Abstract
During patient care simulations, cauliflower mosaic virus DNA and bacteriophage MS2 performed similarly as surrogate markers of pathogen dissemination. These markers disseminated to the environment in a manner similar to Clostridium difficile spores but were more frequently detected on skin and clothing of personnel after personal protective equipment removal.
Keywords: bacteriophage MS-2, cauliflower mosaic virus, DNA, environment, transmission
Benign surrogate markers such as viral DNA and live nonpathogenic viruses provide useful tools to study routes of pathogen transmission [1–6]. Cauliflower mosaic virus DNA placed on toys in childcare centers or on telephone handles in a neonatal intensive care unit disseminated widely to hands and environmental surfaces [1, 2]. Bacteriophage MS2, a nonenveloped RNA virus, inoculated on hands or surfaces has similarly been shown to disseminate widely in a variety of settings, including hotels, households, schools, and healthcare facilities [3–6]. Nonpathogenic viruses have also provided information on the contamination of healthcare personnel during removal of personal protective equipment (PPE) [7–9]. In many of these studies, reductions in contamination have been achieved through interventions such as hand hygiene, environmental cleaning, and education on correct PPE removal [1,3,4,6,7].
Although benign surrogate markers are valuable tools to study pathogen dissemination, it is not known if results are comparable for viral DNA markers and live nonpathogenic viruses. In addition, it is not known how well findings using these surrogates correlate with dissemination of bacterial pathogens. Therefore, we compared dissemination of cauliflower mosaic virus DNA, bacteriophage MS2, and nontoxigenic Clostridium difficile spores during simulations of patient care.
METHODS
The protocol was approved by the Louis Stokes Veterans Affairs Medical Center’s Institutional Review Board. Bacteriophage MS2 and nontoxigenic C. difficile spores (American Type Culture Collection 43593) were prepared as previously described [7,10]. A 222 base-pair DNA marker was generated using methods similar to those of Oelberg et al [2]. All 210 nucleotides of the cauliflower mosaic virus 35S promoter region were used (GenBank: S51061.1) [11], with the addition of GAATTC terminal sequences on each end. The marker DNA sequence was synthesized at Celtek Bioscience and cloned into the plasmid pBluescript II SK (Celtek Bioscience). The plasmid was transformed into ElectroMAX electrocompetent DH10B Escherichia coli cells (Invitrogen) by electroporation and cultured in Super Optimal Broth medium (Fisher Scientific) for 1 hour at 37ºC. The culture medium was plated on tryptic soy agar plates containing 100-µg/mL ampicillin. A colony was selected and cultured in tryptic soy broth, and the DNA marker was extracted by a Qiaprep Spin Maxi kit for plasmids (Qiagen). For detection of the DNA marker, polymerase chain reaction (PCR) was performed with forward primer CCTCAGCAATCGCAGCAAA and reverse primer GGAAGATCAATACATAAAGAGTTGAACTTC, and amplicons were visualized on an agarose gel using GreenGlo (Denville Scientific). The PCR reaction was done in a 20-µL reaction mixture containing 10 µL of Choice Taq Mastermix (Denville Scientific), 1 µL of each primer (10 µmol), and 8 µL of sample. Polymerase chain reaction amplification included 35 cycles of denaturation at 95ºC, primer annealing at 55ºC, and extension at 72ºC.
Initial experiments were conducted to assess persistence on dry surfaces and transmissibility by bare hands. Ten-µL aliquots of phosphate-buffered saline (PBS) containing 106 plaque-forming units (PFUs) of bacteriophage MS2, 106 colony-forming units (CFUs) of nontoxigenic C. difficile spores, or 1 µg of cauliflower mosaic virus DNA were inoculated onto steel disks and allowed to air dry. At 1–21-day intervals for 183 days, disks were transferred to tubes containing 200 µL of DNAse free water and vortexed for 2 minutes. Aliquots of solution were plated on selective media and cultured for C. difficile or bacteriophage MS2 as previously described [7,10]. The limit of detection for C. difficile or bacteriophage MS2 was approximately 1 log10 CFUs or PFUs, respectively. To assess transmissibility, bare fingertips were imprinted onto contaminated disks and then the same contaminated fingertip was sequentially imprinted onto sterile steel disks that were processed as described previously.
To assess susceptibility to disinfectants, the steel disk inoculation sites were covered with 50 µL of 70% ethanol, a 1:10 dilution of household bleach, or a quaternary ammonium disinfectant (Virex II 256, Sealed Air) with exposure times of 2, 2, and 10 minutes, respectively. After exposure to the disinfectants, 1 mL of Dey-Engley neutralizing medium (Remel Products) was added, and the disks were processed as described previously.
The simulations of patient care interactions were conducted as previously described in a simulation room using a life-size mannequin in a hospital bed [12]. A 1-mL solution containing 105 PFUs of bacteriophage MS2, 105 CFUs of nontoxigenic C. difficile spores, and 1 µg of cauliflower mosaic virus DNA marker was applied to the mannequin’s anterior chest and abdomen, spread to cover a 10 × 10 cm area, and allowed to dry for 2 hours. The mannequin was clothed from the waist down. Healthcare personnel donned a SafetyPlus Polyethylene Gown (TIDI Products), nitrile gloves (Denville Scientific), and a face mask. The clinical scenario involved drawing a privacy curtain, pressing the nurse call button, moving the bedside table, raising the bed, examining the mannequin, and closing the privacy curtain. After each simulation, sterile Fisherbrand Polyester-Tipped Applicators (Fisher) premoistened with PBS were used to sample the face mask before removal and the hands and wrists and clothing after PPE removal. The hands, wrists, and clothing were cultured because they could be a source for transmission; the face mask was cultured because preliminary observations demonstrated that it was often touched with bare hands during PPE removal. Each subject participated in only 1 simulation. The personnel were not aware of the purpose of the study and were not told that the mannequin was contaminated. After every 5 simulations, swabs were used to sample several environmental sites that were observed to be frequently touched during simulations in a previous study (eg, privacy curtain, call button, bedside table, bed controls, and bed rail) [12]; after cultures were collected, all surfaces in the room except the walls, ceiling, and floor were thoroughly disinfected with bleach. For each day of sampling, 5 negative control swabs that were opened with no surface contact were processed identically.
Statistical Analysis
The frequencies of environmental, personnel skin and/or clothing, and face mask contamination were compared for the viral surrogates and C. difficile spores using a test of proportions with post hoc pairwise tests using a Holm P value adjustment. Data were analyzed using R version 3.1.1.
RESULTS
On steel disks, recovery of bacteriophage MS2 decreased over time from approximately 4.5 PFUs to approximately 1 PFU at day 11 after inoculation and was not detected after day 11. In contrast, nontoxigenic C. difficile spores and cauliflower mosaic virus DNA marker remained detectable at 183 days. After imprinting a bare fingertip on contaminated disks, each of the viral markers and C. difficile spores were recovered after 9 sequential transfers to sterile steel disks.
After a 2-minute exposure to bleach, each of the viral markers and C. difficile spores were undetectable. The 70% ethanol exposure had no impact on C. difficile spores or the cauliflower mosaic virus DNA marker, but resulted in lack of recovery of bacteriophage MS2. The quaternary ammonium disinfectant had no impact on the DNA marker or C. difficile spores; bacteriophage MS2 continued to be recovered, but the level was reduced by approximately 2 logs.
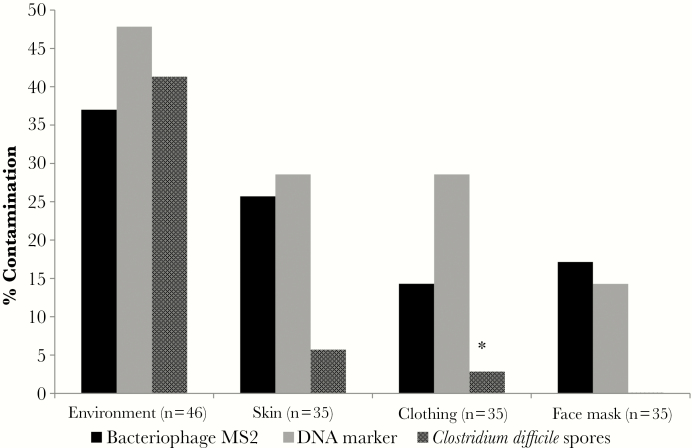
Figure 1 provides a summary of the frequency of contamination of environmental sites, face masks, skin, and clothing for 35 healthcare personnel participating in the simulations. There was no significant difference in the frequency of contamination of environmental sites for the DNA marker, bacteriophage MS2, and C. difficile spores (P ≥ .30). The cauliflower virus DNA marker and bacteriophage MS2 were detected with similar frequency on the face mask and on skin and clothing after PPE removal (P ≥ .32); C. difficile contaminated each of these sites less often, although the difference was only statistically signification for clothing contamination versus the DNA marker (P = .03). All negative control swabs were negative for the viral DNA surrogates and C. difficile.
Figure 1.
Frequency of contamination of environmental sites, face masks, skin, and clothing for 35 healthcare personnel participating in simulations of patient care using a mannequin contaminated with a cauliflower mosaic virus DNA marker, bacteriophage MS2, and nontoxigenic Clostridium difficile spores. *P < .05 for C. difficile spores versus the DNA marker.
DISCUSSION
In laboratory testing, bacteriophage MS2 was detectable on steel disks for up to 11 days, whereas cauliflower mosaic virus remained detectable at 183 days. Bare fingertips were able to transfer each of the viral markers and C. difficile spores to 9 consecutive clean surfaces. In patient care simulations, cauliflower mosaic virus DNA and bacteriophage MS2 performed similarly as surrogate markers of pathogen dissemination. The viral surrogates were recovered from environmental surfaces as frequently as C. difficile spores but more frequently than C. difficile on face masks and on skin and clothing of personnel after PPE removal. Because the limit of detection for recovery of C. difficile spores and MS2 from steel disks was equivalent, less frequent recovery of spores from face masks and from skin and clothing might reflect relatively low pick up of spores from these sites by swabs.
Both cauliflower mosaic virus DNA and bacteriophage MS2 are safe surrogate markers that can be used to study pathogen dissemination in healthcare settings [1–9]. Both are easy to work with but require some level of microbiological and/or molecular biology expertise. One potential advantage of live viruses such as bacteriophage MS2 is that their susceptibility to alcohol and disinfectants approximates that of nonenveloped pathogenic viruses, making it possible to assess the impact of alcohol hand sanitizer and disinfectants on dissemination [1, 3, 4, 6, 7]. We found that bleach reduced recovery of the DNA marker, but a quaternary ammonium disinfectant and 70% ethanol did not. Bacteriophage MS2 can also be detected by culture or PCR, whereas PCR must be used to detect cauliflower mosaic virus DNA [1, 2]. Finally, an advantage of viral DNA markers is it is feasible to produce multiple unique markers that could be used to simultaneously study different sources of transmission.
In addition to common pathogens, the viral surrogate markers can be useful in modeling transmission of potentially fatal viral pathogens such as Ebola virus. For example, Casanova et al [8] used bacteriophage MS2 to assess self-contamination during removal of PPE used for Ebola patient care. Viral surrogate markers can also be used to test the efficacy of interventions to prevent contamination. Koganti et al [13] demonstrated that a spray disinfectant was effective in reducing bacteriophage MS2 contamination on material from gowns designed for care of Ebola virus patients and on cover gowns worn by personnel, but the efficacy was affected by the type of gown material and the correctness of fit. Future studies with viral surrogate markers are needed to design and evaluate interventions to improve environmental cleaning as a strategy to reduce Ebola virus transmission.
Our study has some limitations. We did not assess the impact of interventions such as environmental disinfection on dissemination of the viral surrogate markers. We only included C. difficile spores as a comparator to the viral surrogates. Further studies are needed with other bacterial pathogens. Because the viral surrogate markers were recovered more frequently than C. difficile on face masks and on skin and clothing, it is possible that use of these markers could lead to overestimation of the risk for transmission of bacterial pathogens. Finally, the concentration of the viral surrogates and C. difficile spores was higher than concentrations typically recovered from hospital surfaces [10]. Therefore, our findings may overestimate the risk for dissemination in most clinical settings.
Acknowledgments
Financial support. This work was supported by a Merit Review grant from the Department of Veterans Affairs to C. J. D.
Potential conflicts of interest. C. J. D has received research grants from Merck, GOJO, Clorox, and EcoLab and serves on an advisory board for 3M. All other authors: No reported conflicts of interest.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Jiang X, Dai X, Goldblatt S et al. . Pathogen transmission in child care settings studied by using a cauliflower virus DNA as a surrogate marker. J Infect Dis. 1998;177:881–8. [DOI] [PubMed] [Google Scholar]
- 2. Oelberg DG, Joyner SE, Jiang X et al. . Detection of pathogen transmission in neonatal nurseries using DNA markers as surrogate indicators. Pediatrics. 2000;105:311–5. [DOI] [PubMed] [Google Scholar]
- 3. Reynolds KA, Beamer PI, Plotkin KR et al. . The healthy workplace project: reduced viral exposure in an office setting. Arch Environ Occup Health. 2016;71:157–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sassi HP, Sifuentes LY, Koenig DW et al. . Control of the spread of viruses in a long-term care facility using hygiene protocols. Am J Infect Control. 2015;43:702–6. [DOI] [PubMed] [Google Scholar]
- 5. Koganti S, Alhmidi H, Tomas ME et al. . Evaluation of hospital floors as a potential source of pathogen dissemination using a nonpathogenic virus as a surrogate marker. Infect Control Hosp Epidemiol. 2016;37:1374–7. [DOI] [PubMed] [Google Scholar]
- 6. Sifuentes LY, Koenig DW, Phillips RL et al. . Use of hygiene protocols to control the spread of viruses in a hotel. Food Environ Virol. 2014;6:175–81. [DOI] [PubMed] [Google Scholar]
- 7. Tomas ME, Kundrapu S, Thota P et al. . Contamination of health care personnel during removal of personal protective equipment. JAMA Intern Med. 2015;175:1904–10. [DOI] [PubMed] [Google Scholar]
- 8. Casanova LM, Teal LJ, Sickbert-Bennett EE et al. ; CDC Prevention Epicenters Program Assessment of self-contamination during removal of personal protective equipment for ebola patient care. Infect Control Hosp Epidemiol. 2016;37:1156–61. [DOI] [PubMed] [Google Scholar]
- 9. John A, Tomas ME, Hari A et al. . Do medical students receive training in correct use of personal protective equipment? Med Educ Online. 2017;22:1264125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nerandzic MM, Cadnum JL, Pultz MJ, Donskey CJ. Evaluation of an automated ultraviolet radiation device for decontamination of Clostridium difficile and other healthcare-associated pathogens in hospital rooms. BMC Infect Dis. 2010;10:197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cooke R, Penon P. In vitro transcription from cauliflower mosaic virus promoters by a cell-free extract from tobacco cells. Plant Mol Biol. 1990;14:391–405. [DOI] [PubMed] [Google Scholar]
- 12. Alhmidi H, Koganti S, Tomas ME et al. . A pilot study to assess use of fluorescent lotion in patient care simulations to illustrate pathogen dissemination and train personnel in correct use of personal protective equipment. Antimicrob Resist Infect Control. 2016;5:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Koganti S, Alhmidi H, Tomas ME et al. . Evaluation of an ethanol-based spray disinfectant for decontamination of cover gowns prior to removal. Infect Control Hosp Epidemiol. 2017;38:364–6. [DOI] [PubMed] [Google Scholar]