ABSTRACT
We previously described Salmonella enterica serovar Heidelberg isolates harboring a chromosomal gene cluster similar to the glutathione S-transferase gene, a putative fosA gene conferring resistance to fosfomycin. Here, we show that this new gene, named fosA7, confers resistance to fosfomycin. The introduction of fosA7 into the fosfomycin-susceptible Salmonella enterica serovar Enteritidis resulted in a substantial increase in the fosfomycin MIC. This finding increases the awareness of antibiotic resistance in Salmonella Heidelberg from broilers as related to the food safety and public health.
KEYWORDS: fosfomycin resistance, fosA7 gene, Salmonella Heidelberg, broiler chicken
TEXT
Many pathogenic bacteria have shown resistance to fosfomycin. Recently, a high prevalence of plasmid-mediated fosfomycin resistance among CTX-M-producing Escherichia coli strains in clinical settings (1–5) and in companion animals (6, 7) has been reported in several countries. Most bacteria are inherently resistant to fosfomycin and carry chromosomal mutations that impair its transport, while others possess fosfomycin-modifying enzymes. Several of these enzymes have been reported in a wide range of Gram-negative bacteria and in some Gram-positive pathogenic bacteria. Fosfomycin resistance was first reported in the early 1980s in a clinical Serratia marcescens strain carrying a plasmid-mediated transposable element, Tn2921, which harbored the fosA gene flanked by two terminal copies of an identical insertion sequence (8). The fosA gene product is a glutathione S-transferase, a metalloenzyme transferred through plasmids to Enterobacteriaceae, with two other variants found in Pseudomonas aeruginosa (9). New subtypes of fosA with similar structures have been described (fosA2, fosA3, fosA4, fosA5 fosA6, fosB, fosC, fosC2, fosX, and fosK); the mechanism of resistance associated with each of them has been reviewed elsewhere (10–14).
We previously described Salmonella enterica serovar Heidelberg isolates harboring a glutathione S-transferase gene cluster having 76 to 80% amino acid sequence similarity with fosA (15). Since such similarities do not translate into a function for an antimicrobial resistance phenotype, and due to the clinical relevance of fosfomycin, the objective of this study was to investigate the ability of this new gene to confer a fosfomycin resistance phenotype in S. enterica serovars. This gene was named fosA7 to remain consistent with the nomenclature after the latest fosA6 gene recently described by Guo et al. (14).
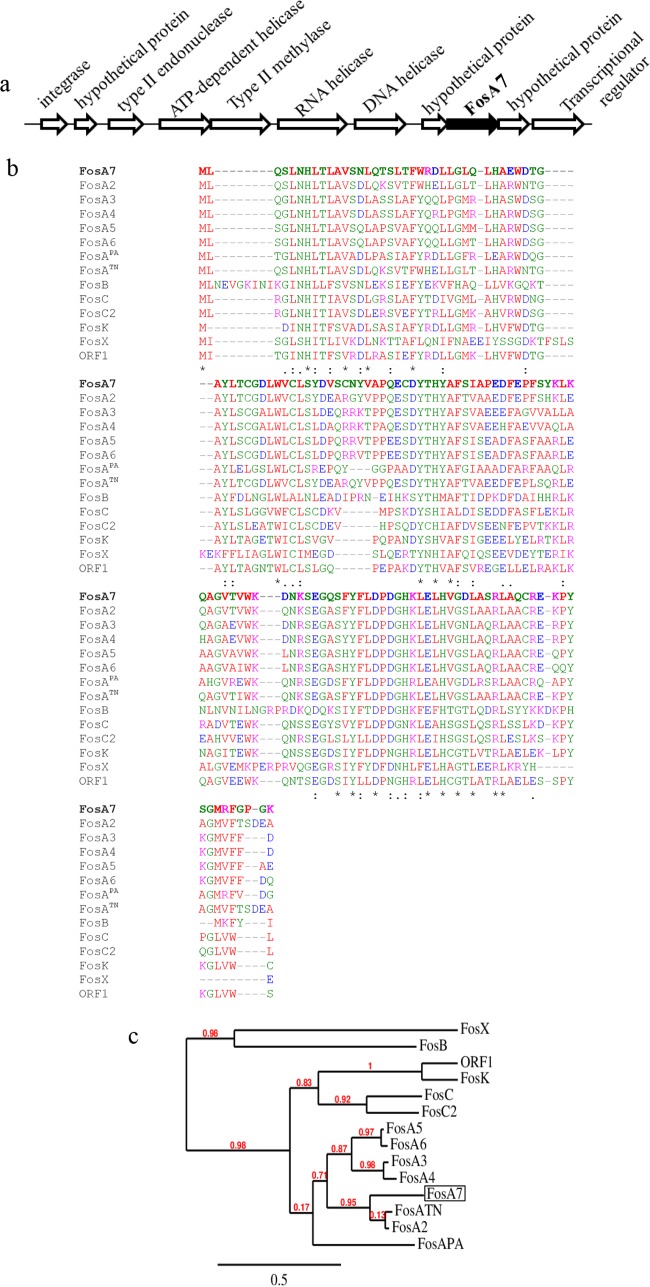
The fosA7 gene was identified on the chromosomes of all four Salmonella Heidelberg strains, surrounded by two hypothetical genes of unknown functions (Fig. 1a). The fosA7 gene has a coding sequence of 423 bp encoding a predicted FosA7 protein of 140 amino acid residues. NCBI-BLAST (16) analysis revealed that the FosA7 protein is highly conserved among different bacteria. Overall, FosA7 shared 30% to 78% amino acid sequence identity with other reported fosfomycin resistance gene subtype products (Fig. 1b). This suggests phylogenetic relationships and a common ancestry with FosA2, FosA4, FosA5, FosA6, FosATN (found in Serratia marcescens), and FosAPA (found in Pseudomonas aeruginosa) (Fig. 1c). FosA7 shares 78.6%, 63.8%, 64.0%, and 62.6% sequence identities with FosA2, FosA4, FosA5, and FosA6, respectively, and 76.4% and 60.3% identities with FosATN and FosAPA, respectively. Interestingly, no fosA7 homolog was found in the closely related Salmonella enterica Enteritidis serovar.
FIG 1.
(a) Genetic environment of a chromosomal fosfomycin resistance gene, fosA7, and its neighboring region (∼19.7 kb) in Salmonella Heidelberg ABB07-SB3031. The gene cassettes are not drawn to scale. (b) Alignment generated in T-Coffee of amino acid sequences of the newly identified FosA7 (in bold) with 13 known fosfomycin resistance proteins found in different bacterial species. Asterisk, amino acid residues conserved among the 14 fosfomycin resistance determinants; colon and dots, amino acid substitutions that result in homologous amino acid residues. The protein GenBank accession numbers are FosA7, KKE03230; FosA2, ACC85616; FosA3, AB522970; FosA4, AB908992; FosA5, AJE60855; FosA6, NG051497; FosAPA, AAT49669; FosATN, AAA98399; FosB, ABS73480; FosC, AAZ14834; FosC2, AB522969; FosK, AB917040; FosX, CWV56762; and open reading frame 1 (ORF1), AAP50248. (c) Phylogenetic relationships between 14 fosfomycin determinants, including the newly described FosA7 (boxed) calculated at http://www.phylogeny.fr.
To assess the fosfomycin resistance phenotype conferred by the chromosomal fosA7 gene, the corresponding 720-bp region derived from the genomic DNA of donor Salmonella Heidelberg strain ABB07-SB3031 was cloned into a high-copy-number vector and transformed into the fosfomycin-susceptible Salmonella Enteritidis ABB07-SB3071. This fosA7-transformed S. Enteritidis strain exhibited complete resistance to fosfomycin (Fig. 2b), in comparison to the recipient transformed with the empty vector, which showed susceptibility to fosfomycin, with an inhibitory zone size greater than 35 mm (Fig. 2a). As shown on Fig. 2c, the contribution of fosA7 in the resistance to fosfomycin was further confirmed in Salmonella Enteritidis ABB07-SB3071 by performing a disk potentiation test, using phosphonoformate (PPF), a specific inhibitor of the fosA gene product, as described by Wachino et al. (17).
FIG 2.
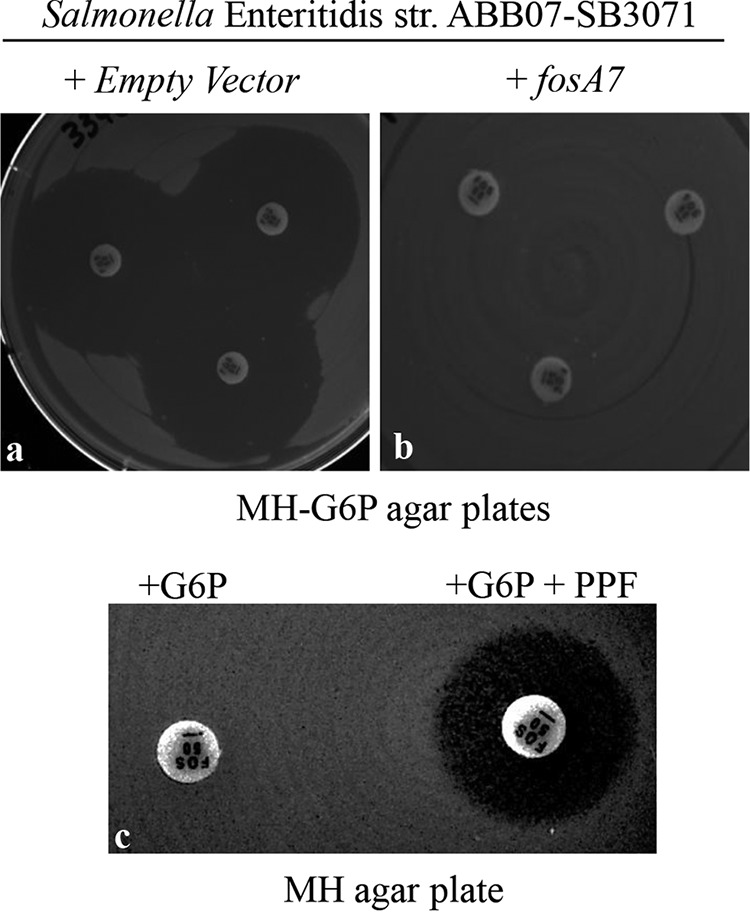
Inhibition zone sizes by fosfomycin (50 μg/disk) in the Kirby-Bauer drug susceptibility test (KBDST) on MH-G6p agar plates (n = 3). (a and b) Salmonella Enteritidis ABB07-SB3071 containing the empty vector (a) and containing the fosA7 gene from Salmonella Heidelberg ABB07-SB3031 (b). (c) Salmonella Enteritidis ABB07-SB3071 transformed with fosA7 on Mueller-Hinton (MH) agar plate; the fosfomycin disk on the left contained 50 μg glucose-6-phosphate (G6P), and the disk on the right contained 50 μg of G6P in addition to 1 mg of phosphonoformate (PPF).
The susceptibility results obtained from the disk diffusion assay were validated using serial broth and agar dilution methods, according to CLSI guidelines (18, 19). The results were interpreted according to the current European Committee on Antimicrobial Susceptibility Testing (26) criteria (susceptible, ≤32 μg/ml; resistant, >32 μg/ml). The MIC values and the Kirby-Bauer test results of four fosA7-positive Salmonella Heidelberg strains and a fosA7-negative Salmonella Enteritidis strain used in this study are presented in Table 1.
TABLE 1.
List of Salmonella strains used in this study and their fosfomycin MIC values with their interpretations
| Inventory no. | Collection date (day-mo-yr) | Organism | Strain or isolate | Zone of inhibition (mm) | Fosfomycin MIC (μg/ml) (resistance)a |
|---|---|---|---|---|---|
| 1768 | 04-Oct-04 | Salmonella Heidelberg | SALB-47-2 | 26 | >32 (R) |
| 1770 | 04-Oct-04 | Salmonella Heidelberg | SALB-46 | 26 | >32 (R) |
| 1773 | 25-Oct-04 | Salmonella Heidelberg | SALB-159-4 | 26 | >32 (R) |
| 3342 | 28-Jun-05 | Salmonella Heidelberg | ABB07-SB3031 | 26 | >32 (R) |
| 3346 | 28-Jun-05 | Salmonella Enteritidis | ABB07-SB3071 | 35 | <2 (S) |
| 3346-1 | 28-Jun-05 | Salmonella Enteritidis | ABB07-SB3071 + empty vector | 35 | <2 (S) |
| 3346-2 | 28-Jun-05 | Salmonella Enteritidis | ABB07-SB3071 + fosA7 | 0 | >512 (R) |
| 12b | NAc | Escherichia coli | ATCC 25922 | 27 | <2 (S) |
MICs were measured by both the broth and agar dilution method recommended by the CLSI and interpreted by EUCAST standard; the interpretation is designated I (intermediate), S (susceptible), or R (resistant).
Quality control strain.
NA, not applicable.
The recipient Salmonella Enteritidis ABB07-SB3071 (containing the fosA7 gene) showed a >256-fold increase in fosfomycin MIC (≥512 μg/ml), thus demonstrating an increased resistance compared to the parent donor strain (Table 1). These results further suggest that fosA7 is responsible for fosfomycin resistance, and if transferred on plasmids, it can induce a high level of resistance in the recipient bacterial strain.
Fosfomycin was recently reintroduced in Canada for the treatment of acute uncomplicated urinary tract infections (UTIs) in adult women caused by E. coli and Enterococcus faecalis (20). Several studies have reported the presence of fosfomycin resistance genes, along with various β-lactamases, including the AmpC-like and extended-spectrum β-lactamases (6, 10). In this study, we characterized a newly identified fosA7 gene that confers resistance to fosfomycin in Salmonella Heidelberg strains isolated from broiler chickens. This fosA7 gene was expressed on a plasmid in a recipient S. Enteritidis strain to induce a >256-fold increase in the fosfomycin MIC value (MIC, ≥512 μg/ml) compared to that of the parental nontransformed strain (MIC, ≥2 μg/ml). The increase in fosfomycin resistance when fosA7 was transferred from the chromosome onto a plasmid suggests that the genetic background may play a role in the expression of this gene. A similar phenomenon was observed for several well-characterized resistance genes, such as chromosomal β-lactamases (21).
Fosfomycin resistance is typically plasmid mediated in most members of Enterobacteriaceae (7). Several published studies have determined the genetic environment of subtypes of this resistance gene, especially fosA3 by PCR mapping and sequencing (1, 22). Two recent studies conducted by Yao et al. (7) and Lin and Chen (23) identified two genetic environments for fosA3 harboring two IS26 transposable elements surrounding the fosA3 gene. The cooccurrence of the fosfomycin resistance gene has often been detected in blaCTX-M-producing and multidrug-resistant bacteria (5, 23). This could further challenge the use of fosfomycin as an alternative treatment approach against UTIs caused by both E. coli and Salmonella (24). Contrary to all previously published data, Hou et al. (6) recently reported that the major type of genes conferring resistance to fosfomycin appears to be chromosomal rather than plasmid mediated. Kitanata et al. (25) identified an integron-mediated chromosomal fosfomycin resistance gene, fosK, in Acinetobacter soli harboring aminoglycoside-modifying enzymes encoded by the aacA4 gene. In the present study, to further explore the prevalence of fosA7, we performed a PCR in 15 Salmonella Heidelberg isolates in our collection, and all were found positive for the fosA7 gene. Additionally, we also performed a BLAST analysis of the FosA7 protein sequence (query length, 140 amino acids) among Salmonella serotypes (∼2,610 known to date) against the microbial protein database. Only 11 matches were found among Salmonella enterica strains. However, in our nucleotide BLAST search (query length, 423 nucleotides) against both the complete and draft genomes (41,831 total genomes available from NCBI RefSeq on 17 February 2017) of Salmonella species available in GenBank, the fosA7 gene was detected in only 35 Salmonella genomes. Of these, 26 genomes (74.3%) were Salmonella Heidelberg, with 100% sequence identity to the query sequence. Others include four Salmonella enterica serovar Agona genomes, three S. enterica serovar Montevideo genomes, and two S. enterica serovar Tennessee genomes, with sequence similarities ranging from 94 to 97%. The nucleotide BLAST against the plasmids (2,417 available in total in the GenBank database on 17 February 2017) showed no significant match. These findings reveal a limited prevalence of fosA7 among the Salmonella serotypes, with Salmonella Heidelberg being the most common carrier of this gene. Unlike E. coli and other pathogens where plasmid-mediated fosfomycin has been detected, the fosA7 gene is exclusively located on the Salmonella chromosome.
In conclusion, we report for the first time a new fosfomycin resistance gene, fosA7, in Salmonella Heidelberg isolates recovered from broiler chickens in British Columbia, Canada. Currently, the presence of fosfomycin resistance among Salmonella species appears to be limited to a few serotypes. Because the fosA7 gene can confer a high level of resistance and is potentially transferable, it is of great concern that fosA7 could further spread via horizontal gene transfer within bacterial communities due to the increased use of fosfomycin in both clinical and veterinary settings. Vigilant monitoring for the spread of fosfomycin resistance in bacteria isolated from humans and animals is needed.
Accession number(s).
The fosfomycin resistance gene (fosA7) sequence is available in GenBank under the accession number KKE03230.1.
ACKNOWLEDGMENTS
This study was supported by a grant from the Genomics Research and Development Initiative on antimicrobial resistance (GRDI-AMR) of the Government of Canada to M.S.D.
We are grateful to Daniel Haft and Mike Feldgarden (NCBI/NIH, Bethesda, MD, USA), for helpful suggestions. We are also thankful to Dion Lepp, Elena Rose Mastin Wood, and Joshua Gong (AAFC) for their assistance.
M.S.D. and M.A.R. designed the study; M.A.R., M.S.D., M.P.-L., and X.Y. wrote the paper; and M.S.D. and M.A.R. reedited the paper.
We declare no conflicts of interest.
REFERENCES
- 1.Wachino J, Yamane K, Suzuki S, Kimura K, Arakawa Y. 2010. Prevalence of fosfomycin resistance among CTX-M-producing Escherichia coli clinical isolates in Japan and identification of novel plasmid-mediated fosfomycin-modifying enzymes. Antimicrob Agents Chemother 54:3061–3064. doi: 10.1128/AAC.01834-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cao XL, Shen H, Xu YY, Xu XJ, Zhang ZF, Cheng L, Chen JH, Arakawa Y. 2016. High prevalence of fosfomycin resistance gene fosA3 in blaCTX-M-harbouring Escherichia coli from urine in a Chinese tertiary hospital during 2010-2014. Epidemiol Infect 145:818–824. doi: 10.1017/S0950268816002879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaase M, Szabados F, Anders A, Gatermann SG. 2014. Fosfomycin susceptibility in carbapenem-resistant Enterobacteriaceae from Germany. J Clin Microbiol 52:1893–1897. doi: 10.1128/JCM.03484-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alrowais H, McElheny CL, Spychala CN, Sastry S, Guo Q, Butt AA, Doi Y. 2015. Fosfomycin resistance in Escherichia coli, Pennsylvania, USA. Emerg Infect Dis 21:2045–2047. doi: 10.3201/eid2111.150750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sato N, Kawamura K, Nakane K, Wachino J, Arakawa Y. 2013. First detection of fosfomycin resistance gene fosA3 in CTX-M-producing Escherichia coli isolates from healthy individuals in Japan. Microb Drug Resist 19:477–482. doi: 10.1089/mdr.2013.0061. [DOI] [PubMed] [Google Scholar]
- 6.Hou J, Huang X, Deng Y, He L, Yang T, Zeng Z, Chen Z, Liu JH. 2012. Dissemination of the fosfomycin resistance gene fosA3 with CTX-M beta-lactamase genes and rmtB carried on IncFII plasmids among Escherichia coli isolates from pets in China. Antimicrob Agents Chemother 56:2135–2138. doi: 10.1128/AAC.05104-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yao H, Wu D, Lei L, Shen Z, Wang Y, Liao K. 2016. The detection of fosfomycin resistance genes in Enterobacteriaceae from pets and their owners. Vet Microbiol 193:67–71. doi: 10.1016/j.vetmic.2016.07.019. [DOI] [PubMed] [Google Scholar]
- 8.Mendoza C, Garcia JM, Llaneza J, Mendez FJ, Hardisson C, Ortiz JM. 1980. Plasmid-determined resistance to fosfomycin in Serratia marcescens. Antimicrob Agents Chemother 18:215–219. doi: 10.1128/AAC.18.2.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beharry Z, Palzkill T. 2005. Functional analysis of active site residues of the fosfomycin resistance enzyme FosA from Pseudomonas aeruginosa. J Biol Chem 280:17786–17791. doi: 10.1074/jbc.M501052200. [DOI] [PubMed] [Google Scholar]
- 10.Falagas ME, Kastoris AC, Kapaskelis AM, Karageorgopoulos DE. 2010. Fosfomycin for the treatment of multidrug-resistant, including extended-spectrum beta-lactamase producing, Enterobacteriaceae infections: a systematic review. Lancet Infect Dis 10:43–50. doi: 10.1016/S1473-3099(09)70325-1. [DOI] [PubMed] [Google Scholar]
- 11.Falagas ME, Vouloumanou EK, Samonis G, Vardakas KZ. 2016. Fosfomycin. Clin Microbiol Rev 29:321–347. doi: 10.1128/CMR.00068-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pérez DS, Tapia MO, Soraci AL. 2014. Fosfomycin: uses and potentialities in veterinary medicine. Open Vet J 4:26–43. [PMC free article] [PubMed] [Google Scholar]
- 13.Xu H, Miao V, Kwong W, Xia R, Davies J. 2011. Identification of a novel fosfomycin resistance gene (fosA2) in Enterobacter cloacae from the Salmon River, Canada. Lett Appl Microbiol 52:427–429. doi: 10.1111/j.1472-765X.2011.03016.x. [DOI] [PubMed] [Google Scholar]
- 14.Guo Q, Tomich AD, McElheny CL, Cooper VS, Stoesser N, Wang M, Sluis-Cremer N, Doi Y. 2016. Glutathione-S-transferase FosA6 of Klebsiella pneumoniae origin conferring fosfomycin resistance in ESBL-producing Escherichia coli. J Antimicrob Chemother 71:2460–2465. doi: 10.1093/jac/dkw177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dhanani AS, Block G, Dewar K, Forgetta V, Topp E, Beiko RG, Diarra MS. 2015. Genomic comparison of non-typhoidal Salmonella enterica serovars Typhimurium, Enteritidis, Heidelberg, Hadar and Kentucky Isolates from broiler chickens. PLoS One 10:e0128773. doi: 10.1371/journal.pone.0128773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 17.Wachino J, Kimura K, Yamada K, Jin W, Arakawa Y. 2014. Evaluation of disk potentiation test using Kirby-Bauer disks containing high-dosage fosfomycin and glucose-6-phosphate to detect production of glutathione S-transferase responsible for fosfomycin resistance. J Clin Microbiol 52:3827–3828. doi: 10.1128/JCM.01805-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.CLSI. 2015. Performance standards for antimicrobial susceptibility testing; 25th informational supplement. CLSI document M100-S25 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 19.Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing; 19th ed, approved standard CLSI document M100-S19 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 20.Forest Laboratories. 1997. Monurol (fosfomycin tromethamine). Forest Laboratories, Inc., St. Louis, MO: https://www.accessdata.fda.gov/drugsatfda_docs/label/2011/050717s007lbl.pdf. [Google Scholar]
- 21.Reisbig MD, Hossain A, Hanson ND. 2003. Factors influencing gene expression and resistance for Gram-negative organisms expressing plasmid-encoded ampC genes of Enterobacter origin. J Antimicrob Chemother 51:1141–1151. doi: 10.1093/jac/dkg204. [DOI] [PubMed] [Google Scholar]
- 22.Fu Z, Liu Y, Chen C, Guo Y, Ma Y, Yang Y, Hu F, Xu X, Wang M. 2016. Characterization of fosfomycin resistance gene, fosB, in methicillin-resisatnt Staphylococcus aureus isolates. PLoS One 11:e0154829. doi: 10.1371/journal.pone.0154829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin D, Chen S. 2015. First detection of conjugative plasmid-borne fosfomycin resistance gene fosA3 in Salmonella isolates of food origin. Antimicrob Agents Chemother 59:1381–1383. doi: 10.1128/AAC.04750-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Allerberger FJ, Dierich MP, Ebner A, Keating MR, Steckelberg JM, Yu PK, Anhalt JP. 1992. Urinary tract infection caused by nontyphoidal Salmonella: report of 30 cases. Urol Int 48:395–400. doi: 10.1159/000282362. [DOI] [PubMed] [Google Scholar]
- 25.Kitanaka H, Wachino J, Jin W, Yokoyama S, Sasano MA, Hori M, Yamada K, Kimura K, Arakawa Y. 2014. Novel integron-mediated fosfomycin resistance gene fosK. Antimicrob Agents Chemother 58:4978–4979. doi: 10.1128/AAC.03131-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.European Committee on Antimicrobial Susceptibility Testing. 2017. Breakpoint tables for interpretation of MICs and zone diameters, v 7.0. http://www.eucast.org. [Google Scholar]