Abstract
While treatments for childhood osteosarcoma have improved, the overall survival for this common type of bone cancer has not changed for three decades, and thus, new targets for therapeutic development are needed. To identify tumor‐related proteins in osteosarcoma, we used isobaric tags in a relative and absolute quantitation proteomic approach to analyze the differentially expressed proteins between osteosarcoma cells and human osteoblastic cells. Through clinical screening and functional evaluation, CCR4–NOT transcription complex subunit 1 (CNOT1) correlated with the growth of osteosarcoma cells. To date, the mechanisms and regulatory roles of CNOT1 in tumors, including osteosarcoma, remain largely elusive. Here, we present evidence that knockdown of CNOT1 inhibits the growth of osteosarcoma in vitro and in vivo. Mechanistically, we observed that CNOT1 interacted with LMNA (lamin A) and functioned as a positive regulator of this intermediate filament protein. The RNA‐seq analysis revealed that CNOT1 depletion inhibited the Hedgehog signaling pathway in osteosarcoma cells. A rescue study showed that the decreased growth of osteosarcoma cells and inhibition of the Hedgehog signaling pathway by CNOT1 depletion were reversed by LMNA overexpression, indicating that the activity of CNOT1 was LMNA dependent. Notably, the CNOT1 expression was significantly associated with tumor recurrence, Enneking stage, and poor survival in patients with osteosarcoma. Examination of clinical samples confirmed that CNOT1 expression positively correlated with LMNA protein expression. Taken together, these results suggest that the CNOT1–LMNA–Hedgehog signaling pathway axis exerts an oncogenic role in osteosarcoma progression, which could be a potential target for gene therapy.
Keywords: CNOT1, Hedgehog signaling pathway, LMNA, osteosarcoma, proliferation
Abbreviations
- CCK‐8
Cell Counting Kit‐8
- CHX
cycloheximide
- CNOT1
CCR4–NOT transcription complex subunit 1
- Co‐IP
co‐immunoprecipitation
- GO
gene ontology
- GSEA
Gene Set Enrichment Analysis
- IHC
immunohistochemistry
- iTRAQ
isobaric tags for relative and absolute quantification
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- LMNA
lamin A
- OS
overall survival
- TFS
tumor‐free survival
1. Introduction
Osteosarcoma is the most common primary malignant bone tumor in children and adolescents. In the past, amputation was the standard treatment, with a 5‐year survival rate of 20% (Berlanga et al., 2016; Coventry and Dahlin, 1957). With the application of neoadjuvant chemotherapy combined with limb salvage surgery, the 5‐year survival rate has risen to 70% (Sampo et al., 2008). However, overall survival (OS) has remained unchanged for over three decades despite attempts to improve outcomes via modification of chemotherapy drugs (Yan et al., 2016). Currently, many patients are not sensitive to chemotherapy and have a poor prognosis (Yao et al., 2012). Therefore, there is an urgent need to discover novel biomarkers and therapeutic targets for the diagnosis and treatment of osteosarcoma.
Quantitative proteomics provides an approach for the rapid identification of new protein targets and helps to elucidate the underlying molecular events associated with the development and progression of cancer (Parker and Borchers, 2014; Ren et al., 2010). Quantitative proteomic analysis has been reported in various tumors, including oral squamous cell carcinoma (Jou et al., 2014), colon cancer (Li et al., 2013), and gastric carcinoma (Subbannayya et al., 2015). Currently, osteosarcoma cell lines as osteoblastic cell models have been extensively used to investigate bone biology (Bernardini et al., 2014). Previously, a comparative proteomic analysis of plasma membrane proteins was performed between human osteosarcoma cells and osteoblastic cells (Zhang et al., 2010). A total of 342 proteins were identified in that study, 68 of which were differentially expressed and one protein CD151 was verified by immunohistochemistry. In addition, proteomic studies have been carried out between osteosarcoma tissues and normal tissues to identify a group of proteins potentially involved in osteosarcoma malignancy (Folio et al., 2009; Kubota et al., 2013; Li et al., 2010).
In the present study, comparative proteomic analysis was performed between osteosarcoma cell lines and an osteoblastic cell line. Through clinical analysis and functional screening, CNOT1, a member of the human CCR4–NOT deadenylase complex, was found to be related to osteosarcoma proliferation. Accumulating evidence suggests that the CCR4–NOT complex is involved in the regulation of mRNA deadenylation, mRNA decay, and cell viability (Ito et al., 2011). This study demonstrated that knockdown of CNOT1 inhibited osteosarcoma cell growth in vitro and in vivo. In addition, CNOT1 was found to interact with lamin A (LMNA) and affect its protein stability in osteosarcoma. Knockdown of CNOT1 could inhibit Hedgehog signaling pathway. Activation of the Hedgehog signaling pathway has been implicated in the development of many human malignancies, including prostate cancer (Xu et al., 2014), ovarian cancer (Li et al., 2016), and breast cancer (Zhou et al., 2016). Overexpression of LMNA could rescue CNOT1 knockdown‐mediated tumor inhibition and Hedgehog signaling pathway inhibition. Notably, we found that CNOT1 was an independent prognostic factor for tumor‐free survival (TFS) and OS in patients with osteosarcoma. A positive correlation between CNOT1 and LMNA expression was detected in osteosarcoma tissues. Taken together, knockdown of CNOT1 inhibited osteosarcoma growth through the Hedgehog signaling pathway via its association with LMNA, suggesting a candidate orientation for osteosarcoma treatment.
2. Materials and methods
2.1. Cell lines and cell culture
Three osteosarcoma cell lines (MNNG/HOS, MG63, and U2OS) and one human osteoblastic cell line (hFOB 1.19) were used. The cells were maintained at 37 °C in a humidified air atmosphere containing 5% CO2. They were cultured in Dulbecco's modified Eagle's medium (MNNG/HOS and MG63) or RPMI‐1640 medium (U2OS) supplemented with 10% fetal bovine serum (Biowest, South America Origin), 100 U·mL−1 penicillin (Sigma‐Aldrich, St Louis, MO, USA), and 100 mg·mL−1 streptomycin (Sigma‐Aldrich). The hFOB 1.19 cell line was cultured according to ATCC protocols.
2.2. Proteomic analysis
The cells were cultured to 80% confluence and collected. The whole‐cell protein was extracted. The proteomic analysis was performed as described previously (Lin et al., 2013). Briefly, 100 μg of the hFOB 1.19, MNNG/HOS, MG63, or U2OS cell lysate was labeled with iTRAQ labeling reagents 114, 115, 116, or 117 (Applied Biosystems, Foster City, CA, USA). After strong cation exchange and NanoLC‐MS/MS analysis, protein identification and iTRAQ quantitation were performed using proteinpilot4.1 software (AB SCIEX, Toronto, OH, USA). Two physical replicates were performed. Figure 1A shows the flow chart of the proteomic analysis. The differentially expressed proteins underwent gene ontology (GO) analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis (Lin et al., 2013).
Figure 1.
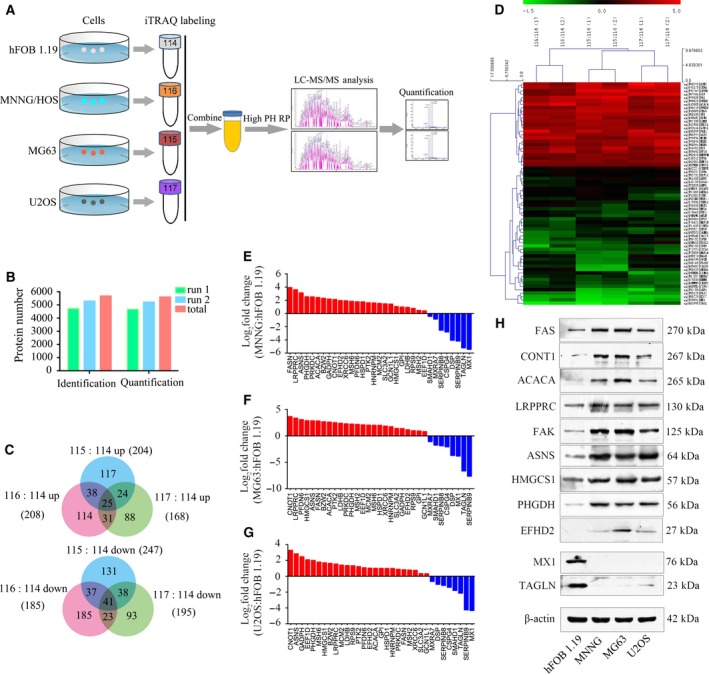
Experimental workflow and results of the quantitative proteomic analysis. (A) The flow chart of the proteomic analysis. (B) The number of identified and quantified proteins in run 1 and run 2. (C) The number of up‐regulated and down‐regulated proteins in osteosarcoma cells. (D) A heat map of dysregulated proteins in osteosarcoma. (E–G) The mRNA expression levels of 25 up‐regulated and eight down‐regulated proteins in osteosarcoma cells compared with hFOB 1.19 cells were verified by qRT‐PCR. (H) Representative blots display the protein expression of nine up‐regulated and two down‐regulated proteins. β‐Actin was used as an internal control.
2.3. RNA isolation and quantitative Real‐time PCR assays
Total RNA from human tissue samples and cultured cells was extracted with TRIzol reagent (Invitrogen, Carlsbad, CA, USA) and quantified with a Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) following the manufacturer's protocol. First‐strand cDNA was synthesized with a PrimeScript RT Reagent Kit (TaKaRa, Shiga, Japan). The cDNA templates were combined with SYBR Green premix with RoxII (Takara, Dalian, Liaoning, China) to perform quantitative PCR reactions. All reactions were performed in a 10‐μL reaction volume in triplicate. The expression level of genes was measured using the comparative Ct method (Yang et al., 2016). Information on the primer sequences used is presented in Table S2.
2.4. Western blot analysis
Equal amounts of protein samples extracted from the harvested cells were separated using 6% or 8% SDS/PAGE and were then transferred to NC membranes (Millipore, Billerica, MA, USA). After blocking in 5% (w/v) nonfat milk, the membranes were incubated with primary antibodies overnight. Information on the primary antibodies used is presented in Table S3. The secondary antibody was anti‐rabbit IgG (1 : 5000; Sigma‐Aldrich) or anti‐mouse IgG (1 : 5000; Sigma‐Aldrich). Subsequent visualization was performed with SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Fisher Scientific).
2.5. Oligonucleotide transfection and stable cell line generation
Predesigned siRNA (Ribobio, Guangzhou, China) and plasmids (Public Protein/Plasmid Library, Nanjing, China) were transfected into cells using Lipofectamine 2000 Reagent (Invitrogen) following the manufacturer's protocol. To construct a stable cell line, scramble control or CNOT1‐specific shRNA particles (Biotend, Shanghai, China) were infected into osteosarcoma cells according to the manufacturer's protocol.
2.6. Cell proliferation assays and cell cycle analysis
2.6.1. Cell proliferation
The cell proliferation assay was performed using a Cell Counting Kit‐8 (CCK‐8; Dojindo, Mashikimachi, Japan). Briefly, a mixture of 10 μL of CCK‐8 and 100 μL of DMEM was added to each well followed by incubation for 2 h. The absorbance at 450 nm was then measured. Each measurement was taken in triplicate, and the experiments were repeated twice.
2.6.2. Cell cycle
The cell cycle was analyzed using a Cell Cycle Detection Kit (Kaiji, Nanjing, China) and a FACSCalibur flow cytometer (BD Biosciences, San Jose, CA, USA). The results were analyzed with modfit software (BD Biosciences). The assays were independently conducted three times.
2.7. Colony forming assay
Forty‐eight hours after siRNA transfection, 1 × 103 MNNG/HOS, MG63, or U2OS cells were seeded in six‐well plates. After 10 d, cells in each well were fixed with 100% methanol for 30 min and stained with 0.1% crystal violet for 30 min. Finally, the cell colonies were counted. The assays were independently conducted three times.
2.8. Migration and invasion assays
Cell migration and invasion assays were performed in a 24‐well plate with 8‐mm pore size chamber inserts (Corning, New York, NY, USA). For the migration assays, 48 h after siRNA transfection, 5 × 104 cells per well were placed in the upper chamber on an uncoated membrane. For the invasion assays, 1 × 105 cells per well were placed in the upper chamber on a Matrigel‐coated membrane. In both assays, cells were diluted with serum‐free culture medium. The lower chambers contained 800 μL of medium with 10% FBS. The cells were incubated for 12 and 16 h for the migration and invasion assays, respectively. Then, the cells that had migrated through the membrane inserts were stained and counted. The assays were independently conducted three times.
2.9. Animal experiments
All animal experiments were approved by the Ethics Committee of the Shanghai Jiao Tong University Affiliated Sixth People's Hospital (YS‐2016‐064, 24 February 2016). For tumor growth assays, MNNG/HOS cells stably expressing sh‐control or sh‐CNOT1 were injected subcutaneously into the left scapulas of nude mice (6‐week‐old BALB/c‐nu/nu, 10 per group, 2 × 106 cells per mouse). The tumor volume was monitored twice a week and was calculated using the formula V = 1/2 × length × width2. After four weeks, the mice were euthanized. For histological analysis, the primary tumors were harvested at necropsy and fixed in 10% formalin.
2.10. Co‐immunoprecipitation (Co‐IP) and mass spectrometry protein identification
Co‐IP was performed in MNNG, MG63, and U2OS cells. Equal amounts of protein (3000 μg) were incubated with antibodies at 4 °C overnight, and the mixture was incubated with protein A/G magnetic beads at 4 °C for 3 h. The beads were washed using phosphate‐buffered saline containing 1‰ Triton X‐100 and eluted using 2× protein loading buffer at 100 °C for 10 min. For silver staining, IgG‐bound or CNOT1‐bound proteins were resolved by SDS/PAGE and stained with a silver staining kit (Beyotime Biotechnology, Shanghai, China). For mass spectrometry analysis, CNOT1‐bound proteins were resolved by SDS/PAGE and stained with Coomassie Blue R250 (Solarbio, Beijing, China). After destaining, reduction, and trypsin digestion for 12 h, the peptides were extracted using acetonitrile. The peptides were analyzed using a NanoLC system (NanoLC‐2D Ultra; Eksigent, Dublin, CA, USA) equipped with a Triple TOF 5600 mass spectrometer (AB SCIEX). Protein identification was performed with proteinpilot4.1 software (AB SCIEX). For this study, a strict unused confidence cutoff > 1.3 and peptides ≥ 2 were used for protein identification.
2.11. Confocal immunofluorescence
Confocal immunofluorescence microscopy was performed on MNNG, MG63, and U2OS cells. Briefly, cells were fixed and incubated with rabbit polyclonal anti‐CNOT1 antibody and mouse monoclonal anti‐LMNA antibody at 4 °C overnight and then incubated with secondary antibodies (Invitrogen) for 1 h. Finally, the cells were incubated with DAPI (Sigma‐Aldrich) for 5 min and viewed with a Fluoview FV1000 microscope (Olympus, Tokyo, Japan).
2.12. Clinical samples and immunohistochemistry (IHC)
Twenty human tissue sample pairs in which each pair consisted of an osteosarcoma sample and a corresponding nontumor tissue sample were obtained at Shanghai Sixth People's Hospital. Total RNA was extracted for clinical screening. The tissue microarray contains a total of 151 patients who had been diagnosed with osteosarcoma at the Shanghai Sixth People's Hospital. All osteosarcoma samples were confirmed by postoperative pathology. They received primary surgical treatment and preoperative and postoperative neoadjuvant therapy. The median age of the patients was 20 years old (range: 6–84 years). The follow‐up period ranged from 24 to 68 months, and the median time was 40 months. Ethics approval was obtained from the local hospital ethic committees, and written informed consent was obtained from each patient before sample collection. A standard IHC staining procedure was followed. Paraffin‐embedded sections were cut at a 4 μm thickness and then dewaxed in xylene and treated with microwave heating at 60 °C for 20 min in an EDTA buffer (pH 9.0) for antigen retrieval. Each slide was blocked for endogenous peroxidase activity by incubation in 0.3% H2O2 for 10 min and then incubated at 37 °C with a 1 : 100 dilution of primary antibodies against CNOT1 (Proteintech, Wuhan, China), LMNA (1 : 200; Abcam, Cambridge, MA, USA), or Ki‐67 (1 : 200; Dako, Glostrup, Denmark). Slides were then rinsed three times in PBS, incubated for 30 min with an EnVision staining kit (Dako), washed three times in PBS, and stained for 3–10 min in a moist chamber at room temperature using DAB. Slides were counterstained with hematoxylin and dehydrated in a graded ethyl alcohol series (70%, 90%, 100%). The primary antibody was substituted with PBS in sections used as negative controls. Assessment of IHC staining was made independently by two expert pathologists. Any discordance was solved through consensus discussion. The IHC signal intensities were scored as negative, weak, positive, or strong. For Ki‐67, the percentage of Ki‐67‐positive cells was calculated.
2.13. Statistical evaluation
The data were compiled and analyzed using spss version 21.0 (Chicago, IL, USA). Comparisons between different groups were made using chi‐square tests. The independent prognostic significance of the parameters was estimated using the Cox proportional hazards model. The correlation analysis between CNOT1 and LMNA was performed by the Spearman correlation test. Survival curves were generated by the Kaplan–Meier method (Bell et al., 2016). The TFS time is the period from surgery to the presence of new local lesions. The OS time was defined as the length of time between the surgery and death. A significant result was considered at P < 0.05.
3. Results
3.1. Comparative proteomic results of osteoblastic cells versus osteosarcoma cells
In the present study, iTRAQ combined with NanoLC‐MS/MS analysis was used to determine differentially expressed proteins between osteoblastic cells (hFOB 1.19) and osteosarcoma cells (MNNG/HOS, MG63, and U2OS). After filtering for an unused protein score > 1.3 and peptide number ≥ 2, 4719 of 5290 proteins were identified and 4663 of 5218 proteins were quantified in the first and second runs, respectively. In total, we identified 5677 proteins and quantified 5601 proteins in the two replicates (Fig. 1B). Twenty‐five up‐regulated proteins and 41 down‐regulated proteins were identified and quantified in all osteosarcoma cell lines compared to the osteoblastic cell line (Fig. 1C,D). Table S1 lists the proteins that were up‐regulated and down‐regulated in osteosarcoma cells. GO analysis and KEGG pathway analysis were performed on the differentially expressed proteins (Fig. S1). Twenty‐five up‐regulated proteins and eight down‐regulated proteins were selected to further validate the proteomic results. The mRNA expression levels of these proteins observed in qRT‐PCR were consistent with the proteomic analysis (Fig. 1E–G). Then, nine up‐regulated proteins – FAS, CNOT1, ACACA, LRPPRC, FAK, ASNS, HMGCS1, PHGDH, and EFHD2 – and two down‐regulated proteins – MX1 and TAGLN – were chosen for protein expression verification. The expression levels of these proteins were consistent with the proteomic analysis (Fig. 1H). These results collectively support the feasibility of our approach for identifying osteosarcoma‐associated proteins.
3.2. The role of CNOT1 in osteosarcoma cells in vitro
Clinical analysis of nine up‐regulated proteins was conducted in 20 pairs of human tissue samples; each pair comprised an osteosarcoma sample and a corresponding nontumor tissue sample. The results revealed that the mRNA expression of CNOT1, ASNS, or EFHD2 was higher in osteosarcoma tissues than in the corresponding nontumor tissues (Fig. S2), whereas no difference was found for the six other proteins. siRNA knockdown and CCK‐8 assay were used for functional screening. The results showed that knockdown of CNOT1 dramatically inhibited osteosarcoma cell proliferation in MNNG/HOS cells (Fig. S3). These results showed that CNOT1 played an important role in the tumorigenesis of osteosarcoma and deserved further research. To explore the functional significance of CNOT1 in osteosarcoma, cell proliferation analysis, cell cycle assay, and colony forming assay were performed. CNOT1 knockdown was validated using qRT‐PCR and western blotting. The mRNA and protein expression of CNOT1 significantly decreased after transfection with CNOT1‐specific siRNA in osteosarcoma cells (Fig. 2A–C). CCK‐8 assay revealed that knockdown of CNOT1 significantly inhibited the proliferation of osteosarcoma cells (Fig. 2D–F). The cell cycle assay showed that knockdown of CNOT1 delayed cell cycle progression by inhibiting S phase transition (Fig. 2G–I). Similarly, colony forming assay showed that knockdown of CNOT1 attenuated cell colony formation (Fig. 2J–L). However, the migration and invasion assays showed that CNOT1 knockdown did not influence the migration or invasion of osteosarcoma, as shown in Fig. S4. Thus, these results indicate that CNOT1 has an oncogenic role in osteosarcoma.
Figure 2.
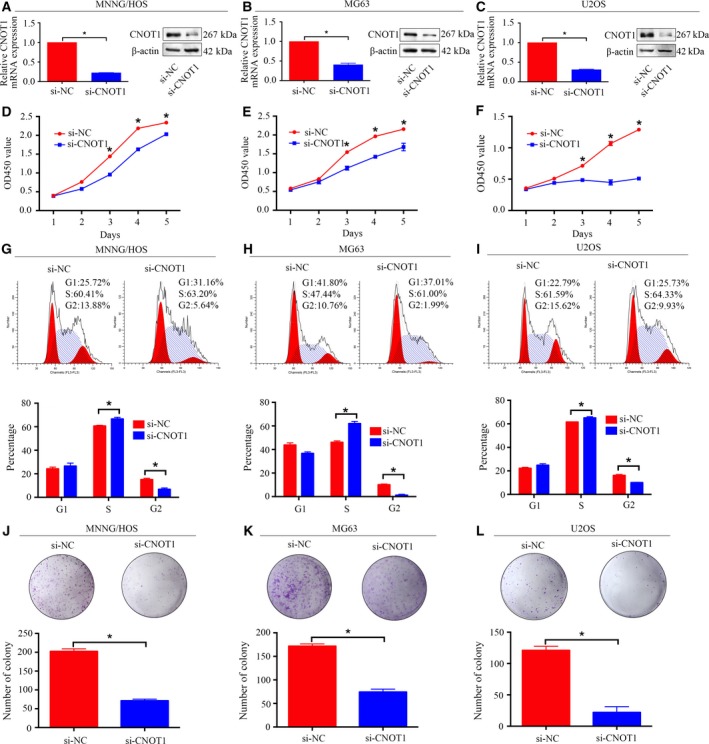
Knockdown of CNOT1 inhibits osteosarcoma cell proliferation in vitro. (A–C) The mRNA and protein expression levels were validated after si‐CNOT1 and si‐NC transfection by qRT‐PCR and western blotting in MNNG/HOS, MG63, and U2OS cells. (D–F) CCK‐8 assay was performed after siRNA transfection. (G–I) Cell cycle assays were performed for CNOT1‐silenced osteosarcoma cells and control cells. (J–L) colony forming assay for CNOT1‐silenced osteosarcoma cells and control cells. Data are representative of the results from three independent experiments. *P < 0.05.
3.3. Knockdown of CNOT1 inhibits tumor growth in vivo
To further determine the effect of CNOT1 on osteosarcoma growth in vivo, MNNG/HOS cells stably expressing sh‐control or sh‐CNOT1 were established. Protein expression was validated by western blotting (Fig. 3A). Then, the cells were subcutaneously injected into the left scapulas of nude mice, and the animals were closely monitored for tumor growth for 4 weeks. The tumor growth curve demonstrated that knockdown of CNOT1 significantly inhibited tumor growth in vivo. The tumor volume and weight were smaller in the CNOT1‐knockdown group than in the scramble group (Fig. 3B–D). The immunohistochemical analysis results shown in Fig. 3E and 3F indicated that the staining of the Ki‐67, a tissue proliferation marker, was lower in the sh‐CNOT1 tumors than in the sh‐control tumors (Fig. 3E,F). Taken together, these results indicated that CNOT1 knockdown hindered the tumorigenesis of osteosarcoma cells in vivo.
Figure 3.
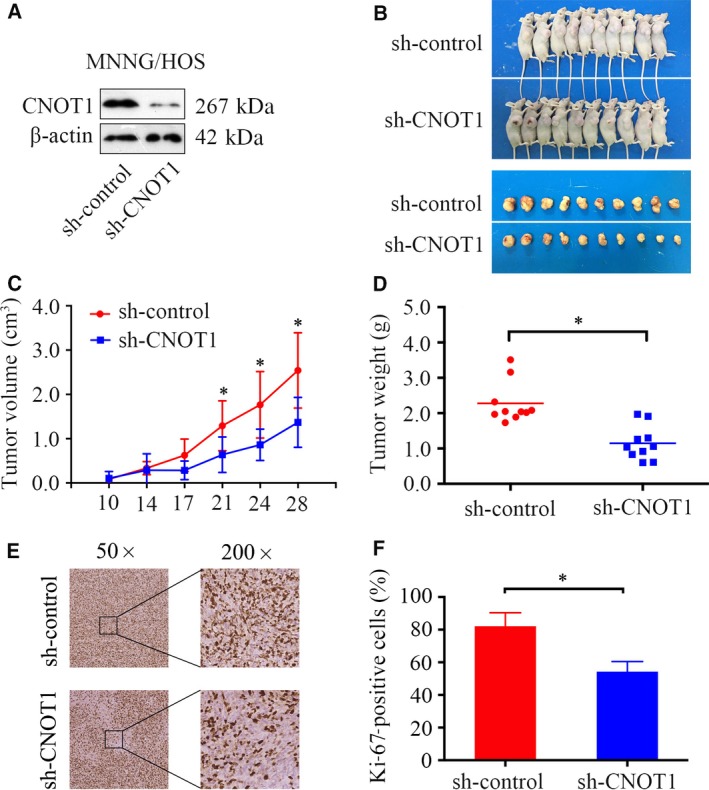
Knockdown of CNOT1 inhibits osteosarcoma cells growth in vivo. (A) Representative blots display the protein expression of CNOT1 in MNNG/HOS cells stably expressing sh‐control or sh‐CNOT1. β‐Actin was used as an internal control. (B) The upper diagram shows a photograph of tumor‐bearing mice, and the lower shows a photograph of tumors when the mice were euthanized. (C) Growth curve drawn by measuring tumor volumes on the indicated days. Error bars represent the SEM. (D) Diagram showing the tumor weights in the sh‐control group and sh‐CNOT1 group. (E,F) Representative images of Ki‐67 staining in the sh‐control group and sh‐CNOT1 group. Magnification, ×50, ×200.
3.4. CNOT1 interacts with LMNA in osteosarcoma
In order to determine the underlying mechanism of CNOT1 in osteosarcoma, Co‐IP followed by mass spectrometry identification was performed in MNNG/HOS cells. The silver staining of SDS/PAGE is shown in Fig. 4A. The identified proteins by mass spectrometry analysis (unused score > 1.3; peptides ≥ 2) are shown in Table S4. To assess the interaction between CNOT1 and other proteins, we pulled down CNOT1. The results showed that CNOT1 specifically interacted with LMNA in osteosarcoma cells as indicated by the Co‐IP assay (Fig. 4B). In addition, we also pulled down LMNA and probed with CNOT1 and found that LMNA interacted with CNOT1 (Fig. 4C). Similarly, we also confirmed the colocalization of LMNA and CNOT1 in osteosarcoma cells by confocal analysis (Fig. 4D). Thus, CNOT1 associates with LMNA in osteosarcoma cells. Next, we determined whether the mRNA and protein levels of LMNA were affected by CNOT1. Knockdown of CNOT1 in MNNG/HOS cells did not alter LMNA mRNA expression but led to a significant decrease in LMNA protein expression (Fig. 4E,F). To further clarify whether CNOT1 affects LMNA protein stability, we measured the LMNA protein half‐lives using cycloheximide (CHX) chase assay. The CHX chase assay results showed that knockdown of CNOT1 significantly decreased the LMNA protein half‐life (Fig. 4G,H). Then the expressions of CNOT1 and LMNA in the tumor tissues of tumor‐bearing mice were detected by IHC analysis. The results showed that CNOT1 expression was decreased in sh‐CNOT1 mice compared to the sh‐control mice (Fig. 4I). Similarly, the expression of LMNA was decreased in sh‐CNOT1 mice compared to sh‐NC mice (Fig. 4J). Taken together, these results strengthen the conclusion that CNOT1 interacts with LMNA and functions as a positive regulator of LMNA protein in osteosarcoma.
Figure 4.
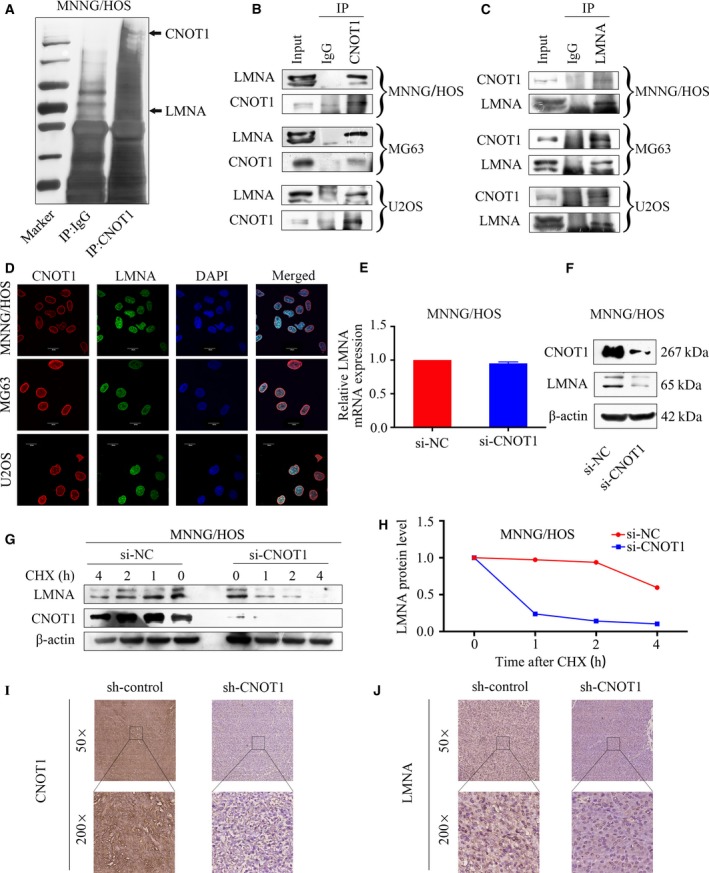
CNOT1 interacts with LMNA and affects its stability in osteosarcoma cells. (A) Silver staining of the SDS/PAGE after IgG or CNOT1 pulldown in MNNG/HOS cell line. (B) Whole‐cell lysates were immunoprecipitated with anti‐CNOT1 antibody followed by immunoblotting (IB) with anti‐LMNA and anti‐CNOT1 antibodies in the indicated osteosarcoma cell lines. IgG was used as a negative control. (C) Whole‐cell lysates were immunoprecipitated with anti‐LMNA antibody followed by IB with anti‐CNOT1 and anti‐LMNA antibodies in the indicated osteosarcoma cell lines. IgG was used as a negative control. (D) Immunofluorescence analysis performed using anti‐CNOT1 and anti‐LMNA antibodies. DAPI was used as a control for nuclear staining. (E,F) MNNG/HOS cells were transfected with si‐NC or si‐CNOT1 for 48 h. The effect of CNOT1 knockdown on LMNA mRNA and protein expression in MNNG/HOS cells was evaluated by qRT‐PCR and western blotting. β‐Actin was used as an internal control. (G,H) MNNG/HOS cells were transfected with si‐NC or si‐CNOT1 for 48 h, followed by treatment with cycloheximide (CHX; 100 mg·mL−1) for 0, 1, 2, and 4 h. Lysates were immunoblotted with anti‐LMNA or anti‐CNOT1. LMNA band intensity was normalized to β‐actin and then normalized to the time = 0 controls. (I) The expression of CNOT1 was detected in tumor tissue of sh‐control mice and sh‐CNOT1 mice by IHC. Magnification, × 50, × 200. (J) The expression of LMNA was detected in tumor tissue of sh‐control mice and sh‐CNOT1 mice by IHC. Magnification, × 50, × 200.
3.5. Knockdown of CNOT1 inhibits the Hedgehog signaling pathway in osteosarcoma
To further clarify the underlying mechanism of CNOT1, RNA‐seq analysis was performed to compare the global difference in gene expression between the si‐NC group and si‐CNOT1 group in MNNG/HOS cells and U2OS cells. The analysis revealed that 798 genes were commonly deregulated in MNNG/HOS and U2OS cells after transfection with si‐CNOT1 (Fig. 5A). Firstly, seven up‐regulated genes and 11 down‐regulated genes were chosen to verify the results of the analysis (Fig. 5B,C). The KEGG pathway analysis and Gene Set Enrichment analysis (GSEA) revealed an enrichment of the Hedgehog signaling pathway (Fig. 5D,E). Figure 5F shows the changes in the genes enriched in the Hedgehog signaling pathway. Then, three key proteins in the Hedgehog signaling pathway, including GLI1, PTCH1, and PTCH2, were chosen to detect the effect of CNOT1 on the pathway. The results showed that knockdown of CNOT1 significantly inhibited the expression of GLI1, PTCH1, and PTCH2 in osteosarcoma cells. Therefore, knockdown of CNOT1 inhibits the Hedgehog signaling pathway in osteosarcoma.
Figure 5.
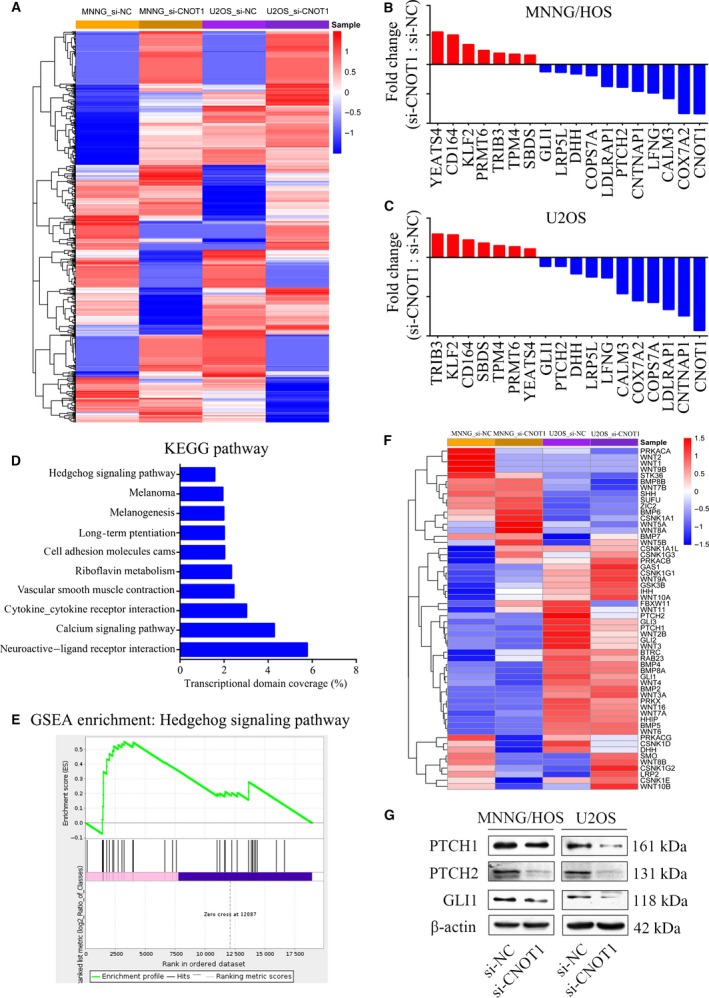
Knockdown of CNOT1 inhibits the Hedgehog signaling pathway in osteosarcoma cells. (A) Heat map of the genes that were differentially expressed between the si‐NC‐treated group and the si‐CNOT1‐treated group in MNNG/HOS and U2OS cells. (B,C) The mRNA expression results of seven up‐regulated genes and 11 down‐regulated genes were chosen for validation by qRT‐PCR. (D) The KEGG pathway analysis of the differentially expressed genes. (E) GSEA demonstrated an enrichment of gene signatures associated with the Hedgehog signaling pathway. (F) Heat map showing the change in genes enriched in the Hedgehog signaling pathway. (G) Representative blots showing the protein expression of GLI1, PTCH1, and PTCH2 in MNNG/HOS and U2OS cells after si‐CNOT1 transfection. β‐Actin was used as an internal control.
3.6. Knockdown of CNOT1 inhibits osteosarcoma cell proliferation through LMNA
Our data demonstrated that CNOT1 interacted with LMNA and affected its protein stability in osteosarcoma cells. We hypothesized that CNOT1 functioned through LMNA. We overexpressed LMNA in CNOT1‐knockdown MNNG/HOS and U2OS cells. Western blotting confirmed that the LMNA protein level was up‐regulated in CNOT1‐knockdown cells after pLVX‐Pruo‐Flag‐LMNA plasmid transfection (Fig. 6A,C). As expected, the CCK‐8 assay and colony forming assay revealed that LMNA overexpression attenuated the CNOT1 knockdown‐induced cell growth inhibition (Fig. 6B,D,E–H). Then, we determined whether the change in the Hedgehog signaling pathway was attenuated in CNOT1‐knockdown cells after LMNA overexpression. The results showed that increasing the expression of LMNA in MNNG/HOS and U2OS cells reversed the down‐regulation of GLI1, PTCH1, and PTCH2 induced by CNOT1 knockdown (Fig. 6I,J). Taken together, knockdown of CNOT1 inhibits osteosarcoma cell proliferation by inhibition of the Hedgehog signaling pathway via LMNA.
Figure 6.
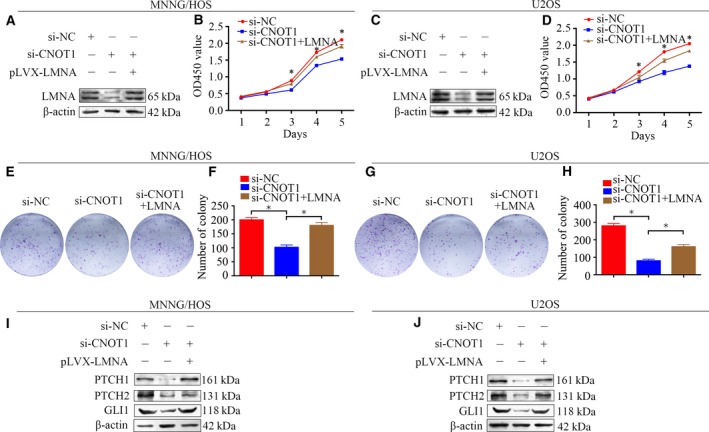
Knockdown of CNOT1 inhibits osteosarcoma cell proliferation through LMNA. (A,C) Representative blots of LMNA after transfection with pLVX‐Pruo‐Flag‐LMNA plasmid plus si‐CNOT1 or si‐NC. β‐Actin was used as an internal control. (B,D) CCK‐8 assay was used to detect tumor cell proliferation after transfection with pLVX‐Pruo‐Flag‐LMNA plasmid plus si‐CNOT1 or si‐NC. (E, F, G, H) colony forming assay was performed after transfection with pLVX‐Pruo‐Flag‐LMNA plasmid plus si‐CNOT1 or si‐NC. (I, J) Representative blots of GLI1, PTCH1, and PTCH2 after transfection with pLVX‐Pruo‐Flag‐LMNA plasmid plus si‐CNOT1 or si‐NC. β‐Actin was used as an internal control.
3.7. CNOT1 is a prognostic marker for osteosarcoma
To further determine the clinicopathologic significance of CNOT1 in osteosarcoma, we performed IHC analysis of CNOT1 in a tissue microarray that included an independent set of 151 cases of osteosarcoma. Representative images of CNOT1 are shown in Fig. 7A. The correlations between the CNOT1 expression level and the clinicopathologic characteristics of the patients with osteosarcoma are summarized in Table 1. The expression level of CNOT1 was higher in patients with a clinically advanced Enneking stage than in those with an early stage (P = 0.000). Further analysis indicated that CNOT1 levels were correlated positively with recurrence (P = 0.005), indicating that CNOT1 expression played an important role in osteosarcoma recurrence. Univariate analysis showed that TFS was related to CNOT1 (P = 0.001) and Enneking stage (P = 0.001). OS was related to CNOT1 (P = 0.000) and Enneking stage (P = 0.000) (Table 2). Variables that were identified as significantly different in univariate analysis were used for multivariate analysis. The Cox proportional hazards model showed that CNOT1 (χ2 = 4.415, HR = 1.306, P = 0.036) and Enneking stage (χ2 = 3.867, HR = 1.722, P = 0.049) were independent prognostic variables for TFS. In addition, the Cox proportional hazards model showed that CNOT1 (χ2 = 5.518, HR = 1.425, P = 0.023) and Enneking stage (χ2 = 5.011, HR = 2.052, P = 0.025) were independent prognostic variables for OS (Table 3). The TFS and OS curves for CNOT1 are presented in Fig. 7B,C. The TFS and OS curves for Enneking stage are presented in Fig. S5. All other factors, including gender, age, tumor location, tumor necrosis rate, and cortical destruction, had no significant influence on prognosis. We also performed Kaplan–Meier survival analyses using microarray data (http://www.kmplot.com) from lung cancer and gastric cancer patients. We found that CNOT1 expression also correlated negatively with patient OS in lung cancer and gastric cancer (Fig. S6). Finally, we detected the expression of LMNA in osteosarcoma tissues. Representative images of LMNA are shown in Fig. 7D. A Spearman correlation analysis revealed a significant correlation between the expression levels of CNOT1 and LMNA (R = 0.473, P = 0.000) in osteosarcoma tissues (Fig. 7E). In summary, CNOT1 was correlated with LMNA in tumor samples and associated with poor prognosis in patients with osteosarcoma.
Figure 7.
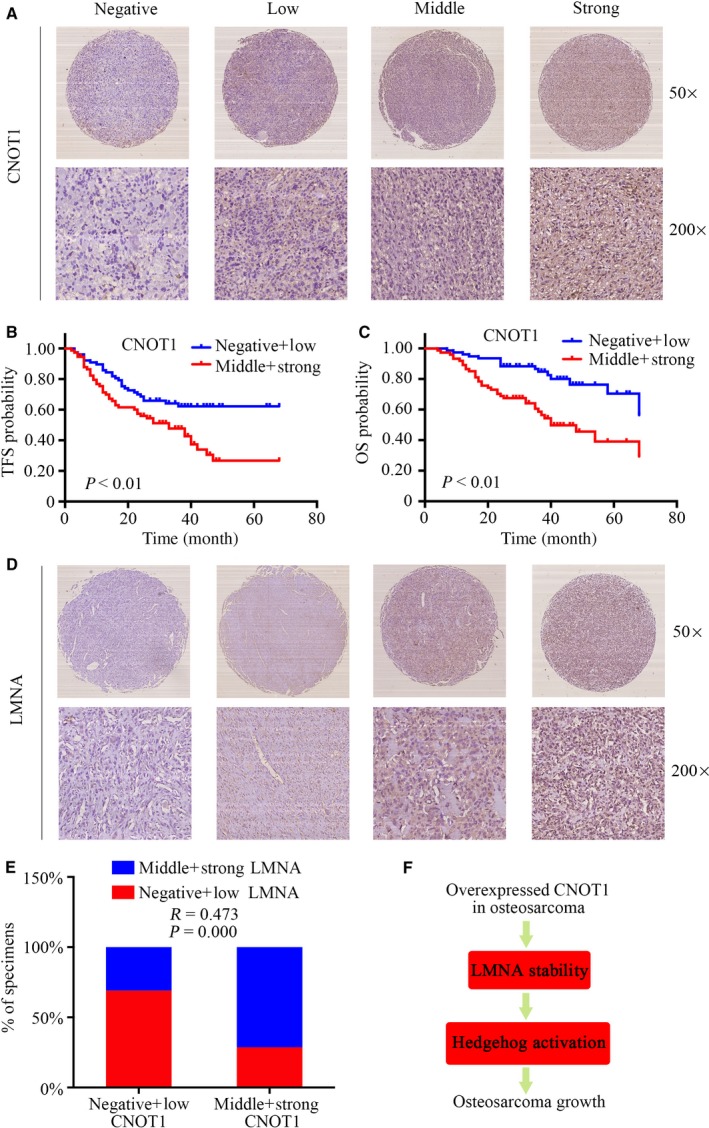
Clinical significance of CNOT1 in patients with osteosarcoma. (A) Representative IHC images of the expression levels of CNOT1 in osteosarcoma tissues. Original magnification: 50 ×, 200 ×. (B,C) The impact of CNOT1 level on TFS and OS of patients with osteosarcoma. (D) Representative IHC images of the expression levels of LMNA in osteosarcoma tissues. Original magnification: 50 ×, 200 ×. (E) Spearman's correlation analysis between CNOT1 protein levels and LMNA protein levels in 151 cases of osteosarcoma tissues. (F) Schematic representation of the function and potential mechanism of CNOT1 in osteosarcoma.
Table 1.
Correlation analyses of CNOT1 protein expression in relation to clinicopathologic variables of 151 patients with osteosarcoma
| Clinicopathologic parameters | CNOT1 expression level | P value | |||
|---|---|---|---|---|---|
| Negative | Low | Middle | Strong | ||
| Gender | |||||
| Male | 13 | 25 | 22 | 25 | 0.193 |
| Female | 18 | 21 | 15 | 12 | |
| Age (years) | |||||
| 6–20 | 16 | 24 | 20 | 21 | 0.071 |
| 20–40 | 13 | 10 | 9 | 14 | |
| 40–60 | 1 | 10 | 4 | 1 | |
| 60–84 | 1 | 2 | 4 | 1 | |
| Location | |||||
| Femur | 13 | 24 | 18 | 26 | 0.234 |
| Tibia | 10 | 14 | 11 | 9 | |
| Elsewhere | 8 | 8 | 8 | 2 | |
| Tumor necrosis rate (%) | |||||
| < 90 | 23 | 38 | 30 | 30 | 0.824 |
| ≥90 | 8 | 8 | 7 | 7 | |
| Cortical destruction | |||||
| Yes | 24 | 42 | 30 | 31 | 0.380 |
| No | 7 | 4 | 7 | 6 | |
| Recurrence | |||||
| Yes | 9 | 19 | 18 | 26 | 0.005a |
| No | 22 | 27 | 19 | 11 | |
| Metastasis | |||||
| Yes | 20 | 22 | 20 | 23 | 0.426 |
| No | 11 | 24 | 17 | 14 | |
| Enneking stage | |||||
| II | 28 | 43 | 26 | 16 | 0.000a |
| III | 3 | 3 | 11 | 21 | |
P < 0.05.
Table 2.
Impact of prognostic factors on TFS and OS by univariate analysis in osteosarcoma
| Clinicopathologic parameters | Tumor‐free survival | Overall survival | ||||||
|---|---|---|---|---|---|---|---|---|
| NO. | HR | 95% CI | P | NO. | HR | 95% CI | P | |
| Gender | ||||||||
| Male | 81 | 0.780 | 0.489–1.244 | 0.297 | 81 | 0.586 | 0.333–1.032 | 0.064 |
| Female | 70 | 70 | ||||||
| Age (years) | ||||||||
| 6–20 | 82 | 1.227 | 0.948–1.586 | 0.120 | 82 | 1.239 | 0.911–1.685 | 0.172 |
| 20–40 | 46 | 46 | ||||||
| 40–60 | 16 | 16 | ||||||
| 60–84 | 7 | 7 | ||||||
| Location | ||||||||
| Femur | 81 | 0.983 | 0.724–1.334 | 0.912 | 81 | 0.859 | 0.585–1.262 | 0.439 |
| Tibia | 44 | 44 | ||||||
| Elsewhere | 26 | 26 | ||||||
| Tumor necrosis rate (%) | ||||||||
| ≥ 90 | 121 | 0.595 | 0.313–1.133 | 0.114 | 121 | 0.636 | 0.299–1.352 | 0.239 |
| ≥ 90 | 30 | 30 | ||||||
| Cortical destruction | ||||||||
| Yes | 127 | 1.215 | 0.622–2.371 | 0.569 | 127 | 1.301 | 0.586–2.887 | 0.518 |
| No | 24 | 24 | ||||||
| CNOT1 | ||||||||
| Negative | 31 | 1.454 | 1.161–1.821 | 0.001a | 31 | 1.676 | 1.277–2.199 | 0.000a |
| Low | 46 | 46 | ||||||
| Middle | 37 | 37 | ||||||
| Strong | 37 | 37 | ||||||
| Enneking stage | ||||||||
| II | 113 | 2.268 | 1.402–3.668 | 0.001a | 113 | 2.985 | 1.726–5.161 | 0.000a |
| III | 38 | 38 | ||||||
P < 0.05.
Table 3.
Variables predictive of survival by COX proportional hazards model in osteosarcoma
| Parameters | Wald χ2 | Risk Ratio | 95% CI | P | |
|---|---|---|---|---|---|
| TFS | CNOT1 | 4.415 | 1.306 | 1.018–1.675 | 0.036 |
| Enneking stage | 3.867 | 1.722 | 1.002–2.960 | 0.049 | |
| OS | CNOT1 | 5.158 | 1.425 | 1.050–1.935 | 0.023 |
| Enneking stage | 5.011 | 2.052 | 1.094–3.849 | 0.025 |
4. Discussion
The 5‐year survival rate for osteosarcoma has remained almost unchanged over the last three decades. Thus, there is an urgent need to explore new therapeutic targets. In this study, quantitative proteomics was used to compare the proteome profiles of osteosarcoma cells and human osteoblastic cells. Twenty‐five up‐regulated proteins and 41 down‐regulated were identified and quantified in osteosarcoma cells compared with osteoblastic cells. Then, 33 potential proteins associated with the tumorigenesis of osteosarcoma were selected for proteomic validation by qRT‐PCR, 11 of which were verified by western blotting. Many of the verified proteins have been reported to be closely associated with carcinogenesis. Zhou et al. (2014) reported that LRPPRC played a critical role in the development of prostate cancer and that its inhibition could present a potential molecular approach for the treatment of prostate cancer. Yu et al. (2016) demonstrated that knockdown of ASNS suppressed gastric cancer cell proliferation and inhibited tumor growth in vivo. FAK and PHGDH were reported to be novel prognostic biomarkers in gastric cancer (Du et al., 2014; Xian et al., 2016). In the present study, we detected the expression of nine verified up‐regulated proteins by qRT‐PCR in 20 pairs of osteosarcoma tissues and their corresponding nontumor tissues. The results showed that CNOT1, ASNS, and EFHD2 were overexpressed in osteosarcoma tissues compared with normal tissues, suggesting that they might play an important role in the tumorigenesis of osteosarcoma. Functional screening was conducted, and the results showed that knockdown of CNOT1 significantly inhibited osteosarcoma cell proliferation, suggesting an important role of CNOT1 in osteosarcoma carcinogenesis.
Currently, the role of CNOT1 in carcinogenesis remains unclear. In a previous study, CNOT1 was reported to act as a transcriptional regulator of nuclear receptor signaling by interacting with RXR in a ligand‐dependent manner (Winkler et al., 2006). Temme et al. (2010) investigated the role of CNOT1 in mRNA deadenylation. They found that CONT1 was required for bulk poly (A) shortening and hsp70 mRNA deadenylation. In addition, it was reported that CNOT1 depletion induced apoptosis by destroying the CCR4–NOT‐associated deadenylase activity. In the present study, we found that knockdown of CNOT1 resulted in significant cell growth inhibition in osteosarcoma cells in vitro. To further clarify the role of CNOT1 in osteosarcoma proliferation in vivo, MNNG/HOS cells with stable CNOT1 depletion were injected into nude mice. The results showed that CNOT1 knockdown could also dramatically impede tumor growth potential in vivo.
To identify the underlying mechanism of CNOT1, we screened CNOT1‐associated proteins using Co‐IP followed by mass spectrometry analysis. In the present study, we found that CNOT1 and LMNA interacted with each other in osteosarcoma cells. LMNA has been implicated in DNA damage response pathways by facilitating the rapid recruitment of 53BP1, a nucleoskeleton protein, to the sites of DNA damage (Gibbs‐Seymour et al., 2015). Depletion of LMNA enhances DNA damage‐induced replication fork arrest, indicating the requirement of LMNA in maintaining genomic stability (Singh et al., 2013). Willis et al. (2008) found that the expression of LMNA was strongly correlated with poor prognosis in colorectal cancer patients. This study, for the first time, demonstrates the interaction of CNOT1 with LMNA in osteosarcoma cells. Knockdown of CNOT1 affected the protein expression level of LMNA in osteosarcoma cells. To determine whether CNOT1 functioned through LMNA, a rescue experiment was performed. Restoration of the protein expression of LMNA rescued tumor cell growth, showing that the control of cell proliferation by CNOT1 was largely dependent on LMNA.
To further determine the underlying mechanism of CNOT1, RNA‐seq analysis was performed to identify the differentially expressed genes following si‐CNOT1 transfection. The KEGG pathway and GSEA analysis showed an enrichment of the Hedgehog signaling pathway. The Hedgehog signaling pathway has been shown to contribute to the growth and chemoresistance of osteosarcoma and other tumors (Hirotsu et al., 2010; Shahi et al., 2008, 2014). Emerging data suggest that interference with the Hedgehog signaling pathway by inhibitors may reduce osteosarcoma cell proliferation and tumor growth, thereby potentially improving the survival of patients with osteosarcoma (Kumar and Fuchs, 2015). We found that CNOT1 depletion inhibited the expression of GLI1, PTCH1, and PTCH2, which were key proteins in the Hedgehog signaling pathway. Then, we clarified the role of LMNA in CNOT1‐mediated Hedgehog signaling pathway regulation. The results showed that overexpression of LMNA rescued the inhibition of the Hedgehog signaling pathway induced by CNOT1 depletion.
5. Conclusions
Collectively, knockdown of CNOT1 dramatically inhibits osteosarcoma cell growth in vitro and in vivo. More importantly, this report provides, for the first time, experimental evidence and implicates the significance of the interaction between CNOT1 and LMNA in osteosarcoma cells. Knockdown of CNOT1 inhibits osteosarcoma cell proliferation by inhibition of the Hedgehog signaling pathway via LMNA, as shown in Fig. 6E. Notably, the expression of CNOT1 is an independent biomarker for TFS and OS in patients with osteosarcoma. Therefore, interventions targeting the CNOT1–LMNA–Hedgehog signaling pathway axis may be a feasible and effective strategy for the treatment of osteosarcoma.
Author contributions
D‐DC and JL wrote the manuscript and designed this experiment. S‐JL helped to conduct the experiment. Q‐CY and C‐YF are in charge of the whole experiment conduction and paper writing. All of the authors reviewed the manuscript before submission and approved the final manuscript.
Supporting information
Fig. S1. GO analysis and KEGG pathway analysis of the differentially expressed proteins between osteosarcoma cells and osteoblastic cells.
Fig. S2. The expression of these 9 up‐regulated proteins was measured using qRT‐PCR in 20 human tissue sample pairs in which each pair consisted of an osteosarcoma sample and a corresponding nontumor tissue sample.
Fig. S3. SiRNA knockdown and CCK‐8 assay was performed to search for osteosarcoma‐related protein in MNNG/HOS cell line.
Fig. S4. Transwell assays were used to detect the migration and invasion capability of each cell line after transfection with si‐CSE1L.
Fig. S5. The impact of Enneking stage on TFS and OS of patients with osteosarcoma.
Fig. S6. Kaplan–Meier survival analyses were performed using microarray data (http://www.kmplot.com) in lung cancer and gastric cancer patients.
Table S1. Lists of proteins that were up‐regulated and down‐regulated in osteosarcoma cells compared with osteoblast.
Table S2. The primer sequences for qRT‐PCR.
Table S3. The primary antibody for western blotting.
Table S4. The identified proteins by mass spectrometry after CNOT1 pulldown.
Acknowledgements
We thank Professor Hui‐zhen Zhang for help with experiments. This work was funded by Shanghai Jiaotong University School of Medicine Doctoral Innovation Foundation (BXJ201634).
Contributor Information
Qing‐cheng Yang, Email: tjyqc@163.com.
Cun‐yi Fan, Email: fancunyi888@hotmail.com.
References
- Bell EH, Chakraborty AR, Mo X, Liu Z, Shilo K, Kirste S, Stegmaier P, McNulty M, Karachaliou N, Rosell R et al (2016) SMARCA4/BRG1 is a novel prognostic biomarker predictive of cisplatin‐based chemotherapy outcomes in resected non‐small cell lung cancer. Clin Cancer Res 22, 2396–2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlanga P, Muñoz L, Piqueras M, Sirerol JA, Sánchez‐Izquierdo MD, Hervás D, Hernández M, Llavador M, Machado I, Llombart‐Bosch A et al (2016) miR‐200c and phospho‐AKT as prognostic factors and mediators of osteosarcoma progression and lung metastasis. Mol Oncol 10, 1043–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardini G, Laschi M, Geminiani M and Santucci A (2014) Proteomics of osteosarcoma. Expert Rev Proteomics 11, 331–343. [DOI] [PubMed] [Google Scholar]
- Coventry MB and Dahlin DC (1957) Osteogenic sarcoma; a critical analysis of 430 cases. J Bone Joint Surg Am 39, 741–757. [PubMed] [Google Scholar]
- Du T, Qu Y, Li J, Li H, Su L, Zhou Q, Yan M, Li C, Zhu Z and Liu B (2014) Maternal embryonic leucine zipper kinase enhances gastric cancer progression via the FAK/Paxillin pathway. Mol Cancer 13, 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folio C, Mora MI, Zalacain M, Corrales FJ, Segura V, Sierrasesúmaga L, Toledo G, San‐Julián M and Patiño‐García A (2009) Proteomic analysis of chemonaive pediatric osteosarcomas and corresponding normal bone reveals multiple altered molecular targets. J Proteome Res 8, 3882–3888. [DOI] [PubMed] [Google Scholar]
- Gibbs‐Seymour I, Markiewicz E, Bekker‐Jensen S, Mailand N and Hutchison CJ (2015) Lamin A/C‐dependent interaction with 53BP1 promotes cellular responses to DNA damage. Aging Cell 14, 162–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirotsu M, Setoguchi T, Sasaki H, Matsunoshita Y, Gao H, Nagao H, Kunigou O and Komiya S (2010) Smoothened as a new therapeutic target for human osteosarcoma. Mol Cancer 9, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K, Takahashi A, Morita M, Suzuki T and Yamamoto T (2011) The role of the CNOT1 subunit of the CCR4‐NOT complex in mRNA deadenylation and cell viability. Protein Cell 2, 755–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jou YJ, Hua CH, Lin CD, Lai CH, Huang SH, Tsai MH, Kao JY and Lin CW (2014) S100A8 as potential salivary biomarker of oral squamous cell carcinoma using nanoLC‐MS/MS. Clin Chim Acta 436, 121–129. [DOI] [PubMed] [Google Scholar]
- Kubota D, Mukaihara K, Yoshida A, Tsuda H, Kawai A and Kondo T (2013) Proteomics study of open biopsy samples identifies peroxiredoxin 2 as a predictive biomarker of response to induction chemotherapy in osteosarcoma. J Proteomics 91, 393–404. [DOI] [PubMed] [Google Scholar]
- Kumar RM and Fuchs B (2015) Hedgehog signaling inhibitors as anti‐cancer agents in osteosarcoma. Cancers (Basel) 7, 784–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Li J and Feng L (2016) Hedgehog signaling pathway as a therapeutic target for ovarian cancer. Cancer Epidemiol 40, 152–157. [DOI] [PubMed] [Google Scholar]
- Li Y, Liang Q, Wen YQ, Chen LL, Wang LT, Liu YL, Luo CQ, Liang HZ, Li MT and Li Z (2010) Comparative proteomics analysis of human osteosarcomas and benign tumor of bone. Cancer Genet Cytogenet 198, 97–106. [DOI] [PubMed] [Google Scholar]
- Li Y, Wen T, Zhu M, Li L, Wei J, Wu X, Guo M, Liu S, Zhao H, Xia S et al (2013) Glycoproteomic analysis of tissues from patients with colon cancer using lectin microarrays and nanoLC‐MS/MS. Mol BioSyst 9, 1877–1887. [DOI] [PubMed] [Google Scholar]
- Lin HC, Zhang FL, Geng Q, Yu T, Cui YQ, Liu XH, Li J, Yan MX, Liu L, He XH et al (2013) Quantitative proteomic analysis identifies CPNE3 as a novel metastasis‐promoting gene in NSCLC. J Proteome Res 12, 3423–3433. [DOI] [PubMed] [Google Scholar]
- Parker CE and Borchers CH (2014) Mass spectrometry based biomarker discovery, verification, and validation–quality assurance and control of protein biomarker assays. Mol Oncol 8, 840–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren F, Wu H, Lei Y, Zhang H, Liu R, Zhao Y, Chen X, Zeng D, Tong A, Chen L et al (2010) Quantitative proteomics identification of phosphoglycerate mutase 1 as a novel therapeutic target in hepatocellular carcinoma. Mol Cancer 9, 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampo MM, Tarkkanen M, Kivioja AH, Taskinen MH, Sankila R and Böhling TO (2008) Osteosarcoma in Finland from 1971 through 1990: a nationwide study of epidemiology and outcome. Acta Orthop 79, 861–866. [DOI] [PubMed] [Google Scholar]
- Shahi MH, Holt R and Rebhun RB (2014) Blocking signaling at the level of GLI regulates downstream gene expression and inhibits proliferation of canine osteosarcoma cells. PLoS One 9, e96593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahi MH, Lorente A and Castresana JS (2008) Hedgehog signalling in medulloblastoma, glioblastoma and neuroblastoma. Oncol Rep 19, 681–688. [PubMed] [Google Scholar]
- Singh M, Hunt CR, Pandita RK, Kumar R, Yang CR, Horikoshi N, Bachoo R, Serag S, Story MD, Shay JW et al (2013) Lamin A/C depletion enhances DNA damage‐induced stalled replication fork arrest. Mol Cell Biol 33, 1210–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbannayya Y, Mir SA, Renuse S, Manda SS, Pinto SM, Puttamallesh VN, Solanki HS, Manju HC, Syed N, Sharma R et al (2015) Identification of differentially expressed serum proteins in gastric adenocarcinoma. J Proteomics 127, 80–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temme C, Zhang L, Kremmer E, Ihling C, Chartier A, Sinz A, Simonelig M and Wahle E (2010) Subunits of the Drosophila CCR4‐NOT complex and their roles in mRNA deadenylation. RNA 16, 1356–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis ND, Cox TR, Rahman‐Casañs SF, Smits K, Przyborski SA, van den Brandt P, van Engeland M, Weijenberg M, Wilson RG, de Bruïne A et al (2008) Lamin A/C is a risk biomarker in colorectal cancer. PLoS One 3, e2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler GS, Mulder KW, Bardwell VJ, Kalkhoven E and Timmers HT (2006) Human Ccr4‐Not complex is a ligand‐dependent repressor of nuclear receptor‐mediated transcription. EMBO J 25, 3089–3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xian Y, Zhang S, Wang X, Qin J, Wang W and Wu H (2016) Phosphoglycerate dehydrogenase is a novel predictor for poor prognosis in gastric cancer. Onco Targets Ther 9, 5553–5560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Cai F, Liu X and Guo L (2014) LKB1 suppresses proliferation and invasion of prostate cancer through hedgehog signaling pathway. Int J Clin Exp Pathol 7, 8480–8488. [PMC free article] [PubMed] [Google Scholar]
- Yan GN, Lv YF and Guo QN (2016) Advances in osteosarcoma stem cell research and opportunities for novel therapeutic targets. Cancer Lett 370, 268–274. [DOI] [PubMed] [Google Scholar]
- Yang J, Cheng D, Zhu B, Zhou S, Ying T and Yang Q (2016) Chromobox homolog 4 is positively correlated to tumor growth, survival and activation of HIF‐1α signaling in human osteosarcoma under normoxic condition. J Cancer 7, 427–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao C, Wu S, Li D, Ding H, Wang Z, Yang Y, Yan S and Gu Z (2012) Co‐administration phenoxodiol with doxorubicin synergistically inhibit the activity of sphingosine kinase‐1 (SphK1), a potential oncogene of osteosarcoma, to suppress osteosarcoma cell growth both in vivo and in vitro. Mol Oncol 6, 392–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q, Wang X, Wang L, Zheng J, Wang J and Wang B (2016) Knockdown of asparagine synthetase (ASNS) suppresses cell proliferation and inhibits tumor growth in gastric cancer cells. Scand J Gastroenterol 51, 1220–1226. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Zhang L, Hua Y, Jia X, Li J, Hu S, Peng X, Yang P, Sun M, Ma F et al (2010) Comparative proteomic analysis of plasma membrane proteins between human osteosarcoma and normal osteoblastic cell lines. BMC Cancer 10, 206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Hou Y, Yang G, Zhang H, Tu G, Du YE, Wen S, Xu L, Tang X, Tang S et al (2016) LncRNA‐Hh strengthen cancer stem cells generation in twist‐positive breast cancer via activation of hedgehog signaling pathway. Stem Cells 34, 55–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Zhang F, Hou X and Zhang N (2014) Downregulation of LRPPRC induces apoptosis in prostate cancer cells through the mitochondria‐mediated pathway. Cancer Biother Radiopharm 29, 345–350. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. GO analysis and KEGG pathway analysis of the differentially expressed proteins between osteosarcoma cells and osteoblastic cells.
Fig. S2. The expression of these 9 up‐regulated proteins was measured using qRT‐PCR in 20 human tissue sample pairs in which each pair consisted of an osteosarcoma sample and a corresponding nontumor tissue sample.
Fig. S3. SiRNA knockdown and CCK‐8 assay was performed to search for osteosarcoma‐related protein in MNNG/HOS cell line.
Fig. S4. Transwell assays were used to detect the migration and invasion capability of each cell line after transfection with si‐CSE1L.
Fig. S5. The impact of Enneking stage on TFS and OS of patients with osteosarcoma.
Fig. S6. Kaplan–Meier survival analyses were performed using microarray data (http://www.kmplot.com) in lung cancer and gastric cancer patients.
Table S1. Lists of proteins that were up‐regulated and down‐regulated in osteosarcoma cells compared with osteoblast.
Table S2. The primer sequences for qRT‐PCR.
Table S3. The primary antibody for western blotting.
Table S4. The identified proteins by mass spectrometry after CNOT1 pulldown.


