ABSTRACT
In the Netherlands, the number of cases of infection with New Delhi metallo-beta-lactamase (NDM)-positive Enterobacteriaceae is low. Here, we report an outbreak of NDM-1-producing Klebsiella pneumoniae infection in a Dutch hospital with interspecies transfer of the resistance plasmid and unexpected occurrence in other unrelated health care centers (HCCs). Next-generation sequencing was performed on 250 carbapenemase-producing Enterobacteriaceae isolates, including 42 NDM-positive isolates obtained from 29 persons at the outbreak site. Most outbreak isolates were K. pneumoniae (n = 26) and Escherichia coli (n = 11), but 5 isolates comprising three other Enterobacteriaceae species were also cultured. The 26 K. pneumoniae isolates had sequence type 873 (ST873), as did 7 unrelated K. pneumoniae isolates originating from five geographically dispersed HCCs. The 33 ST873 isolates that clustered closely together using whole-genome multilocus sequence typing (wgMLST) carried the same plasmids and had limited differences in the resistome. The 11 E. coli outbreak isolates showed great variety in STs, did not cluster using wgMLST, and showed considerable diversity in resistome and plasmid profiles. The blaNDM-1 gene-carrying plasmid present in the ST873 K. pneumoniae isolates was found in all the other Enterobacteriaceae species cultured at the outbreak location and in a single E. coli isolate from another HCC. We describe a hospital outbreak with an NDM-1-producing K. pneumoniae strain from an unknown source that was also found in patients from five other Dutch HCCs in the same time frame without an epidemiological link. Interspecies transfer of the resistance plasmid was observed in other Enterobacteriaceae species isolated at the outbreak location and in another HCC.
KEYWORDS: Klebsiella, NDM, carbapenems, outbreak
INTRODUCTION
In 2008, a new carbapenemase, designated New Delhi metallo-beta-lactamase 1 (NDM-1), was detected in a Klebsiella pneumoniae isolate originating from a Swedish patient who had been hospitalized in India (1). NDM-1 belongs to the Ambler class B beta-lactamases and hydrolyzes virtually all beta-lactam antibiotics, including carbapenems (2). Besides NDM-1, 13 other variants of this metallo-beta-lactamase (NDM-2 to NDM-14) have currently been described, and most of the variants contain either single- or two-amino-acid substitutions compared to NDM-1 (3). The NDM gene has been isolated from a wide range of bacterial hosts, including Vibrio cholerae, and has been located on a great variety of plasmids (4, 5).
After the first report, NDM-positive bacteria were shown to be widely present among hospitalized patients in many countries of the Indian subcontinent and in the United Kingdom in 2008 and 2009 (6). At present, NDM-positive bacteria have been isolated from many countries worldwide, but many isolates have travel-related epidemiological links, with or without hospitalization, to the Indian subcontinent (7). However, some reports showed the presence of NDM-positive isolates in patients with no history of foreign travel, suggesting that the bacteria or the NDM gene had been acquired through local transmission (8, 9).
In the Netherlands, until now, the number of NDM-positive cases has been low, with only sporadic cases reported (10). The first NDM-positive bacteria were detected in 2010 in two Dutch patients who had recently traveled to India (11). More recently, in 2015, an NDM-5-producing K. pneumoniae isolate was isolated from a Dutch patient without a history of travel abroad (12).
Here, we describe a large outbreak of NDM-1-producing Enterobacteriaceae in the Netherlands that occurred in a large teaching hospital. Unexpectedly, the same strain was also found in other health care centers with no detectable epidemiological link.
RESULTS
Transmission of NDM-1-producing K. pneumoniae.
The K. pneumoniae isolates collected from 26 cases at the outbreak location (Table 1) were NDM positive, and analysis of the next-generation sequencing (NGS) data showed that they all harbored the blaNDM-1 gene. Three NDM-positive K. pneumoniae isolates originating from the outbreak location were not epidemiologically linked to the outbreak but also carried the blaNDM-1 gene. The top three carbapenemase genes carried by the 133 K. pneumoniae isolates from the Dutch carbapenemase-producing Enterobacteriaceae (CPE) surveillance, which were used as context in this study, were OXA-48-like (n = 75), KPC (n = 27), and NDM (n = 26) genes (Table 2). Twenty-three of the NDM-positive surveillance isolates carried the NDM-1 variant. Two other isolates carried the blaNDM-5 gene, and the remaining isolate harbored the blaNDM-7 gene.
TABLE 1.
Species isolated from the outbreak location during the NDM-1 outbreak
| Species isolated during the outbreak | No. of cases |
|---|---|
| K. pneumoniae | 17 |
| E. coli | 2 |
| C. freundii | 1 |
| K. pneumoniae and E. coli | 7 |
| K. pneumoniae, E. coli, K. oxytoca, and C. freundii | 1 |
| K. pneumoniae, E. coli, K. oxytoca, and P. mirabilis | 1 |
| Total no. of cases | 29 |
TABLE 2.
Isolates collected from January 2015 to December 2016, used as context in the study (one isolate per case per year)
| Species | Carbapenemase allele | No. of cases |
|---|---|---|
| K. pneumoniae | OXA-48 | 69 |
| OXA-232 | 4 | |
| OXA-181 | 1 | |
| OXA-162 | 1 | |
| KPC-2 | 19 | |
| KPC-3 | 8 | |
| NDM-1 | 23 | |
| NDM-5 | 2 | |
| NDM-7 | 1 | |
| NDM-1, OXA-48 | 3 | |
| NDM-1, OXA-232 | 1 | |
| VIM-1 | 1 | |
| Total | 133 | |
| E. coli | OXA-48 | 31 |
| OXA-181 | 7 | |
| OXA-244 | 4 | |
| KPC-2 | 1 | |
| NDM-5 | 15 | |
| NDM-1 | 7 | |
| NDM-7 | 2 | |
| NDM-6 | 1 | |
| NDM-5, OXA-181 | 1 | |
| VIM-2 | 1 | |
| Total | 70 |
The 26 presumed outbreak cases had meropenem MICs ranging from 0.75 to 12 μg/ml, but the majority of the isolates (n = 20) had MICs below 2 μg/ml. In contrast, 21 of the 26 NDM-positive context isolates from the CPE surveillance had MICs greater than 6 μg/ml, and only 2 isolates had MICs below 2 μg/ml.
Based on the NGS data on the K. pneumoniae isolates, all 26 presumed outbreak cases had sequence type 873 (ST873). Seven of the 133 K. pneumoniae context isolates from the Dutch CPE surveillance also were ST873, and these isolates originated from five other health care centers (HCCs). The HCCs were geographically dispersed throughout the Netherlands, and the ST873 isolates were submitted between December 2015 and October 2016. Six of the seven isolates were submitted after the outbreak period, but a single isolate was submitted during the outbreak period (Fig. 1). The other 126 isolates and the 3 unrelated isolates from the outbreak site submitted between 2015 and 2016 had various other STs.
FIG 1.
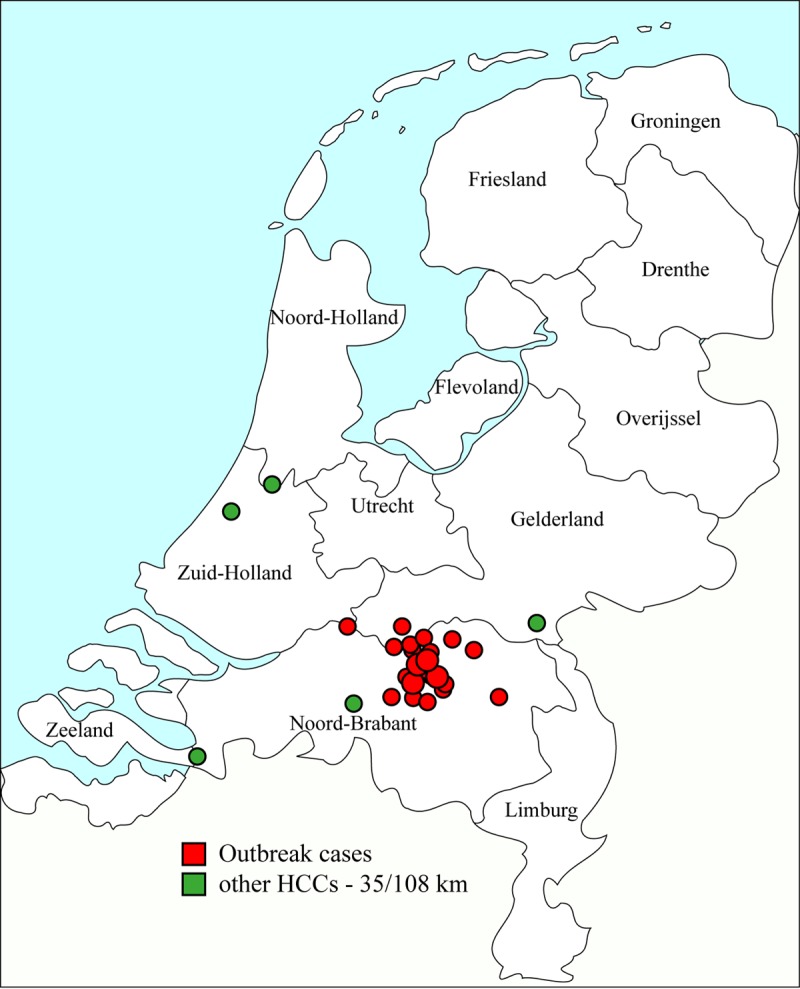
Geographic origins of the 26 K. pneumoniae isolates from outbreak cases and locations of the other five health care centers in the Netherlands. The outbreak cases were plotted based on the postal codes of the NDM carriers, and the sizes of the circles indicate the numbers of carriers with the same postal code.
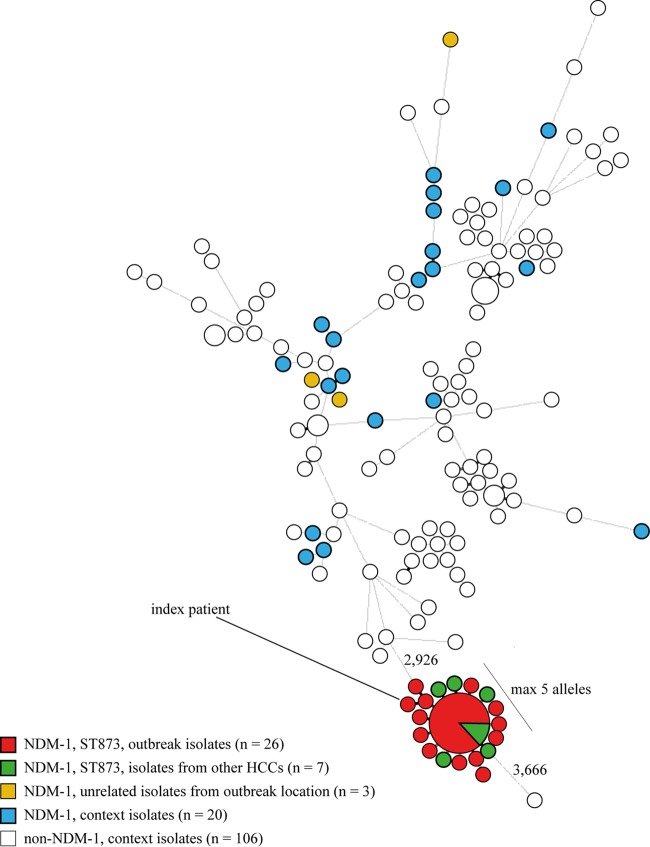
The whole-genome multilocus sequence typing (wgMLST) was based on 4,978 genes, and the resulting minimum spanning tree (MST) showed that the NDM-positive isolates were scattered over the tree, with the exception of a group of 33 ST873 isolates that clustered closely together with a maximum of five genes difference between neighboring isolates (Fig. 2). The distances between this group and the nearest neighbors ranged from 2,926 to 3,666 genes. The distance from the other three epidemiologically unrelated K. pneumoniae isolates that were isolated from the outbreak location ranged from 3,781 to 3,837 genes, indicating that they did not belong to the outbreak.
FIG 2.
Minimum spanning tree of the 162 K. pneumoniae isolates. The tree was based on 4,978 genes of the K. pneumoniae whole-genome MLST scheme. Clustering was done using a categorical coefficient, and the sizes of the circles indicate the numbers of isolates with identical wgMLST profiles. The lengths of the lines between isolates represent the numbers of different alleles.
Resistome analysis of the 33 ST873 NDM-1 K. pneumoniae isolates that clustered closely together in the wgMLST analysis showed that 25 isolates had identical resistomes (Table 3). Of these, 20 were obtained from the outbreak location and 5 isolates originated from four other HCCs. The remaining 8 isolates differed in their resistance gene profiles, each lacking one or more resistance genes compared to the group of 25 isolates.
TABLE 3.
Resistance genes among the outbreak isolates identified by ResFinder software
| Phenotype | Resistance gene | Presencea |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
K. pneumoniae (n = 33) |
E. coli (n = 11) |
||||||||||||
| V1 (n = 25) | V2 (n = 2) | V3 (n = 2) | V4 (n = 1) | V5 (n = 1) | V6 (n = 1) | V7 (n = 1) | V1 (n = 5) | V2 (n = 3) | V3 (n = 1) | V4 (n = 1) | V5 (n = 1) | ||
| Beta-lactam resistance | blaNDM-1 | + | + | + | + | + | + | + | + | + | + | + | + |
| blaCMY-6 | + | + | + | + | + | + | + | + | + | + | + | + | |
| blaOXA-1 | + | + | + | + | + | + | + | ||||||
| blaSHV-27 | + | + | + | + | + | + | + | ||||||
| blaCTX-M-15 | + | + | + | + | + | + | |||||||
| blaTEM-1B | + | + | + | + | + | + | |||||||
| Aminoglycoside resistance | aac(3)-IId | + | |||||||||||
| aadA1 | + | ||||||||||||
| aadA2 | + | ||||||||||||
| aadA5 | + | ||||||||||||
| Fluoroquinolone and aminoglycoside resistance | aac(6′)Ib-cr | + | + | + | + | + | + | + | |||||
| Aminoglycoside resistance | aph(3′)-Ia | + | + | + | + | + | + | + | + | + | + | + | + |
| rmtC | + | + | + | + | + | + | + | + | + | + | + | + | |
| strA | + | + | + | + | + | + | |||||||
| strB | + | + | + | + | + | + | |||||||
| aph(3′)-VIa | + | + | + | + | + | + | + | + | + | + | + | + | |
| Rifampin resistance | arr-2 | + | + | + | + | + | + | + | + | + | + | + | + |
| Phenicol resistance | catB3 | + | + | + | + | + | + | + | |||||
| cmlA1 | + | + | + | + | + | + | + | + | + | + | + | + | |
| Trimethoprim resistance | dfrA12 | + | |||||||||||
| dfrA14 | + | + | + | + | + | + | + | + | + | ||||
| dfrA17 | + | ||||||||||||
| Fosfomycin resistance | fosA | + | + | + | + | + | + | + | |||||
| Macrolide resistance | mph(A) | + | |||||||||||
| Quinolone resistance | oqxA | + | + | + | + | + | + | + | |||||
| oqxB | + | + | + | + | + | + | + | ||||||
| qnrB66 | + | + | + | + | |||||||||
| qnrS1 | + | + | |||||||||||
| Sulfonamide resistance | sul1 | + | + | + | + | + | + | + | + | + | + | + | + |
| sul2 | + | + | + | + | + | + | + | ||||||
| sul3 | + | ||||||||||||
| Tetracycline resistance | tet(A) | + | + | + | + | + | + | ||||||
+, present.
Plasmid analyses showed that the cluster of 33 ST873 K. pneumoniae isolates all had identical plasmid compositions. The blaNDM-1 gene was located on a variant of the pNDM-US plasmid (CP006661), a 140-kb plasmid belonging to the IncA/C2 group (13). Compared to the pNDM-US reference genome, the plasmid in the 33 ST873 K. pneumoniae isolates lacked a 740-bp segment that carried the aac(6′)-1B-cr (fluoroquinolone and aminoglycoside resistance) gene. This finding was corroborated by electroporation experiments, since the transformants originating from two ST873 K. pneumoniae isolates, one from the outbreak location and one from another HCC, did not carry the gene. Besides blaNDM-1, the plasmid also harbored the beta-lactam resistance gene blaCMY-6 and the resistance genes rmtc (aminoglycoside resistance) and sul-1 (sulfonamide resistance). In addition to the variant of the pNDM-US plasmid, all 33 ST873 K. pneumoniae isolates also carried two plasmids belonging to the IncFIB(K) and IncFII(K) groups. The three unrelated K. pneumoniae isolates from the outbreak location and the NDM-positive K. pneumoniae context isolates obtained from the national CPE surveillance did not carry the plasmid found in the ST873 isolates.
Transmission of the pNDM-US variant to E. coli.
During the outbreak, Escherichia coli isolates producing NDM-1 carbapenemase were cultured from 11 patients at the outbreak location. Nine of these patients also carried an ST873 K. pneumoniae isolate. An epidemiologically unrelated E. coli isolate submitted from the outbreak location in June 2015, 5 months prior to the outbreak, also produced NDM-1, whereas the other unrelated isolate carried a blaNDM-5 gene. PCR of the 70 E. coli isolates originating from the national CPE surveillance that served as context showed that they carried the blaOXA-48 (n = 42), blaNDM (n = 26), blaKPC (n = 1), and blaVIM (n = 1) carbapenemase genes (Table 2). Only 7 of the 26 E. coli NDM-positive context isolates produced the NDM-1 variant, whereas 15 other isolates produced NDM-5 and the remaining isolates produced NDM-7 (n = 2) and NDM-6 (n = 1). Similar to the K. pneumoniae outbreak isolates, the meropemen MICs of the 11 E. coli isolates ranged from 0.75 to 6 μg/ml and were considerably lower than those of the E. coli context isolates.
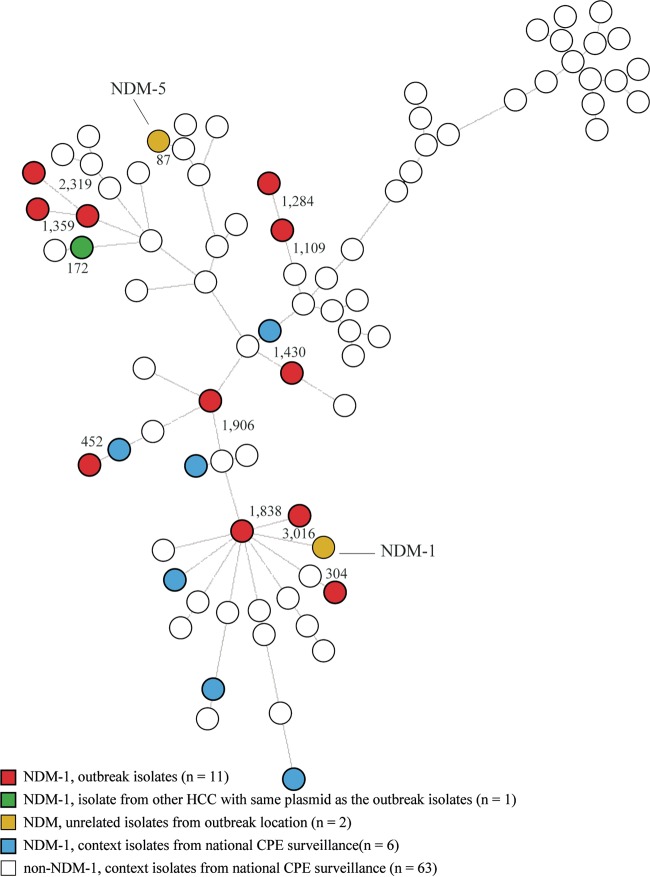
The STs, inferred from the NGS data, revealed that each of the 11 E. coli isolates from the outbreak location had a unique ST. The two unrelated E. coli isolates submitted from the outbreak location and the 70 E. coli isolates obtained from the national CPE surveillance also showed great diversity in STs. The wgMLST analysis, based on 4,503 genes, showed that the 11 E. coli outbreak isolates were scattered over the tree and that no clustering occurred (Fig. 3). The average distance from one of the outbreak E. coli isolates to the closest neighboring E. coli isolate was 1,091 genes, with a range between 304 and 2,319 genes. The unrelated blaNDM-1-positive E. coli isolate from the outbreak location did not cluster with any other isolate, and the distance from the closest neighbor was 3,016 genes. The other unrelated blaNDM-5-positive E. coli isolate clustered closely with another E. coli isolate carrying blaOXA-181, with a distance of 87 alleles.
FIG 3.
Minimum spanning tree based on whole-genome MLST of 83 E. coli isolates. The tree was based on 4,503 genes of the E. coli scheme, and clustering was done using a categorical coefficient. Each isolate in the tree is displayed as a circle. The lines between the isolates denote the distance in numbers of alleles.
Compared to the K. pneumoniae isolates, the E. coli isolates from the outbreak location showed more diversity in the resistome (Table 3). Plasmid analysis showed that all 11 E. coli outbreak isolates and the NDM-1-positive E. coli isolate, submitted in June 2015, harbored the same plasmid as the K. pneumoniae isolates, namely, the Inc A/C2 group plasmid variant of pNDM-US. A transformant of an E. coli isolate from the outbreak location confirmed that the same plasmid was present in both species. Apart from this plasmid, the E. coli isolates showed considerable diversity in their plasmid profiles, and the compositions were different in all the isolates (data not shown).
Four of the seven NDM-1-positive E. coli isolates from the national CPE surveillance also carried a plasmid belonging to the Inc A/C2 group. A single isolate carried the same plasmid variant of pNDM-US as the K. pneumoniae and E. coli outbreak isolates. This isolate originated from a patient from another HCC, and the patient was one of the seven persons who also carried an ST873 K. pneumoniae isolate (Fig. 1 and 2). The plasmid compositions of the other three NDM-1-positive E. coli surveillance isolates differed from the plasmid present in the K. pneumoniae and E. coli outbreak isolates, indicating that another plasmid from the Inc A/C2 group carried the NDM-1 gene.
Multiple NDM-1-producing species in the same patient.
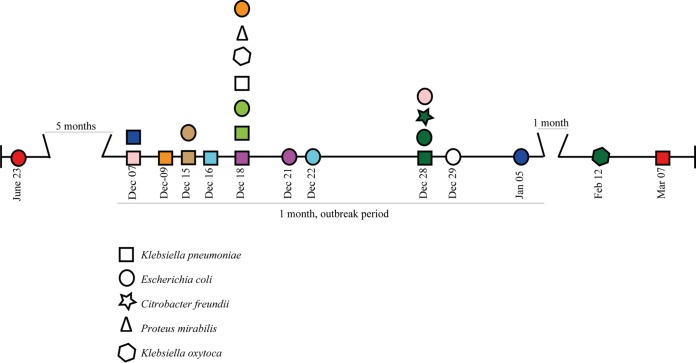
From 9 of the 31 outbreak patients, two to four different NDM-1-positive Enterobacteriaceae species were isolated during the outbreak period. Seven patients carried K. pneumoniae and E. coli, one also carried Citrobacter freundii and Klebsiella oxytoca, and one patient carried Proteus mirabilis and K. oxytoca, in addition to K. pneumoniae and E. coli (Table 1 and Fig. 4). In all nine cases, the K. pneumoniae isolate was cultured before or on the same day as the other species. However, from a patient not related to the outbreak whose E. coli isolate was submitted in June 2015, prior to the outbreak, a ST873 K. pneumoniae isolate was also cultured after the outbreak in March 2016. Similar to K. pneumoniae and E. coli, all the other species carried the variant of plasmid pNDM-US lacking the aac(6′)-1B-cr gene. An NDM-1-positive C. freundii isolate was cultured from a single patient, and this isolate had the same ST and resistome as the C. freundii isolate cultured from the patient who also carried NDM-1-positive K. pneumoniae and E. coli isolates.
FIG 4.
Timeline of the outbreak for 10 patients who carried multiple NDM-1-producing Enterobacteriaceae species. The colors indicate isolates originating from the same patient. The dates below the isolates represent the sampling dates.
DISCUSSION
To date, only a few NDM-positive isolates have been described in the Netherlands, indicating a limited presence of strains producing this metallo-beta-lactamase (11, 12). Here, we report a large outbreak in a Dutch hospital comprising 26 patients colonized with NDM-1-producing K. pneumoniae. In addition, 16 NDM-1-positive isolates comprising five other Enterobacteriaceae species harboring the same plasmid were also cultured at the outbreak location.
All K. pneumoniae outbreak isolates had ST873, were identical or very closely related in the wgMLST, and contained a variant of the IncA/C2 plasmid pNDM-US (CP006661) that harbors the blaNDM-1 gene. This indicates that the outbreak was caused by transmission of the same NDM-1-producing K. pneumoniae strain. Three epidemiologically unrelated blaNDM-1-positive K. pneumoniae isolates collected at the outbreak location were also genetically unrelated according to wgMLST, had a completely different resistome, and did not carry the variant of plasmid pNDM-US. Consequently, they did not belong to the outbreak.
The NDM-1-positive K. pneumoniae outbreak strain was also isolated from seven other persons originating from five other HCCs geographically dispersed throughout the Netherlands. These seven isolates were submitted for the Dutch CPE surveillance between December 2015 and October 2016. Despite extensive epidemiological investigations, none of the seven cases could be connected to the cases from the hospital where the outbreak occurred. Considering their very limited genetic variation, these strains most likely come from a common source. This conclusion was supported when the observed genetic variation was compared to a recently defined cutoff for relatedness between K. pneumoniae isolates (14). Although no evidence was found for this outbreak, NDM-1-producing K. pneumoniae strains could have been introduced via food supplied to various HCCs, as exemplified by the recent study from Egypt, where 12 of 100 tested carcasses were contaminated with CPE isolates, 11 of which were K. pneumoniae (15). The contamination of meat with CPE has not been described in the Netherlands, but several reports have shown high prevalence of extended-spectrum beta-lactamase (ESBL)-producing Enterobacteriaceae (16, 17). Furthermore, several of the health care centers, including the hospital with the outbreak, received poultry from the same supplier. Unfortunately, an attempt to reconstruct the diets of the patients involved in the outbreak was unsuccessful at the time that food was considered a possible source.
Another scenario could be a prolonged, unnoticed circulation of this K. pneumoniae strain or of other NDM-1 strain-carrying Enterobacteriaceae in the Dutch population or in patients or health care workers at the outbreak location. This scenario is supported by the fact that an E. coli strain carrying the same plasmid as the outbreak isolates was isolated at the outbreak location in June 2015, 5 months prior to the outbreak. Moreover, an NDM-1-positive ST873 K. pneumoniae isolate was cultured from the same patient after the outbreak in March 2016. However, given the short outbreak period and the fact that no further transmission was noticed after the outbreak, the latter suggestion seems less likely.
The blaNDM-1 gene was also present in 11 E. coli isolates collected at the outbreak location during the outbreak. The wgMLST and resistome analyses showed that the isolates were genetically unrelated. Like the K. pneumoniae outbreak isolates, all 11 E. coli isolates carried the same variant of plasmid pNDM-US, indicating multiple occurrences of horizontal gene transfer rather than transmission of the same E. coli strain. This was further supported by the fact that the transfer of the plasmid was also observed in five other isolates, comprising three other Enterobacteriaceae species, at the outbreak location. In addition, the transfer of the plasmid was also seen in another HCC where an ST873 K. pneumoniae isolate and an E. coli isolate from the same patient harbored the same plasmid as the outbreak isolates.
Our study has a number of limitations. First, our study has fully focused on bacterial isolates that harbored carbapenemase genes. The genetic background and potential spread of noncarbapenemase isolates from the different HCCs is therefore unknown. Second, our analysis comprised only isolates from the Dutch CPE surveillance. NGS data from publicly available databases containing international carbapenemase-producing K. pneumoniae isolates were not assessed and used. However, we did notice that ST873 K. pneumoniae isolates were occasionally found in recent studies from Switzerland and China, but these isolates did not produce carbapenemase (18, 19).
In conclusion, we report a hospital outbreak with an NDM-1-producing K. pneumoniae strain from an unknown source that was also found in patients from five other HCCs in the Netherlands in the same time frame. The plasmid carrying the blaNDM-1 gene was also found in other Enterobacteriaceae species isolated at the outbreak location and in another HCC. The fact that the outbreak strain was also found in other HCCs without a clear epidemiological link shows that analysis of the NGS data on CPE isolates collected during national CPE surveillance is important to reveal the spread of carbapenemase-producing pathogens in the Netherlands.
MATERIALS AND METHODS
Hospital setting and outbreak description.
The outbreak was first noted during screening for a possible ESBL transmission in the surgical ward of a 683-bed tertiary teaching hospital in the southern Netherlands. On 23 November 2015, a carbapenemase-producing blaNDM-positive, ESBL-positive K. pneumoniae strain was isolated during this screening. At the time of identification, the patient had already been discharged. Shortly after, ESBL screening of cultures from long-term-admitted surgical patients revealed two additional patients with NDM-producing K. pneumoniae. Contact tracing and weekly screening rounds identified additional NDM carriers. There were two wards with a single NDM carrier and seven wards with two or more NDM carriers. All the patients admitted to one of these wards from the 1October 2015 on were defined as at risk for carrying NDM. The outbreak period was defined as the date 2 weeks prior (1 October) to the admission of the initial patient until the date when all suspected NDM carriers were identified and put into contact isolation (30 December). Weekly screening rounds were continued during the first 4 weeks of January 2016 on risk wards to confirm that the outbreak was successfully controlled. All 2,964 patients admitted to one of the risk wards during the outbreak period were tested for NDM carriage. Six months after the end of the outbreak, over 95% of the patients at risk had been screened, and in total, 29 NDM carriers (1%) had been identified.
Bacterial isolates.
The first three isolates were obtained from an ESBL screening using an ESwab on ChromID ESBL agar (bioMérieux Inc., Marcy l'Etoile, France). All subsequent isolates were obtained by screening for carbapenem resistance using enrichment in tryptone soya broth supplemented with ertapenem and vancomycin. During the outbreak period, 42 isolates carrying a blaNDM gene detected by PCR were obtained from 29 persons at the outbreak site and included in the study (Table 1). In most cases, a single NDM-producing species was found per patient. However, in nine patients, two to four distinct NDM-producing species were isolated. Most isolates were K. pneumoniae (n = 26) and E. coli (n = 11), but two C. freundii isolates, two K. oxytoca isolates, and one P. mirabilis isolate were also cultured. Retrospective analysis showed that five more isolates carrying a blaNDM gene had been isolated at the outbreak site between 2015 and 2016 and subsequently submitted to the national reference center. These isolates (three K. pneumoniae and two E. coli) were also included in the study. For the Dutch national surveillance of CPE, medical microbiology laboratories are requested to submit Enterobacteriaceae isolates with a meropenem MIC of >0.25 μg/ml and/or an imipenem MIC of >1 μg/ml to the National Institute for Public Health and the Environment for further molecular characterization. From the Dutch CPE surveillance, NGS data from all K. pneumoniae (n = 133) and all E. coli (n = 70) carbapenemase-producing isolates obtained between January 2014 and December 2016 were included for analysis to provide genetic context. All isolates were tested for the production of carbapenemase using the carbapenem inactivation method (CIM) and subjected to PCR to detect the presence of carbapenemase-encoding genes, as previously described (20). Isolates that were shown by PCR to carry a blaNDM gene and by CIM to produce carbapenemase are referred to as NDM positive.
Antimicrobial susceptibility testing.
At the outbreak location, all isolates were cultured and analyzed for resistance using Vitek (bioMérieux Inc., Marcy l'Etoile, France). Resistance to carbapenem was confirmed by assessing the meropenem MICs for all 250 isolates using Etest (bioMérieux Inc., Marcy l'Etoile, France).
Next-generation sequencing.
All 250 isolates were subjected to NGS using the Illumina HiSeq 2500 (BaseClear, Leiden, the Netherlands). The NGS data on the K. pneumoniae and E. coli isolates were used for MLST and wgMLST analyses using the available wgMLST schemes in SeqSphere software version 2.3.0 (Ridom GmbH, Münster, Germany). The resulting data were imported into Bionumerics version 7.6 for comparative analyses (Applied Maths, Sint-Martens-Latem, Belgium).
The resistomes and plasmid compositions of all the isolates were determined by uploading the assembled contigs into the online tools ResFinder (version 2.1) and PlasmidFinder (version 1.3) via the Center for Genomic Epidemiology website (21, 22). For ResFinder, a 90% identity threshold and a minimum length of 60% were used as criteria, whereas for PlasmidFinder, an identity of 95% was utilized. The NGS data on the presumed outbreak isolates were mapped against known NDM plasmids present in the NCBI database in May 2016.
Electroporation experiments.
To confirm that the same plasmid was transferred between species, an NDM-positive K. pneumoniae isolate and an NDM-positive E. coli isolate from the outbreak location were used to transfer the plasmid carrying the blaNDM gene to a susceptible E. coli (DH10B) isolate by electroporation. The plasmid of another NDM-positive K. pneumoniae isolate from another health care center was also transferred to show that the same plasmid was present in multiple locations. For this, the electroporation protocol of the manufacturer was used (Invitrogen, Carlsbad, CA, USA). Transformants were subjected to PCR to confirm the presence of the blaNDM gene, and NGS was performed on each transformant. The resulting NGS data were analyzed in CLC Genomics Workbench version 9.5.3 (Qiagen, Hilden, Germany).
ACKNOWLEDGMENTS
We thank all the Dutch medical microbiology laboratories for submitting their CPE isolates for the national CPE surveillance. We particularly thank Karen Heemstra for searching for epidemiological links between the related NDM-1-positive K. pneumoniae cases from another health care center and the outbreak cases.
REFERENCES
- 1.Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, Lee K, Walsh TR. 2009. Characterization of a new metallo-beta-lactamase gene, blaNDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother 53:5046–5054. doi: 10.1128/AAC.00774-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nordmann P. 2014. Carbapenemase-producing Enterobacteriaceae: overview of a major public health challenge. Med Mal Infect 44:51–56. doi: 10.1016/j.medmal.2013.11.007. [DOI] [PubMed] [Google Scholar]
- 3.Zou D, Huang Y, Zhao X, Liu W, Dong D, Li H, Wang X, Huang S, Wei X, Yan X, Yang Z, Tong Y, Huang L, Yuan J. 2015. A novel New Delhi metallo-beta-lactamase variant, NDM-14, isolated in a Chinese hospital possesses increased enzymatic activity against carbapenems. Antimicrob Agents Chemother 59:2450–2453. doi: 10.1128/AAC.05168-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khong WX, Xia E, Marimuthu K, Xu W, Teo YY, Tan EL, Neo S, Krishnan PU, Ang BS, Lye DC, Chow AL, Ong RT, Ng OT. 2016. Local transmission and global dissemination of New Delhi metallo-beta-lactamase (NDM): a whole genome analysis. BMC Genomics 17:452. doi: 10.1186/s12864-016-2740-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walsh TR, Weeks J, Livermore DM, Toleman MA. 2011. Dissemination of NDM-1 positive bacteria in the New Delhi environment and its implications for human health: an environmental point prevalence study. Lancet Infect Dis 11:355–362. doi: 10.1016/S1473-3099(11)70059-7. [DOI] [PubMed] [Google Scholar]
- 6.Kumarasamy KK, Toleman MA, Walsh TR, Bagaria J, Butt F, Balakrishnan R, Chaudhary U, Doumith M, Giske CG, Irfan S, Krishnan P, Kumar AV, Maharjan S, Mushtaq S, Noorie T, Paterson DL, Pearson A, Perry C, Pike R, Rao B, Ray U, Sarma JB, Sharma M, Sheridan E, Thirunarayan MA, Turton J, Upadhyay S, Warner M, Welfare W, Livermore DM, Woodford N. 2010. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect Dis 10:597–602. doi: 10.1016/S1473-3099(10)70143-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson AP, Woodford N. 2013. Global spread of antibiotic resistance: the example of New Delhi metallo-beta-lactamase (NDM)-mediated carbapenem resistance. J Med Microbiol 62:499–513. doi: 10.1099/jmm.0.052555-0. [DOI] [PubMed] [Google Scholar]
- 8.Denis C, Poirel L, Carricajo A, Grattard F, Fascia P, Verhoeven P, Gay P, Nuti C, Nordmann P, Pozzetto B, Berthelot P. 2012. Nosocomial transmission of NDM-1-producing Escherichia coli within a non-endemic area in France. Clin Microbiol Infect 18:E128–E130. doi: 10.1111/j.1469-0691.2012.03761.x. [DOI] [PubMed] [Google Scholar]
- 9.Jovcic B, Lepsanovic Z, Suljagic V, Rackov G, Begovic J, Topisirovic L, Kojic M. 2011. Emergence of NDM-1 metallo-beta-lactamase in Pseudomonas aeruginosa clinical isolates from Serbia. Antimicrob Agents Chemother 55:3929–3931. doi: 10.1128/AAC.00226-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.European Centre for Disease Prevention and Control. 2013. Carbapenemase-producing bacteria in Europe: interim results from the European Survey on carbapenemase-producing Enterobacteriaceae (EuSCAPE) project. European Centre for Disease Prevention and Control, Stockholm, Sweden. [Google Scholar]
- 11.Leverstein-van Hall MA, Stuart JC, Voets GM, Versteeg D, Roelofsen E, Fluit AC. 2010. Carbapenem-resistant Klebsiella pneumoniae following foreign travel. Ned Tijdschr Geneeskd 154:A2013 (In Dutch.) [PubMed] [Google Scholar]
- 12.Bathoorn E, Rossen JW, Lokate M, Friedrich AW, Hammerum AM. 2015. Isolation of an NDM-5-producing ST16 Klebsiella pneumoniae from a Dutch patient without travel history abroad, August 2015. Euro Surveill 20. doi: 10.2807/1560-7917.ES.2015.20.41.30040. [DOI] [PubMed] [Google Scholar]
- 13.Hudson CM, Bent ZW, Meagher RJ, Williams KP. 2014. Resistance determinants and mobile genetic elements of an NDM-1-encoding Klebsiella pneumoniae strain. PLoS One 9:e99209. doi: 10.1371/journal.pone.0099209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kluytmans-van den Bergh MF, Rossen JW, Bruijning-Verhagen PC, Bonten MJ, Friedrich AW, Vandenbroucke-Grauls CM, Willems RJ, Kluytmans JA. 2016. Whole-genome multilocus sequence typing of extended-spectrum-beta-lactamase-producing Enterobacteriaceae. J Clin Microbiol 54:2919–2927. doi: 10.1128/JCM.01648-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abdallah HM, Reuland EA, Wintermans BB, Al Naiemi N, Koek A, Abdelwahab AM, Ammar AM, Mohamed AA, Vandenbroucke-Grauls CM. 2015. Extended-spectrum beta-lactamases and/or carbapenemases-producing Enterobacteriaceae isolated from retail chicken meat in Zagazig, Egypt. PLoS One 10:e0136052. doi: 10.1371/journal.pone.0136052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leverstein-van Hall MA, Dierikx CM, Cohen Stuart J, Voets GM, van den Munckhof MP, van Essen-Zandbergen A, Platteel T, Fluit AC, van de Sande-Bruinsma N, Scharinga J, Bonten MJ, Mevius DJ, National ESBL Surveillance Group. 2011. Dutch patients, retail chicken meat and poultry share the same ESBL genes, plasmids and strains. Clin Microbiol Infect 17:873–880. doi: 10.1111/j.1469-0691.2011.03497.x. [DOI] [PubMed] [Google Scholar]
- 17.Overdevest I, Willemsen I, Rijnsburger M, Eustace A, Xu L, Hawkey P, Heck M, Savelkoul P, Vandenbroucke-Grauls C, van der Zwaluw K, Huijsdens X, Kluytmans J. 2011. Extended-spectrum beta-lactamase genes of Escherichia coli in chicken meat and humans, The Netherlands. Emerg Infect Dis 17:1216–1222. doi: 10.3201/eid1707.110209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wohlwend N, Endimiani A, Francey T, Perreten V. 2015. Third-generation-cephalosporin-resistant Klebsiella pneumoniae isolates from humans and companion animals in Switzerland: spread of a DHA-producing sequence type 11 clone in a veterinary setting. Antimicrob Agents Chemother 59:2949–2955. doi: 10.1128/AAC.04408-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang J, Zhou K, Zheng B, Zhao L, Shen P, Ji J, Wei Z, Li L, Zhou J, Xiao Y. 2016. High prevalence of ESBL-producing Klebsiella pneumoniae causing community-onset infections in China. Front Microbiol 7:1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van der Zwaluw K, de Haan A, Pluister GN, Bootsma HJ, de Neeling AJ, Schouls LM. 2015. The carbapenem inactivation method (CIM), a simple and low-cost alternative for the Carba NP test to assess phenotypic carbapenemase activity in gram-negative rods. PLoS One 10:e0123690. doi: 10.1371/journal.pone.0123690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carattoli A, Zankari E, Garcia-Fernandez A, Voldby Larsen M, Lund O, Villa L, Moller Aarestrup F, Hasman H. 2014. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]