ABSTRACT
The ability of clinical microbiology laboratories to reliably detect carbapenemase-producing carbapenem-resistant Enterobacteriaceae (CP-CRE) is an important element of the effort to prevent and contain the spread of these pathogens and an integral part of antimicrobial stewardship. All existing methods have limitations. A new, straightforward, inexpensive, and specific phenotypic method for the detection of carbapenemase production, the carbapenem inactivation method (CIM), was recently described. Here we describe a two-stage evaluation of a modified carbapenem inactivation method (mCIM), in which tryptic soy broth was substituted for water during the inactivation step and the length of this incubation was extended. A validation study was performed in a single clinical laboratory to determine the accuracy of the mCIM, followed by a nine-laboratory study to verify the reproducibility of these results and define the zone size cutoff that best discriminated between CP-CRE and members of the family Enterobacteriaceae that do not produce carbapenemases. Bacterial isolates previously characterized through whole-genome sequencing or targeted PCR as to the presence or absence of carbapenemase genes were tested for carbapenemase production using the mCIM; isolates with Ambler class A, B, and D carbapenemases, non-CP-CRE isolates, and carbapenem-susceptible isolates were included. The sensitivity of the mCIM observed in the validation study was 99% (95% confidence interval [95% CI], 93% to 100%), and the specificity was 100% (95% CI, 82% to 100%). In the second stage of the study, the range of sensitivities observed across nine laboratories was 93% to 100%, with a mean of 97%; the range of specificities was 97% to 100%, with a mean of 99%. The mCIM was easy to perform and interpret for Enterobacteriaceae, with results in less than 24 h and excellent reproducibility across laboratories.
KEYWORDS: Enterobacteriaceae, antimicrobial susceptibility testing, bacterial antibiotic resistance, bacteriological techniques, carbapenemase, carbapenems
INTRODUCTION
The proportion of members of the family Enterobacteriaceae resistant to multiple antimicrobial classes has grown, and clinicians increasingly turn to agents from the broad-spectrum carbapenem class as options of last resort for the effective treatment of serious infections caused by these pathogens. Accordingly, the emergence and spread of carbapenem-resistant Enterobacteriaceae (CRE) is an issue of great clinical and public health concern (1–4).
The mechanisms underlying carbapenem resistance in Enterobacteriaceae are complex and include both the production of carbapenem-hydrolyzing β-lactamases (carbapenemase-producing CRE [CP-CRE]) and resistance due to the presence of a combination of other factors (non-CP-CRE), such as hyperproduction of AmpC β-lactamases or extended-spectrum β-lactamases (ESBLs) combined with altered membrane permeability (5–7). Characterization of the mechanism of carbapenem resistance is currently not recommended for the guidance of therapeutic decisions (8, 9) and is not routine in most clinical laboratories; however, this distinction between CP-CRE and non-CP-CRE is important for infection control and epidemiologic purposes because many carbapenemases are carried on mobile genetic elements that facilitate horizontal transfer of resistance between Gram-negative organisms. CP-CRE can spread rapidly, and their detection may warrant implementation of more-intensive infection control interventions than would be employed for non-CP-CRE (10). Additionally, as novel antimicrobial agents with activity against CP-CRE are introduced, distinguishing CP-CRE from non-CP-CRE will be increasingly important for antimicrobial stewardship programs seeking to rationally prioritize the use of these new drugs (11). Furthermore, a recent report suggested that CP-CRE might be more virulent than non-CP-CRE (12); if this finding is confirmed, routine delineation of resistance mechanisms in CRE may become important for clinical care. Unfortunately, the phenotypic antimicrobial susceptibility testing (AST) profiles of CP-CRE and non-CP-CRE overlap (13, 14). Therefore, the Centers for Disease Control and Prevention (CDC) currently recommend that clinical laboratories consider actively screening for carbapenemase production in isolates that meet the CDC surveillance definition for CRE (15).
Each method currently recommended by the Clinical and Laboratory Standards Institute (CLSI) for identification of CP-CRE has limitations (7, 8). The modified Hodge test (MHT) employs reagents readily available in most laboratories but involves subjective result interpretation and suffers from both false-positive results (particularly with Enterobacter spp. that have AmpC enzymes and porin alterations) and false-negative results (including with New Delhi metallo-β-lactamase [NDM]-producing isolates) (16–19). The Carba NP test and its variants require the acquisition of dedicated reagents (with associated costs and training needs), are interpreted subjectively, and are poorly sensitive for the detection of OXA-48-type carbapenemases (20, 21). OXA-48-type enzymes are more difficult to phenotypically detect than many other carbapenemases because on their own they can result in relatively low carbapenem MICs and spare the third-generation cephalosporins (22). Genotypic assays (such as PCR and DNA microarray tests) for the detection of carbapenemase genes are limited in their scope because only known targets are detected and mutations within targets could compromise assay performance. In addition, these molecular methods are expensive, require special equipment and expertise to perform, and are not in widespread use. Other commercially available reagents (none of which are currently approved for clinical use in the United States) include a combination of antimicrobial-containing disks with inhibitors (Neo-Sensitabs; Rosco Diagnostica, Tasstrup, Denmark), a kit-based version of the Carba NP test with lyophilized reagents in a plastic strip—the RAPIDEC Carba NP (bioMérieux, Inc., Marcy l'Etoile, France), and a metallo-β-lactamase Etest (bioMérieux, Inc., Marcy l'Etoile, France), all of which have also been reported to have limitations (21, 23).
A new phenotypic method for the detection of carbapenemase production, the carbapenem inactivation method (CIM), was first described in 2015 (24). This test is based on the principle that when a 10-μg meropenem (MEM) disk is incubated for 2 h in an aqueous suspension of a carbapenemase-producing microorganism, the carbapenem in the disk is degraded by the carbapenemase; in contrast, if the test microorganism does not produce carbapenemase, MEM retains its antimicrobial activity after incubation in the bacterial suspension. The disk is removed from the suspension and placed onto a Mueller-Hinton agar (MHA) plate seeded with a suspension of a carbapenem-susceptible indicator organism; following overnight incubation, the zone of inhibition is measured to determine whether the MEM had been hydrolyzed (growth of the indicator organism close to the disk), or is still active (a large zone of inhibition around the disk). The initial description of the CIM reported very promising results, including high sensitivity for the detection of a variety of carbapenemases and excellent specificity (24). The test was straightforward to perform and interpret and involved low-cost materials already readily available in clinical laboratories (24). Three early independent comparisons of the CIM to the Carba NP test found the CIM to be an accurate method for the detection of CP-CRE, with equal or better sensitivity for the detection of OXA-48-type carbapenemases in Enterobacteriaceae (25–27). However, other investigations have found the CIM to have lower detection rates of the OXA-48-type carbapenemases, with reported sensitivities of 80%, 50%, and 91% (21; B. M. Willey, S. Rajadurai, D. N. Grohn, R. Ioboni, X. Trimi, G. Ricci, P. Lo, T. Mazzulli, and S. M. Poutanen, presented at the 26th European Congress of Clinical Microbiology and Infectious Diseases, Amsterdam, The Netherlands, 9 to 12 April 2016; A. Aguirre, D. Gamal, M. E. Cano, J. Calvo, and L. Martinez-Martinez, presented at the 26th European Congress of Clinical Microbiology and Infectious Diseases, Amsterdam, The Netherlands, 9 to 12 April 2016). Pilot data suggested that modifying the carbapenem inactivation step by preparing the bacterial suspension in tryptic soy broth (TSB) and extending the time of incubation to 4 h could improve the detection of some carbapenemases (S. D. Das, P. Patel, L. R. Peterson, K. Mangold, and R. B. Thomson, unpublished data). We, a CLSI working group, were charged with validating the performance of this modified carbapenem inactivation method (mCIM) in both a single-laboratory and multilaboratory evaluation.
RESULTS
Single-laboratory mCIM validation study.
In the first stage of the study, 91 of the 92 isolates previously characterized as carrying carbapenemase genes gave positive results by mCIM, and all 23 isolates characterized as not carrying carbapenemase genes gave negative results by mCIM; there were no indeterminate results (Table 1). The single isolate with a false-negative result was an Enterobacter cloacae carrying blaKPC-4 for which the carbapenem MICs were low (FDA-CDC Antimicrobial Resistance Isolate Bank [AR Isolate Bank] no. 0050; Table 2). The sensitivity of the mCIM observed in this first stage of the study was 91/92 = 99% (95% confidence interval [95% CI], 93% to 100%), and the specificity was 23/23 = 100% (95% CI, 82% to 100%) compared to the genotype.
TABLE 1.
mCIM results for isolates tested during the stage 1 single-laboratory studya
| Isolate, carbapenemase class, and carbapenemase gene | Species (n) | MIC (μg/ml)b |
mCIM result(s)c | |||
|---|---|---|---|---|---|---|
| ETP | IPM | MEM | DOR | |||
| Isolates carrying carbapenemase genes (n = 92) | ||||||
| Ambler class A (41) | ||||||
| KPC (31) | Citrobacter freundii (1) | ≥16 | 16 | ≥16 | ≥16 | Pos |
| Enterobacter cloacae (6) | 0.5 to ≥16 | 1 to 8 | 0.5 to ≥16 | 0.5 to 8 | Pos (5); neg (1)d | |
| Escherichia coli (3) | 1 to 8 | 4 | 1 to 4 | 1 to 4 | Pos | |
| Klebsiella oxytoca (1) | 0.5 | 4 | 1 | 0.5 | Pos | |
| Klebsiella ozaenae (1) | ≥16 | ≥16 | ≥16 | ≥16 | Pos | |
| Klebsiella pneumoniae (14) | ≥4 to ≥16 | 8 to ≥16 | 8 to ≥16 | 8 to ≥16e | Pos | |
| Kluyvera ascorbata (1) | 8 | 4 | 8 | 4 | Pos | |
| Morganella morganii (1) | 8 | 8 | 4 | 4 | Pos | |
| Proteus mirabilis (2) | 1 to 2 | ≥16 | 0.5 to 2 | 2 to 4 | Pos | |
| Raoultella ornithinolytica (1) | 1 | 4 | 1 | 2 | Pos | |
| NMC (2) | Enterobacter cloacae complex (2) | ≥16 | ≥16 | ≥16 | ≥16f | Pos |
| SME (8) | Serratia marcescens (8) | ≥16 | ≥16 | ≥16 | ≥16g | Pos |
| Ambler class B (40) | ||||||
| NDM (29) | Citrobacter species (1) | ≥16 | ≥16 | ≥16 | ≥16 | Pos |
| Escherichia coli (12) | 4 to ≥16 | 8 to ≥16 | 8 to ≥16 | 8 to ≥16h | Pos | |
| Klebsiella pneumoniae (13)i | 4 to ≥16 | 4 to ≥16 | ≥16 | ≥16j | Pos | |
| Morganella morganii (1) | 2 | 8 | 4 | 8 | Pos | |
| Proteus mirabilis (1) | 4 | ≥16 | 4 | ≥16 | Pos | |
| Providencia rettgeri (1) | 8 | ≥16 | 8 | ≥16 | Pos | |
| Salmonella enterica serotype Senftenberg (1) | ≥16 | 4 | 8 | 8 | Pos | |
| VIM (5) | Enterobacter cloacae (1) | 2 | 4 | 2 | 4 | Pos |
| Klebsiella pneumoniae (4) | 1 to ≥16 | 4 to ≥16 | 4 to ≥16 | 4 to ≥16 | Pos | |
| IMP (5) | Enterobacter aerogenes (1) | 2 | 2 | 2 | 2 | Pos |
| Enterobacter cloacae (1) | ≥4 | 2 | 2 | Unspecified | Pos | |
| Klebsiella pneumoniae (3) | 2 to ≥4 | 2 to ≥16 | 1 to 16 | 4 to 8k | Pos | |
| Ambler class D (11) | ||||||
| OXA-48-type | Enterobacter aerogenes (1) | 2 | 4 | 2 | 2 | Pos |
| Klebsiella ozaenae (1) | ≥16 | 4 | 4 | 4 | Pos | |
| Klebsiella pneumoniae (9) | 4 to ≥16 | 2 to ≥16 | 2 to ≥16 | 2 to ≥16l | Pos | |
| Isolates not carrying carbapenemase genes (n = 23) | ||||||
| Escherichia coli (8) | ≤0.12 to 2 | ≤0.5 | ≤0.5 to 1 | ≤0.12 to 0.25 | Neg | |
| Klebsiella oxytoca (1) | 8 | 1 | 8 | 2 | Neg | |
| Enterobacter aerogenes (1) | 1 | ≤0.5 | ≤0.12 | ≤0.12 | Neg | |
| Enterobacter cloacae (4) | ≤0.12 to 1 | ≤0.5 | ≤0.12 | ≤0.12 | Neg | |
| Klebsiella pneumoniae (8) | 0.25 to ≥16 | ≤0.5 to 16 | ≤0.12 to ≥16 | ≤0.12 to ≥16 | Neg | |
| Proteus mirabilis (1) | 0.5 | 4 | 1 | 0.5 | Neg | |
mCIM, modified carbapenem inactivation method. Information about specific resistance mechanisms and carbapenem MICs for each isolate is provided in Table S1 in the supplemental material.
ETP, ertapenem; IPM, imipenem; MEM, meropenem; DOR, doripenem.
All mCIM results in this stage of the study were the same regardless of whether the original, provisional mCIM interpretive zone diameters (6 to 10 mm, positive [pos]; 11 to 19 mm, indeterminate; ≥20 mm, negative [neg]) or the revised, optimized criteria (6 to 15 mm, pos; 16 to 18 mm, indeterminate; ≥19 mm, neg) were applied.
Information about the single discrepant result is provided in Table 2.
DOR MICs were unspecified for eight isolates.
The DOR MIC was unspecified for one isolate.
DOR MICs were unspecified for six isolates.
DOR MICs were unspecified for four isolates.
Two Klebsiella pneumoniae isolates with blaNDM also carried blaOXA-48-type carbapenemase genes.
The DOR MIC was unspecified for one isolate.
The DOR MIC was unspecified for one isolate.
DOR MICs were unspecified for two isolates.
TABLE 2.
Isolates with discrepant results
| Stage, result, and isolate (AR Isolate Bank no.)a | Key known resistance determinant | Carbapenem MIC (μg/ml)b |
No. of labs with discrepant result | mCIM zone diam (mm) (interpretation)c | Repeat mCIM zone diam (mm) (interpretation)c,d |
||||
|---|---|---|---|---|---|---|---|---|---|
| ETP | IPM | MEM | DOR | MEM disk included during first subculture | MEM disk not included during subculture | ||||
| Stage 1 single-laboratory study | |||||||||
| False-negative results | |||||||||
| E. cloacae (0050) | KPC-4 | 0.5 | 1 | 0.5 | 0.5 | 1/1 | 22 (neg) | ND | ND |
| Stage 2 nine-laboratory study | |||||||||
| False-negative results | |||||||||
| E. coli (0104) | KPC-4 | 1 | 4 | 1 | 1 | 1/9 | 30 (neg) | 6 (pos) | ND |
| K. pneumoniae (0080) | IMP-4 | 4 | 4 | 4 | 8 | 1/9 | 18 (ind) | 22 (neg) | ND |
| K. pneumoniae (0066) | OXA-232 | >8 | 4 | >8 | >8 | 1/9 | 21 (neg) | C (pos) | ND |
| K. pneumoniae (0075) | OXA-232 | >8 | 8 | >8 | >8 | 4/9 | 20 (neg) | 6 (pos) | 24 (neg) |
| 24 (neg) | ND | ND | |||||||
| 22 (neg) | C (pos) | ND | |||||||
| 21 (neg) | 21 (neg) | ND | |||||||
| False-positive results | |||||||||
| E. coli (0058) | TEM-52 | 1 | ≤0.5 | 0.25 | 0.25 | 1/9 | 6 (pos) | ND | ND |
| E. coli (0084) | TEM-1 | ≤0.12 | ≤0.5 | ≤0.12 | ≤0.12 | 1/9 | 18 (ind) | ND | 22 (neg) |
AR Isolate Bank, FDA-CDC Antimicrobial Resistance Isolate Bank.
ETP, ertapenem; IPM, imipenem; MEM, meropenem; DOR, doripenem.
Modified carbapenem inactivation method (mCIM) zone diameters were classified as follows: 6 to 15 mm, positive (pos); 16 to 18 mm, indeterminate (ind); ≥19 mm, negative (neg). When multiple small bacterial colonies were observed growing within the zone of inhibition around the disk (C), the mCIM results were classified as positive when the outer zone of inhibition measured ≤15 mm in diameter and as indeterminate when the outer zone of inhibition measured ≥16 mm in diameter.
For isolates yielding discrepant results, a fresh subculture plate was created from the frozen stock, and the organism was subcultured twice before repeating mCIM testing. For those isolates with initially false-negative results, a MEM disk was placed between the first and second streak quadrants of the first subculture plate. Growth from around the disk was selected for the second subculture, and then the mCIM was repeated. ND, not done.
Receiver operating characteristic analysis to establish mCIM interpretive criteria.
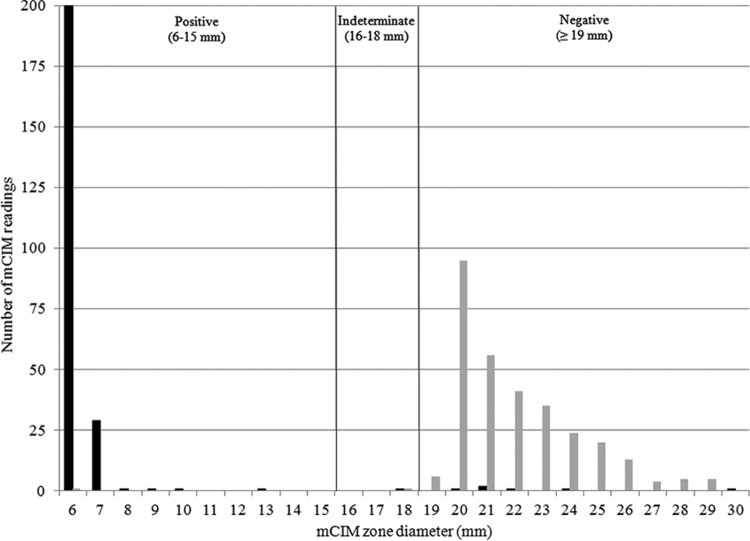
Using the zone diameter data from the multilaboratory study, the average sensitivity and specificity across all testing sites and the sum of average sensitivity and specificity that would be achieved by setting the positive mCIM cutoff criterion at all possible zone diameter values were calculated. The maximum sum of average sensitivity and specificity was found when defining a positive mCIM result as ≤18 mm and a negative mCIM result as ≥19 mm (see Table S2 in the supplemental material). Although this cutoff provided excellent separation between CP-CRE and those Enterobacteriaceae not producing carbapenemases, the CLSI working group recommended that an indeterminate interpretive range should be included in the testing procedure to prevent small, uncontrolled technical factors inherent to disk diffusion testing from causing major or very major errors. Therefore, a zone diameter of 6 to 15 mm was considered a positive result, a zone diameter of 16 to 18 mm was considered an indeterminate result (requiring further testing to establish the presence or absence of carbapenemase production), and a zone diameter of ≥19 mm was considered a negative result (Fig. 1). When multiple small bacterial colonies were observed growing within the zone of inhibition around the disk, the mCIM results were classified as positive when the zone of confluent growth inhibition was ≤18 mm in diameter and as indeterminate when the zone was ≥19 mm in diameter (Fig. 2). Analyzing the data from the first stage of the study using the original, provisional cutoff values and these revised criteria gave identical sensitivity and specificity results.
FIG 1.
Distribution of mCIM zone diameter measurements. Displayed are the numbers of mCIM readings with specific zone diameter measurements during the second stage, nine-laboratory study, in which 27 isolates with carbapenemase genes and 34 isolates without carbapenemase genes were tested, generating a total of 549 mCIM results. Among the 243 mCIM results for isolates with carbapenemase genes, a total of 3 exhibited multiple small bacterial colonies within the zone of inhibition around the disk; zone size data for these 3 readings are not displayed. Isolates with carbapenemase genes (black bars) and isolates without carbapenemase genes (gray bars) are indicated.
FIG 2.

Example of pinpoints within mCIM inhibition zone. During mCIM testing of some isolates, multiple small colonies were observed growing throughout the zone of inhibition of the 10-μg meropenem (MEM) disk; results were interpreted as positive when the zone of confluent growth inhibition measured ≤18 mm in diameter and as indeterminate when the zone measured ≥19 mm in diameter.
Nine-laboratory study of mCIM accuracy and reproducibility.
The mCIM results from the second stage of the study are shown in Table 3. These results include the mCIM results for the isolates tested during the multilaboratory accuracy and reproducibility study, and for comparison, the CIM results from the single laboratory that performed both CIM and mCIM testing on each isolate concurrently. All quality control (QC) results were within the ranges at all sites. Out of the 27 isolates genetically characterized as carrying carbapenemase genes, 23 yielded mCIM-positive results in all nine laboratories and an additional 3 gave mCIM-positive results in eight of the nine laboratories; for the one remaining isolate, a Klebsiella pneumoniae carrying blaOXA-232, a false-negative result was obtained by four laboratories. Among the 34 isolates genetically characterized as lacking carbapenemase genes, 32 had mCIM-negative results in all nine laboratories and the remaining two had mCIM-negative results in eight of the nine laboratories. The range of sensitivities observed across the nine laboratories (classifying indeterminate results as false-negative results when observed during the testing of isolates with known carbapenemase genes) was 93% to 100%, with a mean of 97%. The range of specificities across the nine laboratories (classifying indeterminate results as false-positive results when observed during the testing of isolates without carbapenemase genes) was 97% to 100%, with a mean of 99%. The laboratory that performed CIM and mCIM testing in parallel found the methods to be equally specific (100% [95% CI, 87% to 100%]), but the CIM to have a lower sensitivity (82% [95% CI, 61% to 93%]) than the mCIM (93% [95% CI, 74% to 99%]).
TABLE 3.
mCIM results for isolates tested during the stage 2 nine-laboratory studya
| Isolate, carbapenemase class, and carbapenemase gene | Species (n) | No. of labs with the following mCIM result (no. of isolates)b |
CIM result in one lab (no. of isolates)c | ||
|---|---|---|---|---|---|
| Pos | Ind | Neg | |||
| Isolates carrying carbapenemase genes (27) | |||||
| Ambler class A (11) | |||||
| KPC (8) | Citrobacter freundii (1) | 9 | Pos | ||
| Enterobacter cloacae (2) | 9 | Pos (2) | |||
| Escherichia coli (2) | 9 (1); 8 (1) | 1 (1)d | Pos (2) | ||
| Morganella morganii (1) | 9 | Pos | |||
| Proteus mirabilis (1) | 9 | Pos | |||
| Raoultella ornithinolytica (1) | 9 | Pos | |||
| NMC (1) | Enterobacter cloacae complex (1) | 9 | Pos | ||
| SME (2) | Serratia marcescens (2) | 9 | Pos (2) | ||
| Ambler class B (10) | |||||
| NDM (7) | Citrobacter species (1) | 9 | Pos | ||
| Escherichia coli (2) | 9 | Pos (2) | |||
| Morganella morganii (1) | 9 | Pos | |||
| Proteus mirabilis (1) | 9 | Pos | |||
| Providencia rettgeri (1) | 9 | Neg | |||
| Salmonella enterica serotype Senftenberg (1) | 9 | Pos | |||
| VIM (2) | Enterobacter cloacae (1) | 9 | Pos | ||
| Klebsiella pneumoniae (1) | 9 | Pos | |||
| IMP (1) | Klebsiella pneumoniae (1) | 8 | 1d | Neg | |
| Ambler class D (6) | |||||
| OXA-48-type | Enterobacter aerogenes (1) | 9 | Pos | ||
| Klebsiella ozaenae (1) | 9 | Pos | |||
| Klebsiella pneumoniae (4) | 9 (2); 8 (1); 5 (1) | 1 (1)d; 4 (1)d | Pos (1); neg (3) | ||
| Isolates not carrying carbapenemase genes (34) | |||||
| Enterobacter aerogenes (4) | 9 | Neg (4) | |||
| Enterobacter cloacae (5) | 9 | Neg (5) | |||
| Escherichia coli (12) | 1 (1)d | 1 (1)d | 9 (10); 8 (2) | Neg (12) | |
| Klebsiella oxytoca (1) | 9 | Neg (1) | |||
| Klebsiella pneumoniae (11) | 9 | Neg (11) | |||
| Proteus mirabilis (1) | 9 | Neg (1) | |||
Information about specific resistance mechanisms and carbapenem MICs for each isolate is provided in Table S1 in the supplemental material.
The modified carbapenem inactivation method (mCIM) zone diameters were classified as follows: 6 to 15 mm, positive (Pos); 16 to 18 mm, indeterminate (Ind); ≥19 mm, negative (Neg). When multiple small bacterial colonies were observed growing within the zone of inhibition around the disk, the mCIM results were classified as positive when the outer zone of inhibition measured ≤15 mm in diameter and as indeterminate when the outer zone of inhibition measured ≥16 mm in diameter.
The carbapenem inactivation method (CIM) results were classified as follows: no zone of inhibition present, positive (pos); any zone of inhibition present, negative (neg).
Details about the isolates with discrepant results (and about repeat testing, where applicable) are shown in Table 2.
With the exception of a single isolate, discrepant results occurred in only one out of nine laboratories during the second stage of the study. Details about the isolates with discrepant results (and about repeat testing, where applicable) are shown in Table 2. In the single laboratory that performed both CIM and mCIM testing, five isolates characterized as carrying carbapenemase genes had negative CIM results but positive mCIM results. These isolates with discrepant results included the following isolates: K. pneumoniae with blaOXA-232 (n = 2; AR Isolate Bank no. 0066 and 0075), K. pneumoniae with blaOXA-181 (n = 1; AR Isolate Bank no. 0039), K. pneumoniae with blaIMP-4 (n = 1; AR Isolate Bank no. 0080), and Providencia rettgeri with blaNDM-1 (n = 1; AR Isolate Bank no. 0082).
DISCUSSION
In our study, the mCIM exhibited excellent sensitivity for the detection of carbapenemase production among Enterobacteriaceae carrying a variety of different carbapenemase genes, including those belonging to Ambler classes A, B, and D. The assay was also highly specific, including when testing isolates meeting the 2015 CDC surveillance definition for CRE on the basis of mechanisms other than expression of carbapenemase genes (non-CP-CRE), such as ESBL production in combination with decreased outer membrane permeability, and when testing isolates with Ambler class C (AmpC-type) β-lactamases (both chromosomal and plasmid mediated). The mCIM was easy to perform and interpret for these enteric organisms, supported by overall excellent reproducibility of the results across nine testing sites. The mCIM uses inexpensive materials readily available in clinical microbiology laboratories at a cost of less than $1 per test, which is similar to the cost of MHT (<$1 per test), less than the cost of the Carba NP test ($2 to $10 per test) (21), and substantially less than molecular methods (e.g., $55 per test [list price] for the Cepheid Xpert Carba-R, the only FDA-cleared test). After reviewing the data generated during this evaluation, the CLSI Subcommittee on Antimicrobial Susceptibility Testing voted to add the mCIM to the 27th edition of the CLSI Performance Standards for Antimicrobial Susceptibility Testing M100 Supplement (M100) as a reliable, standardized method available to laboratories that aim to identify CP-CRE for epidemiologic or infection control purposes or as additional testing for isolates with carbapenem MICs of ≥2 μg/ml in laboratories that have not yet implemented the current CLSI or FDA carbapenem breakpoints (28).
The two modifications of the original CIM that we implemented in our study were both changes to the carbapenem inactivation step: (i) using TSB instead of water, and (ii) extending the incubation time from 2 to 4 h. While our study did not directly compare the performance of the mCIM to the previously published CIM in all nine laboratories, the single site that did directly compare the two tests found the mCIM to be more sensitive (93% versus 82%) and equally specific (100%) to the CIM. In addition, we observed the mCIM to be more sensitive for the detection of OXA-48-type carbapenemases than what has recently been reported for the CIM by several other groups (21; B. M. Willey, S. Rajadurai, D. N. Grohn, R. Ioboni, X. Trimi, G. Ricci, P. Lo, T. Mazzulli, and S. M. Poutanen, presented at the 26th European Congress of Clinical Microbiology and Infectious Diseases, Amsterdam, The Netherlands, 9 to 12 April 2016; A. Aguirre, D. Gamal, M. E. Cano, J. Calvo, and L. Martinez-Martinez, presented at the 26th European Congress of Clinical Microbiology and Infectious Diseases, Amsterdam, The Netherlands, 9 to 12 April 2016). These data suggest that incubating the isolate and MEM disk in TSB and for 4 h enhance the detection of carbapenemases with either weaker hydrolytic activities, lower levels of expression, or metallo-β-lactamases that require divalent cations for activity. Further studies would be necessary to separate the contributions of the growth medium versus water from the extended incubation time. Although we did not directly compare the performance of the mCIM to the MHT or the Carba NP test, we found the sensitivity of the mCIM for the detection of NDM carbapenemases and OXA-48-type carbapenemases to be higher than that reported in some evaluations of the MHT and Carba NP test, respectively (16–21). In addition, we saw very few false-positive mCIM results, including among Enterobacter spp. expressing AmpC β-lactamases, in contrast to what has been reported for the MHT (17–19). Furthermore, at test sites where laboratory staff also had experience performing either the MHT, the Carba NP test, or both, the interpretation of mCIM results was thought to be less subjective and the mCIM to be simpler to perform.
One consideration for laboratories deciding which method to employ for screening CRE for carbapenemase production is that the mCIM, similar to the MHT, requires an overnight incubation with the indicator organism; this is in contrast to molecular methods and the Carba NP test, with which results can be generated within a single work shift. Additionally, a positive mCIM result does not provide information about the specific carbapenemase gene present in a given bacterial isolate, such as might be gleaned by the performance of a molecular test; nonetheless, the distinction between CP-CRE and non-CP-CRE may be sufficient to guide the stratification of infection control and antimicrobial stewardship interventions in many facilities. Furthermore, the results of mCIM testing might allow laboratories to consider restricting the utilization of molecular tests for specific carbapenemase genes to the subset of CRE that are shown to be CP-CRE by mCIM testing.
Our investigation has several limitations. We utilized isolates from an organism repository, rather than prospectively collected clinical isolates, such as those that would be tested by laboratories that implement the mCIM in routine clinical practice. However, this study design allowed for the inclusion of isolates with a representative spectrum of resistance mechanisms of interest, including those that might be encountered only rarely by clinical laboratories. If we had instead performed the mCIM on consecutively clinically recovered CRE and performed whole-genome sequencing to characterize them as to the presence or absence of carbapenemase genes, we would likely have included more non-CP-CRE and KPC-producing CRE and fewer NDM-, OXA-48-type-, VIM-, and IMP-producing CRE, which are currently less commonly isolated in U.S. clinical laboratories (29). In addition, testing consecutive clinical isolates would have raised the likelihood that our evaluation would have included clonal, rather than unique, isolates, potentially limiting the generalizability of our results. Second, we did not systematically confirm preservation of resistance (either through antimicrobial susceptibility testing or genotypic testing) after thawing and subculturing isolates during the second stage of our study. Doing so would have aided in our ability to draw conclusions about the isolates that had false-negative mCIM results in one or more laboratories. A third limitation is that the specific individuals performing the testing may not have been blind to the whole-genome sequencing and PCR results of specific isolates. However, while measuring the size of zones of inhibition is subject to a small amount of variability, such measurements are sufficiently objective that the categorical interpretation of the mCIM should not be altered from positive to negative (or vice versa) by interreader differences, particularly with the inclusion of an indeterminate range in the method's interpretive criteria.
In the future, it will be important to evaluate the performance of the mCIM with additional Enterobacteriaceae isolates, including larger numbers with AmpC β-lactamases combined with porin alterations and larger numbers with OXA-48-type carbapenemases, since such isolates have challenged the performance of other phenotypic carbapenemase detection methods. Additionally, differentiation between carbapenem resistance mediated by carbapenemase production and that imparted by other mechanisms may be desirable for other bacteria (such as Pseudomonas aeruginosa and Acinetobacter baumannii complex) for epidemiologic or infection control purposes. The CLSI mCIM ad hoc working group is currently undertaking a study to explore this issue. While the current overnight incubation with the indicator organism prior to reading mCIM results is likely to be practical for many laboratories, determining whether results can be accurately interpreted at an earlier time point (such as after 6 h, as was reported in the original description of the CIM) would be of interest to laboratories in which the workflow would accommodate the performance of test steps on multiple work shifts. Finally, it would be interesting to further explore the impact of the presence of a carbapenem disk during the subculture of organisms prior to mCIM testing. We did not routinely include this step during our study because we sought to avoid extending the test procedure by an additional day, which would lengthen turnaround time in clinical laboratories. In addition, we thought that the inclusion of a carbapenem disk during subculture would be unlikely to improve the sensitivity of the method for the majority of CP-CRE. However, limited data (Table 2) generated during repeat testing of the three mCIM false-negative strains in the second stage of our study suggest that inclusion of a carbapenem disk during subculture might improve the already excellent sensitivity of the test for a subset of isolates. The mechanism by which this would occur (e.g., prevention of plasmid loss, selection of colonies with increased carbapenemase production, or even induction of carbapenemase production) is unknown, as is whether or not the inclusion of a carbapenem disk during subculture might adversely impact the specificity of the mCIM.
In conclusion, we found the mCIM to be a simple, inexpensive, accurate, and reproducible method for the identification of carbapenemase production among Enterobacteriaceae. The inclusion of the standardized mCIM procedure developed during this study in the widely used CLSI M100 document has the potential to facilitate the identification of CP-CRE in clinical laboratories, which may in turn aid facilities in understanding the local epidemiology of carbapenem resistance and serve as an important component of the critical, multifaceted effort to combat the spread of these pathogens.
MATERIALS AND METHODS
Bacterial isolates tested.
During the first stage of the study, which was performed in a single laboratory (NorthShore University HealthSystem, Evanston, IL), a total of 117 members of the family Enterobacteriaceae representing 11 genera were used, including 107 from either the Gram-Negative Carbapenemase Detection Panel or the Enterobacteriaceae Carbapenemase Diversity Panel of the FDA-CDC Antimicrobial Resistance Isolate Bank (AR Isolate Bank; http://www.cdc.gov/drugresistance/resistance-bank) and 10 from the UCLA Clinical Microbiology Laboratory (UCLA Medical Center, Los Angeles, CA). Each isolate had been previously characterized as to the presence or absence of carbapenemase genes, either by targeted PCR (n = 6) or whole-genome sequencing (n = 4) at UCLA as previously described (30–34) or by whole-genome sequencing at the CDC. At the CDC, antimicrobial resistance genes were detected using c-SSTAR (https://github.com/chrisgulvik/c-SSTAR), and a command line version of SSTAR (35) was used with a compatible ResFinder database (36) (accessed 25 October 2016). Reported resistance genes needed to meet a 99% sequence identity and 100% coverage threshold. Reported truncated outer membrane porin (OMP) genes, due to the presence of premature stop codons, had a ≥80% sequence identity and 100% coverage threshold. OMP genes were manually added to the ResFinder database (35). Overall, 94 isolates were classified as carrying a variety of carbapenemase genes, and 23 isolates were characterized as lacking such genes (Table 1 and Table S1 in the supplemental material). Of the 117 isolates evaluated, 115 were ultimately included in the analysis because the previously identified carbapenemase gene (blaNDM in both cases) was not detected when molecular testing of two isolates was repeated at the CDC, possibly reflecting plasmid loss during subculture.
During the second stage of the study, which was conducted in nine different laboratories, a total of 61 Enterobacteriaceae representing 10 genera were used, all of which were obtained from the AR Isolate Bank; 27 had been classified as carrying carbapenemase genes, and 34 as lacking such genes (Table 3 and Table S1). Among these 61 isolates, 50 had also been included in the first stage of the study and 11 had not; these 11 additional isolates, which were members of the Enterobacteriaceae Carbapenem Breakpoint Panel, were negative for carbapenemase genes. On the basis of phenotypic determination of reference broth microdilution MICs at the CDC (37), 14 of the 34 isolates without carbapenemase genes met the 2015 CDC surveillance definition of CRE, and 20 did not (15).
The positive and negative quality control (QC) strains used in this study were Klebsiella pneumoniae ATCC BAA-1705 (blaKPC-positive by PCR) and Klebsiella pneumoniae ATCC BAA-1706, respectively (American Type Culture Collection, Manassas, VA).
From frozen (−80°C) stock, each isolate was subcultured twice on tryptic soy agar with 5% sheep blood (Trypticase soy agar with 5% sheep blood [TSA with 5% SB]; Becton, Dickinson and Company, Sparks, MD), incubating each subculture in ambient air at 35°C ± 2°C for 18 to 24 h to ensure purity and viability before performance of the mCIM test.
Algorithm used for evaluation of mCIM.
During the first stage of the study, mCIM testing was performed on each isolate. During the second stage, each of the nine participating laboratories performed mCIM testing on every isolate; in addition, one site simultaneously performed unmodified CIM testing (24), with inoculum for both CIM and mCIM testing taken from the same subculture plate. All laboratories also performed mCIM testing on each QC strain on each day of testing.
mCIM testing.
Using a sterile inoculating loop, 1 μl of test organism was added into a tube containing 2 ml of tryptic soy broth (TSB) (BD BBL tryptic soy broth or BD Bacto tryptic soy broth [Becton, Dickinson and Company, Sparks, MD] or Remel tryptic soy broth [Thermo Fisher Scientific, Inc., Waltham, MA]); the bacterial suspension was vortexed for 10 to 15 s. Next, a 10-μg MEM disk (BD BBL Sensi-Disc susceptibility test disc) was aseptically added into the bacterial suspension. The tube was then incubated for 4 h ± 15 min at 35°C ± 2°C in ambient air. Just prior to completion of the 4-h carbapenem inactivation step, a suspension of the mCIM indicator organism (Escherichia coli ATCC 25922, a carbapenem-susceptible strain) with turbidity equivalent to a 0.5 McFarland standard was prepared, and the surface of a MHA plate (BD BBL Mueller-Hinton agar or Remel Mueller-Hinton agar) was inoculated using the procedure for standard disk diffusion susceptibility testing (38). The meropenem (MEM) disk was then removed from the TSB bacterial suspension using a 10-μl inoculating loop; the loop was dragged along the edge of the tube during removal to remove excess liquid, and the disk was placed on the inoculated MHA plate, which was then incubated in an inverted position for 18 to 24 h at 35°C ± 2°C in ambient air.
mCIM result interpretation.
The diameter of the zone of inhibition around each MEM disk was measured (Fig. 3A). During the first phase of the study, a zone diameter of 6 to 10 mm was considered a positive result (i.e., carbapenemase production detected), a zone diameter of 11 to 19 mm was considered an indeterminate result, and a zone diameter of ≥20 mm was considered a negative result (i.e., no carbapenemase production detected); these provisional cutoffs were chosen on the basis of the results of previously published studies that reported “uninhibited growth” of the indicator strain for CP-CRE (24, 25) and zone diameters of ≥20 mm for Enterobacteriaceae not producing carbapenemases (25). A narrow ring of growth abutting the MEM disk, representing carryover of the test organism from the TSB, was ignored (Fig. 3B). Analysis of the zone diameter data generated during the second phase of the study (which included results of testing performed by multiple individuals in multiple laboratories) informed the establishment of the ultimate, standardized interpretive criteria for the mCIM.
FIG 3.
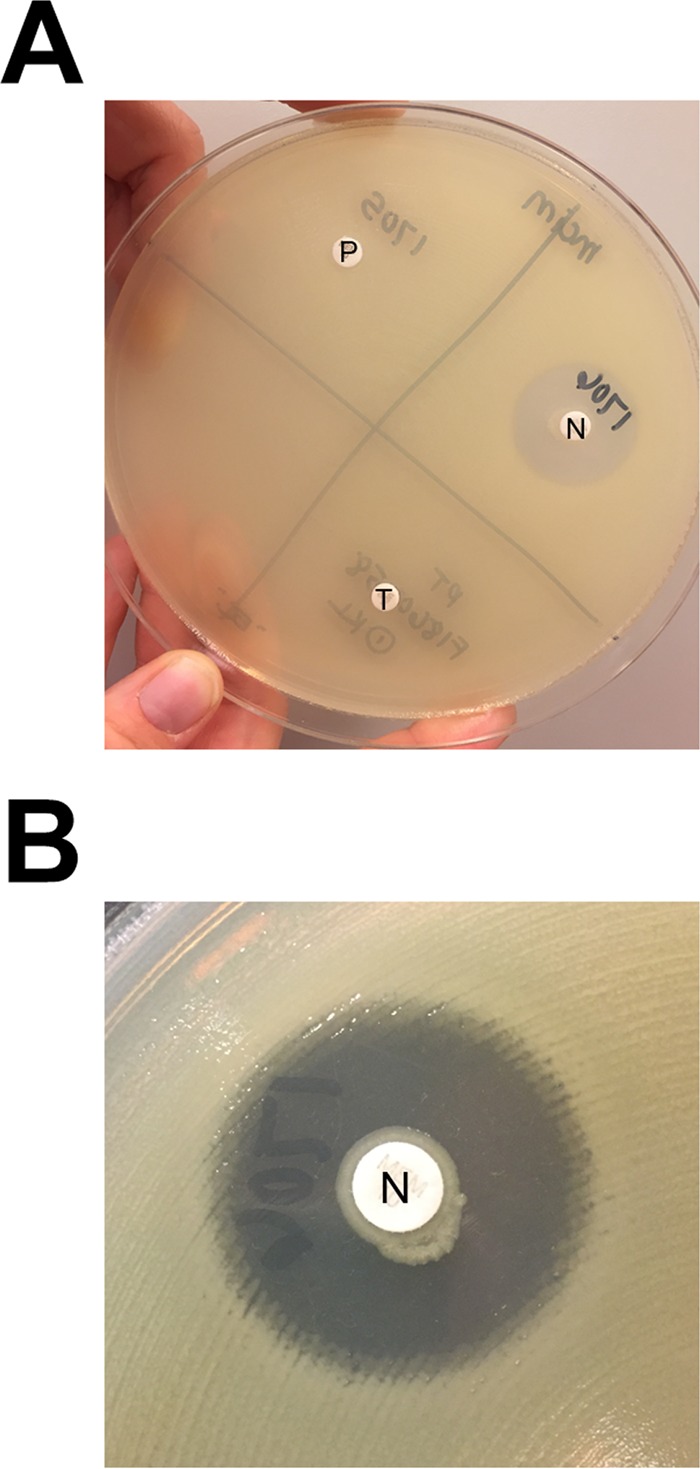
Reading and interpretation of mCIM results. (A) A 22-mm zone of inhibition of growth of the carbapenem-susceptible indicator organism is present around the meropenem (MEM) disk that was incubated with the negative-control organism (N), while no zone of inhibition is present around the MEM disks that were incubated with the positive-control organism (P) or the test organism (T). (B) Closer examination of the zone around the MEM disk incubated with the negative-control organism reveals a narrow ring of growth abutting the disk, which represents carryover of the test organism from the tryptic soy broth; this growth is ignored when interpreting mCIM results.
Statistical analysis.
Data from the first stage of the study were used to calculate the sensitivity, specificity, and associated 95% confidence intervals for the mCIM, using genotypic testing (whole-genome sequencing or targeted PCR) as the reference method to which the performance of the mCIM was compared. Data from the second stage were used to calculate the sensitivity and specificity of the mCIM in each of the nine participating laboratories and the proportion of laboratories that observed the expected categorical result (positive or negative) for each isolate. Indeterminate mCIM results were classified as false-negative results when they occurred for isolates that carried carbapenemase genes and as false-positive results when they occurred for isolates that did not carry carbapenemase genes. Finally, the average overall sensitivity and average overall specificity generated by the application of each possible zone diameter cutoff value were calculated and used to determine an optimized cutoff value for the test through receiver operating characteristic analysis.
Supplementary Material
ACKNOWLEDGMENTS
We gratefully acknowledge Cindy Bethel, Krizia Chambers, Jekia J. Cox, Kristianne Dawa, Nicole Karikari, Susan M. Kircher, Maria Jose Machado, Rachelle Markham, Belita N. A. Opene, Qinfang Qian, Monica Raposo, James K. Rasheed, Jennifer Rivers, Parul Patel, Dena Shibib, Jean Spargo, Corey Sparkes, Vanda White, and Brian B. Yoo for technical assistance and thank Janet Hindler and Romney Humphries for providing a subset of the study isolates.
This work was conducted with support from Harvard Catalyst (The Harvard Clinical and Translational Science Center) (National Center for Research Resources and the National Center for Advancing Translational Sciences, National Institutes of Health Award UL1 TR001102) and financial contributions from Harvard University and its affiliated academic health care centers.
The findings and conclusions in this report are those of the authors and do not necessarily represent the official views of Harvard Catalyst, Harvard University and its affiliated academic health care centers, or the National Institutes of Health or the Centers for Disease Control and Prevention. Use of trade names and commercial sources is for identification purposes and does not constitute endorsement by the Public Health Service or the U.S. Department of Health and Human Services.
We have no conflicts of interest relevant to this report to disclose.
Footnotes
For a commentary on this article, see https://doi.org/10.1128/JCM.00538-17.
Supplemental material for this article may be found at https://doi.org/10.1128/JCM.00193-17.
REFERENCES
- 1.Nordmann P, Naas T, Poirel L. 2011. Global spread of carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis 17:1791–1798. doi: 10.3201/eid1710.110655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention. 2013. Vital signs: carbapenem-resistant Enterobacteriaceae. MMWR Morb Mortal Wkly Rep 62:165–170. [PMC free article] [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention. 2013. Antibiotic resistance threats in the United States, 2013. Centers for Disease Control and Prevention, Atlanta, GA: http://www.cdc.gov/drugresistance/threat-report-2013/. [Google Scholar]
- 4.World Health Organization. 2014. Antimicrobial resistance: global report on surveillance 2014. World Health Organization, Geneva, Switzerland: http://www.who.int/drugresistance/documents/surveillancereport/en/. [Google Scholar]
- 5.Vasoo S, Barreto JN, Tosh PK. 2015. Emerging issues in gram-negative bacterial resistance: an update for the practicing clinician. Mayo Clin Proc 90:395–403. doi: 10.1016/j.mayocp.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 6.Peleg AY, Hooper DC. 2010. Hospital-acquired infections due to gram-negative bacteria. N Engl J Med 362:1804–1813. doi: 10.1056/NEJMra0904124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lutgring JD, Limbago BM. 2016. The problem of carbapenemase-producing-carbapenem-resistant-Enterobacteriaceae detection. J Clin Microbiol 54:529–534. doi: 10.1128/JCM.02771-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clinical and Laboratory Standards Institute. 2016. Performance standards for antimicrobial susceptibility testing, 26th ed CLSI supplement M100S. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 9.European Committee on Antimicrobial Susceptibility Testing. 2016. Breakpoint tables for interpretation of MICs and zone diameters, version 6.0. http://www.eucast.org/clinical_breakpoints/.
- 10.Centers for Disease Control and Prevention. 2015. Facility guidance for control of carbapenem-resistant Enterobactericeae (CRE): November 2015 update–CRE toolkit. Centers for Disease Control and Prevention, Atlanta, GA: http://www.cdc.gov/hai/organisms/cre/cre-toolkit/. [Google Scholar]
- 11.Boucher HW, Talbot GH, Benjamin DK Jr, Bradley J, Guidos RJ, Jones RN, Murray BE, Bonomo RA, Gilbert D, Infectious Diseases Society of America. 2013. 10 × ′20 Progress–development of new drugs active against gram-negative bacilli: an update from the Infectious Diseases Society of America. Clin Infect Dis 56:1685–1694. doi: 10.1093/cid/cit152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tamma PD, Goodman KE, Harris AD, Tekle T, Roberts A, Taiwo A, Simner PJ. 2017. Comparing the outcomes of patients with carbapenemase-producing and non-carbapenemase-producing carbapenem-resistant Enterobacteriaceae bacteremia. Clin Infect Dis 64:257–264. doi: 10.1093/cid/ciw741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chea N, Bulens SN, Kongphet-Tran T, Lynfield R, Shaw KM, Vagnone PS, Kainer MA, Muleta DB, Wilson L, Vaeth E, Dumyati G, Concannon C, Phipps EC, Culbreath K, Janelle SJ, Bamberg WM, Guh AY, Limbago B, Kallen AJ. 2015. Improved phenotype-based definition for identifying carbapenemase producers among carbapenem-resistant Enterobacteriaceae. Emerg Infect Dis 21:1611–1616. doi: 10.3201/eid2109.150198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tamma PD, Huang Y, Opene BN, Simner PJ. 2016. Determining the optimal carbapenem MIC that distinguishes carbapenemase-producing and non-carbapenemase-producing carbapenem-resistant Enterobacteriaceae. Antimicrob Agents Chemother 60:6425–6429. doi: 10.1128/AAC.00838-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Centers for Disease Control and Prevention. 2015. FAQs about choosing and implementing a CRE definition. Centers for Disease Control and Prevention, Atlanta, GA: http://www.cdc.gov/hai/organisms/cre/definition.html. [Google Scholar]
- 16.Girlich D, Poirel L, Nordmann P. 2012. Value of the modified Hodge test for detection of emerging carbapenemases in Enterobacteriaceae. J Clin Microbiol 50:477–479. doi: 10.1128/JCM.05247-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anderson KF, Lonsway DR, Rasheed JK, Biddle J, Jensen B, McDougal LK, Carey RB, Thompson A, Stocker S, Limbago B, Patel JB. 2007. Evaluation of methods to identify the Klebsiella pneumoniae carbapenemase in Enterobacteriaceae. J Clin Microbiol 45:2723–2725. doi: 10.1128/JCM.00015-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carvalhaes CG, Picao RC, Nicoletti AG, Xavier DE, Gales AC. 2010. Cloverleaf test (modified Hodge test) for detecting carbapenemase production in Klebsiella pneumoniae: be aware of false positive results. J Antimicrob Chemother 65:249–251. doi: 10.1093/jac/dkp431. [DOI] [PubMed] [Google Scholar]
- 19.Mochon AB, Garner OB, Hindler JA, Krogstad P, Ward KW, Lewinski MA, Rasheed JK, Anderson KF, Limbago BM, Humphries RM. 2011. New Delhi metallo-beta-lactamase (NDM-1)-producing Klebsiella pneumoniae: case report and laboratory detection strategies. J Clin Microbiol 49:1667–1670. doi: 10.1128/JCM.00183-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Papagiannitsis CC, Studentova V, Izdebski R, Oikonomou O, Pfeifer Y, Petinaki E, Hrabak J. 2015. Matrix-assisted laser desorption ionization-time of flight mass spectrometry meropenem hydrolysis assay with NH4HCO3, a reliable tool for direct detection of carbapenemase activity. J Clin Microbiol 53:1731–1735. doi: 10.1128/JCM.03094-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tamma PD, Opene BNA, Gluck A, Chambers KK, Carroll KC, Simner PJ. 11 January 2017. A comparison of eleven phenotypic assays for the accurate detection of carbapenemase-producing Enterobacteriaceae. J Clin Microbiol. doi: 10.1128/JCM.02338-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poirel L, Potron A, Nordmann P. 2012. OXA-48-like carbapenemases: the phantom menace. J Antimicrob Chemother 67:1597–1606. doi: 10.1093/jac/dks121. [DOI] [PubMed] [Google Scholar]
- 23.Doyle D, Peirano G, Lascols C, Lloyd T, Church DL, Pitout JD. 2012. Laboratory detection of Enterobacteriaceae that produce carbapenemases. J Clin Microbiol 50:3877–3880. doi: 10.1128/JCM.02117-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van der Zwaluw K, de Haan A, Pluister GN, Bootsma HJ, de Neeling AJ, Schouls LM. 2015. The carbapenem inactivation method (CIM), a simple and low-cost alternative for the Carba NP test to assess phenotypic carbapenemase activity in gram-negative rods. PLoS One 10:e0123690. doi: 10.1371/journal.pone.0123690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tijet N, Patel SN, Melano RG. 2016. Detection of carbapenemase activity in Enterobacteriaceae: comparison of the carbapenem inactivation method versus the Carba NP test. J Antimicrob Chemother 71:274–276. doi: 10.1093/jac/dkv283. [DOI] [PubMed] [Google Scholar]
- 26.Yamada K, Kashiwa M, Arai K, Nagano N, Saito R. 2016. Comparison of the Modified-Hodge test, Carba NP test, and carbapenem inactivation method as screening methods for carbapenemase-producing Enterobacteriaceae. J Microbiol Methods 128:48–51. doi: 10.1016/j.mimet.2016.06.019. [DOI] [PubMed] [Google Scholar]
- 27.Aktas E, Malkocoglu G, Otlu B, Copur Cicek A, Kulah C, Comert F, Sandalli C, Gursoy NC, Erdemir D, Bulut ME. 30 August 2016. Evaluation of the carbapenem inactivation method for detection of carbapenemase-producing Gram-negative bacteria in comparison with the RAPIDEC CARBA NP. Microb Drug Resist doi: 10.1089/mdr.2016.0092. [DOI] [PubMed] [Google Scholar]
- 28.Clinical and Laboratory Standards Institute. 2017. Performance standards for antimicrobial susceptibility testing, 27th ed CLSI supplement M100. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 29.Centers for Disease Control and Prevention. 2015. Tracking CRE. Centers for Disease Control and Prevention, Atlanta, GA: https://www.cdc.gov/hai/organisms/cre/TrackingCRE.html#CREmapKPC. [Google Scholar]
- 30.Pollett S, Miller S, Hindler J, Uslan D, Carvalho M, Humphries RM. 2014. Phenotypic and molecular characteristics of carbapenem-resistant Enterobacteriaceae in a health care system in Los Angeles, California, from 2011 to 2013. J Clin Microbiol 52:4003–4009. doi: 10.1128/JCM.01397-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hemarajata P, Yang S, Hindler JA, Humphries RM. 2015. Development of a novel real-time PCR assay with high-resolution melt analysis to detect and differentiate OXA-48-like beta-lactamases in carbapenem-resistant Enterobacteriaceae. Antimicrob Agents Chemother 59:5574–5580. doi: 10.1128/AAC.00425-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang S, Hemarajata P, Hindler J, Ward K, Adisetiyo H, Li F, Aldrovandi GM, Green NM, Russell D, Rubin Z, Humphries RM. 2016. Investigation of a suspected nosocomial transmission of blaKPC3-mediated carbapenem-resistant Klebsiella pneumoniae by whole genome sequencing. Diagn Microbiol Infect Dis 84:337–342. doi: 10.1016/j.diagmicrobio.2015.12.019. [DOI] [PubMed] [Google Scholar]
- 33.Humphries RM, Yang S, Hemarajata P, Ward KW, Hindler JA, Miller SA, Gregson A. 2015. First report of ceftazidime-avibactam resistance in a KPC-3-expressing Klebsiella pneumoniae isolate. Antimicrob Agents Chemother 59:6605–6607. doi: 10.1128/AAC.01165-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang S, Hemarajata P, Hindler J, Li F, Adisetiyo H, Aldrovandi GM, Sebra R, Kasarskis A, MacConnell D, Didelot X, Russell D, Rubin Z, Humphries RM. 1 April 2017. Evolution and transmission of carbapenem-resistant Klebsiella pneumoniae expressing the blaoxa-232 gene during an institutional outbreak associated with endoscopic retrograde cholangiopancreatography. Clin Infect Dis doi: 10.1093/cid/ciw876. [DOI] [PubMed] [Google Scholar]
- 35.de Man TJ, Limbago BM. 2016. SSTAR, a stand-alone easy-to-use antimicrobial resistance gene predictor. mSphere 1:e00050-15. doi: 10.1128/mSphere.00050-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Clinical and Laboratory Standards Institute. 2015. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard–tenth edition. CLSI document M07-A10. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 38.Clinical and Laboratory Standards Institute. 2015. Performance standards for antimicrobial disk susceptibility tests; approved standard–twelfth edition. CLSI document M02-A12. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.