Abstract
Purpose
We propose a systematic methodology to quantify incidentally identified pulmonary nodules based on observed radiological traits (semantics) quantified on a point scale and a machine learning method using these data to predict cancer status.
Materials and Methods
We investigated 172 patients who had low-dose computed tomography (LDCT) images, with 102 and 70 patients grouped into training and validation cohorts, respectively. On the images, 24 radiological traits were systematically scored and a linear classifier was built to relate the traits to malignant status. The model was formed both with and without size descriptors to remove bias due to nodule size. The multivariate pairs formed on the training set was tested on an independent validation data set to evaluate its performance.
Results
The best four feature set that included a size measurement (Set 1), was short axis, contour, concavity, and texture, which had an area under the receiver operator characteristic curve (AUROC) of 0.88 (Accuracy= 81%, Sensitivity= 76.2%, Specificity= 91.7%). If size measures were excluded, the four best features (Set 2) were: location, fissure attachment, lobulation, and spiculation which had an AUROC of 0.83 (Accuracy= 73.2%, Sensitivity= 73.8%, Specificity= 81.7%) in predicting malignancy in primary nodules. The validation test AUROC was 0.8 (Accuracy=74.3%, Sensitivity =66.7%, Specificity= 75.6%) and 0.74 (Accuracy=71.4%, Sensitivity = 61.9%, Specificity = 75.5%) for Sets 1 and 2, respectively.
Conclusions
Radiological image traits are useful in predicting malignancy in lung nodules. These semantic traits can be used in combination with size-based measures to enhance prediction accuracy and reducing false positives.
Keywords: semantic features, radiological image traits, lung nodules, CT
INTRODUCTION
Lung cancer is the leading cause of cancer-related deaths globally and in the U.S. (1). Low-dose computed tomography (LDCT) has evolved to be a sensitive imaging modality to detect pulmonary nodules. The National Lung Screening Trial (NLST), which compared LDCT and standard chest radiography (CXR) for three annual screens, found a 20% reduction in lung cancer mortality for CT compared to CXR(2). In the NLST trial, at least 39% of LDCT study participants had a nodule-positive scan during the study, and 96.4% of these were non-cancerous (i.e., false positive). The CXR arm had a lower rate of positive detection (16%) with a comparatively lower rate of false positives (2, 3). In the NLST, it is estimated that about 18.5% (95% CI, 5.4% – 30.6%) of the lung cancers detected in the study population were clinically insignificant and hence, over-diagnosed (4). Other studies have shown that over-diagnosis of lung cancer can be as high as 96% (5–7). Despite the high false positive rate, the United States Preventive Services Task Force (USPSTF) recommended lung cancer screening for high-risk individuals (8). However, debate on the effectiveness of LDCT screening still continues. The availability of abundant data has helped the development of clinical assessment models to predict probabilities of malignancy (9, 10).
Identification of malignancy continues to be a challenge even in the screening setting, patients with indeterminate pulmonary nodules (IPN) are typically monitored with scheduled follow-up scans (11). Advancements in image acquisition and improved computer-aided diagnostic tools coupled with effective treatment strategy have shown to improve patient survival (12). In recent work, nodule characteristics coupled with clinical risk factors have been widely used to differentiate malignancy (13–15). Although the NLST shows < 4% of the subjects with non-calcified nodules (NCN) were diagnosed with lung cancer within a year (17), at present, there is limited ability to provide individual patient-level risk (16),
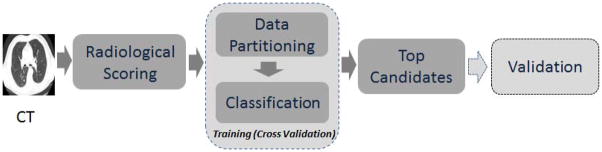
In this work we focus on quantifying radiological imaging characteristics of nodules (shape, location, texture) including associated structures of the lung and relate them to cancer status. We followed a rigorous approach to find the optimal imaging characteristics in a training cohort using cross-validation methodology and validated in a test cohort data set. These quantitative predictive scores (accuracy and or AUROC) obtained from images were used to develop classifier models to identify a risk of malignancy (see Figure 1).
Figure 1.
Study design to find discriminant Semantic features. The blocks describe the methodology followed in the manuscript. The observed radiological trait by an expert was related to outcome with a train and validation setting.
MATERIAL AND METHODS
Patient cohort
In this study we collected two cohorts (training and validation) from the Vanderbilt University Medical Center (VUMC), Nashville and Veterans Affairs (VA) Medical Center, Nashville. The training cohort had 102 patients consisting of 42 with lung cancer and 60 patients with a positive scan that was not lung cancer (i.e., benign nodule). Of the 102 patients in the training set, there were 206 nodules (84 malignant, 122 benign). While the validation set had 70 patients 21 with lung cancer and 49 patients with positive scan that was non lung cancer. Of the 70 patients in validation cohort, there were 102 nodules (26 malignant, 76 benign). The patients had 2 years of follow up from the time of CT scans and the biopsy results confirmed their cancer status. The median patient age for the training set was 67 years (σ = 8.6) while the validation set had 65 years (σ = 9.3). The patient samples were retrospectively curated, first batch of patients were used in the training and later batch was used in the validation set, the cohorts had a collection time difference of 6 to 9 months.
Table 1 describes the patient demographics, and Supplemental Table-S1 describes the nodule dimensions in the two cohorts. This study was approved by the Institutional Review Board (IRB) at the collecting institution (VUMC/VA Hospital) and as a retrospective study to review de-identified patient records at the collaborating institution (Moffitt Cancer Center). The requirement for patients’ informed consent was waived.
Table 1.
Patient demographics and tumor stage for the training and testing samples.
| Training Set | Validation Set | ||||||
|---|---|---|---|---|---|---|---|
| Overall | Lung Cancer Cases | Patients with Benign Nodules | Overall | Lung Cancer Cases | Patients with Benign Nodules | ||
| Demographics patients | 102 | 42 | 60 | 70 | 21 | 49 | |
| Nodules | 206 | 84 | 122 | 102 | 26 | 76 | |
| Age: median, mean (σ) | 67, 66.7 (8.6) | 69, 68.4 (8.7) | 65, 65.6, (8.3) | 65, 65.5 (9.3) | 69, 70.1 (7.4) | 64, 63.5 (9.5) | |
| Gender n (%) (Male/Female) | 102 | 95.2%/4.7% | 98%/1.6% | 70 | 28.6%/0% | 67.1%/4.3% | |
| Male | 99 | 40 | 59 | 68 | 20 | 47 | |
| Female | 3 | 2 | 1 | 3 | 0 | 3 | |
| Smoking: Pack Yearsmedian, mean (σ) | 46, 58.22, (42.15) | 50, 68, (53.53) | 51.3654530.56934781 | 45, 48.1 (37.5) | 57, 61.4 (34.4) | 40, 41.9 (37.5) | |
| Race (Caucasian/African American/Native/Others) | 92/9/0/1 | 35/7/0/0 | 57/2/0/1 | 65/2/3/1 | 21/0/0/0 | 44/2/3/1 | |
LDCT protocol
Chest LDCT scans were performed with a single deep breath hold by using a Discovery VCT (GE Healthcare, Waukesha, WI, USA) from the base of the neck to the posterior lung gutters. The patients were scanned without IV contrast and images obtained by filtered back projection image reconstruction with a soft tissue filter to obtain a 512 × 512 matrix. CT energy was 120 KVp, with variable mAs, (range 30 – 400) to minimize radiation. Helical data were acquired using collimation of 40 mm, pitch 1.375:1, table speed 55 cm/sec, reconstructed as 1.25 mm pixels at contiguous 1.25 mm intervals, producing isotropic voxels. The field of view (FOV) was based on patient body habitus, typically 35 to 43 cm.
Image analysis and reader agreement
All LDCT images were reviewed by a clinical radiologist (Y.L.) with more than 6 years of experience in LDCT imaging of thoracic malignancies, who was blind to the clinical details and final diagnosis for the nodules at the time of image interpretation. Thin-section LDT images were displayed using both standard mediastinal (width, 350 HU; level, 40 HU) and lung (width, 1500 HU; level, −600 HU) window-width and window-level settings. Totally, 24 CT image descriptors were developed to characterize the pulmonary nodules and these were classified into eight categories: (1) location; (2) size; (3) shape; (4) margin; (5) density; (6) internal features; (7) external features; and (8) associated findings, example cases are shown in Supplemental Figure 1. Each CT descriptor was rated using either an ordinal scale or a binary categorical variable (See Supplemental Table S-2 and Supplemental Table-S-3).
To measure the reproducibility of semantic scoring metric, we randomly selected 80 patients (40 malignant and 40 benign) from the cohort and provided the scoring sheet with approximate anatomical location of the nodules to two different radiologists (Y.L. and Q.L. who is a resident radiologist with 3 years of clinical experience). The scoring sheet was used to compute the concordance of these discrete scoring between readers using kappa statistics (18, 19). In radiological observations, a value of kappa coefficient greater than 0.8 is considered perfect agreement, 0.61 to 0.8 is considered substantial agreement, 0.41 to 0.6 moderate agreement; 0.21 to 0.4 fair agreement and below 0.2 is considered poor agreement (20). The Supplemental Table S-4 shows the kappa coefficient and the confidence limits in different sampling cohorts. Out of the 24 features, 10 semantic features had kappa > 0.95 of which 9 features exhibited a perfect score. Eight semantic features had kappa coefficient between 0.85 – 0.95, while three were between 0.7 to 0.8. One feature (distribution) could not be scored due to limited examples in the sample population. The two size-based features (long and short axis) repeatability was evaluated by computing Interclass Correlation Coefficient (ICC) which was 0.94 (0.89–0.968) and 0.96 (0.834–0.985) respectively.
Reader Variability and Prediction Outcome
We evaluated reader variability on the classification outcome in a subset of 80 patients, which was further divided into training and testing (40 patients in each) with equal number of patients with benign and malignant nodules. Two radiologist independently score the semantics metrics as described in the previous section. We compared prediction results (AUC) of the classifier by using semantic scoring for test and train samples coming from the same radiologist to the prediction testing carried out using semantic scoring coming from different radiologist. Supplemental Table S-5 shows the results of the AUC (Sensitivity and Specificity) of the inter-reader classification carried out in both ways. We find semantic metrics of contour and concavity showed differences of 10.2% and 6.6%, respectively, in the AUC derived from different radiologists. Notably, other semantic metrics showed less than 5% difference in the AUC.
Statistical analysis
Discriminatory analysis was conducted using a liner classifier to find the best predictive feature of cancer status. The error of classification was estimated using the hold-out cross validation method, where 80% of the sample was selected for training and 20% for testing. The process was randomized and repeated for a large number of times (over 200) and the average test accuracy (or error) was reported. For each combination, the AUROC was computed and compared with the clinical model proposed by Gould et.al (9, 21). To make a comparison to the cross validation method, clinical data were resampled using a bootstrap method and the clinical model prediction was computed for each random partition (22–24). The average AUROC with deviation across multiple runs was reported, along with sensitivity and specificity. The classifier model was first built on the training cohort using the cross validation method described and independently applied to the validation cohort to find the most promising feature combination.
The feature combination that exhibited the highest sensitivity and specificity (Youden J index) (25, 26) in the training set was then selected to be tested for performance on the test cohort. The final lists of top candidate features were selected based on their performance on both cohorts (training and test). This approach provides an additional validation step to overcome typical considerations of cross-validation methods (27–29). As such, the larger cohort sample size provided better performance capabilities and hence the elevated AUCs.
Integrity of training and testing samples was independently maintained without mixing the samples. We evaluated the overall survival difference between the classifier discriminated patient population using Kaplan Meier survival plots and p-value the log-rank test.
Finding the best set of features is a challenge. Various feature reduction methods have been proposed in the past, most often these methods have a range of performance, typically dependent on the complexity of the datasets(30). We used an exhaustive search to find the best performing feature finding all possible feature combinations (up to four dimensions, over 12,650 combinations). The top discriminating features are reported in Tables 2 and Supplemental Table S-6 (all nodules in S-7 & S-8) for the training data along with clinical comparator. Accuracy (1-Error), AUROC, sensitivity, and specificity, were all considered in identifying the best discriminant combinations. We then used the top discriminating feature pairs short listed from the training and applied the discriminator blindly on the validation data. The error rate with sensitivity and specificity is reported in Table 3.
Table 2.
Prediction error rate for test and train set using four best semantic features with primary nodule. a) Including Size based features b) without size based features.
| 2. a) Including Size Based Features | ||||||
|---|---|---|---|---|---|---|
| Features | Training | |||||
| Accuracy(Error), % | Sensitivity | Specificity | E[AUC] (μ,σ, [95% CI]) | E[AUC] Gould* (μ,σ, [95% CI]) | ||
| 1 | short-axis-(cm), contour, concavity, texture | 81.02 (18.98) | 0.762 | 0.917 | 0.88,0.08 [0.69,0.98] | 0.57,0.04 [0.49,0.65] |
| 2 | long-axis-(cm), border-definition, vascular-convergence, lymphadenopathy | 79.6 (20.4) | 0.762 | 0.95 | 0.87,0.1 [0.53,0.98] | 0.58,0.05 [0.48,0.69] |
| 3 | short-axis-(cm), contour, concavity, nodules-in-nontumor-lobes | 79.02 (20.98) | 0.786 | 0.917 | 0.86,0.09 [0.7,0.98] | 0.58,0.06 [0.42,0.66] |
| 4 | short-axis-(cm), contour, concavity, spiculation | 82.42 (17.58) | 0.762 | 0.917 | 0.86,0.08 [0.66,0.98] | 0.58,0.05 [0.47,0.66] |
| 5 | short-axis-(cm), contour, spiculation, nodules-in-nontumor-lobes | 80.9 (19.1) | 0.762 | 0.917 | 0.81,0.1 [0.62,0.98] | 0.59,0.05 [0.5,0.67] |
| 2. b) No Size Based Feature | ||||||
|---|---|---|---|---|---|---|
| Features | Training | |||||
| Accuracy (Error),% | Sensitivity | Specificity | E[AUC] (μ,σ, [95% CI]) | E[AUC] Gould* (μ,σ, [95% CI]) | ||
| 1 | location fissure-attachment lobulation spiculation | 73.2 (26.8) | 0.738 | 0.817 | 0.83,0.09 [0.68,0.98] | 0.59,0.049 [0.48,0.66] |
| 2 | location fissure-attachment spiculation vascular-convergence | 73.6 (26.4) | 0.738 | 0.817 | 0.82,0.09 [0.65,0.96] | 0.578,0.06 [0.46,0.69] |
| 3 | concavity border-definition spiculation peri-nodule-fibrosis- | 69.3 (30.7) | 0.714 | 0.8 | 0.81,0.09 [0.57,0.95] | 0.586,0.056 [0.47,0.7] |
| 4 | concavity border-definition spiculation texture | 67.3 (32.7) | 0.738 | 0.733 | 0.8,0.1 [0.57,0.98] | 0.567,0.05 [0.49,0.67] |
| 5 | location pleural-attachment spiculation vascular-convergence | 71.5 (28.5) | 0.714 | 0.817 | 0.8,0.08 [0.7,0.97] | 0.587,0.047 [0.51,0.69] |
Table 3.
Validation using 70 patients with 49 being normal and 21 identified cancerous. a) Including Size based features b) without size based features.
| 3. a) Validation Data Set (With Size Based Features) | ||||||
|---|---|---|---|---|---|---|
| Features | Coefficient (a0 to a5) | Testing | ||||
| Accuracy (Error), % | AUC | Sensitivity | Specificity | |||
| 1 | short-axis-(cm), contour, concavity, texture | (0.401033,0.141517,0.436219,0.042036,) | 74.3 (25.7) | 0.8 | 0.667 | 0.776 |
| 2 | long-axis-(cm), border-definition, vascular-convergence, lymphadenopathy | (0.319822,0.035229,0.552036,0.595986,) | 64.3 (35.7) | 0.73 | 0.667 | 0.633 |
| 3 | short-axis-(cm), contour, concavity, nodules-in-nontumor-lobes | (0.396477,0.144428,0.455541, −0.028359,) | 74.3 (25.7) | 0.80 | 0.667 | 0.776 |
| 4 | short-axis-(cm), contour, concavity, spiculation | (0.372755,0.100117,0.363608,0.206085,) | 78.6 (21.4) | 0.81 | 0.714 | 0.816 |
| 5 | short-axis-(cm), contour, spiculation, nodules-in-nontumor-lobes | (0.414338,0.203542,0.283427, −0.031426,) | 80 (20) | 0.81 | 0.714 | 0.837 |
| Combined Voting (all five combinations) | Accuracy: 77.2% | 0.714 | 0.796 | |||
| 3. b) Validation Data Set (No Size Based Features) | ||||||
|---|---|---|---|---|---|---|
| Features | Coefficient (a0 to a5) | Testing | ||||
| Accuracy (Error),% | AUC | Sensitivity | Specificity | |||
| 1 | location fissure-attachment lobulation spiculation | (−2.106962,0.108965, 0.635492,0.425675, 0.429703) | 71.4 (28.6) | 0.74 | 0.619 | 0.755 |
| 2 | location fissure-attachment spiculation vascular-convergence | (−1.515249,0.094082, 0.768446,0.428057, 0.698817) | 65.7 (34.3) | 0.68 | 0.619 | 0.673 |
| 3 | concavity border-definition spiculation peri-nodule-fibrosis- | (−1.892709,0.557655, 0.033625,0.258149, 0.103441) | 71.4 (28.6) | 0.78 | 0.714 | 0.714 |
| 4 | concavity border-definition spiculation texture | (−1.88911,0.564026, 0.062267,0.286365, 0.025501) | 68.6 (31.4) | 0.75 | 0.81 | 0.633 |
| 5 | Location pleural-attachment spiculation vascular-convergence | (−1.359628,0.07856, 0.240703,0.393194, 0.632097) | 64.3 (35.7) | 0.68 | 0.571 | 0.673 |
| 6 | Combined Voting (all five combinations) | Accuracy: 77.46 % | 0.619 | 0.694 | ||
Clinical Predictor (Gould Model)
Clinical patient characteristic including size of the nodules has been widely used as prognostic factors. There are several models proposed in the past; we used clinical model proposed by Gould et.al (9, 21). In the model, clinical malignancy of a nodule was predicted based on smoking status, age of the patient, diameter of the nodule and number of years since the patient quit smoking. These factors in the logistic regression model showed a high accuracy of malignancy prediction. We used this model as a baseline to compare the semantic based predictors.
RESULTS
Prediction of cancer status
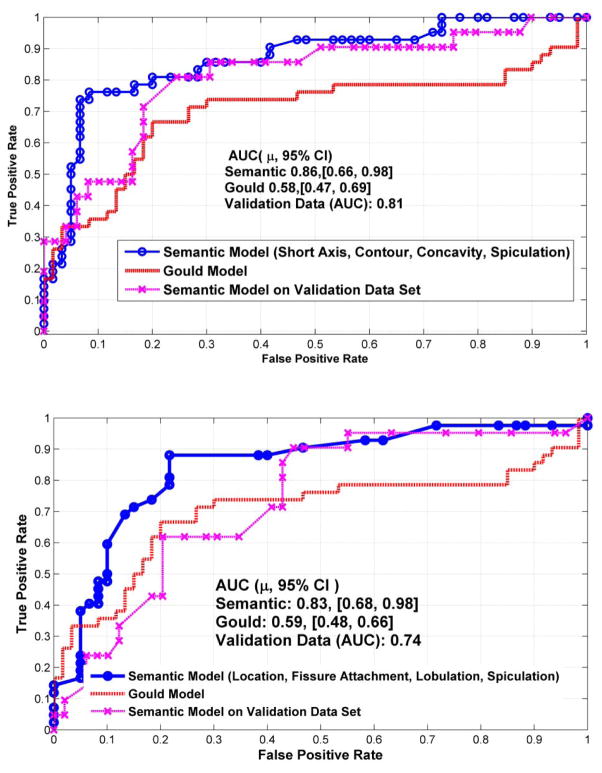
We investigated combinations of up to four features, with and without size (long axis, short axis and size category) based descriptors. Figure 2 shows an example of cancerous and benign nodule across different slices. As expected, size-based features by themselves were good predictors for cancerous nodules, providing an average accuracy of 73% (AUC of 0.89, CI [0.69, 0.98]), with a low sensitivity (0.476) and high specificity (0.93), reported in Supplemental Table S-6. Individual traits, including lymphadenopathy or vascular convergence, provided accuracies of 70 and 72% (AUC range: 0.71, CI [0.58, 0.84] to 0.72, CI [0.55,0.9]), respectively. Using all the nodules identified in the patients shows varied prediction accuracy (see Supplemental Tables-S7 and S8). Multivariate analyses of image features were shown to improve the accuracy of prediction; as shown in Table 2. For example, using size based short axis with contour, concavity and texture improved prediction accuracy to 81% (AUC of 0.88, CI [0.68, 0.98]) using the primary largest nodule. The size-based features are conventionally known to be informative of malignancy and hence we removed size measurements to avoid bias in the predictions and repeated the process to find best non-size-based predictors of cancer status. The accuracy for the best non-size based features (4-dimentions) was in the range of 67.2% to 73.6%, the average AUROC was in the order of 0.8 (CI [0.57, 0.98]) to 0.83 (CI [0.68, 0.98]), with sensitivity in the range of 0.71 to 0.74, and specificity of 0.73 to 0.82. Figure 3 shows receiver operator curves (ROC) with four semantic features (both with and without size) to predict malignancy. These were compared against the Gould model and a validation data set. The best semantic model was based on size, concavity, contour and spiculation. The non-size based predictor was based location, fissure attachment, lobulation and spiculation, which are known to be related to malignancy (31–36).
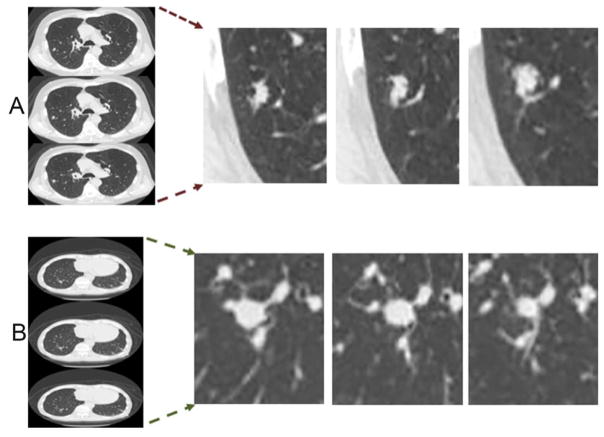
Figure 2.
Representative slices selected based on four radiological traits (Lobulation, Border definition, Texture and Nodules in primary tumor lobe) which was found to be one of the best discriminant pairs to predict malignant nodules. The slices in panel A) correspond to malignant case (Lobulation:3, border definition:2, Texture: 3, Nodules-in-Primary-Tumor:0) and B) Benign case (Lobulation:1, border definition:1, Texture: 3, Nodules-in-Primary-Tumor:0).
Figure 3.
Receiver operator characteristics (ROC) for semantic feature based predictors (blue) compared to conventional clinical parameters using Gould model (red) and on the independent validation data set. The panels below uses pairs (a) with size feature and (b) without size based features.
Prediction of Overall-survival (OS)
These semantic based predictor models were used to partition the samples into two groups, which showed significant differences in survival based on the Kaplan-Meier survival curves. As expected, the model that included a size-based feature was significantly associated with overall survival (Supplemental Figure 2a; P = 0.013) while the model without size-based features was borderline significantly associated with overall survival (Supplemental Figure 2b; P =0.048).
The models were then blindly applied to the validation data set to assess the ability to predict malignancy and the predictor’s discrimination ability was measured using accuracy, sensitivity and specificity. As noted in Table 3 (and Supplemental Table S-9), accuracy in predicting cancer status in the validation set was in the range of 64.3 to 80% (with AUROC 0.73 to 0.80, sensitivity 66.7 to 71.4%, specificity 63.3 to 83.7%) using a combination of size-based and semantic features. Semantic features by themselves had prediction accuracy in the range of 64.3 to 71.4 % (AUC 0.68 to 0.78, sensitivity 57 to 81%, specificity 67 to 75.5%).
To improve reliability of the predictors, the top five discriminating combinations were used to obtain an ensemble decision to predict cancer status. The voting-based top multidimensional feature predictor should improve the sensitivity and specificity. The accuracy in blindly predicting cancer status in the validation data was 77.2% (sensitivity 71.4%, specificity 79.6%) using primary nodules. In contrast, non-size based features provided a comparable accuracy of 77.4% (sensitivity 61.9%, specificity 69.4%).
DISCUSSION
In this study, we used observed radiological traits to systematically characterize the size, shape and location of indeterminate pulmonary nodules and quantitatively represented these traits on a point scale. Traditionally, these semantics have been used to prognosticate malignancy in lung cancer(37). A linear classification model was applied on these quantified observed image traits to predict malignancy. The training data set was used to find feature combinations and estimate the accuracy of the predictor in a cross validation setting, graded based on the accuracy, sensitivity, specificity, and the AUROC. The ability of the predictor was blindly evaluated by applying it on validation set. The top five, four-dimensional features were determined, and it was interesting to observe that seven unique features appeared as candidates in both size- and non-size-based models: border definition, vascular convergence, concavity, lobulation, texture, spiculation and nodules in non-tumor lobe (see Supplemental Table S-10). The non-size based feature categories had four additional features selected by the top combinations, namely: location, fissure attachment, pleural attachment, peri-nodule fibrosis. Although linear methods may not discriminate non-linearly separable cases, the limitations are mitigated by using multiple linear fits to derived an ensemble decision (38, 39).
Comparison of various CT features such as contour, shape, and margin, can be helpful for distinguishing between malignant and benign nodules (31, 40–42). The positive relationship of lesion size to likelihood of malignancy has been clearly demonstrated (32, 33). Zerhouni et al. (33) found that more than 80% of benign solitary pulmonary nodules were less than 2 cm in diameter; by contrast, diameters of malignant nodules were nearly uniformly distributed in the range of 1–6 cm, and 50% of the malignant nodules were larger than 2 cm in diameter. We similarly observed that nodules of bigger size were more likely to be malignant. Importantly, we also found some top semantic features are good predictors of malignancy even after size based features were removed to avoid size based bias. A spiculated contour occurs significantly more often in malignant lung nodules (31, 35). This was supported by our results in which the majority of malignant lesions exhibited spiculation. In pathological studies, spiculated contours were shown to be due to thickened interlobular septa, fibrosis caused by obstruction of peripheral vessels, or lymphatic channels filled with tumor cells (43). Nevertheless, in benign lung nodules, especially in inflammatory pseudo tumors or tuberculomas, spiculated edges may also be found (31). Our results agree with other publications (31, 44) that reported similar morphological appearances for the differentiation of benign and malignant IPNs, such as regular shape for benign lesions and lobular for malignant lesions. In a recent study, it is confirmed that the prevalence of lung cancer among current smokers increased from 1.1% for those without emphysema to 2.3% for those with emphysema; among former smokers, the prevalence increased from 0.9% to 1.8%, and for never smokers it increased from 0.4% to 2.6%. Thus, there was a little more than 2-fold increase for current and former smokers while a 6-fold increase among never smokers (45). This could be verified in the current study as we observed severe peri-nodule emphysema has a high frequency to be seen in malignant nodules.
Size (WHO, RECIST: long and short axis), rate of change in size and volume are largely the most important prognostic metrics that have been widely used (46). In response to community’s need to converge on a standard, the American College of Radiology (ACR) created the guidelines to define a positive scan, Lung-Rads (47). Current clinical guidelines relies heavily on the nodule size (11, 47)(cite NCCN and LungRads). Based on nodule size, a wide range of false positives has been reported, 96.4% by the CT arm of the NLST, and 25% by others.(4, 48, 49) Our semantic model in addition to size based predictors will aid the clinical decision support system, including monitoring of the nodule growth.
Designing a predictive method poses several challenges; most often the cohort population has larger number of benign nodules compared to cancerous with range of nodule sizes. Image traits observed by the expert radiologist has the ability to adjust with the system variations (CT parameters) and nodule size differences. It then becomes critical for predictive models to balance positive and negative (sensitivity and specificity) findings rather than relying on single figure of merit. The approach followed by us allows grading the feature pairs according to the predictive performance. We believe our approach of radiological semantics is novel that uses the observed traits on a quantitative scale and apply machine learning classification approach in a systematic cross validation setting. Our results show better performance compared to one of the widely used clinical model (9).
Semantic approach has practical relevance in nodule classification. In a recent Lung nodule classification challenge, our team proposed semantic based approaches to classify indeterminate pulmonary nodules. The challenge had about 10 samples for calibration or training (known outcome) and about 60 samples for blinded testing. The semantic based approach came second with a test AUC of 0.66, while the computerized CAD feature based method was first with AUC of 0.68 with over 15 international participants (50–52).
Due to the retrospective study design and small sample size, to avoid over-fitting the data we collected two independent cohorts from two institutions (train and validation from Vanderbilt Medical Center and Veteran Affairs Hospital in Nashville, samples randomly mixed in the cohorts). Further multi-Institutional studies with large number of patients are warranted to replicate these novel results.
Study Limitations
Our study has some limitations. First, the number of patients was not large. We have taken effort to reduce false discovery by using training and test cohort. Despite this approach, it is possible there could be biases in the patient population, as the current cohorts were predominantly male, derived as they were from a VA population. This could be mitigated by collecting samples in a larger multi-institutional cohort study. Radiologist training and preferences will influence the semantic scoring. While this is less a concern in a research setting, it may be an issue in a clinical practice. Efforts have been made to standardize the scoring sheet with a descriptive atlas (e.g. Supplemental Figure 1 & Supplemental Table S-2) in a way that will be acceptable by the community at large.
CONCLUSION
We have shown radiological image traits are useful in differentiating malignant from non-benign nodules. These semantic features along with size measurement, certainly enhances the prediction accuracy and reduces false positives. The usefulness of radiological imaging traits (semantics) in predicting cancer status shows ability to reduce diagnostic errors compared to clinical models. These, along with conventional measure based on the size, could be collectively used in clinical workflow to better diagnose malignancy.
TRANSLATIONAL / CLINICAL RELEVANCE.
Radiological image traits have been investigated extensively to prognosticate in lung CT both in the context of lesions and nodules. Most of the studies relied on patient survival based models using single or multiple traits.
In this study we have taken a systematic approach to describe and traits on a point scale and score the patient scans for the appearance of a trait. Agnostic learning method was used in a cross validation setting to find the relationships of the traits to the malignancy status. The combination pairs were tested for reliability on a validation cohort.
These pairs of radiological traits (semantics) could be readily be used by the practicing clinician to provide risk assessment for pulmonary nodules, which will help to standardize radiologist inference and improve patient care.
Certainly any inference on biomarkers need to be used with a caution, one needs to account for system level variability at the clinic, parameter settings of the scanner and the operator precision needs to brought to perspective.
Acknowledgments
Author Contributions: Dr. Gillies and Dr. Massion had full access to all the data in the study at respective institutions and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Dr. Liu: contributed to the study design, semantic scoring and manuscript writing.
Dr. Balagurunathan: contributed to study design, statistical analysis and manuscript writing.
Dr. Atwater: contributed to collection of cohort data and patient level information.
Dr. Antic: contributed to collection of cohort data and patient level information.
Dr. Li: contributed to semantic scoring and manuscript writing.
Dr. Walker: contributed in study design and manuscript writing.
Dr. Smith: contributed in the study design and manuscript writing.
Dr. Massion: contributed in the study design, data inference and manuscript writing.
Dr. Schabath: contributed in the study design and manuscript writing.
Dr. Gillies: contributed in the study design, data inference and manuscript writing.
Role of Sponsors: The National Institute of Health (NIH) grant (CA 143062-01) and State of Florida Department of Health (2KT01) grant provided protected time for Drs. Gillies, Balagurunathan, Liu and Qian to work on the research project. The content of the article is solely the responsibility of the authors.
Footnotes
Conflict of Interest
RJG is a consultant and shareholder in HealthMyne, Inc. and oncology-specific PACS system. No other authors of this manuscript have relationships with any companies, whose products or services may be related to the subject matter of the article.
References
- 1.Siegel R, Ma J, Zou Z, AJ Cancer statistics. CA: a cancer journal for clinicians. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Aberle DR, Adams AM, Berg CD, Black WC, Clapp JD, Fagerstrom RM, et al. Reduced lung-cancer mortality with low-dose computed tomographic screening. The New England journal of medicine. 2011;365:395–409. doi: 10.1056/NEJMoa1102873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aberle DR, Adams AM, Berg CD, Clapp JD, Clingan KL, Gareen IF, et al. Baseline characteristics of participants in the randomized national lung screening trial. Journal of the National Cancer Institute. 2010;102:1771–9. doi: 10.1093/jnci/djq434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Patz EF, Jr, Pinsky P, Gatsonis C, Sicks JD, Kramer BS, Tammemagi MC, et al. Overdiagnosis in low-dose computed tomography screening for lung cancer. JAMA internal medicine. 2014;174:269–74. doi: 10.1001/jamainternmed.2013.12738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Swensen SJ, Jett JR, Hartman TE, Midthun DE, Mandrekar SJ, Hillman SL, et al. CT screening for lung cancer: five-year prospective experience. Radiology. 2005;235:259–65. doi: 10.1148/radiol.2351041662. [DOI] [PubMed] [Google Scholar]
- 6.Croswell JM, Baker SG, Marcus PM, Clapp JD, Kramer BS. Cumulative incidence of false-positive test results in lung cancer screening: a randomized trial. Annals of internal medicine. 2010;152:505–12. doi: 10.7326/0003-4819-152-8-201004200-00007. [DOI] [PubMed] [Google Scholar]
- 7.Henschke CI, Yankelevitz DF, Mirtcheva R, McGuinness G, McCauley D, Miettinen OS. CT screening for lung cancer: frequency and significance of part-solid and nonsolid nodules. American Journal of Roentgenology. 2002;178:1053–7. doi: 10.2214/ajr.178.5.1781053. [DOI] [PubMed] [Google Scholar]
- 8.Boiselle PM, Chiles C, Partz E, Tammemagi M, DEW Expert opinion:United States preventive services task force recommendation on screening for lung cancer. J Thorac Imaging. 2014:4. doi: 10.1097/RTI.0000000000000094. [DOI] [PubMed] [Google Scholar]
- 9.Gould MK, Ananth L, PGB A clinical model to estimate the pretest probability of lung cancer in patients with solitary pulmonary nodules. CHEST Journal. 2007:131. doi: 10.1378/chest.06-1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Patel VK, Naik SK, Naidich DP, Travis WD, Weingarten JA, Lazzaro R, et al. A practical algorithmic approach to the diagnosis and management of solitary pulmonary nodules: part 2: pretest probability and algorithm. CHEST Journal. 2013;143:840–6. doi: 10.1378/chest.12-1487. [DOI] [PubMed] [Google Scholar]
- 11.NCCN. NCCN Guidelines for Lung cancer screening. 2015 http://www.nccn.org/patients/guidelines/lung_screening/
- 12.El-Baz A, Suri J. Lung Imaging and Computer Aided Diagnosis. Taylor & Francis; 2011. [Google Scholar]
- 13.Brandman S, Ko JP. Pulmonary nodule detection, characterization, and management with multidetector computed tomography. Journal of thoracic imaging. 2011;26:90–105. doi: 10.1097/RTI.0b013e31821639a9. [DOI] [PubMed] [Google Scholar]
- 14.Sayyouh M, Vummidi DR, Kazerooni EA. Evaluation and management of pulmonary nodules: state-of-the-art and future perspectives. Expert opinion on medical diagnostics. 2013;7:629–44. doi: 10.1517/17530059.2013.858117. [DOI] [PubMed] [Google Scholar]
- 15.Matsuguma H, Mori K, Nakahara R, Suzuki H, Kasai T, Kamiyama Y, et al. Characteristics of subsolid pulmonary nodules showing growth during follow-up with CT scanning. CHEST Journal. 2013;143:436–43. doi: 10.1378/chest.11-3306. [DOI] [PubMed] [Google Scholar]
- 16.Massion P, Walker R. Indeterminate pulmonary nodules: risk for having or for developing lung cancer? Cancer prevention research. 2014:7. doi: 10.1158/1940-6207.CAPR-14-0364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pinsky PF, Nath PH, Gierada DS, Sonavane S, ES Short-and Long-term lung cancer risk associated with noncalcified nodules observed on low-dose CT. Cancer prevention research. 2014:7. doi: 10.1158/1940-6207.CAPR-13-0438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.JC Assessing agreement on classification tasks: The kappa statistic. Computational Linguistics. 1996;22:249–54. [Google Scholar]
- 19.Sim J, CW The Kappa Statistic in Reliability Studies: Use, Interpretation, and Sample Size Requirements. Physical Therapy. 2005;85:257–68. [PubMed] [Google Scholar]
- 20.Kundel HL, MP Measurement of observer agreement. Radiology. 2003;228:303–8. doi: 10.1148/radiol.2282011860. [DOI] [PubMed] [Google Scholar]
- 21.Gould MK, Ananth L, Barnett PG. A clinical model to estimate the pretest probability of lung cancer in patients with solitary pulmonary nodules. CHEST Journal. 2007;131:383–8. doi: 10.1378/chest.06-1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Efron B. Nonparametric estimates of standard error: The jackknife, the bootstrap and other methods. Biometrika. 68:589–99. [Google Scholar]
- 23.Efron BT, Robert J. An introduction to the bootstrap. New York: Chapman & Hall; 1993. [Google Scholar]
- 24.Good P. Permutation, Parametric and Bootstrap Tests of Hypotheses. Springer; 2005. [Google Scholar]
- 25.Ruopp MD, Perkins NJ, Whitcomb BW, Schisterman EF. Youden Index and Optimal Cut-Point Estimated from Observations Affected by a Lower Limit of Detection. Biometrical journal Biometrische Zeitschrift. 2008;50:419–30. doi: 10.1002/bimj.200710415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.WJY Index for rating diagnostic tests. Cancer. 1950;3:32–5. doi: 10.1002/1097-0142(1950)3:1<32::aid-cncr2820030106>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 27.Smialowski P, Frishman D, Kramer S. Pitfalls of supervised feature selection. Bioinformatics (Oxford, England) 2010;26:440–3. doi: 10.1093/bioinformatics/btp621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.GCC, NLC T. Preventing Over-Fitting during Model Selection via Bayesian Regularisation of the Hyper-Parameters. J Mach Learn Res. 2007;8:841–61. [Google Scholar]
- 29.Hemphill E, Lindsay J, Lee C, Mandoiu II, Nelson CE. Feature selection and classifier performance on diverse bio-logical datasets. BMC bioinformatics. 2014;15(Suppl 13):S4. doi: 10.1186/1471-2105-15-S13-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.James G, Witten D, Hastie T, RT . A introduction to statistical learning. Springer; 2013. [Google Scholar]
- 31.Zwirewich C, Vedal S, Miller R, Müller N. Solitary pulmonary nodule: high-resolution CT and radiologic-pathologic correlation. Radiology. 1991;179:469–76. doi: 10.1148/radiology.179.2.2014294. [DOI] [PubMed] [Google Scholar]
- 32.Henschke CI, Yankelevitz DF, Naidich DP, McCauley DI, McGuinness G, Libby DM, et al. CT Screening for Lung Cancer: Suspiciousness of Nodules according to Size on Baseline Scans 1. Radiology. 2004;231:164–8. doi: 10.1148/radiol.2311030634. [DOI] [PubMed] [Google Scholar]
- 33.Zerhouni EA, Stitik F, Siegelman S, Naidich D, Sagel S, Proto A, et al. CT of the pulmonary nodule: a cooperative study. Radiology. 1986;160:319–27. doi: 10.1148/radiology.160.2.3726107. [DOI] [PubMed] [Google Scholar]
- 34.Hu H, Wang Q, Tang H, Xiong L, Lin Q. Multi-slice computed tomography characteristics of solitary pulmonary ground-glass nodules: Differences between malignant and benign. Thoracic cancer. 2016;7:80–7. doi: 10.1111/1759-7714.12280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fan L, Liu SY, Li QC, Yu H, Xiao XS. Multidetector CT features of pulmonary focal ground-glass opacity: differences between benign and malignant. The British journal of radiology. 2012;85:897–904. doi: 10.1259/bjr/33150223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gomez-Saez N, Hernandez-Aguado I, Vilar J, Gonzalez-Alvarez I, Lorente MF, Domingo ML, et al. Lung cancer risk and cancer-specific mortality in subjects undergoing routine imaging test when stratified with and without identified lung nodule on imaging study. Eur Radiol. 2015;25:3518–27. doi: 10.1007/s00330-015-3775-3. [DOI] [PubMed] [Google Scholar]
- 37.Wang H, Schabath M, Liu Y, Berglund A, Bloom A, Kim J, et al. Semiquantitative Computed Tomography Characteristics for Lung Adenocarcinoma and Their Association With Lung Cancer Survival. 2015:1–11. doi: 10.1016/j.cllc.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Herman GT, KTDY On Piecewise-Linear Classification. IEEE Trans Pattern Anal Mach Intell. 1992:14. [Google Scholar]
- 39.Sklansky J, LM Locally trained piecewise linear classifiers. IEEE Trans Pattern Anal Mach Intell. 1980:2. doi: 10.1109/tpami.1980.4766988. [DOI] [PubMed] [Google Scholar]
- 40.Yang Z-G, Sone S, Takashima S, Li F, Honda T, Maruyama Y, et al. High-resolution CT analysis of small peripheral lung adenocarcinomas revealed on screening helical CT. American Journal of Roentgenology. 2001;176:1399–407. doi: 10.2214/ajr.176.6.1761399. [DOI] [PubMed] [Google Scholar]
- 41.Li F, Sone S, Maruyama Y, Takashima S, Yang Z-G, Hasegawa M, et al. Correlation between high-resolution computed tomographic, magnetic resonance and pathological findings in cases with non-cancerous but suspicious lung nodules. European radiology. 2000;10:1782–91. doi: 10.1007/s003300000426. [DOI] [PubMed] [Google Scholar]
- 42.Li F, Sone S, Abe H, MacMahon H, Doi K. Malignant versus Benign Nodules at CT Screening for Lung Cancer: Comparison of Thin-Section CT Findings 1. Radiology. 2004;233:793–8. doi: 10.1148/radiol.2333031018. [DOI] [PubMed] [Google Scholar]
- 43.Kuriyama K, Tateishi R, Doi O, Kodama K, Tatsuta M, Matsuda M, et al. CT-pathologic correlation in small peripheral lung cancers. American Journal of Roentgenology. 1987;149:1139–43. doi: 10.2214/ajr.149.6.1139. [DOI] [PubMed] [Google Scholar]
- 44.Webb WR. Radiologic evaluation of the solitary pulmonary nodule. AJR American journal of roentgenology. 1990;154:701–8. doi: 10.2214/ajr.154.4.2107661. [DOI] [PubMed] [Google Scholar]
- 45.Henschke CI, Yip R, Boffetta P, Markowitz S, Miller A, Hanaoka T, et al. CT screening for lung cancer: Importance of emphysema for never smokers and smokers. Lung Cancer. 2015;88:42–7. doi: 10.1016/j.lungcan.2015.01.014. [DOI] [PubMed] [Google Scholar]
- 46.Prosch H. Implementation of lung cancer screening: promises and hurdles. Trans Lung Canc Res. 2014;3:286–90. doi: 10.3978/j.issn.2218-6751.2014.09.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pinsky PF, Gierada DS, Black W, Munden R, Nath H, Aberle D, et al. Performance of Lung-RADS in the National Lung Screening Trial: a retrospective assessment. Ann Intern Med. 2015;162:485–91. doi: 10.7326/M14-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yankelevitz DF, Kostis WJ, Henschke CI, Heelan RT, Libby DM, Pasmantier MW, et al. Overdiagnosis in chest radiographic screening for lung carcinoma: frequency. Cancer. 2003;97:1271–5. doi: 10.1002/cncr.11185. [DOI] [PubMed] [Google Scholar]
- 49.Veronesi G, Maisonneuve P, Bellomi M, Rampinelli C, Durli I, Bertolotti R, et al. Estimating overdiagnosis in low-dose computed tomography screening for lung cancer: a cohort study. Ann Intern Med. 2012;157:776–84. doi: 10.7326/0003-4819-157-11-201212040-00005. [DOI] [PubMed] [Google Scholar]
- 50.SPIE-Medical-Imaging-2015. Challenge Press Release. 2015 [cited 2015 May 2016]; Available from: http://spie.org/about-spie/press-room/press-releases/mi15--lungx-wrap-3-24-2015.
- 51.Armato SG, 3rd, Hadjiiski L, Tourassi GD, Drukker K, Giger ML, Li F, et al. LUNGx Challenge for computerized lung nodule classification: reflections and lessons learned. Journal of medical imaging (Bellingham, Wash) 2015;2:020103. doi: 10.1117/1.JMI.2.2.020103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Armato SG, 3rd, et al. SPIE-AAPM-NCI Lung Nodule Classification Challenge Dataset. 2015 [cited 2016 May]; Available from: https://wiki.cancerimagingarchive.net/display/Public/LUNGx+SPIE-AAPM-NCI+Lung+Nodule+Classification+Challenge.