected by qRT-PCR after silencing of H19. Data are representative of three independent experiments and represent the mean ± SD. #P<0.05, ##P<0.01.
Association between EZH2 expression and clinical pathological characteristics in TSCC patients
Our results implied that H19 modulated EZH2 expression in TSCC cells; we therefore examined the expression of EZH2 in TSCC tissue with and without metastasis, as well as in adjacent normal tissue. Results from IHC staining showed that EZH2 expression levels were much higher in TSCC tissues than in adjacent normal tongue tissue. Moreover, TSCC tissue with metastasis exhibited higher EZH2 expression levels than did TSCC tissues without metastasis (Figure 4A). Furthermore, 50 matched pairs of fresh adjacent non-cancerous and TSCC tissue samples were used to measure the expression of EZH2 by qRT-PCR. We found that EZH2 expression levels was notably elevated in cancer tissues compared to levels in matched adjacent non-cancerous tissues (P=0.0051, Figure 4B). Furthermore, EZH2 expression was elevated in TSCC patients in line with WHO stage, and was predominantly upregulated in late-stage tumor tissues (P=0.028, P=0.037, Figure 4C). Additionally, EZH2 expression level was closely associated with lymph node metastasis (P=0.019, Figure 4D). We also analyzed the correlation between EZH2 expression and clinicopathological features using Kaplan-Meier analysis and the logrank test in 123 TSCC tissues. As shown in Figure 4E, the length of overall survival time varied significantly different between patients with high and low EZH2 expression (P<0.001), with the low EZH2 expression group having a longer overall survival time, compared to the high-level EZH2 expression group.
Figure 4.
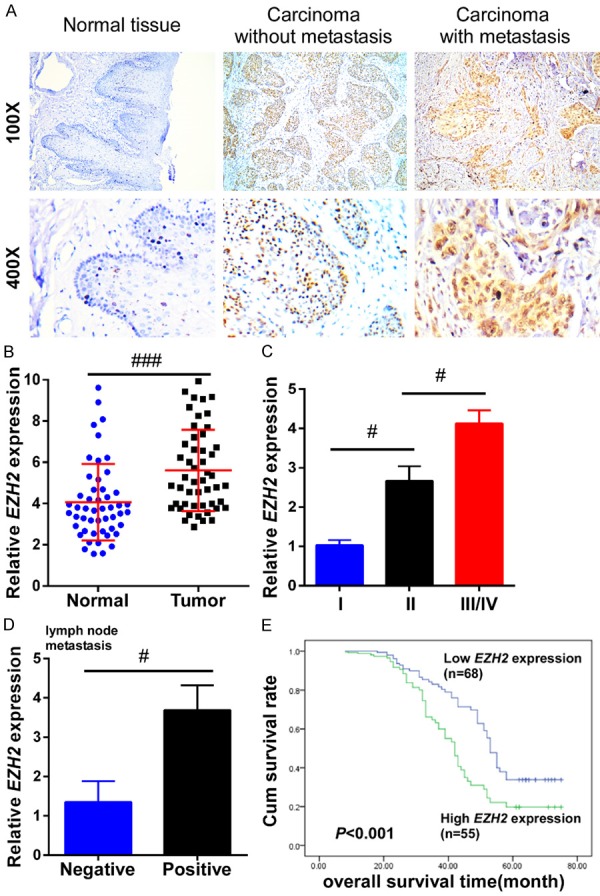
EZH2 was upregulated in TSCC tissues and closely associated with poor prognosis. A. Immunohistochemistry was performed to detect the expression of EZH2 in TSCC tissues with and without metastasis, and in adjacent normal tongue tissues. The carcinoma with metastasis had a higher expression of EZH2 than that in the carcinoma without metastasis. B. The relative expression of EZH2 was examined by qRT-PCR in 50 TSCC tissues and matched adjacent non-cancerous tissues. The relative gene expression was normalized to GAPDH. C. The relative expression of EZH2 was much higher at advanced pathological stages in TSCC patients. D. The relative expression of EZH2 was analyzed between TSCC patients with or without lymph node metastasis. E. Kaplan-Meier overall survival curves were generated for patients with different EZH2 expression levels. The higher expression of EZH2 had a much shorter lifespan. Data represent the mean ± SD. #P<0.05, ##P<0.01, ###P<0.001.
Correlation among H19, β-catenin, EZH2, and EMT marker expression in TSCC patients
To confirm the correlation among H19, EZH2, β-catenin and the progression of tongue cancer, the expression of β-catenin and EMT markers, E-cadherin and vimentin were examined in TSCC tissues with and without metastasis and in adjacent normal tissue using IHC. As shown in Figure 5A, TSCC tissue with metastasis expressed much higher levels of β-catenin and vimentin, but lower levels of E-cadherin, than TSCC tissue without metastasis, compared to the normal tongue tissues (Figure 5A). To further illustrate the relationship between H19, EZH2, and E-cadherin, the correlations above were analyzed by Person analysis; we found a strong positive correlation between H19 and EZH2 (P=0.0005, R=0.553, Figure 5B), negative correlation between H19 and E-cadherin (P=0.0038, R=-0.402, Figure 5C), and negative correlation between EZH2 and E-cadherin (P=0.0016, R=-0.435, Figure 5D).
Figure 5.
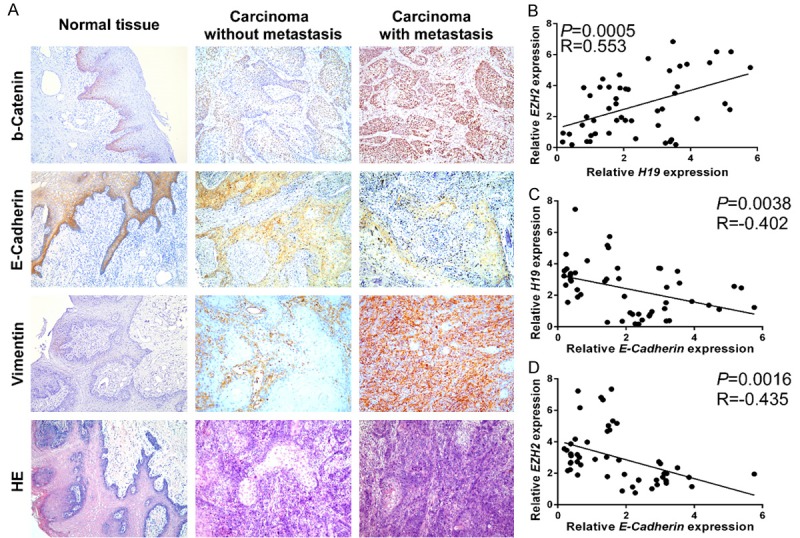
Overexpression of H19 was positively associated with EZH2/β-catenin up-regulation and promoted TSCC EMT. (A) The expression levels of β-catenin, E-cadherin, and vimentin were measured by immunohistochemistry in TSCC tissues with and without metastasis, and in adjacent normal tongue tissue. The carcinoma with metastasis had a higher expression of β-catenin and vimentin, but lower expression of E-Cadherin than that in the carcinoma without metastasis. Correlation analysis showing (B) positive correlation between H19 and EZH2; (C) Negative correlation between H19 and E-cadherin; (D) Negative correlation between EZH2 and E-cadherin in TSCC patients.
Silencing H19 expression attenuates tumor growth and metastasis of TSCC in vivo
Because knockdown of H19 expression inhibits the proliferation and invasion of TSCC cells in vitro, we further assessed its effect on tumor growth and metastasis in vivo. H19 was stably inhibited in UM1 cells by infection with lentivirus containing shRNA against H19. As shown in Figure 6A and 6B, knockdown of H19 expression significantly decreased tumor weight and tumor volume in BALB/c-nu mice subcutaneously inoculated with UM1-shH19 cells. When the mice were xenografted, the lungs were harvested and H&E staining was performed to detect tumor metastasis status. We found that silencing H19 expression significantly attenuated metastasis in the lungs of mice bearing UM1-shH19 xenografts, as compared with UM1-sh-NC mice (Figure 6C). These findings suggest that H19 promotes the metastasis of TSCC in vivo.
Figure 6.
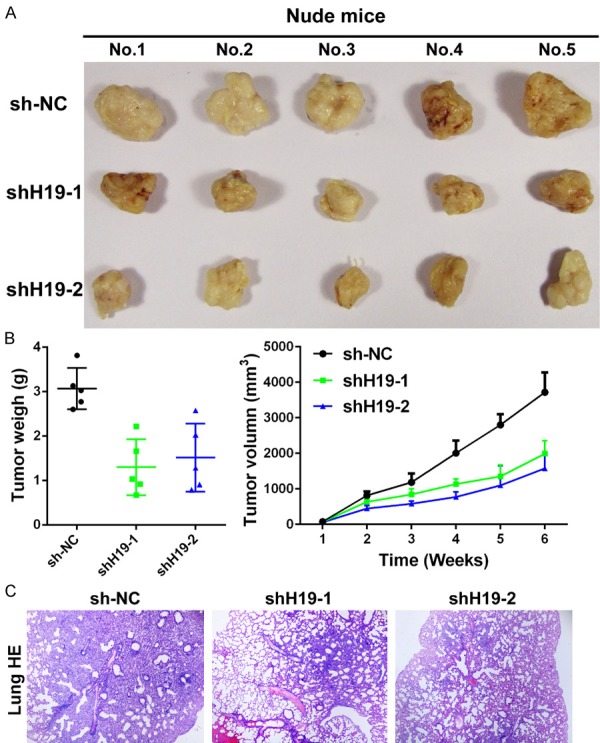
Knockdown of H19 inhibits UM1 xenograft lung metastasis in BALB/c-nu mice. UM1-sh-NC and UM1-sh19s cells were injected into nude mice (n=5/group). (A) Images of tumors from all mice in each group. Knockdown of H19 expression by lentivirus can remarkably attenuate the tumor growth and formation. (B) Mean tumor weight (left panel), tumor volume (right panel), and (C) lung metastasis stained by H&E were analyzed.
Discussion
TSCC is one of the most prevalent malignant tumors in the world, influencing millions of patients worldwide each year, especially in South Asia [38]. Through improvements have been achieved in past decades, the 5-year survival rate still remains at about 50% [39,40]. Previous research has suggested that the progression of TSCC is dependent on tumor cell proliferation, apoptosis, invasion, metastasis and recurrence [41,42]. Therefore, unveiling and understanding the underlying regulatory mechanism of this malignancy is crucial to the development of novel and effective therapeutic strategies for TSCC.
The discovery of long non-coding RNA was a significant conceptual breakthrough in the post-genome era. LncRNAs represented a new class of non-coding RNA (>200 nt) that do not exhibit protein coding potential. The available data indicate that lncRNAs are involved in a variety of vital biological activities, including embryonic development, organofaction, tumorigenesis and cancer metastasis [11,43,44]. It was reported that Malat1 was associated with larger tumor size, advanced TNM stage, more lymph node metastases, and distant metastases in multiple cancer types [45-47]. The abnormal upregulation of Hotair was found to be significantly correlated with tumor invasion, advanced clinical stage, lymph node metastasis, and poor prognosis in several tumors [48,49]. In addition, the downregulation of GAS5 was also found to be closely associated with aberrant cell cycle, proliferation, apoptosis, and tumor metastasis in cancer [50]. Although significant progress has been achieved in understanding the function and mechanism of lncRNAs, the expression pattern of H19, EZH2, and EMT and molecular mechanism involved in TSCC remain largely dismal.
The β-catenin pathway is involved in regulating a wide variety of physiological and pathological processes, including embryogenesis, differentiation, and carcinogenesis [51]. Furthermore, β-catenin participates in cell-cell adhesion and signal transduction as a downstream hinge of the Wnt/β-catenin pathway [34,35,52]. Although much effort has gone into understanding the molecular mechanism of the β-catenin pathway, little is known about the correlation between H19 and β-catenin in TSCC. In the present study, we observe that downregulation of H19 caused a significant decrease in the expression of β-catenin, EZH2, cyclin D1, c-Myc and mesenchymal genes, such as N-cadherin and vimentin, while increasing E-cadherin and ZO-1 expression levels.
It has been reported that H19 can regulate EZH2 expression by interacting with miRNAs and promotes cell invasion and metastasis through EMT signaling in a variety of carcinomas. However, there is no evidence of direct interaction between H19 and EZH2. H19 may function as an miRNA sponge to suppress miRNA activity by sequestering miRNAs away from their target genes [53]. Notably, in a bladder cancer model, H19 was verified to increases the binding of EZH2, which accelerates Wnt/β-catenin activation and results in decreased E-cadherin expression and tumor invasion [14].
We found that H19 and EZH2 expression levels were significantly higher in TSCC tissues compared with adjacent normal tissues, and there was an obvious correlation between H19 and EZH2 expression in TSCC. Additionally, increased H19 and EZH2 expression levels accelerated the progress of TSCC, which was reflected not only in the gradual elevation of H19 and EZH2 expression from normal tongue tissue to cancer tissue, but also in the relationship of H19 and EZH2 expression with WHO grade and lymph node metastasis. Lastly, overexpressed EZH2 combined with H19 downregulation completely reversed the sh-H19 mediated repression of β-catenin and GSK-3β activation in tongue cancer cells. Based on these findings, we conclude that EZH2 is not only involved in the carcinogenic process, but is also activated in tumor progression.
Conclusions
In conclusion, overexpression of H19 promotes tongue cancer cell invasion and apoptosis resistance in vitro. In addition, the upregulation of both H19 and EZH1 is closely associated with poor prognosis in tongue cancer patients. H19 plays a crucial role in the progression of TSCC by regulating the expression of β-catenin and GSK-3β via EZH2, indicating that inhibition of H19 might be a potential target for the treatment of TSCC.
Acknowledgements
This project was supported by the China Postdoctoral Science Foundation 2016M590839 (Z. Lin).
Disclosure of conflict of interest
None.
References
- 1.Neville BW, Day TA. Oral Cancer and PrecancerousLesions. CA Cancer J Clin. 2002;52:195–215. doi: 10.3322/canjclin.52.4.195. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 4.Bello IO, Soini Y, Salo T. Prognostic evaluation of oral tongue cancer: means, markers and perspectives (I) Oral Oncol. 2010;46:630–635. doi: 10.1016/j.oraloncology.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 5.Bello IO, Soini Y, Salo T. Prognostic evaluation of oral tongue cancer: means, markers and perspectives (II) Oral Oncol. 2010;46:636–643. doi: 10.1016/j.oraloncology.2010.06.008. [DOI] [PubMed] [Google Scholar]
- 6.Leemans CR, Braakhuis BJ, Brakenhoff RH. The molecular biology of head and neck cancer. Nat Rev Cancer. 2011;11:9–22. doi: 10.1038/nrc2982. [DOI] [PubMed] [Google Scholar]
- 7.Gold KA, Lee HY, Kim ES. Targeted therapies in squamous cell carcinoma of the head and neck. Cancer. 2009;115:922–935. doi: 10.1002/cncr.24123. [DOI] [PubMed] [Google Scholar]
- 8.Evans JR, Feng FY, Chinnaiyan AM. The bright side of dark matter: lncRNAs in cancer. J Clin Invest. 2016;126:2775–2782. doi: 10.1172/JCI84421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21:354–361. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 10.Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152:1298–1307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29:452–463. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1:391–407. doi: 10.1158/2159-8290.CD-11-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matouk IJ, Halle D, Raveh E, Gilon M, Sorin V, Hochberg A. The role of the oncofetal H19 lncRNA in tumor metastasis: orchestrating the EMT-MET decision. Oncotarget. 2015;7:3748–3765. doi: 10.18632/oncotarget.6387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luo M, Li Z, Wang W, Zeng Y, Liu Z, Qiu J. Long non-coding RNA H19 increases bladder cancer metastasis by associating with EZH2 and inhibiting E-cadherin expression. Cancer Lett. 2013;333:213–221. doi: 10.1016/j.canlet.2013.01.033. [DOI] [PubMed] [Google Scholar]
- 15.Byun HM, Wong HL, Birnstein EA, Wolff EM, Liang G, Yang AS. Examination of IGF2 and H19 loss of imprinting in bladder cancer. Cancer Res. 2007;67:10753–10758. doi: 10.1158/0008-5472.CAN-07-0329. [DOI] [PubMed] [Google Scholar]
- 16.Vennin C, Spruyt N, Dahmani F, Julien S, Bertucci F, Finetti P, Chassat T, Bourette RP, Le Bourhis X, Adriaenssens E. H19 non coding RNA-derived miR-675 enhances tumorigenesis and metastasis of breast cancer cells by downregulating c-Cbl and Cbl-b. Oncotarget. 2015;6:29209–29223. doi: 10.18632/oncotarget.4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han D, Gao X, Wang M, Qiao Y, Xu Y, Yang J, Dong N, He J, Sun Q, Lv G, Xu C, Tao J, Ma N. Long noncoding RNA H19 indicates a poor prognosis of colorectal cancer and promotes tumor growth by recruiting and binding to eIF4A3. Oncotarget. 2016;7:22159–22173. doi: 10.18632/oncotarget.8063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liang WC, Fu WM, Wong CW, Wang Y, Wang WM, Hu GX, Zhang L, Xiao LJ, Wan DC, Zhang JF, Waye MM. The lncRNA H19 promotes epithelial to mesenchymal transition by functioning as miRNA sponges in colorectal cancer. Oncotarget. 2015;6:22513–22525. doi: 10.18632/oncotarget.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao T, He B, Pan Y, Gu L, Chen L, Nie Z, Xu Y, Li R, Wang S. H19 DMR methylation correlates to the progression of esophageal squamous cell carcinoma through IGF2 imprinting pathway. Clin Transl Oncol. 2014;16:410–417. doi: 10.1007/s12094-013-1098-x. [DOI] [PubMed] [Google Scholar]
- 20.Li H, Yu B, Li J, Su L, Yan M, Zhu Z, Liu B. Overexpression of lncRNA H19 enhances carcinogenesis and metastasis of gastric cancer. Oncotarget. 2014;5:2318–2329. doi: 10.18632/oncotarget.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Conigliaro A, Costa V, Lo Dico A, Saieva L, Buccheri S, Dieli F, Manno M, Raccosta S, Mancone C, Tripodi M, De Leo G, Alessandro R. CD90+ liver cancer cells modulate endothelial cell phenotype through the release of exosomes containing H19 lncRNA. Mol Cancer. 2015;14:155. doi: 10.1186/s12943-015-0426-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsang WP, Kwok TT. Riboregulator H19 induction of MDR1-associated drug resistance in human hepatocellular carcinoma cells. Oncogene. 2007;26:4877–4881. doi: 10.1038/sj.onc.1210266. [DOI] [PubMed] [Google Scholar]
- 23.Mizrahi A, Czerniak A, Levy T, Amiur S, Gallula J, Matouk I, Abu-lail R, Sorin V, Birman T, de Groot N, Hochberg A, Ohana P. Development of targeted therapy for ovarian cancer mediated by a plasmid expressing diphtheria toxin under the control of H19 regulatory sequences. J Transl Med. 2009;7:69. doi: 10.1186/1479-5876-7-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Medrzycki M, Zhang Y, Zhang W, Cao K, Pan C, Lailler N, McDonald JF, Bouhassira EE, Fan Y. Histone h1.3 suppresses h19 noncoding RNA expression and cell growth of ovarian cancer cells. Cancer Res. 2014;74:6463–6473. doi: 10.1158/0008-5472.CAN-13-2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma C, Nong K, Zhu H, Wang W, Huang X, Yuan Z, Ai K. H19 promotes pancreatic cancer metastasis by derepressing let-7’s suppression on its target HMGA2-mediated EMT. Tumour Biol. 2014;35:9163–9169. doi: 10.1007/s13277-014-2185-5. [DOI] [PubMed] [Google Scholar]
- 26.Zhu M, Chen Q, Liu X, Sun Q, Zhao X, Deng R, Wang Y, Huang J, Xu M, Yan J, Yu J. lncRNA H19/miR-675 axis represses prostate cancer metastasis by targeting TGFBI. FEBS J. 2014;281:3766–3775. doi: 10.1111/febs.12902. [DOI] [PubMed] [Google Scholar]
- 27.Kim KH, Roberts CW. Targeting EZH2 in cancer. Nat Med. 2016;22:128–134. doi: 10.1038/nm.4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chase A, Cross NC. Aberrations of EZH2 in cancer. Clin Cancer Res. 2011;17:2613–2618. doi: 10.1158/1078-0432.CCR-10-2156. [DOI] [PubMed] [Google Scholar]
- 29.Behrens C, Solis LM, Lin H, Yuan P, Tang X, Kadara H, Riquelme E, Galindo H, Moran CA, Kalhor N, Swisher SG, Simon GR, Stewart DJ, Lee JJ, Wistuba II. EZH2 protein expression associates with the early pathogenesis, tumor progression, and prognosis of non-small cell lung carcinoma. Clin Cancer Res. 2013;19:6556–6565. doi: 10.1158/1078-0432.CCR-12-3946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Vries NA, Hulsman D, Akhtar W, de Jong J, Miles DC, Blom M, van Tellingen O, Jonkers J, van Lohuizen M. Prolonged Ezh2 depletion in glioblastoma causes a robust switch in cell fate resulting in tumor progression. Cell Rep. 2015 doi: 10.1016/j.celrep.2014.12.028. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 31.Feng H, Yu Z, Tian Y, Lee YY, Li MS, Go MY, Cheung YS, Lai PB, Chan AM, To KF, Chan HL, Sung JJ, Cheng AS. A CCRK-EZH2 epigenetic circuitry drives hepatocarcinogenesis and associates with tumor recurrence and poor survival of patients. J Hepatol. 2015;62:1100–1111. doi: 10.1016/j.jhep.2014.11.040. [DOI] [PubMed] [Google Scholar]
- 32.He LR, Liu MZ, Li BK, Jia WH, Zhang Y, Liao YJ, Chen YC, Zhang LJ, Guan XY, Zeng YX, Kung HF, Xie D. High expression of EZH2 is associated with tumor aggressiveness and poor prognosis in patients with esophageal squamous cell carcinoma treated with definitive chemoradiotherapy. Int J Cancer. 2010;127:138–147. doi: 10.1002/ijc.25031. [DOI] [PubMed] [Google Scholar]
- 33.Lee ST, Li Z, Wu Z, Aau M, Guan P, Karuturi RK, Liou YC, Yu Q. Context-specific regulation of NF-kappaB target gene expression by EZH2 in breast cancers. Mol Cell. 2011;43:798–810. doi: 10.1016/j.molcel.2011.08.011. [DOI] [PubMed] [Google Scholar]
- 34.Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 35.MacDonald BT, Tamai K, He X. Wnt/betacatenin signaling: components, mechanisms, and diseases. Dev Cell. 2009;17:9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jung HY, Jun S, Lee M, Kim HC, Wang X, Ji H, McCrea PD, Park JI. PAF and EZH2 induce Wnt/beta-catenin signaling hyperactivation. Mol Cell. 2013;52:193–205. doi: 10.1016/j.molcel.2013.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu P, Wang Y, Huang G, Ye B, Liu B, Wu J, Du Y, He L, Fan Z. lnc-beta-Catm elicits EZH2-dependent beta-catenin stabilization and sustains liver CSC self-renewal. Nat Struct Mol Biol. 2016;23:631–639. doi: 10.1038/nsmb.3235. [DOI] [PubMed] [Google Scholar]
- 38.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–32. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 39.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 40.Neville BW, Day TA. Oral cancer and precancerous lesions. CA Cancer J Clin. 2002;52:195–215. doi: 10.3322/canjclin.52.4.195. [DOI] [PubMed] [Google Scholar]
- 41.Miyazawa J, Mitoro A, Kawashiri S, Chada KK, Imai K. Expression of mesenchyme-specific gene HMGA2 in squamous cell carcinomas of the oral cavity. Cancer Res. 2004;64:2024–2029. doi: 10.1158/0008-5472.can-03-1855. [DOI] [PubMed] [Google Scholar]
- 42.Civantos FJ, Zitsch RP, Schuller DE, Agrawal A, Smith RB, Nason R, Petruzelli G, Gourin CG, Wong RJ, Ferris RL, El Naggar A, Ridge JA, Paniello RC, Owzar K, McCall L, Chepeha DB, Yarbrough WG, Myers JN. Sentinel lymph node biopsy accurately stages the regional lymph nodes for T1-T2 oral squamous cell carcinomas: results of a prospective multi-institutional trial. J. Clin. Oncol. 2010;28:1395–1400. doi: 10.1200/JCO.2008.20.8777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qi P, Du X. The long non-coding RNAs, a new cancer diagnostic and therapeutic gold mine. Mod Pathol. 2013;26:155–165. doi: 10.1038/modpathol.2012.160. [DOI] [PubMed] [Google Scholar]
- 44.Zhang H, Chen Z, Wang X, Huang Z, He Z, Chen Y. Long non-coding RNA: a new player in cancer. J Hematol Oncol. 2013;6:37. doi: 10.1186/1756-8722-6-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou X, Liu S, Cai G, Kong L, Zhang T, Ren Y, Wu Y, Mei M, Zhang L, Wang X. Long non coding RNA MALAT1 promotes tumor growth and metastasis by inducing epithelial-mesenchymal transition in oral squamous cell carcinoma. Sci Rep. 2015;5:15972. doi: 10.1038/srep15972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tee AE, Liu B, Song R, Li J, Pasquier E, Cheung BB, Jiang C, Marshall GM, Haber M, Norris MD, Fletcher JI, Dinger ME, Liu T. The long noncoding RNA MALAT1 promotes tumor-driven angiogenesis by up-regulating pro-angiogenic gene expression. Oncotarget. 2016;7:8663–8675. doi: 10.18632/oncotarget.6675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jadaliha M, Zong X, Malakar P, Ray T, Singh DK, Freier SM, Jensen T, Prasanth SG, Karni R, Ray PS, Prasanth KV. Functional and prognostic significance of long non-coding RNA MALAT1 as a metastasis driver in ER negative lymph node negative breast cancer. Oncotarget. 2016;7:40418–40436. doi: 10.18632/oncotarget.9622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu Y, Zhang L, Wang Y, Li H, Ren X, Wei F, Yu W, Wang X, Zhang L, Yu J, Hao X. Long noncoding RNA HOTAIR involvement in cancer. Tumour Biol. 2014;35:9531–9538. doi: 10.1007/s13277-014-2523-7. [DOI] [PubMed] [Google Scholar]
- 49.Zhang J, Zhang P, Wang L, Piao HL, Ma L. Long non-coding RNA HOTAIR in carcinogenesis and metastasis. Acta Biochim Biophys Sin (Shanghai) 2014;46:1–5. doi: 10.1093/abbs/gmt117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu X, Li Z. Long non-coding RNA growth arrest-specific transcript 5 in tumor biology. Oncol Lett. 2015;10:1953–1958. doi: 10.3892/ol.2015.3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Valenta T, Hausmann G, Basler K. The many faces and functions of β-catenin. EMBO J. 2012;31:2714–2736. doi: 10.1038/emboj.2012.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Clevers H, Nusse R. Wnt/beta-catenin signaling and disease. Cell. 2012;149:1192–1205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 53.Kallen AN, Zhou XB, Xu J, Qiao C, Ma J, Yan L, Lu L, Liu C, Yi JS, Zhang H, Min W, Bennett AM, Gregory RI, Ding Y, Huang Y. The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol Cell. 2013;52:101–112. doi: 10.1016/j.molcel.2013.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]


