ABSTRACT
The search for a cure for HIV infection has highlighted the need for increasingly sensitive and precise assays to measure viral burden in various tissues and body fluids. We describe the application of a standardized assay for HIV-1 RNA in multiple specimen types. The fully automated Aptima HIV-1 Quant Dx assay (Aptima assay) is FDA cleared for blood plasma HIV-1 RNA quantitation. In this study, the Aptima assay was applied for the quantitation of HIV RNA in peripheral blood mononuclear cells (PBMCs; n = 72), seminal plasma (n = 20), cerebrospinal fluid (CSF; n = 36), dried blood spots (DBS; n = 104), and dried plasma spots (DPS; n = 104). The Aptima assay was equivalent to or better than commercial assays or validated in-house assays for the quantitation of HIV RNA in CSF and seminal plasma. For PBMC specimens, the sensitivity of the Aptima assay in the detection of HIV RNA decayed as background uninfected PBMC counts increased; proteinase K treatment demonstrated some benefit in restoring signal at higher levels of background PBMCs. Finally, the Aptima assay yielded 100% detection rates of DBS in participants with plasma HIV RNA levels of ≥35 copies/ml and 100% detection rates of DPS in participants with plasma HIV RNA levels of ≥394 copies/ml. The Aptima assay can be applied to a variety of specimens from HIV-infected subjects to measure HIV RNA for studies of viral persistence and cure strategies. It can also detect HIV in dried blood and plasma specimens, which may be of benefit in resource-limited settings.
KEYWORDS: assay development, cerebrospinal fluid, dried blood spots, dried plasma spots, human immunodeficiency virus, peripheral blood mononuclear cells, plasma, semen
INTRODUCTION
Combination antiretroviral therapy (ART) has dramatically reduced morbidity and mortality resulting from HIV-1 infection (1). ART restricts HIV replication and thereby suppresses plasma viremia to levels below the limit of detection of standard assays (2), but it is unable to eradicate HIV infection due to the persistence of latent provirus (3, 4). Integrated virus has been identified in multiple tissue types, including peripheral blood, lymph nodes, gut-associated lymphoid tissue, the central nervous system, and the genital tract (5). Attention has therefore turned to developing cure strategies that target latently infected cells in order to eradicate HIV infection. Increasing interest in the latent HIV reservoir has emphasized the need for sensitive and precise assays capable of quantifying residual virus in different tissues (6). However, methods used to measure HIV-1 in specimen types from biological compartments other than blood have not been standardized. Moreover, some specimen types contain substances that can inhibit the quantitation of nucleic acids, posing an additional challenge to the development of HIV RNA assays.
More than 90% of new HIV-1 infections occur in low- and middle-income countries with limited health care resources (7). Management of infection is challenging in these settings because of difficulties in performing diagnostic virology assays. Barriers include limited refrigeration, unreliable electricity, and lack of access to expensive equipment and reagents among other resource limitations. Many treatment programs engaged in the quantitation and sequencing of HIV have therefore resorted to utilizing clinical specimens that can be obtained without venipuncture and are easy to store and transport, such as dried blood or plasma spots (8, 9).
The Aptima HIV-1 Quant Dx assay (Aptima assay) is an FDA-cleared fully automated assay on the Panther system (Hologic, San Diego, CA). The assay is based on target-capture transcription-mediated amplification (TMA) (10) and real-time detection technologies. It uses 0.5 ml of plasma and is intended for monitoring HIV-1 viral load, with a lower limit of quantitation of 30 copies/ml and a specificity of 100% for all HIV-1 groups and group M subtypes (11). The experiments described here were performed to determine whether the assay can be adapted to detect and quantitate HIV-1 RNA in clinical specimens other than blood plasma, such as peripheral blood mononuclear cells (PBMCs), seminal plasma, cerebrospinal fluid (CSF), dried blood spots (DBS), and dried plasma spots (DPS). Detection and quantitation of HIV-1 RNA in these diverse specimen types may have important practical applications for HIV care in low- and middle-income countries as well as for research efforts to eradicate HIV.
RESULTS
PBMC specimens.
The application of the Aptima assay for the detection of cell-associated HIV RNA in PBMCs was examined using samples containing different quantities of background cellular RNA and protein. Cell-associated HIV RNA levels were measured in samples of background uninfected PBMCs spiked with 10 8E5 cells, which contain a single copy of integrated HIV DNA in each cell, prepared in replicates of 6 (n = 72). HIV RNA levels were comparable in samples with 0 background PBMCs and 0.3 × 106 background PBMCs processed using the standard protocol (“no treatment”; Fig. 1). However, as background PBMC concentration increased, significant reductions in the HIV RNA signal with single-phase decay kinetics were noted in the no treatment group (mean decrease of 2.96 logs with every 3-fold increase in background PBMCs; R2 = 0.90). It was hypothesized that cellular proteins were inhibiting nucleic acid extraction by contributing to sample viscosity and RNA degradation. To overcome this potential source of inhibition, a digestion step using proteinase K was added. Samples treated with proteinase K (“proteinase K”) showed significant decreases of 0.24 logs (P = 0.004) for samples with 0.3 × 106 background PBMCs and 0.70 logs (P = 0.002) for samples with 1 × 106 background PBMCs compared to those of samples with 0 background PBMCs. However, when background PBMCs increased to 3 × 106 and 10 × 106 cells/sample, no further significant decreases in signal were seen (Fig. 1). Overall, the two groups showed similar results at 0 and 0.3 × 106 background PBMCs (P = 0.33 and P = 0.57, respectively); no treatment showed higher sensitivity at background PBMC counts of 1 × 106 (P = 0.004); and proteinase K treatment showed higher sensitivity at background PBMC counts of 3 × 106 and 10 × 106 (P = 0.03 and P = 0.008, respectively). There were no false positives detected in 6 negative PBMC controls run in each system.
FIG 1.
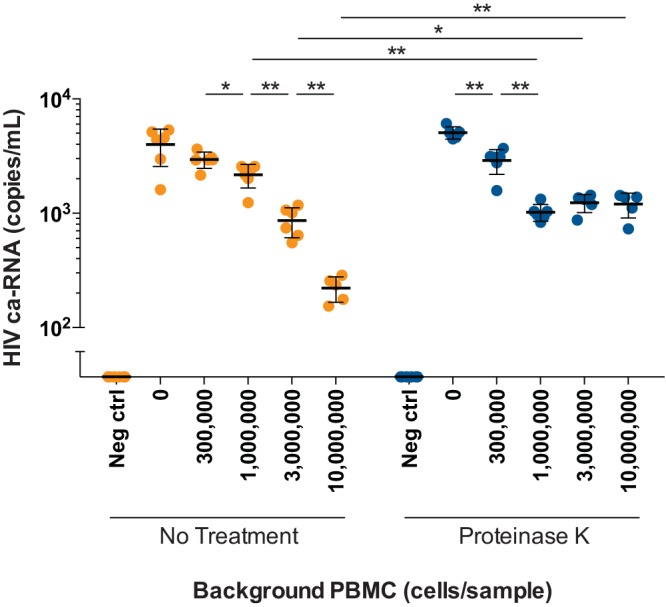
Cell-associated HIV RNA (ca-RNA) in PBMCs spiked with 8E5 cells. Cell-associated HIV RNA levels as measured by the Aptima assay in samples with different background concentrations of uninfected PBMCs (0, 0.3 × 106, 1 × 106, 3 × 106, and 10 × 106 cells/sample), each spiked with 10 8E5 cells. Samples pretreated with proteinase K, indicated by blue circles, were compared with untreated samples (labeled as “No Treatment”), indicated by orange circles. Black lines indicate means with error bars denoting 1 standard deviation. Mann-Whitney t tests were performed with significant results shown (P < 0.05 denoted by a single asterisk; P < 0.01 denoted by a double asterisk).
Seminal plasma specimens.
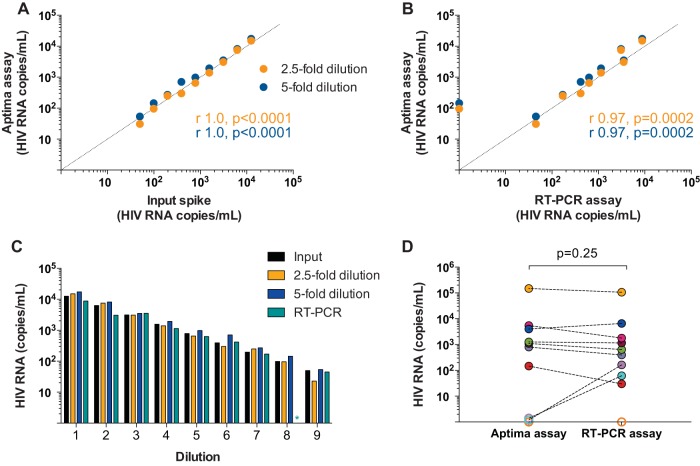
To assess the utility of the Aptima assay for HIV RNA quantitation in the complex composition of semen, we first evaluated assay performance by testing serial 2-fold dilutions of an HIV stock spiked into uninfected seminal plasma (n = 9; range, 50 to 12,500 copies/ml and 1 negative control), with parallel testing using an in-house real-time PCR (RT-PCR) assay. Aptima assay testing was performed in samples diluted 2.5-fold and 5-fold with specimen transfer medium (STM) and compared with previously validated RT-PCR (12) that was performed in an undiluted specimen. Aptima assay measurements of the two dilutions significantly correlated with each other (r = 1.0; P < 0.0001) and with input copy numbers (r = 1.0; P < 0.0001 for both 2.5-fold dilutions and 5-fold dilutions) (Fig. 2A). Aptima assay results also significantly correlated with RT-PCR assay results (r = 0.97; P = 0.0002 for both 2.5-fold and 5-fold dilutions) (Fig. 2B). Values from analysis of samples diluted 2.5-fold were comparable to known input copy numbers (P = 0.91), whereas values from samples diluted 5-fold slightly overestimated input copy numbers (P = 0.004) (Fig. 2C; see also Table S1 in the supplemental material). RT-PCR results were not significantly different from input copy numbers (P = 0.10), and the two were significantly correlated (r = 0.97; P = 0.0002). One negative control was run for each assay, and there were no false positives with either assay. Assay performance was next tested against RT-PCR in paired seminal samples from 10 HIV-infected donors that were not on ART with seminal plasma HIV RNA levels ranging from 0 to 106,408 copies/ml as measured by in-house RT-PCR (n = 10) (Fig. 2D). Samples run using the Aptima assay were diluted 2.5-fold with specimen transfer medium. Results of the two assays were significantly correlated (r = 0.92; P = 0.001). One sample estimated to have 61 copies/ml by RT-PCR was detected but below the quantitative threshold of the Aptima assay (<30 copies/ml), one sample with 163 copies/ml as measured by RT-PCR was undetectable with the Aptima assay, and a final sample had undetectable HIV RNA levels with both assays.
FIG 2.
HIV RNA in prepared seminal plasma spikes and in samples from HIV-infected donors. (A, B) Correlation between Aptima assay measurements and input copy numbers of a prequantified viral stock spiked into seminal plasma from an uninfected donor (A) and RT-PCR results from identical spiked samples (B). The Aptima assay was performed at dilutions of 2.5-fold and 5-fold (orange and blue circles, respectively). Dotted lines indicate an ideal correlation (y = x). Results of correlation analysis (Spearman r and P values) for 2.5-fold (orange text) and 5-fold (blue text) dilutions are shown. (C) Comparison of results of the Aptima (orange and blue bars for 2.5-fold and 5-fold dilutions, respectively) and RT-PCR assays (teal bars) in prepared seminal plasma spikes (input copies denoted by black bars). Teal asterisk denotes undetectable viral copy numbers with the RT-PCR assay. (D) Comparison of results of the Aptima and RT-PCR assays in samples from 10 HIV-infected donors; each donor is depicted with a distinct colored circle. Open circles denote samples with viral copy numbers below the assay limit of quantitation.
Cerebrospinal fluid specimens.
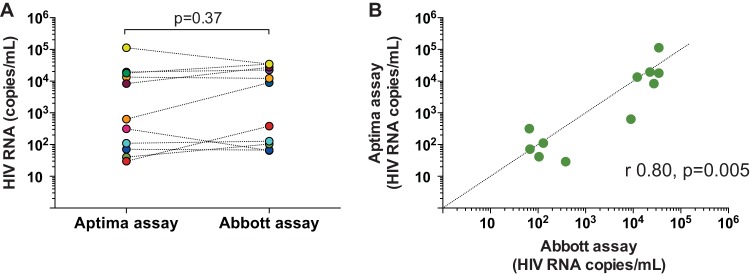
Performance of the Aptima assay was compared to that of the Abbott assay in 12 HIV-infected donors not on ART, with detectable CSF HIV RNA (ranging from 53 to 34,631 copies/ml), and 16 donors (from whom 24 samples were available) on suppressive ART and with undetectable CSF HIV RNA using the Abbott assay, which has a lower limit of quantitation of 40 copies/ml (total n = 36). One sample with 53 copies/ml detected by the Abbott assay had a copy number below the limit of quantitation of the Aptima assay (<30 copies/ml) in both replicates and was therefore excluded from the analysis. In samples from donors with detectable CSF HIV RNA with the Abbott assay, Aptima assay measurements gave comparable results (P = 0.37) (Fig. 3A; see also Table S2 in the supplemental material) that demonstrated significant correlation with the Abbott assay (r = 0.80; P = 0.005) (Fig. 3B). With samples from donors with undetectable CSF HIV RNA with the Abbott assay, all 12 samples tested singly were undetectable and 9 of 12 samples tested in multiple (9, 10) replicates were undetectable (all replicates undetectable) using the Aptima assay. For each of the 3 detectable samples, only 1 out of 10 replicates tested was detectable with copy numbers below the assay limit of quantitation.
FIG 3.
HIV RNA in CSF from HIV-infected donors. (A) Parallel testing of CSF samples from 12 HIV-infected donors with the Aptima (orange circles) and Abbott (blue circles) assays. The Aptima assay was performed in 2 replicates; means are denoted. Each donor is depicted with a distinct colored circle. P value calculated by Wilcoxon t test. (B) Correlation of Aptima and Abbott assays (Spearman r and P value). Dotted line denotes an ideal correlation (y = x).
Dried blood spot and dried plasma spot specimens.
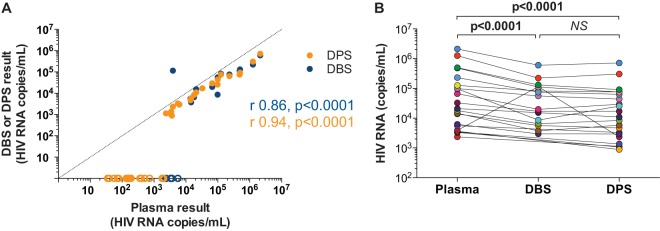
Dried whole blood or plasma on filter paper provides the opportunity to measure HIV RNA from subjects in resource limited settings where storing or processing fresh samples may not be feasible. Parallel testing of plasma samples, DBS and DPS from 104 HIV-infected participants was performed using the Aptima assay (DBS, n = 104; DPS, n = 104). Based on results from plasma testing, 40 participants had plasma HIV RNA ranging from 35 to 2,130,846 copies/ml, 40 had plasma HIV RNA that was detectable below the assay limit of quantitation (<30 copies/ml), and 24 had plasma HIV RNA that was undetectable. For the 40 participants with plasma HIV RNA above the limit of quantitation, DBS and DPS results correlated significantly with each other (r = 0.76; P = 0.001) and with plasma HIV RNA (r = 0.86, P < 0.0001 and r = 0.94, P < 0.0001, respectively) (Fig. 4A; see also Table S3 in the supplemental material). DBS had detectable HIV RNA in at least 1 of 5 replicates for participants with plasma HIV RNA levels of ≥35 copies/ml and in 5 of 5 replicates at plasma HIV RNA levels of ≥2,363 copies/ml. DPS had detectable HIV RNA in at least 1 of 5 replicates for participants with plasma HIV RNA levels of ≥394 copies/ml and in 5 of 5 replicates at plasma HIV RNA levels of ≥724 copies/ml. For samples with detectable HIV RNA in 5 of 5 replicates, DBS and DPS underestimated plasma HIV RNA by a median of 0.46 logs (P = 0.002) and 0.40 logs (P < 0.0001), respectively (Fig. 4B). Of the 40 participants with detectable plasma HIV RNA levels of <30 copies/ml, 4 were undetectable by DBS and 36 were undetectable by DPS. Of the 24 participants with undetectable plasma HIV RNA, 18 participants (75%) had positive DBS while no participants had positive DPS.
FIG 4.
HIV RNA measured in paired plasma samples, dried blood spots, and dried plasma spots. (A) HIV RNA as quantified in participants with detectable HIV RNA using the Aptima assay in paired plasma samples, DBS (blue circles), and DPS (orange circles) from 104 HIV-infected donors. Dotted line indicates an ideal correlation (y = x). Results of correlation analysis (Spearman r and P values) are shown for DBS (blue text) and DPS (orange text). Samples with undetectable HIV RNA in 1 or more replicates are assigned a conservative value of 0 copies/ml denoted by open circles. (B) Comparison of HIV RNA levels in samples with >1,000 copies/ml as measured in paired plasma (teal circles) against levels measured in DBS (blue circles) and DPS (orange circles). Undetectable DBS values are not shown; they are reported in the supplemental data (see Table S3 in the supplemental material).
DISCUSSION
The search for a cure for HIV infection has highlighted the need for increasingly sensitive and precise assays capable of detecting viral burden in various tissue and body fluid types. Useful assays for measuring HIV RNA in blood or plasma on filter paper will also facilitate patient management in resource-limited settings. Here, we describe the application of a standardized assay developed for blood plasma to the measurement of HIV-1 RNA in PBMCs, CSF, semen, and dried blood and plasma spots.
Several commercial assays have been approved by regulatory agencies for quantitation of HIV RNA in plasma. The new Aptima assay on the Panther system is FDA approved for HIV-1 RNA quantitation and CE approved for both diagnosis and monitoring of plasma HIV-1 RNA. Based on extensive replicate analysis of serially diluted HIV standards of diverse genotypes, the assay has a 95% limit of detection of 12 copies/ml and a lower limit of quantitation of 30 copies/ml when performed on single 0.5-ml plasma input volumes (11). Other investigators have reported good agreement between the Aptima assay and the RealTime HIV-1 (Abbott Molecular), COBAS TaqMan HIV-1 test v2.0 (Roche Molecular Diagnostics, Pleasanton, CA), NucliSens v2.0 (bioMérieux Diagnostics, Durham, NC), and Xpert HIV-1 (Cepheid International, Sunnyvale, CA) assays for measurement of plasma HIV RNA (13, 14). Here, we compared the performance of the Aptima assay with that of the commercially available Abbott RealTime HIV-1 assay in CSF samples from 12 HIV-infected participants. Our results demonstrated excellent agreement between the assays with linear association and significant correlation in samples across a wide range of viral loads, consistent with that which has been reported for plasma samples. We therefore suggest that the Aptima assay may be used to detect low levels of HIV RNA in CSF, which may in turn play a role in optimizing ART regimens in patients with CSF viral escape (15).
The measurement of HIV RNA in other clinical specimens, such as genital fluids and PBMCs, is more challenging due to a lack of commercial assays validated for specific use in these sample types and the presence of inhibitors in some of these fluids that prevents accurate quantitation using standard RT-PCR-based assays (16, 17). Most studies have used noncommercial assays for this purpose (12, 18), making comparisons between different studies and across different specimen types in compartmentalization studies problematic. The Aptima assay may be adapted for use in difficult sample types, such as those that are highly viscous or contain inhibitors, with the addition of an elution or dilution step with specimen transfer medium (STM; a detergent-based solution) followed by target capture, which concentrates HIV-RNA and removes potential inhibitors of TMA. Using this method significantly reduces reagents and processing time required for sample preparation compared with alternative methods, such as the Boom extraction technique (19). In this study, we applied this method to quantitation of HIV RNA in seminal plasma and cell-associated HIV RNA in PBMCs. For seminal plasma specimens, we demonstrated the accuracy of the Aptima assay in measurement of HIV RNA in a serial dilution of viral stock into seminal plasma as well as the comparability of the Aptima assay to an extensively validated in-house RT-PCR assay in samples from HIV-infected donors. For PBMC samples, signal diminished exponentially when background cell numbers increased over a certain threshold (approximately 0.3 × 106 cells per sample), suggesting the presence of an additional inhibitor that is not overcome by target capture and STM alone at elevated cell concentrations. We hypothesized that this drop in signal may be due to insufficient cell lysis, nucleic acid release from protein-bound states, or the presence of RNases leading to RNA degradation. We therefore tested the addition of proteinase K to overcome this inhibition. Treatment with proteinase K reduced signal loss at higher background concentrations of uninfected cells; however, proteinase K treatment was associated with significantly lower signal compared to untreated samples at low background concentrations of uninfected cells, possibly due to loss of target nucleic acids in the additional heating and washing steps. Quantitation of cell-associated HIV RNA with the Aptima assay will therefore require meticulous dilution of samples to a maximum concentration of 0.3 × 106 cells per aliquot to prevent oversaturating nucleic acid extraction techniques, which may limit sensitivity of detection of low levels of cell-associated RNA or require replicate testing to achieve higher sensitivity.
The use of advanced nucleic acid testing-based techniques in resource-limited settings is hindered by lack of equipment, such as centrifuges and cooling systems for sample processing and storage; financial constraints that limit access to reagents; and poor infrastructure that limits efficient transportation of precious temperature-sensitive samples. Dried fluid spots have therefore been proposed as a means for HIV RNA detection that bypass costly processing, storage, and transport requirements. Currently available assays that have been evaluated in the literature show a range of sensitivities, with lower limits of detection mostly reported in the 3 to 4 log10 copies/ml range (9, 20). Our findings demonstrate improved sensitivities using the Aptima assay, with 100% detection rates of DBS in participants with plasma HIV RNA levels of ≥35 copies/ml and 100% detection rates of DPS in participants with plasma HIV RNA levels of ≥394 copies/ml. In this study, both DBS and DPS yielded consistently lower values than those of conventional plasma assays. These differences may be attributable to imperfect nucleic acid extraction methods from the DBS and DPS cards, with additional variation introduced by the small volumes of fluid tested. Exceptions to the rule are DBS values at low plasma HIV RNA levels; results here and elsewhere suggest that DBS tend to overestimate viral load at low plasma HIV RNA levels, likely due to cell-associated HIV nucleic acids that are proportionally more apparent at these levels of viremia (9). DNA is not detected using TMA with the Aptima assay, but the assay may detect low copies of cell-associated HIV RNA. DPS showed a more consistent relationship with plasma viral load in our study, with no detection of HIV nucleic acid in DPS when plasma HIV RNA was undetectable, which further supports the proposal that false-positive results in DBS are most likely attributable to the presence of infected CD4 cells. Based on the above findings, we propose that the Aptima DBS and DPS assays may be useful in establishing diagnosis of acute HIV-1 infection when viral load exceeds several log10 copies per milliliter. The improved sensitivity and specificity of DPS may also be beneficial in treatment monitoring in resource-limited settings, in which a plasma HIV RNA cutoff of 1,000 copies/ml is often used to define treatment failure (21). However, the additional requirement of centrifugation for DPS testing may be a constraint. Further work will be necessary to improve DBS specificity for its application in viral load monitoring during ART.
The Aptima assay confers several advantages. (i) Nucleic acid extraction is performed using bead-based HIV RNA-specific target-specific capture technology, dispensing with the need for ultracentrifugation and extraction steps and efficiently removing potential inhibitors. (ii) Specificity of the assay for HIV RNA avoids spurious signal from HIV DNA. (iii) Adaptability of the assay to different tissue types permits the standardized measurement of HIV RNA across anatomic compartments. (iv) Automation of the assay using the Panther system permits high-throughput testing, which is crucial both in the setting of disease screening and in large cohort studies. (v) Multiple replicates may be extracted from one specimen, and the random access feature allows users to run up to 120 samples at any given time (13) enabling large numbers samples as well as replicates of plasma or other sample types to be tested for improved sensitivity and also enabling quantitation when sample is scarce. (vi) The assay is commercially available and has CE certification and FDA clearance for HIV-1 RNA quantitation.
This study has several limitations. We compared the HIV RNA-specific Aptima assay to several commercial and in-house RT-PCR-based assays, which amplify both RNA and DNA. To improve the comparability of assays, CSF and semen samples were processed by centrifugation and cell pellets were discarded to limit PCR contamination with HIV DNA species. Another limitation is that the comparator commercial assays are not validated for use in nonplasma specimens, limiting the strength of comparative studies. This further serves to emphasize the need for standardized, well-validated assays for diverse biologic specimen types.
The Aptima assay has been well validated for the detection of HIV-1 RNA in plasma specimens (11). Extending the application of the Aptima assay to other tissue specimens should prove useful in studies of the latent HIV reservoir and candidate interventions to eradicate this reservoir. The application to filter paper specimens containing dried fluid spots should provide a useful application for patient management in resource-limited settings. Future studies to further develop this technology will include the use of different methods to overcome inhibition in PBMCs and improve nucleic acid extraction in DBS; the application of the assay in other sample types, including saliva, breast milk, and cervical-vaginal lavage fluid; and the direct application of the uses demonstrated here to assays measuring inducible viral expression, such as the viral outgrowth assay.
MATERIALS AND METHODS
Study participants.
Study participants were recruited across two study sites (University of California San Diego [UCSD] and University of Washington). Each study site had a protocol that was approved by the local institution's institutional review board. Informed consent was obtained from study participants. All samples were tested after removal of specific subject identifiers.
The Aptima HIV-1 Quant Dx assay.
The Aptima HIV-1 Quant Dx assay (Hologic Incorporated, Bedford, MA), run on the fully automated Panther platform, uses specific target-capture transcription-mediated amplification (TMA) and real-time detection technologies to achieve high sensitivity and a broad dynamic range for HIV-1 detection and quantitation (11). In line with other commercial HIV-1 nucleic acid tests developed by Hologic Incorporated (formerly Gen-Probe, San Diego, CA) for HIV-1 diagnosis and blood screening, such as the Aptima HIV-1 RNA qualitative assay (19), the Procleix HIV-1/HCV assay (22, 23), and the Procleix Ultrio and Ultrio Elite (HIV-1&2/HCV/HBV) assays (24), this assay also targets highly conserved regions of HIV-1 polymerase (pol) and long terminal repeat (LTR). Target capture is mediated by oligonucleotides directed at conserved regions of the HIV-1 genome to enable specific pulldown of HIV RNA species (11). The multiple primers used in this product amplify HIV-1 groups M, N, O, and P along with an internal control. The primer design unique to TMA and the dual target approach ensure accurate detection and quantitation of all groups and subtypes of HIV-1. According to data in the FDA-cleared package insert, the lower limit of quantitation of the assay is 30 copies/ml and the 95% limit of detection is 12 copies/ml using a single analysis of 0.5 ml (11).
Peripheral blood mononuclear cells.
Whole blood (200 ml) from an HIV-seronegative donor was processed to obtain PBMCs by density gradient separation. A T-cell line (8E5) with each cell containing a single integrated but defective (replication-incompetent) copy of proviral HIV DNA (25) was used for spiking into uninfected PBMCs. The 8E5 cells have a defective reverse transcriptase, thus producing cell-associated and cell-free HIV RNA without the propagation of new infection. PBMCs and 8E5 were counted by flow cytometry (Accuri flow cytometer; BD Biosciences, San Jose, CA). Samples of 10 8E5 cells spiked into 0, 0.3 × 106, 1 × 106, 3 × 106, and 10 × 106 PBMCs were prepared in triplicate, lysed in 1 ml of specimen transfer medium (STM; Hologic Incorporated, Bedford, MA), and stored at −80°C. Cell-associated HIV RNA in PBMC specimens spiked with 8E5 (n = 72) was quantified in two different systems that used no proteinase K treatment (no treatment) or treatment with Aptima transfer solution proteinase K (Hologic Incorporated, Bedford, MA). Samples with no treatment were processed directly with the Aptima assay protocol. For proteinase K treatment, 300 μl Aptima transfer solution containing proteinase K was added to cell lysates and incubated at 95°C for 15 min before the Aptima assay was performed. The Aptima assay was performed in 6 replicates of 0.5 ml each under both conditions.
Seminal plasma.
Quantitation of HIV RNA in seminal plasma was performed using the following two sample types (total n = 20): (i) seminal plasma from an HIV-uninfected donor spiked with a quantified viral stock and (ii) seminal plasma from HIV-infected donors from the extensively characterized San Diego Primary Infection Cohort (26). Spiked seminal samples were prepared by adding a viral stock (NIH AIDS Research and Reference Reagent Program) into uninfected seminal plasma (BioreclamationIVT, Baltimore, MD). A serial 2-fold dilution was set up with sample HIV RNA concentration ranging from 12,500 copies/ml to 50 copies/ml. One sample of uninfected seminal plasma was used as a negative control. Spiked samples were then divided in two and stored at −80°C before testing. Parallel testing was performed with the Aptima assay and an extensively validated in-house real-time PCR (RT-PCR) assay (12). Upon thawing, samples were diluted 5-fold with STM to overcome inhibition of nucleic acid amplification (19) and 2.5-fold to test whether comparable assay sensitivity can be achieved at lower sample dilutions, which would enable testing of larger sample volumes. These dilutions were tested in 1 and 2 replicates of 0.5 ml, respectively. The RT-PCR assay was performed as previously described (12). Briefly, 0.5 ml of seminal samples were diluted 1:1 with phosphate-buffered saline (PBS) and ultracentrifuged (23,500 × g at 4°C for 1 h). HIV RNA was extracted using the High Pure viral RNA kit (Roche Molecular Diagnostics, Pleasanton, CA) according to the manufacturer's recommendations and quantified by reverse transcriptase RT-PCR using primers for HIV pol (mf302 and mf299).
Ten HIV-infected subjects not on ART donated semen samples. Seminal plasma was separated from seminal cells by centrifugation at 700 × g for 12 min within 4 h of collection and stored at −80°C. HIV RNA levels had been previously quantified in 0.5-ml seminal plasma samples using the in-house real-time PCR (RT-PCR) assay described above. For Aptima assay testing, a second replicate of seminal plasma (0.5 ml) from the same time point was diluted 1:2.5 with STM and processed as described above.
Cerebrospinal fluid.
Quantitation of HIV RNA in CSF was performed using samples from HIV-infected subjects selected from two distinct cohorts (total n = 36). First, Aptima assay performance was tested against that of the quantitative Abbott RealTime HIV-1 assay (Abbott Molecular, Des Plaines, IL) in 12 HIV-infected donors not on ART recruited at the UCSD HIV Neurobehavioral Research Center. These donors had undergone CSF testing with the Abbott assay and were known to have detectable CSF HIV RNA. Paired CSF samples collected at the same time point were used for Aptima assay testing. Briefly, CSF from lumbar punctures was collected, divided into batches of 5 ml, and centrifuged at 300 × g for 15 min. Supernatant was carefully isolated using a transfer pipette, leaving behind 50 μl of sample containing any pelleted cells. Cell-free CSF samples were divided into multiple replicates of 0.5 or 1 ml. All samples were stored at −80°C before testing. One milliliter of sample from each participant was tested with the Abbott assay. The Aptima assay was performed in duplicate using two replicates (0.5 ml each) from each donor. Mean copy number was used for analysis, except in cases when one of two replicates had a copy number below the limit of quantitation, in which case that replicate was excluded from the final analysis.
To measure low copy numbers in the CSF of well-suppressed subjects, assay performance was evaluated by testing 24 high-volume samples (40 ml) collected from 16 HIV-infected donors on suppressive ART enrolled in the San Diego Primary Infection Cohort known to have undetectable levels of CSF HIV RNA as measured in 1-ml samples with the Abbott assay (limited of detection, 40 copies/ml). Eight of the 16 donors were sampled at two time points. Samples were processed as above; for 12 samples, single replicates of 0.5 ml were tested, and in the remaining 12 samples, 9 to 10 replicates of 0.5 ml were tested.
Dried blood and plasma spots.
Matched sets of dried blood spots (DBS), dried plasma spots (DPS), and plasma were collected from 104 HIV-infected participants and prepared at the Center for AIDS Research (CFAR) at the University of Washington. Samples were from well-characterized participants representing a wide range of pre- and posttreatment HIV loads, determined based on historical results from individual participants (dried blood spots, n = 104; dried plasma spots, n = 104). DBS and DPS samples were prepared as follows. Whole blood samples were used to spot DBS cards before centrifugation. DBS cards were prepared by spotting at least 50 μl of fresh whole blood per spot onto a Whatman 903 protein saver card (VWR Scientific, West Chester, PA). Plasma was then separated from whole blood within 24 h of collection by centrifugation at 1,000 to 3,000 × g for 10 min. DPS cards were prepared by spotting at least 50 μl of plasma onto a Whatman 903 protein saver card. Each card had 5 spots for a total of 5 DBS replicates and 5 DPS replicates per individual donor. DBS and DPS cards were air-dried overnight and then placed into zip-lock bags containing desiccant packs. Specimens tested within 2 weeks were kept at room temperature; those tested later were stored at −20°C prior to testing. The remaining plasma volume after spotting ranged from 1.5 to 3.0 ml and was divided into tubes of approximately 1 ml each. Plasma samples were stored at ≤15°C until shipment. Aptima assay for plasma samples was performed in two replicates of 0.5 ml each. For DBS and DPS samples, spots were punched from the donor card (5 spots per card) and placed into tubes containing 1 ml of STM. Tubes were placed on a rocker platform and incubated at room temperature for 30 min. Following incubation, samples were directly loaded onto the Panther instrument for HIV RNA quantitation. Results where all five DBS or DPS replicates were unquantifiable were assigned a value of “target not detected.” Where all five replicates gave quantifiable results, the average of the 5 replicates was used. All other results were assigned a value of “<30 copies/ml detected.” A conversion factor of 36.4 was applied to DBS results and 20 to both DBS and DPS results to account for hematocrit and dilution, respectively.
Statistical methods.
Statistical analysis was performed with Prism version 6.0 (GraphPad Software, San Diego, CA). Comparisons between groups were performed using Wilcoxon and Mann-Whitney tests for paired and unpaired continuous variables, respectively. Correlations were examined using the Spearman test.
Supplementary Material
ACKNOWLEDGMENTS
We thank the study subjects for their time and participation in this study.
The expert technical support of Caroline Ignacio and Brianna Scott is gratefully acknowledged. We also thank our collaborators at the University of Washington for providing the DBS and DPS samples.
Hologic supplied reagents for RNA extraction from semen and PBMCs, and Aptima assays were performed at Hologic on their equipment by their employees. Hologic did not provide funding for personnel or work performed at UCSD.
This research was supported by the Collaboratory for AIDS Research on Eradication (CARE; U19 AI096113 and 1UM1AI126619), the BEAT-HIV Delaney Collaboratory (1UM1Al126620), the UCSD CFAR (AI306214), the amfAR grant 109316 with support from FAIR, the Department of Veterans Affairs, and the James B. Pendleton Charitable Trust. M.M. is supported by the BP-DGR AGAUR Postdoctoral Fellowship.
D.D.R. has consulted for Chimerix, Gilead, Antiva, Merck, and Monogram.
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JCM.00425-17.
REFERENCES
- 1.Walensky RP, Paltiel AD, Losina E, Mercincavage LM, Schackman BR, Sax PE, Weinstein MC, Freedberg KA. 2006. The survival benefits of AIDS treatment in the United States. J Infect Dis 194:11–19. doi: 10.1086/505147. [DOI] [PubMed] [Google Scholar]
- 2.Perelson AS, Essunger P, Cao Y, Vesanen M, Hurley A, Saksela K, Markowitz M, Ho DD. 1997. Decay characteristics of HIV-1-infected compartments during combination therapy. Nature 387:188–191. doi: 10.1038/387188a0. [DOI] [PubMed] [Google Scholar]
- 3.Finzi D, Blankson J, Siliciano JD, Margolick JB, Chadwick K, Pierson T, Smith K, Lisziewicz J, Lori F, Flexner C, Quinn TC, Chaisson RE, Rosenberg E, Walker B, Gange S, Gallant J, Siliciano RF. 1999. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat Med 5:512–517. doi: 10.1038/8394. [DOI] [PubMed] [Google Scholar]
- 4.Richman DD, Margolis DM, Delaney M, Greene WC, Hazuda D, Pomerantz RJ. 2009. The challenge of finding a cure for HIV infection. Science 323:1304–1307. doi: 10.1126/science.1165706. [DOI] [PubMed] [Google Scholar]
- 5.Eisele E, Siliciano RF. 2012. Redefining the viral reservoirs that prevent HIV-1 eradication. Immunity 37:377–388. doi: 10.1016/j.immuni.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Massanella M, Richman DD. 2016. Measuring the latent reservoir in vivo. J Clin Invest 126:464–472. doi: 10.1172/JCI80567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shao Y, Williamson C. 2012. The HIV-1 epidemic: low- to middle-income countries. Cold Spring Harb Perspect Med 2:a007187. doi: 10.1101/cshperspect.a007187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hamers RL, Smit PW, Stevens W, Schuurman R, Rinke de Wit TF. 2009. Dried fluid spots for HIV type-1 viral load and resistance genotyping: a systematic review. Antivir Ther 14:619–629. [PubMed] [Google Scholar]
- 9.Bertagnolio S, Parkin NT, Jordan M, Brooks J, Garcia-Lerma JG. 2010. Dried blood spots for HIV-1 drug resistance and viral load testing: a review of current knowledge and WHO efforts for global HIV drug resistance surveillance. AIDS Rev 12:195–208. [PubMed] [Google Scholar]
- 10.Kacian DL, Fultz TJ. March 1995. Nucleic acid sequence amplification methods.US patent 5399491.
- 11.Nair SV, Kim HC, Fortunko J, Foote T, Peling T, Tran C, Nugent CT, Joo S, Kang Y, Wilkins B, Lednovich K, Worlock A. 2016. Aptima HIV-1 Quant Dx–a fully automated assay for both diagnosis and quantification of HIV-1. J Clin Virol 77:46–54. doi: 10.1016/j.jcv.2016.02.002. [DOI] [PubMed] [Google Scholar]
- 12.Gianella S, Strain MC, Rought SE, Vargas MV, Little SJ, Richman DD, Spina CA, Smith DM. 2012. Associations between virologic and immunologic dynamics in blood and in the male genital tract. J Virol 86:1307–1315. doi: 10.1128/JVI.06077-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mor O, Gozlan Y, Wax M, Mileguir F, Rakovsky A, Noy B, Mendelson E, Levy I. 2015. Evaluation of the RealTime HIV-1, Xpert HIV-1, and Aptima HIV-1 Quant Dx assays in comparison to the NucliSens EasyQ HIV-1 v20 assay for quantification of HIV-1 viral load. J Clin Microbiol 53:3458–3465. doi: 10.1128/JCM.01806-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schalasta G, Borner A, Speicher A, Enders M. 2016. Comparative evaluation of the Aptima HIV-1 Quant Dx assay and COBAS TaqMan HIV-1 v2.0 assay using the Roche High Pure system for the quantification of HIV-1 RNA in plasma. Clin Chem Lab Med 54:493–499. doi: 10.1515/cclm-2015-0522. [DOI] [PubMed] [Google Scholar]
- 15.Canestri A, Lescure FX, Jaureguiberry S, Moulignier A, Amiel C, Marcelin AG, Peytavin G, Tubiana R, Pialoux G, Katlama C. 2010. Discordance between cerebral spinal fluid and plasma HIV replication in patients with neurological symptoms who are receiving suppressive antiretroviral therapy. Clin Infect Dis 50:773–778. doi: 10.1086/650538. [DOI] [PubMed] [Google Scholar]
- 16.Dyer JR, Gilliam BL, Eron JJ Jr, Grosso L, Cohen MS, Fiscus SA. 1996. Quantitation of human immunodeficiency virus type 1 RNA in cell free seminal plasma: comparison of NASBA with Amplicor reverse transcription-PCR amplification and correlation with quantitative culture. J Virol Methods 60:161–170. doi: 10.1016/0166-0934(96)02063-0. [DOI] [PubMed] [Google Scholar]
- 17.Shepard RN, Schock J, Robertson K, Shugars DC, Dyer J, Vernazza P, Hall C, Cohen MS, Fiscus SA. 2000. Quantitation of human immunodeficiency virus type 1 RNA in different biological compartments. J Clin Microbiol 38:1414–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cillo AR, Sobolewski MD, Bosch RJ, Fyne E, Piatak M Jr, Coffin JM, Mellors JW. 2014. Quantification of HIV-1 latency reversal in resting CD4+ T cells from patients on suppressive antiretroviral therapy. Proc Natl Acad Sci U S A 111:7078–7083. doi: 10.1073/pnas.1402873111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nugent CT, Dockter J, Bernardin F, Hecht R, Smith D, Delwart E, Pilcher C, Richman D, Busch M, Giachetti C. 2009. Detection of HIV-1 in alternative specimen types using the APTIMA HIV-1 RNA qualitative assay. J Virol Methods 159:10–14. doi: 10.1016/j.jviromet.2009.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smit PW, Sollis KA, Fiscus S, Ford N, Vitoria M, Essajee S, Barnett D, Cheng B, Crowe SM, Denny T, Landay A, Stevens W, Habiyambere V, Perriens JH, Peeling RW. 2014. Systematic review of the use of dried blood spots for monitoring HIV viral load and for early infant diagnosis. PLoS One 9:e86461. doi: 10.1371/journal.pone.0086461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Phillips A, Shroufi A, Vojnov L, Cohn J, Roberts T, Ellman T, Bonner K, Rousseau C, Garnett G, Cambiano V, Nakagawa F, Ford D, Bansi-Matharu L, Miners A, Lundgren JD, Eaton JW, Parkes-Ratanshi R, Katz Z, Maman D, Ford N, Vitoria M, Doherty M, Dowdy D, Nichols B, Murtagh M, Wareham M, Palamountain KM, Chakanyuka Musanhu C, Stevens W, Katzenstein D, Ciaranello A, Barnabas R, Braithwaite RS, Bendavid E, Nathoo KJ, van de Vijver D, Wilson DP, Holmes C, Bershteyn A, Walker S, Raizes E, Jani I, Nelson LJ, Peeling R, Terris-Prestholt F, Murungu J, Mutasa-Apollo T, Hallett TB, Revill P. 2015. Sustainable HIV treatment in Africa through viral-load-informed differentiated care. Nature 528:S68–S76. doi: 10.1038/nature16046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jackson JB, Smith K, Knott C, Korpela A, Simmons A, Piwowar-Manning E, McDonough S, Mimms L, Vargo JM. 2002. Sensitivity of the Procleix HIV-1/HCV assay for detection of human immunodeficiency virus type 1 and hepatitis C virus RNA in a high-risk population. J Clin Microbiol 40:2387–2391. doi: 10.1128/JCM.40.7.2387-2391.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Giachetti C, Linnen JM, Kolk DP, Dockter J, Gillotte-Taylor K, Park M, Ho-Sing-Loy M, McCormick MK, Mimms LT, McDonough SH. 2002. Highly sensitive multiplex assay for detection of human immunodeficiency virus type 1 and hepatitis C virus RNA. J Clin Microbiol 40:2408–2419. doi: 10.1128/JCM.40.7.2408-2419.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koppelman MH, Assal A, Chudy M, Torres P, de Villaescusa RG, Reesink HW, Lelie PN, Cuypers HT. 2005. Multicenter performance evaluation of a transcription-mediated amplification assay for screening of human immunodeficiency virus-1 RNA, hepatitis C virus RNA, and hepatitis B virus DNA in blood donations. Transfusion 45:1258–1266. doi: 10.1111/j.1537-2995.2005.00197.x. [DOI] [PubMed] [Google Scholar]
- 25.Rodriguez-Alfageme C, Chen Z, Sonoda G, Testa JR, Damiani RD, Astrin SM. 1998. B cells malignantly transformed by human immunodeficiency virus are polyclonal. Virology 252:34–38. doi: 10.1006/viro.1998.9445. [DOI] [PubMed] [Google Scholar]
- 26.Little SJ, Frost SD, Wong JK, Smith DM, Pond SL, Ignacio CC, Parkin NT, Petropoulos CJ, Richman DD. 2008. Persistence of transmitted drug resistance among subjects with primary human immunodeficiency virus infection. J Virol 82:5510–5518. doi: 10.1128/JVI.02579-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.