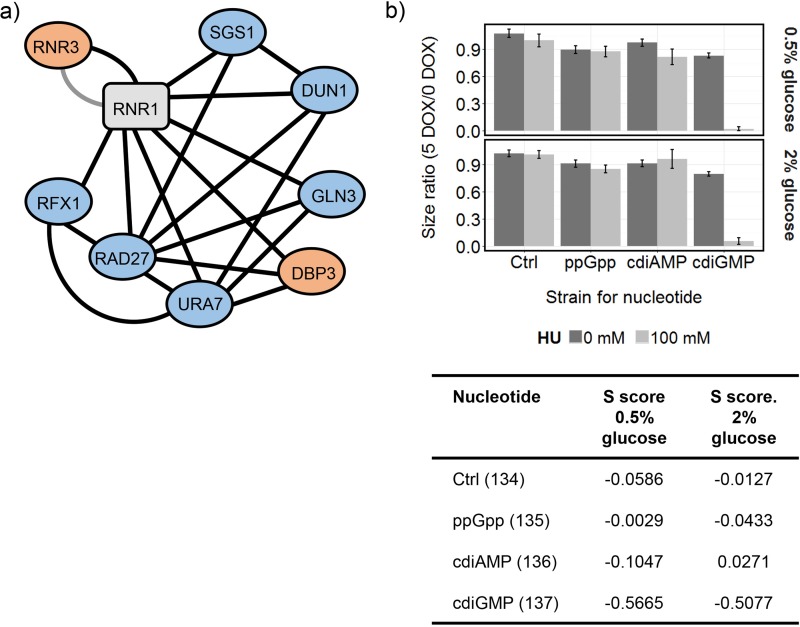
FIG 6 .
Ribonucleotide reductase activity is important for surviving exposure to cdiGMP. (a) Genes known to interact with RNR1 which synthetically alleviated (orange nodes) or aggravated (blue nodes) the effect of cdiGMP on cell fitness in the SGA screens. Edges connecting nodes are shown in black (genetic interaction) or gray (physical interaction) and are based on interaction data for RNR1 collated at http://www.yeastgenome.org. (b) BY4741-112-134 (Ctrl), BY4741-112-135 (ppGpp), BY4741-112-136 (cdiAMP), and BY4741-112-137 (cdiGMP) were arrayed onto YNB minimal medium plates containing 0 or 100 mM HU, with (5 DOX) or without (0 DOX) doxycycline. Strains were spotted 24 times each, and the data were collected and analyzed after 3 days of growth. Mean size changes (± standard errors; n = 24) are shown alongside interaction (S) scores, calculated based on the changes in colony areas. Strongly negative values (less than −0.1) indicate synthetic sick interactions between HU and nucleotide synthesis.