ABSTRACT
The phylogenetically diverse family of methanogenic archaea universally use methyl coenzyme M reductase (MCR) for catalyzing the final methane-forming reaction step of the methanogenic energy metabolism. Some methanogens of the orders Methanobacteriales and Methanococcales contain two isoenzymes. Comprehensive phylogenetic analyses on the basis of all three subunits grouped MCRs from Methanobacteriales and Methanococcales into three distinct types: (i) MCRs from Methanobacteriales, (ii) MCRs from Methanobacteriales and Methanococcales, and (iii) MCRs from Methanococcales. The first and second types contain MCR isoenzymes I and II from Methanothermobacter marburgensis, respectively; therefore, they were designated MCR type I and type II and accordingly; the third one was designated MCR type III. For comparison with the known MCR type I and type II structures, we determined the structure of MCR type III from Methanotorris formicicus and Methanothermococcus thermolithotrophicus. As predicted, the three MCR types revealed highly similar overall structures and virtually identical active site architectures reflecting the chemically challenging mechanism of methane formation. Pronounced differences were found at the protein surface with respect to loop geometries and electrostatic properties, which also involve the entrance of the active-site funnel. In addition, the C-terminal end of the γ-subunit is prolonged by an extra helix after helix γ8 in MCR type II and type III, which is, however, differently arranged in the two MCR types. MCR types I, II, and III share most of the posttranslational modifications which appear to fine-tune the enzymatic catalysis. Interestingly, MCR type III lacks the methyl-cysteine but possesses in subunit α of M. formicicus a 6-hydroxy-tryptophan, which thus far has been found only in the α-amanitin toxin peptide but not in proteins.
IMPORTANCE Methyl coenzyme M reductase (MCR) represents a prime target for the mitigation of methane releases. Phylogenetic analyses of MCRs suggested several distinct sequence clusters; those from Methanobacteriales and Methanococcales were subdivided into three types: MCR type I from Methanobacteriales, MCR type II from Methanobacteriales and Methanococcales, and the newly designated MCR type III exclusively from Methanococcales. We determined the first X-ray structures for an MCR type III. Detailed analyses revealed substantial differences between the three types only in the peripheral region. The subtle modifications identified and electrostatic profiles suggested enhanced substrate binding for MCR type III. In addition, MCR type III from Methanotorris formicicus contains 6-hydroxy-tryptophan, a new posttranslational modification that thus far has been found only in the α-amanitin toxin.
KEYWORDS: methanogenic archaea, X-ray crystallography, crystal structural analysis, methanogenesis, phylogenetic analysis, posttranslational modification
INTRODUCTION
The exponential increase in the methane concentration in the atmosphere correlates well with the onset of the industrial revolution. Atmospheric methane increased by ∼150% between preindustrial and present times (1). Roughly half of the annual methane production is of biogenic origin, essentially produced by methanogenic archaea. These strictly anaerobic euryarchaeota are subdivided into the orders Methanopyrales, Methanococcales, Methanobacteriales, Methanomicrobiales, Methanocellales, Methanosarcinales, and Methanomassiliicoccales and inhabit a broad range of ecological niches (see Table S1 in the supplemental material).
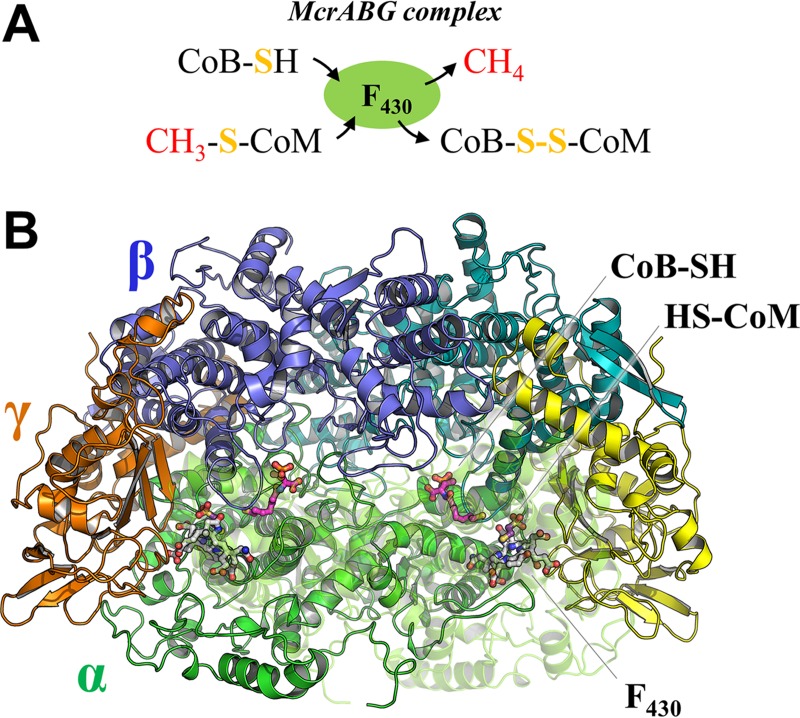
The key enzyme of the methanogenic energy metabolism is methyl coenzyme M reductase (MCR). MCR catalyzes the reduction of methyl coenzyme M (2-methylthioethanesulfonate; methyl-S-CoM) with coenzyme B (CoB-SH) to produce methane and a heterodisulfide (CoB-S-S-CoM) (Fig. 1A) (2). Due to its central role in energy metabolism, this enzyme is an attractive target to slow down methane emissions from biological sources such as livestock. Recently, it was shown that 3-nitrooxypropanol, an analogue of methyl-S-CoM, strongly inhibits MCR (3, 4).
FIG 1.
Chemical reaction and quaternary structure of MCR. (A) Reaction scheme of MCR. Methyl-S-CoM and CoB-SH react to methane and the heterodisulfide of HS-CoM and CoB-SH (CoM-S-S-CoB). (B) The structure of MCR type III from M. thermolithotrophicus is presented in cartoon with each subunit in a different color. F430 (white), CoB-SH (pink), and HS-CoM (purple) are depicted in ball-and-stick models. The chain α′ is drawn for clarity in a transparent mode.
The α-, β-, and γ-subunits of MCR are encoded by three genes: mcrA, mcrB, and mcrG (5). The function of the additional accessory genes, mcrC and mcrD, in many mcr gene clusters remains unclear. MCR is organized as a dimer of the heterotrimers αβγ (Fig. 1B) (5) and contains a unique nickel porphinoid, F430. To compensate for the relatively low catalytic rate of MCR, its cellular content consists of up to 10% of the total cytoplasmic protein (6). Most methanogens belonging to Methanobacteriales and Methanococcales synthesize two MCR isoenzymes (7, 8) with a primary structure identity of 60 to 70%. Production of the isoenzymes I and II in Methanothermobacter marburgensis is regulated by growth conditions, including factors such as temperature, pH, and exponential/linear growth phases (7, 8). Crystal structures of MCR isoenzyme I from M. marburgensis (5, 9) and Methanothermobacter wolfeii (10), of MCR isoenzyme II from M. marburgensis and M. wolfeii (10), and of MCR from Methanosarcina barkeri and Methanopyrus kandleri (11) have been reported previously.
The mcrA gene is used as a phylogenetic marker of methanogenic archaea because the corresponding phylogenetic tree resembles that obtained from 16S RNA gene sequences (12, 13). Exceptions to this strict correlation are sequences encoding Methanobacteriales MCR isoenzyme II, which include MCRs from both Methanococcales and Methanobacteriales (14, 15). Therefore, it was argued that the MCR isoenzyme II of Methanobacteriales is the result of a horizontal gene transfer from Methanococcales (12, 16). In the present work, we performed a precise amino acid sequence-based comparison of the three MCR subunits, allowing the classification of MCRs from Methanobacteriales and Methanococcales into three types tentatively designated MCR types I, II, and III. Next, a comprehensive structural comparison between the three types of MCRs was performed after the determination of the structures of MCRs from Methanotorris formicicus and Methanothermococcus thermolithotrophicus, which belong to the MCR type III.
RESULTS
Phylogenetic analysis of MCR.
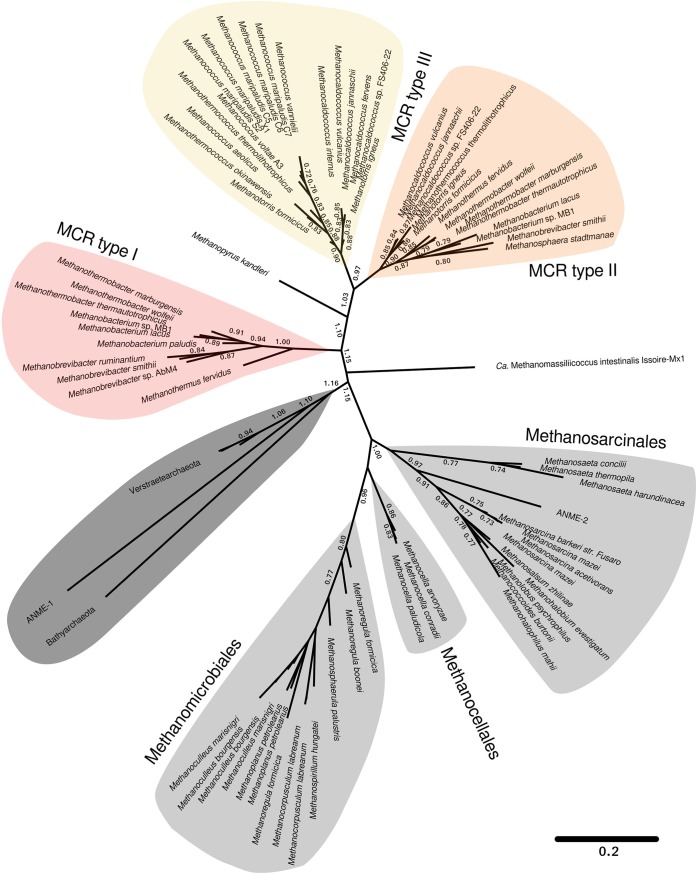
Phylogenetic analyses based on concatenated amino acid sequence alignments of α-, β-, and γ-subunits of MCRs revealed seven distinct clusters of MCRs (Fig. 2). This phylogenetic tree is similar to the trees calculated from the individual subunits McrA, McrB, and McrG (data not shown). MCR sequences from Methanosarcinales, Methanomicrobiales, and Methanocellales were clearly separated from those of Methanococcales and Methanobacteriales, which might be a consequence of their distinct habitats and substrates (see Table S1 in the supplemental material). The MCR of ANME-1 (where ANME is anaerobic methanotrophic archaea) (17) and the recently described Bathyarchaeota and Verstraetearchaeota (14, 18) share a cluster and are rather distinct from all others. The same held true for the Methanomassiliicoccales (19). These sequence comparisons partially contradict data reported by Li and colleagues, which have indicated an uncertainty in the phylogenetic placement of Methanocellales dependent on the marker genes used (20). In addition, the genomes of Methanomicrobiales (20) and Methanosarcinales (21) were found to be severely affected by horizontal gene transfer compared to those of other methanogenic orders. However, the MCR sequences formed distinct clusters.
FIG 2.
Phylogenetic tree based on concatenated alignments of McrA, McrB, and McrG amino acid sequences. Numbers at nodes represent the confidence for resolved branches based on bootstrapping. The scale bar refers to amino acid substitutions per site. The type strains were mostly used for this analysis; otherwise, the strain name is indicated. Ca, Candidatus.
MCRs from Methanococcales and Methanobacteriales branch into three MCR clusters, designated types I, II, and III. Type I is composed of the MCR isoenzyme I from M. marburgensis and its homologues from Methanobacteriales. Type II includes the MCR isoenzyme II from M. marburgensis and its homologues from Methanobacteriales and Methanococcales. Type III contains the MCR from Methanococcales, consisting exclusively of marine organisms, which include the fastest growing thermophilic and hyperthermophilic methanogens, such as M. thermolithotrophicus and Methanocaldococcus jannaschii (22, 23).
The phylogenetic analysis suggested that MCRs in the MCR types II and III feature a common ancestor which branched off from the MCR I cluster. Type II can be subdivided into two subtypes, each containing species from Methanobacteriales and Methanococcales. MCR type II from Methanobacteriales is evidently a result of horizontal gene transfer, as previously proposed (12, 16). Horizontal transfer of genes encoding catabolic enzymes has also been reported for, e.g., dissimilatory sulfite reductase (DsrAB) (24) and dissimilatory adenosine-5′-phosphosulfate reductase (AprBA) (25). Two MCR isoenzymes coexist in most methanogens from Methanobacteriales and Methanococcales, which suggests a functional advantage of maintaining both for the growth and/or survival of the methanogen. Moreover, both isoenzymes are produced in the methanogenic cells and are regulated by specific growth conditions (8, 10). The gene clusters encoding MCR I and III host five mcr genes (mcrBDCGA); however, those of MCR II lack one or two accessory genes (mcrC and/or mcrD).
Structure determination of MCR type III.
To investigate the structural differences between the three MCR types, we determined the X-ray structure of MCR type III from the thermophile M. thermolithotrophicus (optimum growth temperature, 65°C) and the hyperthermophile M. formicicus (80°C), both belonging to Methanococcales. MCR type II and type III from M. formicicus were isolated from cells harvested at the late log phase. The two isoenzymes could not be separated by the three chromatography steps used (see Materials and Methods). Therefore, the mixture of the two isoenzymes was used for crystallization. The MCR from M. thermolithotrophicus obtained from cells harvested in stationary phase was purified. This enzyme contains only MCR type III, which was identified by SDS-PAGE and subsequent sequence analysis using matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS). The applied growth conditions might not have allowed the detection of another isoenzyme, which belongs to MCR type II.
The samples from the two members of the Methanococcales were crystallized under air. Two crystal forms from M. formicicus diffracted to a medium resolution of 2.8 Å. Even though the MCR population from M. formicicus was a mixture of MCR types II and III, X-ray-suitable crystals grew, and the structure was determined by the molecular replacement method and further refined. The resulting electron density contained the amino acid sequence only of MCR type III. This selective power of the crystallization process was also found for MCRs from anaerobic methanotrophic archaea (26). One crystal form of MCR type III from M. thermolithotrophicus was determined at 1.9-Å resolution. Data quality and refinement statistics are listed in Table 1. In the following sections, we mainly used the highest-resolution model of MCR type III from M. thermolithotrophicus for the comparative studies with MCR types I and II.
TABLE 1.
Data collection and refinement statistics
| Parameter | Value for the parameter in: |
||
|---|---|---|---|
| MCR type III of M. thermolithotrophicus | MCR type III of M. formicicus |
||
| P21 form | P3221 form | ||
| Data collection | |||
| Wavelength (Å) | 0.99979 | 0.99998 | 0.99998 |
| Space group | P21 | P21 | P3221 |
| Resolution (Å)a | 46.4–1.9 (2.0–1.9) | 49.4–2.8 (2.95–2.8) | 48.1–2.8 (2.95–2.8) |
| Cell dimensions | |||
| a, b, c (Å) | 111.8, 77.2, 145.5 | 103.7, 81.2, 155.0 | 127.70, 127.70, 160.38 |
| α, β, γ (°) | 90.0, 107.0, 90.0 | 90.0, 107.8, 90.0 | 90.0, 90.0, 120.0 |
| Rmerge (%)a | 8.7 (57.7) | 10.3 (75.5) | 18.8 (73.9) |
| CC1/2a,d | 99.4 (80.4) | 99.7 (31.7) | 98.7 (33.4) |
| I/σI a | 8.1 (1.9) | 10.8 (2.1) | 7.4 (2.3) |
| Completeness (%)a | 99.2 (98.7) | 99.9 (100.0) | 99.9 (100.0) |
| Redundancya | 3.7 (3.7) | 5.0 (5.0) | 5.0 (4.9) |
| Refinement | |||
| Resolution (Å) | 45.2–1.9 | 48.4–2.8 | 39.9–2.8 |
| No. of reflections | 184,823 | 60,657 | 37,735 |
| Rwork/Rfree (%)b | 17.0/19.0 | 19.4/20.4 | 19.9/22.1 |
| No. of atoms | |||
| Protein | 19,426 | 19,370 | 9,714 |
| Ligands/ions | 200 | 181 | 92 |
| Solvent | 1283 | 28 | 0 |
| Avg B factors (Å2) | 35.4 | 94.2 | 61.2 |
| MolProbity clash score, all atoms (nth percentile) | 1.2 (100) | 4.4 (100) | 3.5 (100) |
| Ramachandran plot (no. [%]) | |||
| Favored regions | 2,393 (97.0) | 2,379 (96.2) | 1,187 (96.0) |
| Outlier regions | 3 (0.12) | 2 (0.08) | 1 (0.08) |
| RMSDc | |||
| Bond lengths (Å) | 0.009 | 0.008 | 0.008 |
| Bond angles (°) | 1.04 | 1.05 | 1.01 |
| PDB code | 5N1Q | 5N28 | 5N2A |
Values for the highest-resolution shell are within parentheses.
Rfree was calculated as the Rwork for 5% of the reflections that were not included in the refinement.
RMSD, root mean square deviation.
CC1/2 is the Pearson correlation coefficient between two random half data sets.
Overall structure and active site.
Comparison of the structurally known MCRs from M. marburgensis, M. wolfeii, M. thermolithotrophicus, and M. formicicus resulted in a highly similar overall structure, as shown by the data compiled in Table 2. The primary structures of MCR type III from both organisms are also highly similar (Table 2 and Fig. S1).
TABLE 2.
Sequence identity between the MCR subunits and root mean square deviations of the overall superposition of MCR structures and the active-site residues
| MCR type and parameter | Value for the parameter |
||
|---|---|---|---|
| MCR type II from M. wolfeii | MCR type III from M. thermolithotrophicus | MCR type III from M. formicicus | |
| MCR type I from M. marburgensis | |||
| Subunit sequence identity (%) | |||
| α | 73 | 70 | 73 |
| β | 68 | 65 | 68 |
| γ | 66 | 64 | 66 |
| Structural similarity (Å)a | |||
| All Cα atoms | 0.61 (1,090) | 0.70 (1,108) | 0.57 (1,081) |
| Cα atoms of active-site residues | 0.12 (35) | 0.18 (40) | 0.27 (60) |
| MCR type II from M. wolfeii | |||
| Subunit sequence identity (%) | |||
| α | 79 | 80 | |
| β | 67 | 68 | |
| γ | 76 | 74 | |
| Structural similarity (Å) | |||
| All Cα atoms | 0.49 (1,130) | 0.47 (1,135) | |
| Cα atoms of active-site residues | 0.13 (35) | 0.23 (42) | |
| MCR type III from M. thermolithotrophicus | |||
| Subunit sequence identity (%) | |||
| α | 91 | ||
| β | 86 | ||
| γ | 87 | ||
| Structural similarity (Å) | |||
| All Cα atoms | 0.37 (1,124) | ||
| Cα atoms of active-site residues | 0.25 (44) | ||
Structural similarity, root mean square deviation between the structures of the three MCR types. The number of Cα atoms used is given in parentheses.
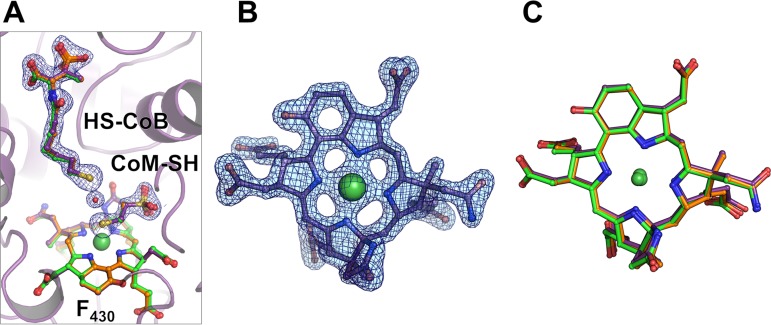
An analysis of the active-site structure of both MCR type III structures indicates clear electron densities for coenzyme M, coenzyme B, and F430 (Fig. 3A and B); their conformation and positions accurately correspond to those found in the MCRox1-silent form of other MCR crystal structures (5, 10, 11, 26). The structure of the F430 of MCR type III is identical to structures from most of the other MCRs (Fig. 3C) except for the methylthio-F430 in the ANME-I MCR (26) and an F430 variant with a circulated 3-mercaptopropionate thioester in M. jannaschii and Methanococcus maripaludis (27). Overall structure and the residues surrounding CoM-SH, HS-CoB, and F430 are strictly conserved among the three types of MCRs, and the conformations of the side chains are also virtually identical (Fig. 4 and Table 2). These results reflect the sensitivity of the MCR reaction to changes in the highly optimized polypeptide during evolution, which does not allow any freedom for neutral amino acid exchanges. This high conservation is indicative of the complexity of the catalytic mechanism and not found in other enzymes of the methanogenic pathway.
FIG 3.
CoM-SH, CoB-SH, and F430 in MCR type III compared to those of MCR types I and II. (A) The active site of MCR type III from M. thermolithotrophicus with CoM-SH, CoB-SH, and F430 represented as a stick model (purple). The cofactor and substrates observed in the structures of MCR type I (green) and MCR type II (orange) are superimposed. The 2Fo − Fc electron density map for the CoM-SH, HS-CoB, and the putative water molecule in between is contoured at 1.0 σ. (B) The 2Fo − Fc electron density of F430 of M. thermolithotrophicus MCR type III contoured at 1.5 σ. (C) Superposition of F430 from M. marburgensis MCR type I (green), from M. wolfeii MCR type II (orange), and from M. thermolithotrophicus MCR type III (purple).
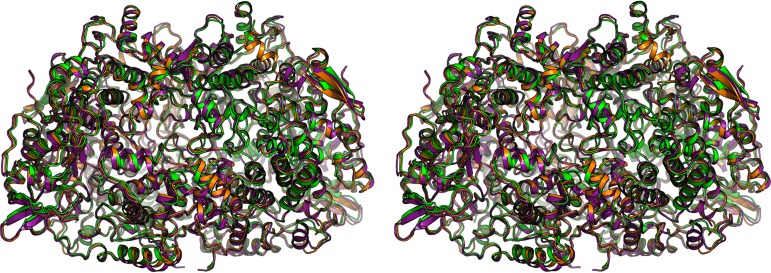
FIG 4.
Structural comparison of the three MCR types. Superimposition of the M. marburgensis MCR type I (green), M. wolfeii MCR type II (orange), and M. thermolithotrophicus MCR type III (purple) presented in stereo.
Structural deviations.
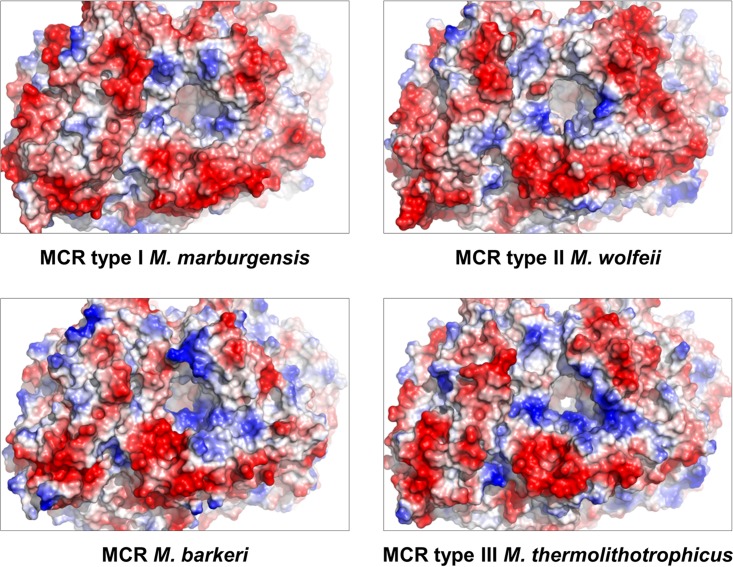
Structural differences between the MCRs are essentially found only at the protein surface where several solvent-exposed loops, such as α40 to α61, α357 to α368, β281 to β291, and γ181 to γ188 (numbering according to the MCR type III from M. thermolithotrophicus), significantly deviate among the three types of MCRs (Fig. 4 and Table 2). Compared to the other two types of MCRs, MCR type III contains a high number of basic residues exposed at the surface near the entrance of the substrate-binding tunnel, which might more efficiently attract the negatively charged methyl-S-CoM and CoB-SH toward the active site (Fig. 5). Related substrate-funneling strategies have already been reported for other enzymes (28). In addition, distinct surface charge profiles might also reflect differences in intracellular salts or temperature optima of different methanogens.
FIG 5.
Comparison of the electrostatic surface potentials of different MCRs focusing on their substrate entrance. The potential is displayed by a gradient from most negative in red to most positive in dark blue.
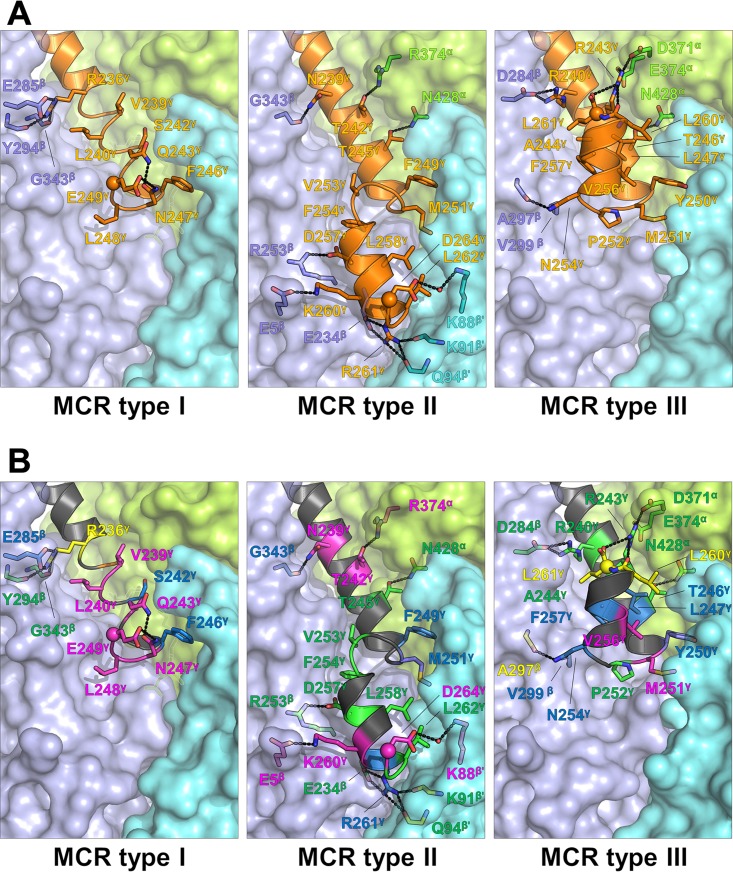
The most obvious difference was observed at the C-terminal end of the γ-subunit, which was partly predictable by primary structure analysis (Fig. S1). Figure 6 highlights the differences between the representative models of MCR type I from M. marburgensis (PDB accession number 5A0Y), MCR type II from M. wolfeii (PDB accession number 5A8W), and MCR type III from M. thermolithotrophicus (PDB accession number 5N1Q); the resolutions are 1.15 Å, 1.8 Å, and 1.9 Å, respectively. In MCR type I, the interactions between the C-terminal helix of the γ subunit (γ8) and the core are relatively weak, consisting of only one salt bridge (Glu285β-Arg236γ) and a few poorly conserved van der Waals interactions (Fig. 6B). MCR type II contains an additional C-terminal helix in the γ subunit (γ9) which prolongs off-center helix γ8 after a short kink. Helix γ9 forms multiple interactions to subunits β and β′, among them two invariant salt bridges (Asp257γ-Arg253β and Arg261γ-Glu234β). MCR type III also possesses helix γ9, but it is rotated about 180° compared to that of MCR type II, mainly due to the invariant Pro252γ. Helix γ9 of MCR type III is attached to helix γ8, subunit β, and subunit α but not to subunit β′. The C-terminal helix γ9 is stabilized by two conserved salt bridges (Arg240γ-Asp284β and Arg243γ-Asp371α plus Asp374α) and several hydrogen bonds. Notably, the C-terminal carboxy group of Leu261γ interacts with the conserved Arg243γ.
FIG 6.
Difference in the C-terminal extensions of the subunits γ between the MCR type I from M. marburgensis, MCR type II from M. wolfeii, and MCR type III from M. thermolithotrophicus. (A) Surface representation of subunits α (light green), β (light purple), and β′ (cyan) and the C-terminal helix of subunit γ: γ8 in MCR type I and γ8 and γ9 in MCR types II and III (orange). The residues involved in hydrogen bonding and salt bridges are highlighted as a stick model; residues involved in hydrophobic/van der Waals interactions are shown only for subunit γ to avoid overcrowding. (B) The same representation as in panel A but with a focus on the conservation of the interacting residues (perfectly conserved, green; highly conserved, dark blue; partially conserved, pink; and not conserved, yellow).
Posttranslational modifications.
A pronounced feature of MCR represents posttranslational modifications located in subunit α near the active site (5, 10, 29, 30). In the high-resolution structure of MCR type III from M. thermolithotrophicus, methyl-histidine 260, methyl-arginine 274, methyl-glutamine 402, and thioglycine 447 were identified in the electron density and confirmed by mass spectrometric analyses. The same modification pattern was also observed in MCR types II and III from M. formicicus by mass spectrometric analysis (data not shown). Posttranslational modifications show a higher variability than amino acid changes of the active site. For example, a didehydroaspartate has been detected in MCR type I and type II from M. marburgensis and in the MCR from M. barkeri but not in the close relative M. wolfeii MCR (10). Moreover, MCRs I and II from M. marburgensis and M. wolfeii contain methyl-cysteine, which is not present in MCRs from M. kandleri and M. jannaschii or in MCR type III from M. thermolithotrophicus and M. formicicus.
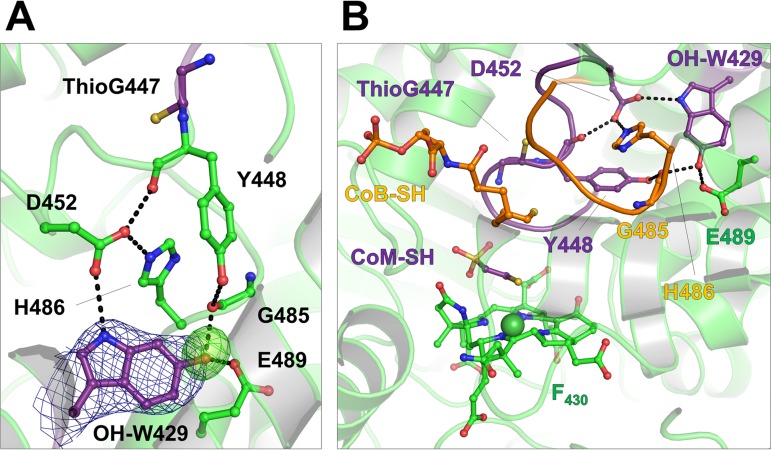
A potentially new posttranslational modification was detected at Trp429α in the M. formicicus MCR type III (Fig. 7) but not in the M. thermolithotrophicus. Extra electron density adjacent to C-6 of the indole group was visible in the heterotrimer αβγ of the asymmetric unit of a trigonal crystal form (resolution of 2.8 Å) and in both protomers from the second monoclinic crystal form (resolution of 2.8 Å). The electron density peak was interpreted as a proton-donating hydroxy group rather than a keto group. The potential hydroxy group at C-6 of Trp429α interacts with the main-chain carbonyl-oxygen of Gly485α (2.7 Å) and the carboxy group of Glu489α (2.6 Å). Peptide fragmentation analysis of MCR type III from M. formicicus using MALDI-TOF MS indicated that residue 429α is 16 Da heavier than tryptophan, in accordance with the mass of hydroxy-Trp429α observed in the crystal structure (Table 3; Fig. S2).
FIG 7.
The presence of a 6-hydroxy-tryptophan in MCR type III from M. formicicus. (A) Extra electron density in the vicinity of Trp429 in subunit α. The 2Fo − Fc (dark blue) and omit (light green) maps are contoured at 1.0 σ and at 3.0 σ, respectively. (B) The putative 6-hydroxy-tryptophan sits at a crucial point between the CoM-SH (purple) and the HS-CoB (orange) binding loops. In the MCR type I and type II of M. marburgensis, Asp452α is posttranslationally modified to a didehydroaspartate.
TABLE 3.
MALDI-TOF MS/MS mass fragmentation data of a trypsin-digested peptide from the α-subunit of MCR from M. formicicus
| Peptide sequencea | Mol mass [M+H]+ (Da) |
||
|---|---|---|---|
| Calculated (a) | Measured (b) | b − a | |
| 422SNAGLNGWYLSQILHK437 | 1,800.939 | 1,816.932 | 15.993 |
| 423NAGLNGWYLSQILHK437 | 1,713.907 | 1,729.886 | 15.979 |
| 424AGLNGWYLSQILHK437 | 1,599.864 | 1,615.866 | 16.002 |
| 425GLNGWYLSQILHK437 | 1,528.827 | 1,544.825 | 15.998 |
| 426LNGWYLSQILHK437 | 1,471.806 | 1,487.792 | 15.986 |
| 427NGWYLSQILHK437 | 1,358.722 | 1,374.719 | 15.997 |
| 428GWYLSQILHK437 | 1,244.679 | 1,260.675 | 15.996 |
| 429WYLSQILHK437 | 1,187.657 | 1,203.651 | 15.994 |
| 430YLSQILHK437 | 1,001.578 | 1,001.579 | 0.001 |
| 431LSQILHK437 | 838.515 | 838.514 | −0.001 |
The sequence of the α-subunit peptide is 403AAVAAAASGISVCMATGNSNAGLNGWYLSQILHK437. The presence of hydroxyl-tryptophan Trp429 (in bold) was not included for the calculation of the mass. The MS/MS spectrum is shown in Fig. S2.
It is worth mentioning that the didehydro-Asp452α of MCR types I and II from M. marburgensis interacts with Trp427α (10), which corresponds to Trp429 modified in MCR type III from M. formicicus. The double bond between the Cα and Cβ of the didehydroaspartate suggests a local backbone distortion, which shortens the hydrogen bond distance to the imidazolium group of His484 (His486α in M. formicicus) and the indole group of Trp427α (OH-Trp429α in M. formicicus). The introduction of a hydroxyl group to Trp429α in MCR type III increases the hydrogen bonding network and might have a similar effect in adjusting thioglycine 447α (Fig. 7B) and the loop involved in CoM-SH/CoB-SH binding.
DISCUSSION
Amino acid-based sequence analyses of concatenated MCR alignments allowed the classification of MCRs from Methanobacteriales and Methanococcales in MCR types I, II, and III, which are clearly distinct from those of Methanomicrobiales, Methanosarcinales, Methanocellales, and ANME-I/Bathyarchaeota/Verstraetearchaeota. Based on the derived phylogenetic placement, MCR type I and MCR types II/III evolved from an ancestral MCR. MCR types II and III formed clear sister lineages, suggesting that either of them is a result of an early horizontal gene transfer event (Fig. 2). A common feature of Methanobacteriales and Methanococcales is the presence of MCR type II, while MCR type I and type III are specific for Methanobacteriales and Methanococcales, respectively.
Despite the phylogenetic diversity of methanogens and the evolution of different MCR types, MCR structures are highly conserved. Even rather distantly related MCRs such as the MCR type III structures from M. formicicus and M. thermolithotrophicus and MCR structures from M. barkeri and ANME-1 are structurally highly similar with respect to overall fold and active-site architecture, including the structure and the binding mode of the F430 and the substrates. The high degree of conservation of the active site points to vertical evolution, originating from an early, already well adapted, ancestral MCR.
However, MCRs significantly differ regarding the electrostatic surface potentials, loop architectures, and, in particular, the C-terminal end of their γ-subunits that interact with α- and β-subunits in a different manner. Although amino acid exchanges at the protein surface appear at first view to be silent with respect to the catalytic process, they might be a response to individual cellular conditions provoked by specific environmental specifications and thus indirectly affect the reaction rate. Likewise, the differential expression of two isoenzymes in Methanobacteriales and Methanococcales under different growth conditions might also be a biological response to specifically cope with various environmental influences. Unfortunately, a verification of these assumptions is not feasible as MCRs purified in the Ni(II) state cannot be activated to Ni(I), with the exception of the MCR I of M. marburgensis (31).
MCRs feature a wide range of posttranslational modifications. We currently think that posttranslational modifications primarily serve to accelerate the enzymatic process beyond the possibility of amino acid exchanges. From an energetic point of view, the costs for posttranslational modifications might be compensated by the reduced amounts of MCR required to produce the same amount of methane produced per time. 6-Hydroxy-Trp429α, a new posttranslational modification in protein, was identified in the electron density map of MCR type III from M. formicicus and confirmed by a MALDI-TOF MS experiment. Its extra hydroxy group is in contact with the substrate-binding loop and might have a role equivalent to that of the didehydroaspartate modification in MCR types I and II from M. marburgensis and the MCR from M. barkeri (10), which contain an unmodified tryptophan. Another example is the extra 7-OH group in Trpα333 in combination with an unmodified arginine in the ANME-I MCR (26). Obviously, different posttranslational modifications can be performed to optimize catalysis dependent on the synthetic repertoire of the individual methanogenic organism. This finding also classifies posttranslational modifications as evolutionarily later than amino acid exchanges.
Hydroxy-tryptophan modifications are already identified, e.g., 5-hydroxy-tryptophan in serotonin from mammals (32) and 7-hydroxy-tryptophan in methylamine dehydrogenases from Paracoccus denitrificans (33) and in the MCR from ANME-I (26). Interestingly, 6-hydroxy-tryptophan has already been reported in the α-amanitin toxin from Amanita genus mushrooms (34). The toxin is produced by the ribosome and targets the RNA polymerase II and III machineries (35). However, to our knowledge, a 6-hydroxy-tryptophan modification has not as yet been detected in proteins. Incorporation of a hydroxy group normally requires O2 as the substrate, as described for serotonin biosynthesis (32), and was not investigated for posttranslational modifications in anaerobic methanogenic and methanotrophic archaea.
MATERIALS AND METHODS
Phylogenetic analysis.
Available methanogen genomes were retrieved from the genome repository of NCBI by using National Center for Biotechnology Information e-utilities implemented in Biopython (version 1.65) (36). Mcr sequences for subunits α, β, and γ were identified in collected genomes by blastp searches (version 2.2.31) (37) of reference sequences from Methanosarcina barkeri strain Fusaro against a BLAST database constructed from collected methanogen genomes (38). Unpublished Mcr sequences from M. wolfeii were obtained from the Institute of Microbiology and Genetics of the Georg August University Göttingen. Verstraetearchaeota Mcr sequences were obtained by collecting available metagenome assembled genomes, followed by gene prediction and in silico translation using prodigal (version 2.6.3) (39). Subunit-specific protein alignments were generated by applying T-Coffee (version 11.00) (40) using the psicoffee setting. The reliability of the individual alignments was validated against other common alignment algorithms (e.g., MAFFT [38], MUSCLE [41], and ProbCons [42]) using M-Coffee. Subunit alignments were concatenated using a python script and subsequently subjected to phylogenetic tree construction using FastTree (version 2.1.8) (43). FastTree was found to be orders of magnitude faster than RaxML (44) without a substantial difference in tree accuracy (45). A maximum-likelihood tree was calculated by applying the WAG model (46), nearest neighbor interchange (NNI) for optimizing tree topology, and the CAT approximation to account for evolutionary rate heterogeneity (44). A bootstrapped tree was calculated in addition (n = 1,000) and used to validate the topology determined based on NNI. Phylogenetic trees were visualized using FigTree (http://tree.bio.ed.ac.uk/software/figtree/) and polished using Inkscape (https://inkscape.org/en/). Determined multiple sequence alignments were also inspected for sequence motifs characteristic for suggesting MCR clades. Corresponding motifs were visualized using WebLogo (version 3.0) (47).
Cultivation of methanogenic archaea.
Methanothermococcus thermolithotrophicus (DSM 2095) and Methanotorris formicicus (DSM 16983) were obtained from the Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ). M. thermolithotrophicus was cultivated in a previously described minimal medium (48) with some modifications. The following components were dissolved in 900 ml of distilled water: 0.306 g of KCl, 0.9 g of NH4Cl, 0.09 g of CaCl2·2H2O, 0.253 g of K2HPO4, 0.252 g of KH2PO4, 22.5 g of NaCl, 7.72 g of MgCl2·6H2O, 0.16 g of Na2CO3, 7.56 g of PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)], and 4.88 g of 2-(N-morpholino)ethanesulfonic acid (MES). The final pH was adjusted to 6.2 by NaOH. A 100-fold-concentrated trace element solution was prepared by dissolving 1,360 mg of nitrilotriacetic acid in 700 ml of distilled water (pH 7.0 by NaOH), to which the different salts were added: 73 mg of MnCl2·4H2O, 135 mg of FeCl2·4H2O, 60 mg of CaCl2, 180 mg of CoCl2·6H2O, 90 mg of ZnCl2, 70 mg of CuSO4, 46 mg of Na2MoO4·2H2O, 90 mg of NiCl2·6H2O, and 15 mg of NaHSeO3. M. thermolithotrophicus was cultivated in 1.5-liter fermentors at 63°C with 800 rotations per minute and a continuous flow of 1.5 liters · min−1 of a gas mixture containing 80% H2–20% CO2–0.01% H2S. When the optical density at 600 nm (OD600) of the culture reached 3, the cells were harvested under anoxic conditions and stored at −80°C.
M. formicicus was cultivated in previously described medium (49) with some modifications: no cysteine was added, the amount of NH4Cl was increased (0.4 g per liter), and pH was adjusted to 6.5. The methanogen grew routinely in 1.5-liter fermentors at 75°C, with 1,000 rotations per minute and with a continuous flow at 1 liter · min−1 of a gas mixture composed of 80% H2–20% CO2–0.01% H2S. When the OD600 of the culture approached 1.0, the gas flow rate was increased to 2 liters · min−1. When the OD600 of the culture reached 2.0, the gas flow rate was increased to 3.5 liters · min−1. During growth, the pH was controlled by several injections of 10% (vol/vol) ammonia solution. When the OD600 reached approximately 2.5, the cultures were cooled down to 4°C, and the cells were harvested under anoxic conditions by centrifugation and stored at −80°C. For both organisms, a small-scale batch culture in anoxic vial bottles was necessary to adapt the cells to large-scale cultivation. For this small-scale batch culture, the medium was reduced by addition of 2 mM Na2S, and the gas phase contained 80% H2–20% CO2. A ratio of 1:10 of medium/gas phase was used, which was optimal for growth. The organisms showed similar growth profiles with a doubling time of roughly 30 min in fermentors.
Purification of MCRs.
Wet cells (30 to 50 g) of the methanogens were suspended in a 9-fold volume of 50 mM 3-(N-morpholino)propanesulfonic acid (MOPS)–NaOH, pH 7.0, containing 10 mM MgCl2 and 2 mM dithiothreitol (DTT) in an anoxic tent (95% N2, 5% H2), which led to cell lysis by osmotic shock. The lysate was centrifuged twice at 10,000 × g for 60 min at 4°C. All MCRs were purified under anoxic conditions at 18°C in the anaerobic tent. The purification methods were similar for the two organisms, with three chromatography steps used for both. For M. thermolithotrophicus the supernatant was applied to a 70-ml DEAE-Sepharose fast flow column (GE Healthcare, Freiburg, Germany) equilibrated with 50 mM Tricine-NaOH, pH 8.0, containing 2 mM DTT, and MCR was eluted by a gradient of NaCl (0 to 1 M) in 5 column volumes at a flow rate of 4 ml · min−1. MCR eluted in a fraction at 225 to 325 mM NaCl. The fractions were pooled and then diluted by the same volume of 50 mM Tricine/NaOH, pH 8.0, containing 2 mM DTT and applied on a Q Sepharose fast flow (45 ml) column equilibrated with the same buffer. Proteins were eluted by a NaCl gradient (0 to 1 M) of 8 column volumes at a flow of 3.5 ml · min−1. MCR eluted between 400 and 450 mM NaCl. The sample was diluted by 3 volumes of a buffer composed of 2 M (NH4)2SO4, 25 mM Tris/HCl, pH 7.6, and 2 mM DTT and applied on a 15-ml Source 15 Phe column equilibrated with the same buffer. The elution was performed at a flow rate of 1 ml · min−1 in 5 column volumes by a decreasing gradient of (NH4)2SO4 (2 to 0 M). MCR eluted in fractions between 1.24 M and 1.05 M (NH4)2SO4. The MCR-containing fractions were pooled and concentrated using a 50-kDa-cutoff filter (Merck Millipore, Darmstadt, Germany). A 0.5-ml sample was then injected onto a 10/300 Superose 6 column (GE Healthcare) equilibrated in 25 mM Tris/HCl, pH 7.6, containing 10% glycerol and 2 mM DTT and then eluted at a flow rate of 0.4 ml · min−1. MCR eluted as a sharp Gaussian peak at 13.6 ml.
For the MCR from M. formicicus, the supernatant was applied to a 45-ml DEAE-Sepharose fast flow column (GE Healthcare, Freiburg) equilibrated with 25 mM Tris/HCl, pH 7.6, and 2 mM DTT. Elution was performed using a gradient of NaCl (0 to 1 M) in 3 column volumes at a flow of 4.5 ml · min−1. Both MCR types II and III eluted between 225 and 500 mM NaCl. The fractions were pooled and diluted by the same volume of 25 mM Tris/HCl, pH 7.6, and 2 mM DTT and applied on a Q Sepharose fast flow (45 ml) column equilibrated with the same buffer. The elution was performed by applying a NaCl gradient (0 to 1 M) of 10 column volumes at a flow of 4 ml · min−1. Both MCRs eluted between 435 and 480 mM NaCl. The MCR sample was diluted by 3 column volumes of a buffer composed of 2 M (NH4)2SO4, 25 mM Tris, pH 7.6, and 2 mM DTT. The eluate was loaded on a phenyl-Sephacryl column of 40 ml equilibrated with the same buffer. The elution was performed at a flow rate of 1 ml · min−1 in 3 column volumes by a decreasing gradient of (NH4)2SO4 (2 to 0 M). MCR type III eluted between 1.1 M and 0.78 M (NH4)2SO4, and MCR type II eluted between 0.95 M and 0.67 M (NH4)2SO4. The MCR type III-containing fractions were concentrated by passing them through a 50-kDa-cutoff filter, injected onto a 10/300 Superose 6 column (GE Healthcare) equilibrated in 25 mM Tris/HCl, pH 7.6, containing 10% glycerol and 2 mM DTT, and eluted at a flow rate of 0.4 ml · min−1. MCR types II and III eluted together at 13.4 ml.
For both organisms the final pooled samples from the gel filtration were concentrated by passing them through a 50-kDa-cutoff filter, and the final concentration was measured by the Bradford method using dye solution (Bio-Rad, Munich, Germany) (50). Each elution profile was systematically controlled by separation on SDS-polyacrylamide gels (15%) to select the purest fractions.
Protein crystallization.
The purified enzymes were concentrated, stored on ice in 25 mM Tris/HCl, pH 7.6, containing 10% glycerol and 2 mM DTT, and used for crystallization within 12 h. Crystals were obtained using the sitting-drop method (CombiClover Junior Plate; Jena Bioscience) under air at 18°C. One microliter of MCR type III from M. thermolithotrophicus at a concentration of 35 mg/ml was mixed with 1 μl of reservoir solution. Best crystals with a yellow-brick morphology appeared after a few days in 19% (wt/vol) polyethylene glycol (PEG) 3350 and 200 mM MgCl2 in the absence of buffer. The crystals were immersed in a solution containing 19% (wt/vol) PEG 3350, 200 mM MgCl2, and 30% glycerol (vol/vol) prior to being frozen in liquid nitrogen. Crystals also grew at 18°C from a mixture of 1 μl of MCR type III/MCR II from M. formicicus (35 mg/ml) and 1 μl of reservoir solution. Yellow rod-shaped crystals of a monoclinic form could be reproduced in 20% PEG 8000 (wt/vol) and 100 mM MES-NaOH, pH 7.0, with 2 μl of protein and 1 μl of reservoir solution. The cryoprotection solution contains 20% PEG 8000 (wt/vol), 100 mM MES-NaOH, pH 7.0, and 25% ethylene glycol (vol/vol). Yellow cubic crystals of a trigonal form appeared 1 year later and could be directly frozen. The reservoir solution contains 200 mM potassium bromide, 200 mM potassium thiocyanate, 100 mM Na cacodylate, pH 6.5, 3% (wt/vol) low-molecular-weight poly-γ-glutamic acid polymers (PGA-LM), and 30% PEG 400 (vol/vol).
X-ray crystallography and structural analysis.
Crystallographic data were collected at 100 K on the Swiss Light Source (SLS) beamline X10SA and processed with XDS (51) and iMOSFLM (52). All data were scaled with SCALA in the CCP4 suite (53). The crystal structures of MCR type III from M. formicicus and M. thermolithotrophicus were determined by molecular replacement with MOLREP (54) using the coordinates of MCR type II from M. marburgensis and of MCR type III from M. formicicus, respectively. The correctly positioned models were first refined with REFMAC5 (55). All models were then manually rebuilt with COOT (56) and further refined with REFMAC5, PHENIX (57), or BUSTER (58). A translation-liberation-screw rotation (TLS) refinement (59) was applied for all three structures; restraints for noncrystallographic symmetry were used only for the MCR type III structure of M. thermolithotrophicus and of the monoclinic form of M. formicicus. Final models were validated through the MolProbity server (http://molprobity.biochem.duke.edu) (60). Data collection and refinement statistics, as well as Protein Data Bank (PDB) codes for the deposited models and structure factors are listed in Table 2. Figures were generated with PyMOL (Schrödinger, LLC).
Peptide analysis using mass spectrometry.
Purified MCRs were subjected to SDS-PAGE. The protein bands in the gel stained with Coomassie brilliant blue G250 were cut out, and the gel pieces were chopped. After destaining with 30% isopropanol (vol/vol) containing 50 mM NH4HCO3 and 30 mM thioglycolic acid (pH 8.3), the gel pieces were dehydrated with 100% isopropanol, rehydrated in 5 mM NH4HCO3 in 10% acetonitrile (vol/vol) containing 10 mM DTT and 2.5 mg/liter sequencing-grade modified trypsin (Promega), and incubated for 10 h at 22°C. The extracted peptide mixture was injected onto a PepMap100 C18 RP nanocolumn (Dionex, Idstein) of an UltiMate 3000 liquid chromatography system. The column was eluted with an acetonitrile gradient consisting of 0 to 40% buffer B (80% [vol/vol] acetonitrile, 0.04% [vol/vol] trifluoroacetic acid) in 40 min and with 40 to 100% buffer B in 10 min at a flow rate of 300 nl/min. We used a Probot microfraction collector (Dionex, Idstein, Germany) to spot the fractionated peptides on a MALDI target plate. Each fraction was mixed with a matrix composed of 3 mg/ml α-cyano-4-hydroxycinnamic acid in 80% (vol/vol) acetonitrile and 0.1% (vol/vol) trifluoroacetic acid. MALDI-TOF MS analysis was performed with a 4800 Proteomics Analyzer (Applied Biosystems/MDS Sciex, Forster City, CA) using the 4800 Series Explorer software (positive-ion reflector mode in a mass range from 840 to 5,000 Da, with an signal/noise minimum set to 80). The data were calibrated with an external standard using the peptide mixture from Bruker (Bremen, Germany), which was spotted onto the same target position. Tandem MS (MS/MS) data were used for searching against the genome sequences using the Mascot program in the GPS Explorer software (MDS Sciex).
Accession number(s).
Structural data are available in the Protein Data Bank under accession numbers 5N1Q for the MCR type III from M. thermolithotrophicus, 5N28 for the monoclinic form of the MCR type III from M. formicicus, and 5N2A for the trigonal form of the MCR type III from M. formicicus.
Supplementary Material
ACKNOWLEDGMENTS
We thank Rolf Thauer for helpful suggestions, Hartmut Michel for continuous support, and the staff of the PXII beamline at the Swiss Light Source (Villigen).
This work was supported by a grant of the Max Planck Society to R. Thauer and S.S. S.S. thanks the Japan Science and Technology Agency, the PRESTO program, for support.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00197-17.
REFERENCES
- 1.Intergovernmental Panel on Climate Change. 2013. Summary for policymakers, p 3–32. In Stocker TF, Qin D, G.-KPlattner G-K, Tignor M, Allen SK, Boschung J, Nauels A, Xia Y, VB, Midgley PM (ed), Climate change 2013: the physical science basis contribution of Working Group I to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change. Cambridge University Press, Cambridge, United Kingdom. [Google Scholar]
- 2.Thauer RK. 1998. Biochemistry of methanogenesis: a tribute to Marjory Stephenson. Microbiology 144:2377–2406. doi: 10.1099/00221287-144-9-2377. [DOI] [PubMed] [Google Scholar]
- 3.Hristov AN, Oh J, Giallongo F, Frederick TW, Harper MT, Weeks HL, Branco AF, Moate PJ, Deighton MH, Williams SRO, Kindermann M, Duval S. 2015. An inhibitor persistently decreased enteric methane emission from dairy cows with no negative effect on milk production. Proc Natl Acad Sci U S A 112:10663–10668. doi: 10.1073/pnas.1504124112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duin EC, Wagner T, Shima S, Prakash D, Cronin B, Yanez-Ruiz DR, Duval S, Rumbeli R, Stemmler RT, Thauer RK, Kindermann M. 2016. Mode of action uncovered for the specific reduction of methane emissions from ruminants by the small molecule 3-nitrooxypropanol. Proc Natl Acad Sci U S A 113:6172–6177. doi: 10.1073/pnas.1600298113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ermler U, Grabarse W, Shima S, Goubeaud M, Thauer RK. 1997. Crystal structure of methyl-coenzyme M reductase: the key enzyme of biological methane formation. Science 278:1457–1462. doi: 10.1126/science.278.5342.1457. [DOI] [PubMed] [Google Scholar]
- 6.Goubeaud M, Schreiner G, Thauer RK. 1997. Purified methyl-coenzyme-M reductase is activated when the enzyme-bound coenzyme F430 is reduced to the nickel(I) oxidation state by titanium(III) citrate. Eur J Biochem 243:110–114. doi: 10.1111/j.1432-1033.1997.00110.x. [DOI] [PubMed] [Google Scholar]
- 7.Bonacker LG, Baudner S, Morschel E, Bocher R, Thauer RK. 1993. Properties of the two isoenzymes of methyl-coenzyme-M reductase in Methanobacterium thermoautotrophicum. Eur J Biochem 217:587–595. doi: 10.1111/j.1432-1033.1993.tb18281.x. [DOI] [PubMed] [Google Scholar]
- 8.Bonacker LG, Baudner S, Thauer RK. 1992. Differential Expression of the two methyl-coenzyme M-reductases in Methanobacterium thermoautotrophicum as determined immunochemically via isoenzyme-specific antisera. Eur J Biochem 206:87–92. doi: 10.1111/j.1432-1033.1992.tb16904.x. [DOI] [PubMed] [Google Scholar]
- 9.Grabarse W, Mahlert F, Duin EC, Goubeaud M, Shima S, Thauer RK, Lamzin V, Ermler U. 2001. On the mechanism of biological methane formation: structural evidence for conformational changes in methyl-coenzyme M reductase upon substrate binding. J Mol Biol 309:315–330. doi: 10.1006/jmbi.2001.4647. [DOI] [PubMed] [Google Scholar]
- 10.Wagner T, Kahnt J, Ermler U, Shima S. 2016. Didehydroaspartate modification in methyl-coenzyme M reductase catalyzing methane formation. Angew Chem Int Ed Engl 55:10630–10633. doi: 10.1002/anie.201603882. [DOI] [PubMed] [Google Scholar]
- 11.Grabarse WG, Mahlert F, Shima S, Thauer RK, Ermler U. 2000. Comparison of three methyl-coenzyme M reductases from phylogenetically distant organisms: unusual amino acid modification, conservation and adaptation. J Mol Biol 303:329–344. doi: 10.1006/jmbi.2000.4136. [DOI] [PubMed] [Google Scholar]
- 12.Luton PE, Wayne JM, Sharp RJ, Riley PW. 2002. The mcrA gene as an alternative to 16S rRNA in the phylogenetic analysis of methanogen populations in landfill. Microbiology 148:3521–3530. doi: 10.1099/00221287-148-11-3521. [DOI] [PubMed] [Google Scholar]
- 13.Paul K, Nonoh JO, Mikulski L, Brune A. 2012. “Methanoplasmatales,” Thermoplasmatales-related archaea in termite guts and other environments, are the seventh order of methanogens. Appl Environ Microbiol 78:8245–8253. doi: 10.1128/AEM.02193-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Evans PN, Parks DH, Chadwick GL, Robbins SJ, Orphan VJ, Golding SD, Tyson GW. 2015. Methane metabolism in the archaeal phylum Bathyarchaeota revealed by genome-centric metagenomics. Science 350:434–438. doi: 10.1126/science.aac7745. [DOI] [PubMed] [Google Scholar]
- 15.Watanabe T, Kimura M, Asakawa S. 2009. Distinct members of a stable methanogenic archaeal community transcribe mcrA genes under flooded and drained conditions in Japanese paddy field soil. Soil Biol Biochem 41:276–285. doi: 10.1016/j.soilbio.2008.10.025. [DOI] [Google Scholar]
- 16.Reeve JN, Nolling J, Morgan RM, Smith DR. 1997. Methanogenesis: genes, genomes, and who's on first? J Bacteriol 179:5975–5986. doi: 10.1128/jb.179.19.5975-5986.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meyerdierks A, Kube M, Kostadinov I, Teeling H, Glockner FO, Reinhardt R, Amann R. 2010. Metagenome and mRNA expression analyses of anaerobic methanotrophic archaea of the ANME-1 group. Environ Microbiol 12:422–439. doi: 10.1111/j.1462-2920.2009.02083.x. [DOI] [PubMed] [Google Scholar]
- 18.Vanwonterghem I, Evans PN, Parks DH, Jensen PD, Woodcroft BJ, Hugenholtz P, Tyson GW. 2016. Methylotrophic methanogenesis discovered in the archaeal phylum Verstraetearchaeota. Nat Microbiol 1:16170. doi: 10.1038/nmicrobiol.2016.170. [DOI] [PubMed] [Google Scholar]
- 19.Borrel G, Harris HM, Parisot N, Gaci N, Tottey W, Mihajlovski A, Deane J, Gribaldo S, Bardot O, Peyretaillade E, Peyret P, O'Toole PW, Brugere JF. 2013. Genome sequence of “Candidatus Methanomassiliicoccus intestinalis” Issoire-Mx1, a third Thermoplasmatales-related methanogenic archaeon from human feces. Genome Announc 1:e00453-13. doi: 10.1128/genomeA.00453-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li J, Wong CF, Wong MT, Huang H, Leung FC. 2014. Modularized evolution in archaeal methanogens phylogenetic forest. Genome Biol Evol 6:3344–3359. doi: 10.1093/gbe/evu259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deppenmeier U, Johann A, Hartsch T, Merkl R, Schmitz RA, Martinez-Arias R, Henne A, Wiezer A, Baumer S, Jacobi C, Bruggemann H, Lienard T, Christmann A, Bomeke M, Steckel S, Bhattacharyya A, Lykidis A, Overbeek R, Klenk HP, Gunsalus RP, Fritz HJ, Gottschalk G. 2002. The genome of Methanosarcina mazei: evidence for lateral gene transfer between bacteria and archaea. J Mol Microbiol Biotechnol 4:453–461. [PubMed] [Google Scholar]
- 22.Huber H, Thomm M, Konig H, Thies G, Stetter KO. 1982. Methanococcus thermolithotrophicus, a novel thermophilic lithotrophic methanogen. Arch Microbiol 132:47–50. doi: 10.1007/BF00690816. [DOI] [Google Scholar]
- 23.Jones WJ, Leigh JA, Mayer F, Woese CR, Wolfe RS. 1983. Methanococcus jannaschii sp. nov., an extremely thermophilic methanogen from a submarine hydrothermal vent. Arch Microbiol 136:254–261. doi: 10.1007/BF00425213. [DOI] [Google Scholar]
- 24.Müller AL, Kjeldsen KU, Rattei T, Pester M, Loy A. 2015. Phylogenetic and environmental diversity of DsrAB-type dissimilatory (bi)sulfite reductases. ISME J 9:1152–1165. doi: 10.1038/ismej.2014.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meyer B, Kuever J. 2007. Molecular analysis of the distribution and phylogeny of dissimilatory adenosine-5′-phosphosulfate reductase-encoding genes (aprBA) among sulfur-oxidizing prokaryotes. Microbiology 153:3478–3498. doi: 10.1099/mic.0.2007/008250-0. [DOI] [PubMed] [Google Scholar]
- 26.Shima S, Krueger M, Weinert T, Demmer U, Kahnt J, Thauer RK, Ermler U. 2012. Structure of a methyl-coenzyme M reductase from Black Sea mats that oxidize methane anaerobically. Nature 481:98–101. doi: 10.1038/nature10663. [DOI] [PubMed] [Google Scholar]
- 27.Allen KD, Wegener G, White RH. 2014. Discovery of multiple modified F430 coenzymes in methanogens and anaerobic methanotrophic archaea suggests possible new roles for F430 in nature. Appl Environ Microbiol 80:6403–6412. doi: 10.1128/AEM.02202-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perry JJ, Shin DS, Getzoff ED, Tainer JA. 2010. The structural biochemistry of the superoxide dismutases. Biochim Biophys Acta 1804:245–262. doi: 10.1016/j.bbapap.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kahnt J, Buchenau B, Mahlert F, Kruger M, Shima S, Thauer RK. 2007. Post-translational modifications in the active site region of methyl-coenzyme M reductase from methanogenic and methanotrophic archaea. FEBS J 274:4913–4921. doi: 10.1111/j.1742-4658.2007.06016.x. [DOI] [PubMed] [Google Scholar]
- 30.Selmer T, Kahnt J, Goubeaud M, Shima S, Grabarse W, Ermler U, Thauer RK. 2000. The biosynthesis of methylated amino acids in the active site region of methyl-coenzyme M reductase. J Biol Chem 275:3755–3760. doi: 10.1074/jbc.275.6.3755. [DOI] [PubMed] [Google Scholar]
- 31.Prakash D, Wu YN, Suh SJ, Duin EC. 2014. Elucidating the process of activation of methyl-coenzyme M reductase. J Bacteriol 196:2491–2498. doi: 10.1128/JB.01658-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fitzpatrick PF. 1999. Tetrahydropterin-dependent amino acid hydroxylases. Annu Rev Biochem 68:355–381. doi: 10.1146/annurev.biochem.68.1.355. [DOI] [PubMed] [Google Scholar]
- 33.Jensen LM, Sanishvili R, Davidson VL, Wilmot CM. 2010. In crystallo posttranslational modification within a MauG/pre-methylamine dehydrogenase complex. Science 327:1392–1394. doi: 10.1126/science.1182492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Belliardo F, Massano G. 1983. Determination of alpha-amanitin in serum by HPLC. J Liq Chromatogr 6:551–558. doi: 10.1080/01483918308076067. [DOI] [Google Scholar]
- 35.Bushnell DA, Cramer P, Kornberg RD. 2002. Structural basis of transcription: α-amanitin-RNA polymerase II cocrystal at 2.8 Å resolution. Proc Natl Acad Sci U S A 99:1218–1222. doi: 10.1073/pnas.251664698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cock PJ, Antao T, Chang JT, Chapman BA, Cox CJ, Dalke A, Friedberg I, Hamelryck T, Kauff F, Wilczynski B, de Hoon MJ. 2009. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 25:1422–1423. doi: 10.1093/bioinformatics/btp163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hyatt D, Chen GL, LoCascio PF, Land ML, Larimer FW, Hauser LJ. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Notredame C, Higgins DG, Heringa J. 2000. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J Mol Biol 302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 41.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Do CB, Mahabhashyam MS, Brudno M, Batzoglou S. 2005. ProbCons: probabilistic consistency-based multiple sequence alignment. Genome Res 15:330–340. doi: 10.1101/gr.2821705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Price MN, Dehal PS, Arkin AP. 2010. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 45.Liu K, Linder CR, Warnow T. 2011. RAxML and FastTree: comparing two methods for large-scale maximum likelihood phylogeny estimation. PLoS One 6:e27731. doi: 10.1371/journal.pone.0027731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Whelan S, Goldman N. 2001. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol Biol Evol 18:691–699. doi: 10.1093/oxfordjournals.molbev.a003851. [DOI] [PubMed] [Google Scholar]
- 47.Crooks GE, Hon G, Chandonia JM, Brenner SE. 2004. WebLogo: a sequence logo generator. Genome Res 14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Belay N, Sparling R, Daniels L. 1986. Relationship of formate to growth and methanogenesis by Methanococcus thermolithotrophicus. Appl Environ Microbiol 52:1080–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mukhopadhyay B, Johnson EF, Wolfe RS. 1999. Reactor-scale cultivation of the hyperthermophilic methanarchaeon Methanococcus jannaschii to high cell densities. Appl Environ Microbiol 65:5059–5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 51.Kabsch W. 2010. XDS. Acta Crystallogr D Biol Crystallogr 66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Battye TGG, Kontogiannis L, Johnson O, Powell HR, Leslie AGW. 2011. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr D Biol Crystallogr 67:271–281. doi: 10.1107/S0907444910048675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AGW, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. 2011. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr 67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vagin A, Teplyakov A. 1997. MOLREP: an automated program for molecular replacement. J Appl Crystallogr 30:1022–1025. doi: 10.1107/S0021889897006766. [DOI] [Google Scholar]
- 55.Murshudov GN, Vagin AA, Dodson EJ. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr 53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 56.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 57.Afonine PV, Grosse-Kunstleve RW, Chen VB, Headd JJ, Moriarty NW, Richardson JS, Richardson DC, Urzhumtsev A, Zwart PH, Adams PD. 2010. phenix.model_vs_data: a high-level tool for the calculation of crystallographic model and data statistics. J Appl Crystallogr 43:669–676. doi: 10.1107/S0021889810015608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bricogne G, Blanc E, Brandl M, Flensburg C, Keller P, Paciorek W, Roversi P, Sharff A, Smart OS, Vonrhein C, Womack T. 2016. BUSTER version 2.10.1. Global Phasing, Ltd., Cambridge, United Kingdom. [Google Scholar]
- 59.Winn MD, Isupov MN, Murshudov GN. 2001. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr D Biol Crystallogr 57:122–133. doi: 10.1107/S0907444900014736. [DOI] [PubMed] [Google Scholar]
- 60.Chen VB, Arendall WB, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. 2010. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.