ABSTRACT
Staphylococcus aureus has acquired resistance to nearly all antibiotics used in clinical practice. Whereas some resistance mechanisms are conferred by uptake of resistance genes, others evolve by mutation. In this study, IS256 has been shown to play a role, e.g., in S. aureus strains displaying intermediate resistance to vancomycin (VISA). To characterize the IS256 insertion sites in the genomes of two closely related sequence type 247 (ST247) VISA strains, all insertions were mapped in both VISA and a susceptible control strain. The results showed that the three ST247 strains contained the highest number so far of IS256 insertions for all sequenced S. aureus strains. Furthermore, in contrast to the case with the other IS elements in these genomes, the IS256 insertion sites were not identical in the closely related strains, indicating a high transposition frequency of IS256. When IS256 was introduced into a laboratory strain which was then cultured in the presence of antibiotics, it was possible to isolate small-colony variants (SCVs) that possessed IS256 insertions in guaA and hemY that displayed increased resistance to vancomycin and aminoglycosides, respectively. For these clones, a very rapid reversion to the wild type that resembled the fast reversion of clinical SCVs was observed. The reversion was caused by excision of IS256 in a small number of fast-growing clones that quickly outcompeted the SCVs in broth cultures. In conclusion, the presence of IS256 confers a strong genomic plasticity that is useful for adaptation to antibiotic stress.
KEYWORDS: Staphylococcus aureus, transposition, phase variation, insertion element IS256, vancomycin, aminoglycosides, hemY, guaA, small-colony variants (SCVs), VISA
INTRODUCTION
IS256 was first described as a part of the transposon Tn4001, which harbors the gene aacA-aphD and mediates resistance to several aminoglycosides in the nosocomial pathogen Staphylococcus aureus (1, 2). It has a length of 1,324 bp, and the single gene tnpA encodes a DDE-transposase that contains two Asp (D) and one Glu (E) residue in its active site. (3). The ends of IS256 are formed by two imperfect inverted repeats (IRL and IRR). In addition, the insertions of IS256 are framed by two 8-bp direct repeats which are formed by duplication of the target sequence during transposition (4). IS256 transposes by a “copy and paste” mechanism (5), thereby leading to an accumulation of copies in the genome, since every transposition will increase the copy number by one. The integration of a single insertion sequence (IS) element into a gene or its promoter may result in an inactivation or overexpression of the affected gene. This way, IS256 confers phase variation phenomena concerning biofilm formation or sigma factor B activity on S. aureus and Staphylococcus epidermidis (4, 6–9). In addition, in S. aureus it has been shown to be involved in modulation of antibiotic resistance (10–13) and virulence; e.g., an insertion into the promoter of rot led to an increased virulence in S. aureus USA500 (14).
Here we present the genome sequences of three methicillin-resistant Staphylococcus aureus (MRSA) strains that contain a very high number of IS256 insertions. All strains belong to sequence type 247 (ST247), which is a multiresistant ancient sublineage of clonal complex 8 (CC8). The very first MRSA strain that was discovered in the United Kingdom in the early 1960s, strain COL (15, 16), also belongs to CC8 (ST250) and is the closest relative of these strains in the database. ST247 was most widespread in the early 1990s, when it caused hospital-associated MRSA outbreaks in several European countries and was referred to as Iberian strain or the Northern German Epidemic type as well as UK-EMRSA-5, -8, and -17, Rome clone, Spanish pulsed-field gel electrophoresis (PFGE) type E1, Irish AR22, and Irish New02 (16). Strain S. aureus SA137/93A was originally isolated in 1993 from a tracheal secretion in Germany, and during a retrospective screening in 1997, it was discovered that this isolate showed intermediate resistance to vancomycin (17). After subculturing of strain SA137/93A in the presence of vancomycin (6 μg/ml), a variant with an even higher resistance to vancomycin was isolated and designated S. aureus SA137/93G (18). Further studies of the vancomycin resistance mechanism showed that the presence of IS256 seemed to be important for the evolution of resistance, since it had inserted upstream of the two-component regulatory system walRK, upregulating its expression in strain SA137/93A, and inactivated the gene tcaA in strain SA137/93G (10, 12).
In this study, in order to learn more about the role of IS256 in resistance development, selection in the presence of antibiotics was performed with plasmids harboring IS256 elements that had been tagged with antibiotic resistance genes. These studies showed that IS256 may be involved in the generation of unstable small-colony variant (SCV)-like phenotypes. SCVs are slowly growing cells that are able to cause chronic infections and often have lost susceptibility to antibiotics such as aminoglycosides or trimethoprim-sulfamethoxazole. Clinical SCV isolates are often formed during intracellular growth (19) or prolonged antibiotic therapy (20); they are often unstable and therefore very difficult to handle, especially in broth culture, and most laboratory experiments have been performed with stable genetically engineered variants that are inactivated in hemB and menD or that possess a deletion in hemH (21–23). Recently it was shown that in infection models, long-term persistence induced SCV phenotypes, which rapidly reverted to the wild-type phenotype when the organisms were subcultured in the laboratory (24). The mechanism of this switch is still unclear (20), but it was hypothesized that the stress factors encountered during infection might activate the transition and that SigB is involved in this mechanism (19, 24). Here we show that the formation and fast reversion of SCV-like phenotypes may also be mediated by transposition and subsequent excision of IS256.
RESULTS
Genomes of three ST247 strains.
Table 1 provides an overview of the mobile and accessory genetic elements of the three ST247 strains. The number of mobile genetic elements and resistance genes was striking, especially in strain SA137/93A, which was susceptible only to netilmicin, fusidic acid, and mupirocin. All three strains contained multiple copies of IS elements (IS1272, IS1272-related elements, IS1181, IS431mec, and IS256) which were all mapped by PCR (Table 1; for all insertion sites, see Tables S1 and S2 in the supplemental material).
TABLE 1.
Overview of mobile genetic elements, pathogenicity islands, and amino acid exchanges conferring antibiotic resistance for 3 ST247 strains
| Feature | Datum for strain: |
||
|---|---|---|---|
| SA1450/94 | SA137/93A | SA137/93G | |
| Total no. of IS elements | 46 | 63 | 53 |
| No. of copies of IS elements | |||
| IS256 | 33 | 44 (chromosome) + 1 (pSA93A) | 39 |
| IS1181 | 5 | 7 | 7 |
| IS431mec | 1 | 2 (chromosome) + 2 (pSA93A) | 1 |
| IS1272 | 1 | 1 | |
| IS1272 related | 6 | 6 | 6 |
| Free plasmid pSA93A | pSA93A: merA, merB, cadA, blaZ, qacA | ||
| Integrated pUB110 | pUB110: aadD, bleO | ||
| Transposons | Tn4001: aac(6′)-aph(2″) Tn5801 variant: tetM Tn554: ermA, ant9-Ia | Tn4001: aac(6′)-aph(2″) Tn5801 variant: tetM Tn554: ermA, ant9-Ia | Tn4001: aac(6′)-aph(2″) Tn5801 variant: tetM Tn554: ermA, ant9-Ia |
| SCCmec cassette | Type I | Type IA | Deleted (18) |
| No. of prophages (complete) | 4 | 3 | 3 |
| No. of prophages (incomplete) | 2 | 2 | 2 |
| No. of pathogenicity islands (νSaα, νSaβ, νSaγ) | 3 | 3 | 3 |
| Exchanges in RpoBa | H481N, L875S, V798A | H481N, L875S, V798A, S529L | H481N, L875S, V798A, S529L |
| Exchange in GyrAb | S84L | S84L | S84L |
| Exchange in ParCb | S80F | S80F | S80F |
Exchanges in RpoB confer resistance to rifampin (H481N, L875S, and V798A) and the S529L exchange was previously observed in a VISA strain (58).
Exchanges in in GyrA and ParC confer resistance to quinolones.
In total, 67 different IS256 insertions were detected in the three strains. Unlike the other IS elements (IS1272, IS1272-related elements, IS1181, and IS431mec), which were mostly located between the coding sequences, 46% of the IS256 insertions were intragenic, inactivating hypothetical genes (58%) and genes involved in processing of genetic information (23%), in metabolism (13%), and in regulatory processes (6%) (Table S2).
In spite of the close relationship between strains SA137/93A and SA137/93G, at least four transpositions of IS256 had occurred (Fig. 1A), indicating that its transposition frequency is rather high in these strains. This is also stressed by the fact that all insertions that were found for the other IS elements were conserved between the strains (Table S2). The inactivation of sarU was among the 13 IS256 insertions detected in all three isolates. Strains SA137/93A and SA137/93G carried 21 common IS256 insertions, among others in the promoter of rot and in the open reading frame (ORF) of comGB. The insertion in the promoter of rot was at nearly the same place as the insertion described previously for S. aureus USA500 (14). However, IS256 is inserted in the opposite direction from that in S. aureus USA500; i.e., in strain SA137/93 the strong C-terminal −35 promoter sequence of IS256 is located in the vicinity of the promoter of rot, which might alleviate any negative effects of this insertion on transcription. Indeed, previous microarray experiments comparing strain S. aureus SA137/93A to strain SA1450/94, which does not contain this insertion, had not indicated any differences concerning transcription of rot or any toxins (25). Although IS256 was integrated in the promoter region of fosB in SA137/93A and SA137/93G, the strains showed a fosfomycin resistance phenotype.
FIG 1.
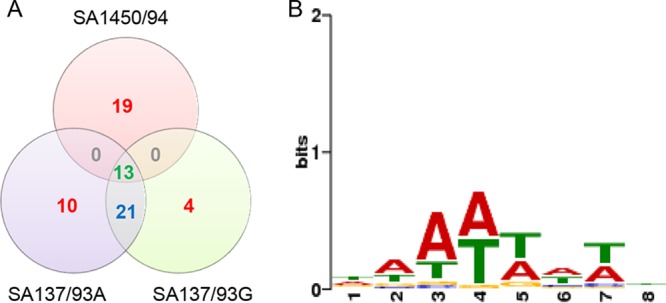
IS256 insertion sites. (A) Distribution of 67 different IS256 insertion sites in three ST247 strains. (B) Weight matrix of 67 different direct repeats of IS256 in ST247 strains.
Three insertions led to recombinations between neighboring IS256 elements and deletion of the sequence between the two copies; e.g., the deletions of SACOL1333 and parts of SACOL1859 were most probably caused by homologous recombination of two IS elements inserted in the same orientation (see Fig. S1A and B in the supplemental material). Most notably, if two IS256 elements are inserted in a head-to-head manner, a 9-bp direct repeat (TGTAAAAGT) is formed. Illegitimate recombination at this direct repeat most probably led to deletion of the largest parts of two IS256 elements inserted in a head-to-head manner (at bp 1754695 in the reference genome of S. aureus COL) (Fig. S1C).
A comparison of all insertion sites showed that AT-rich sequences were dominant, especially in positions 3 to 5 and 7; the median GC content of the insertion sites was 23.5%. Figure 1B shows the weight matrix of all 67 direct repeats.
Insertion into guaA yields an in vitro SCV phenotype with slightly increased resistance to vancomycin.
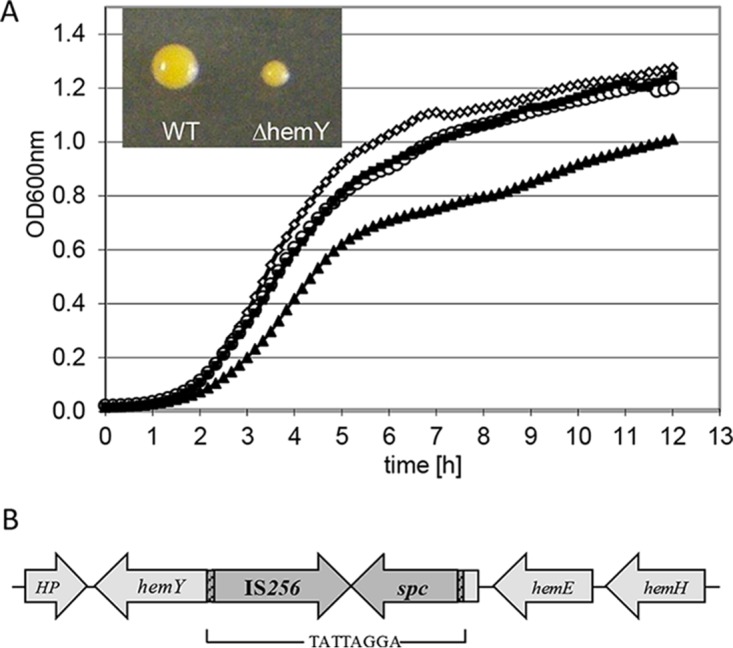
In order to evaluate whether the presence of IS256 confers a selective advantage in the presence of vancomycin, S. aureus HG001 containing plasmid pA3, which contains a composite IS256 (IS256-ermB [26]), and strains that harbored one insertion of IS256-ermB in their genomes (HG001 W5 and HG001 G7) were passaged in the presence of increasing concentrations of vancomycin for at least 10 days. Controls contained the empty vector pBT2, no vector, or pBT2-ermB. After 10 days, all strains were able to grow at vancomycin concentrations of 10 μg/ml. Therefore, we could not show that a selective advantage is always conveyed by the presence of multiple copies of IS256 per se. However, a mutant was isolated from the culture of S. aureus HG001 pA3 that contained an insertion of IS256-ermB in the gene guaA. This mutant was present after day 7 as well as after day 24, showed an MIC of 4 to 8 μg/ml for vancomycin, and formed rather small colonies. guaA encodes the basic metabolic gene GMP synthetase.
In order to evaluate the effect of this insertion on vancomycin susceptibility and exclude the effects of further point mutations in the chromosomal background of the strains, the insertion was transduced into S. aureus HG001 via phage 85 and was verified by PCR analysis. The resulting mutant showed very slow growth with trace amounts of guanine present in tryptic soy broth (TSB) and brain heart infusion (BHI) medium (Fig. 2) and virtually no growth in the absence of guanine in modified RPMI medium (Fig. S2), indicating guanine auxotrophy. On BHI medium, small colonies, reminiscent of SCVs, were formed. Furthermore, there was a slight rise in resistance to vancomycin, from 2 μg/ml (S. aureus HG001) to 3 μg/ml (S. aureus HG001 guaA::IS256-ermB). This increase in the MIC was observed only in the absence of exogenous guanine; the addition of guanine reversed the effect (Fig. 3). In conclusion, the strain displayed the phenotype of a guanine auxotrophic SCV.
FIG 2.
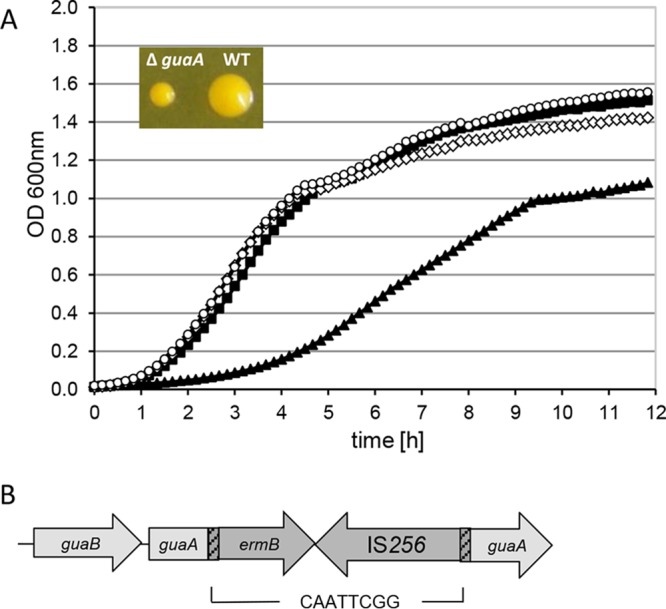
Characterization of the SCV S. aureus HG001 guaA::IS256-ermB. (A) Growth kinetics of S. aureus HG001 guaA::IS256-ermB in the presence (■) and in the absence (▲) of 0.9 mM guanine as well as S. aureus HG001 in the presence (○) and in the absence (♢) of 0.9 mM guanine in BHI medium. The inset shows the colony size after 48 h on BHI agar at 37°C (WT, wild type; ΔguaA, guaA::IS256-ermB). (B) Localization of IS256-ermB in guaA. The inverted repeats are shown in the hatched area, and the 8-bp direct repeat is shown below the diagram.
FIG 3.
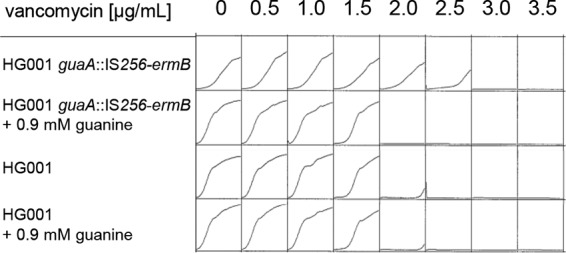
Vancomycin susceptibility of the SCV S. aureus HG001 guaA::IS256-ermB. Growth experiments employed S. aureus HG001 guaA::IS256-ermB and S. aureus HG001 in the presence of different concentrations of vancomycin in BHI medium in the presence and absence of guanine; growth was determined using a microplate reader. Shown are the results of one of three replicates.
Insertion into hemY yields an SCV with increased resistance to aminoglycosides.
In another experiment, plasmid pA6, which does not replicate at 45°C and contains a genetically engineered IS256 element harboring the resistance gene spc or ant(9)-Ia (27), which confers resistance to spectinomycin but not to streptomycin (28), was used for selection of spectinomycin-resistant colonies. After culturing on agar containing spectinomycin (150 μg/ml) at 45°C, one of the colonies contained an insertion of IS256-spc into the 3′ end of hemY (S. aureus HG001 hemY::IS256-spc) as shown in Fig. 4. The colony showed slow growth, and the wild-type phenotype was reconstituted by growth in the presence of hemin (Fig. 4). In the absence of oxygen, the phenotype was indistinguishable from that of the wild type. Hemin auxotrophic SCVs with mutations in the heme biosynthesis genes are often selected in the presence of aminoglycosides (29). This strain also displayed resistance to kanamycin and gentamicin, which is not conveyed by spc and was not present in another mutant, S. aureus HG001 G2, which contained an intergenic insertion of IS256-spc between SAOUHSC_0967 and SAOUHSC_0968, and a revertant that had lost the insertion (Table 2). In conclusion, S. aureus HG001 hemY::IS256-spc showed the phenotype of an aminoglycoside-selected SCV. The insertion hemY::IS256-spc harbored a deletion of the last bases of the direct repeat (GAT). However, a 9-bp direct repeat was still formed, as the first 3 bases of IS256 (GAT) complemented this deletion. Any further mutations in the heme biosynthesis operons, hemEHY and hemAXCDBL, in this clone were excluded by sequencing.
FIG 4.
Characterization of the SCV S. aureus HG001 hemY::IS256-spc. (A) Growth kinetics of S. aureus HG001 hemY::IS256-spc in the presence (■) and in the absence (▲) of 2 μg/ml of hemin as well as S. aureus HG001 G2 in the presence (○) and in the absence (♢) of 2 μg/ml of hemin in TSB medium containing 64 μg/ml of spectinomycin. The inset shows the colony size after 48 h on TSA agar with 150 μg/ml of spectinomycin at 37°C (WT, S. aureus HG001 G2; ΔhemY, hemY::IS256-ermB). (B) Localization of IS256-spc in hemY. The inverted repeats are shown in the hatched area, and the 8-bp direct repeat is shown below the diagram.
TABLE 2.
MICs of drugs for S. aureus HG001 hemY::IS256-spc in comparison to S. aureus HG001 G2 and the revertant S. aureus HG001 R1, which had lost the IS256-spc element, in MH broth
| Strain | MIC (μg/ml) of: |
||
|---|---|---|---|
| Kanamycin | Gentamicin | Spectinomycin | |
| S. aureus HG001 G2 | 3.6 | 0.5 | >128 |
| S. aureus HG001 hemY::IS256-spc | 58 | 4 | >128 |
| S. aureus HG001 R1 | 4 | 0.5 | 64 |
The hemY mutant still produced some staphyloxanthin (Fig. 4), which was particularly noticeable after 40 h of incubation. Typically, clinical SCVs and ΔhemB mutants show no or decreased pigment production compared to that of the wild type (20). However, when CtaB, the enzyme that catalyzes the very last step of the pathway and couples the farnesyl diphosphate chain to protoheme IX, is removed in S. aureus, an increased pigmentation is observed, indicating that farnesyl diphosphate is fed into the pigment production pathway (30). Since the insertion into hemY interrupts the pathway between the steps catalyzed by HemB and CtaB, the hemY mutant was supplemented with hemin on an agar plate to assess staphyloxanthin production in the absence and presence of hemin. The result clearly showed that the hemY mutant produced less color in the absence of hemin (Fig. S3).
SCV phenotypes that are caused by IS256 insertions revert quickly to the normal phenotype.
IS256 replicates by a copy-and-paste mechanism (5). Loss of an insertion is mediated by illegitimate recombination via the 8-bp direct repeats and is a rare event (3). In order to test the stability of the SCV, the guaA and the hemY mutants were passaged in BHI (data not shown) or TSB (Fig. 5A). The selective antibiotic was either absent or present only in the very first culture, which had been inoculated by resuspending an SCV colony (day 0), or present from day 0 to day 4. Every day, aliquots of the cultures were plated on BHI or TSB agar plates (no addition of antibiotics) and SCV colonies and wild-type colonies were counted. In the absence of erythromycin, the reversion of the guaA mutant occurred quickly, with 60% revertants on day 1 after 24 h of incubation (Fig. 5A). The presence of erythromycin (4 μg/ml) in the first culture delayed the reversion (Fig. 5B), and this effect was even more pronounced when the antibiotic was present during all passages (Fig. 5C), indicating that the presence of erythromycin selected for strains that had kept the IS256-ermB element in their genome. Large-colony-forming revertants were checked for the insertion of IS256-ermB in guaA by PCR using the primer pair guaA_forMH and guaA_revMH or guaA_for and guaA_rev. The IS element was lost in most cases, but there were also revertants that showed the large-colony phenotype and still harbored the insertion sequence in guaA. The uptake of guanine or its precursors in S. aureus has not been investigated, but our growth experiments in the presence of guanine in RPMI medium demonstrated that such an uptake must exist. For Escherichia coli, several purine transporters have been identified (purP, ygfO, and yicE) (31) which according to protein BLAST resemble SA2050, pbuX, and SA1042 (pyrP) in S. aureus. However, sequencing (promoters and structural genes of SA1041 [pyrR], SA1042 [pyrP], SA2050, and the riboswitch upstream of xpt-pbuX-guaB-guaA [32]) did not reveal any mutations.
FIG 5.
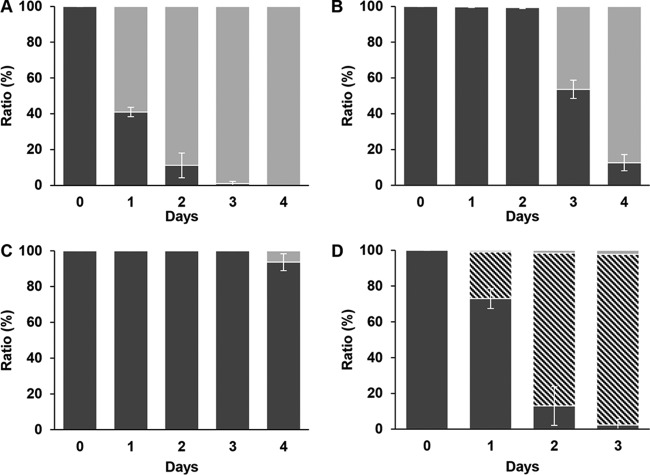
Reversion of S. aureus guaA::IS256-ermB to the wild-type colony phenotype. (A) S. aureus guaA::IS256-ermB (black) reverting to the large-colony phenotype (gray) during daily serial passages in TSB medium without antibiotic. (B and C) Same experiment as for panel A performed in TSB containing antibiotic (4 μg/ml of erythromycin) in the first culture (inoculated on day 0) only (B) and in TSB containing 4 μg/ml of erythromycin from day 0 to day 4 (C). (D) Appearance of surrogate revertants, i.e., blue colonies harboring pMAD (oblique striped) during serial passages in TSB medium containing 25 μg/ml of erythromycin from day 0 to day 3. For this experiment, the initial inoculum was approximately 1 CFU/ml of S. aureus HG001 pMAD and one SCV colony (2 × 105 to 6 × 105 cells/ml). Error bars indicate the standard deviations from at least 3 experiments.
The phenotype of S. aureus HG001 hemY::IS256-spc was also lost very quickly in TSB and BHI (Fig. 6A). As described above, the reversion could be delayed by the addition of spectinomycin (Fig. 6B) and the delay could be prolonged, but not inhibited, if spectinomycin (150 μg/ml) was always present (Fig. 6C). Characterization of 12 randomly chosen revertant colonies from several experiments with S. aureus HG001 hemY::IS256-spc showed that IS256-spc was never retained in hemY; i.e., the reversion was always accompanied by loss of IS256-spc from hemY. Four out of eight hemY revertants chosen on day 4 from experiments that had been conducted in the presence of antibiotics at least at day 0 were still able to grow on antibiotic-containing agar, indicating that the resistance gene was still present in these strains. In order to localize the spc gene in two such mutants, the insertion sites of IS256-spc were analyzed by inverse PCR and sequencing, and it could be shown that IS256-spc had inserted into SAOUHSC_00997 and SAOUHSC_00950, respectively. Both revertants had been isolated on day 4 from the same culture incubated with antibiotic on day 0 only.
FIG 6.
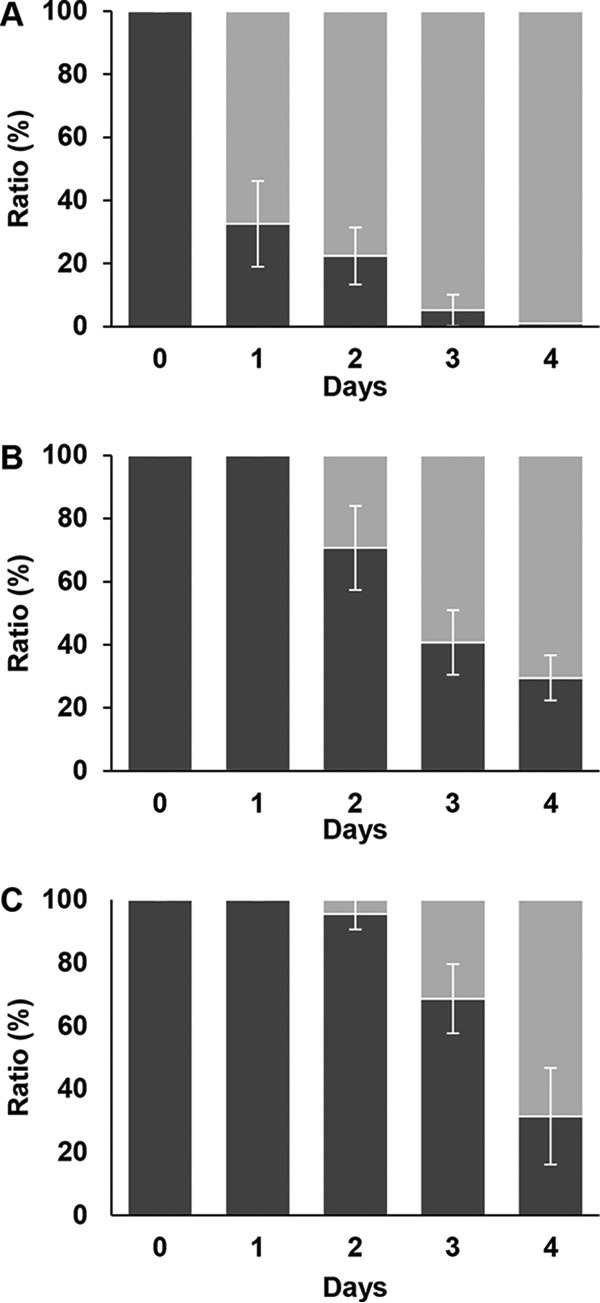
Reversion of S. aureus hemY::IS256-spc to the wild-type colony phenotype. (A) S. aureus hemY::IS256-spc (black) reverting to the large-colony phenotype (gray) during daily serial passages in TSB medium without antibiotic. (B and C) Same experiment as for panel A performed in TSB containing antibiotic (150 μg/ml of spectinomycin) in the first culture on day 0 only (B) and in TSB containing 150 μg/ml of spectinomycin from day 0 to day 4 (C). Error bars indicate the standard deviations from at least 3 experiments.
This result indicated that in the presence of antibiotics, replicative transposition of IS256 with the associated resistance gene into another locus of the chromosome had enabled subsequent loss of the IS element from hemY. This way, reversion of the phenotype even in the presence of the selective antibiotic was possible; however, two successive mutational events, i.e., transposition of IS256 using the copy-and-paste mechanism and the subsequent illegitimate recombination that excised IS256 from hemY, were needed.
Reversion may be mediated by a very low number of quickly growing cells.
As a control, the reversion experiments were repeated with both auxotrophic SCVs in TSB and BHI after supplementation with guanine (0.9 mM) or hemin (2 μg/ml) in the absence of antibiotics, respectively. These conditions conferred a fast-growing phenotype on the SCV cells; therefore, possible revertants could not outcompete the SCV genotype cells. When culture aliquots were plated on guanine-free or hemin-free agar, all cells had retained the SCV phenotype and no revertant colonies were visible after 4 days. This result indicates that the reversion might be mediated by only a few mutational events that occur in a very small subpopulation and is not accomplished by simultaneous mutation of the majority of the cells. We therefore hypothesized that only a few revertant cells would be able to outcompete the SCV by their fast growth. In our media, the doubling time of the SCV cells was twice as long as that of the wild type, and both strains grew more quickly in TSB than in BHI.
In order to visualize the competition between SCV and a fast-growing revertant, we inoculated 5 ml of TSB containing a high concentration of the selective antibiotic (25 μg/ml of erythromycin) with one SCV colony of S. aureus HG001 guaA::IS256-ermB, yielding 2 × 105 to 6 ×105 CFU/ml, and added approximately 1 CFU/ml of the parent strain S. aureus HG001 (displaying the large phenotype) harboring plasmid pMAD. This strain is also resistant to erythromycin at 30°C and forms blue colonies on agar containing 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal). Thus, it served as an easily detectable surrogate “revertant” in this experiment. Figure 5D shows the appearance of blue colonies harboring pMAD in the culture after passages at 30°C. Indeed, the progeny of the plasmid-bearing clone represented the majority of the culture after 2 days, indicating that a very low number of mutants was sufficient for a reversion of the SCV genotype. In BHI the same phenomenon was observed but with a slower appearance of blue colonies.
DISCUSSION
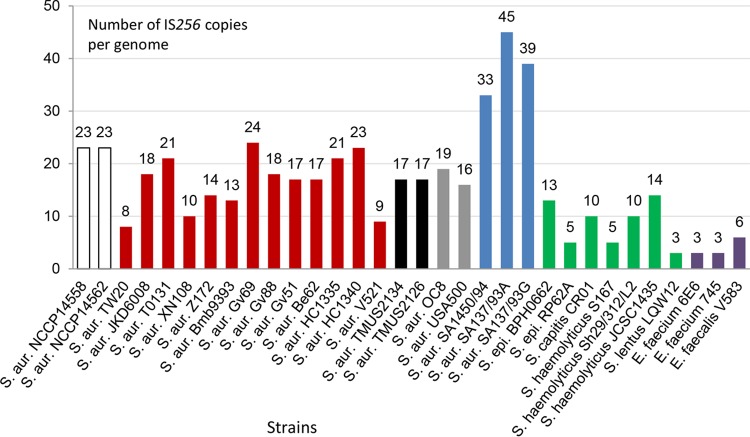
This paper describes the genomes of three strains of the Iberian clone (ST247). The high number of IS256 elements present in the ST247 genomes is striking; it is the highest number found by NCBI BLAST using all complete staphylococcal and enterococcal genomes deposited in the databases so far (Fig. 7). The IS density of S. aureus 137/93A (number of IS elements per kilobase of nucleotides) is 0.021 and is at the upper limit for free-living organisms (33). The highest number of IS elements in prokaryotes has been described for Wolbachia strains (52 to 171 elements per genome); however, more than 70% of these ISs constitute “fossil insertions” that contain inactivating mutations (34). The different insertion sites of IS256 encountered in two closely related isolates indicate a high transposition activity. This might be specific for these isolates, as the copy number of the transposase gene will increase with the number of IS elements; therefore, the transposition activity might increase along with the number of tnpA copies in these strains. In the end, such an “IS expansion” may run out of control and result in genome reduction by favoring homologous recombination (34). Two examples of such deletions are present in the strains described here (Fig. S1), and interestingly, a large IS256-based genomic inversion that contains 19 copies of IS256 has been described for S. aureus OC8 (35). IS256 itself is present only in some successful MRSA lineages, most of which belong to clonal complex 8 (i.e., ST239, ST8, ST247, and ST72 [16]) (Fig. 7). Other staphylococcal IS elements, like IS257/431mec, are found in more lineages and show lower copy numbers and transposition activity. In our hands, IS257/431mec was nearly inactive when we tested its transposition frequency (data not shown). However, IS257/431mec might be involved in recombination events like the integration of pUB into SCCmec in strain SA137/93A or the large deletion present in strain SA137/93G (18) and other VISA strains (36).
FIG 7.
Distribution of IS256 in different cocci. Sequence types (ST) of S. aureus are indicated by colors as follows: ST5, white; ST239, red; ST72, black; ST8, gray; and ST247, blue. Strains with fewer than three IS256 copies are not shown. Coagulase-negative staphylococci are green, and Enterococcus species are purple. IS256 was localized by NCBI BLAST and S. aureus MLST types were determined by MLST 1.8. S. aur., S. aureus; S. epi., Staphylococcus epidermidis.
Plasmids pA3 and pA6 contain the pE194ts replicon, which gives rise to about 14 copies of the plasmid (37), which would be equivalent to 14 copies of IS256 in the genome and is in the range of the average copy number of IS256 in clinical CC8 isolates (Fig. 7). These plasmids were used to introduce a recombinant IS element into S. aureus HG001 in the presence of antibiotics; most intriguingly, unstable SCV-like phenotypes were generated during these experiments.
The insertion into guaA led to slow growth and a slight increase in resistance to vancomycin, if guanine was not added to the BHI serving as culture medium. The absence of GuaA activity elicits the stringent response in staphylococci (38). This phenomenon, which is also triggered by mutations in relA (rsh), relP, or relQ, has already been described in connection with intermediate vancomycin resistance and slow growth in Staphylococcus and Enterococcus (39–41). Indeed, an rsh mutant with an SCV phenotype was also isolated from a patient with a recurrent infection (42). In addition, knockout of guaA leads to thickened cell walls (43). However, guaA deletion mutants are not viable in human serum (43) and therefore represent an in vitro phenotype only. In contrast, inactivation of hemY led to a heme-auxotrophic phenotype, and recently, HemY has been shown to be an essential enzyme in the heme biosynthesis pathway in S. aureus (44). Heme auxotrophy is also observed in vitro after treatment with aminoglycosides (45). Interestingly, recently a clinical S. aureus SCV isolate has been published to possess a truncated hemY gene (46) (called hemG here; this is the E. coli locus designation for hemY).
Both genotypes reverted quickly to the wild-type phenotype in the absence of antibiotics. Whereas IS256 was always lost from the hemY insertion mutants, the guaA mutants seemed also to be rescued by other mutations. The molecular details of this mechanism are still unclear, as sequencing showed no alterations (e.g., insertion of another copy of IS256, forming a stronger promoter) in the promoters of SA2050, pbuX, or SA1042, which represent ORFs with similarity to E. coli purine base transporters (31). However, the regulation of guanine uptake in S. aureus is also unclear, and a mutation in a regulatory gene might still stimulate import of guanine in BHI and TSB medium which still contain trace amounts of this metabolite.
IS256 is excised by illegitimate recombination (slipped-strand mispairing) of the direct repeats (3). The inverted repeat that is flanked by the direct repeats will facilitate such a deletion since it is able to form a hairpin structure that places the direct repeats close to each other (47, 48). The experiment with the pMAD-bearing surrogate revertant clearly demonstrated that outcompetition of the SCV by a revertant may be fast. The doubling times of the SCV in our experiments were about twice as long as those of the wild type. If one revertant was present or arose in a culture containing approximately 105 SCVs, a calculation of the cell numbers indicates that under these conditions (with assumed mean doubling times of 44 min for the wild type and 88 min for the guaA insertion mutant, as seen, for example, in TSB), 24.44 h of exponential growth would be needed to reach equal cell numbers of SCVs and the wild type in the culture. However, the very high cell numbers at this time (about 2 × 1010 CFU per ml) would probably not be supported by the growth medium, and therefore, a second passage is normally needed. This confirms the in vitro data shown in Fig. 5. The speed of the reversion of the hemY mutants in BHI reproducibly exceeded the speed that can be reached after one mutational event. Here several, possibly preexisting, mutants would be required for the observed high reversion rate. Indeed, such revertants already arose during the storage of the SCV colonies on agar plates, because in the first experiments, large colonies were sometimes detected during inspection of agar plates that had been plated directly after resuspension of a hemY::IS256-spc SCV colony, when the colony had been taken from an agar plate stored in the refrigerator for over 1 week. These experiments were not evaluated for this study but served to demonstrate the high excision frequency, which might in part depend on the fact that a 9-bp direct repeat could be used for excision of this insertion.
So far, there seem to be several mechanisms that generate the SCV phenotype in vivo and in vitro (20). There is sound evidence that indicates the presence of regulatory mechanisms, since SigB seems to be essential if SCVs are formed intracellularly in vivo (19) and a high percentage of intracellular cells seems to be able to switch to the SCV phenotype (24). On the other hand, mutations may also play a role since stable hemH mutants have been isolated after treatment with gentamicin in vitro from S. aureus NCTC 8325, which is an rsbU-negative strain and therefore displays very low SigB activity (23), and stable thyA mutants were isolated after long-term exposure to trimethoprim-sulfamethoxazole in vivo (49). Here, we show for the first time that via insertions of IS elements, mutations may also give rise to unstable auxotrophic SCVs. Other genetic mechanisms that can confer fast phenotypic switching, like insertion or deletion of single bases or short repeats by slipped-strand mispairing, have also been discussed previously for phase variation in E. coli and biofilm formation of S. aureus (50, 51).
In conclusion, the isolates of the ST247 lineage sequenced in this study are burdened by a huge number of IS256 insertion elements, which are actively transposing and confer a strong genomic plasticity on these strains. As an example, the presence of IS256 may give rise to unstable SCV phenotypes that are reminiscent of phase variation phenomena but which are nonetheless caused by mutations.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table 3.
TABLE 3.
Strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Reference |
|---|---|---|
| Staphylococcus aureus strains | ||
| HG001 | rsbU-repaired laboratory strain NCTC 8325 | 59 |
| SA137/93A | Clinical VISA/MRSA isolate | 18 |
| SA137/93G | Spontaneous mutant of clinical VISA isolate SA137/93A; ΔSCCmec (Mets) | 18 |
| SA1450/94 | MRSA, Northern German epidemic strain; mutS::IS256 | 18 |
| HG001 hemY::IS256-spc | hemY::IS256-spc insertion mutant; DR: TATTAGGAT | This study |
| HG001 G2 | Intergenic IS256-spc insertion mutant (SAOUHSC_0967/SAOUHSC_0968); DR: TTTTTAGC | 7 |
| HG001 G7 | Intergenic IS256-ermB insertion mutant (SACOL0573/gltX) | This study |
| HG001 W5 | rsbU::IS256-spc insertion mutant; DR: TTTAATTA | 7 |
| HG001 guaA::IS256-ermB | guaA::IS256-ermB insertion mutant; DR: CAATTCGG | This study |
| HG001 R1 | Revertant of HG001 hemY::IS256-spc | This study |
| Plasmids | ||
| pBT2 | E. coli-S. aureus shuttle vector with thermosensitive origin of replication for Gram-positive bacteria; cat | 7 |
| pBT2-ermB | pBT2 harboring the erythromycin resistance-mediating cassette ermB as insert | 26 |
| pA3 | pBT2 carrying a recombinant IS256 element containing an erythromycin resistance-mediating cassette | 26 |
| pA6 | pBT2 carrying a recombinant IS256 element containing a spectinomycin resistance-mediating cassette, spc from S. aureus Mu50 (GenBank accession no. BA000017.4) | 7 |
| pMAD | E. coli-S. aureus shuttle vector with a thermosensitive origin of replication for Gram-positive bacteria; ermC bgaB | 60 |
VISA, vancomycin-intermediate S. aureus; DR, direct repeat.
S. aureus strains were shaken in tryptic soy broth (TSB) (Merck KGaA, Darmstadt, Germany), brain heart infusion (BHI) medium (Oxoid, Wesel, Germany), Mueller-Hinton (MH) broth (Oxoid, Wesel, Germany), or lysogenic broth (LB [52]) at 37°C or 45°C. For solid media, tryptic soy agar (TSA) or BHI agar was used. Growth experiments under anaerobic conditions were performed at 37°C using the GasPak EZ container system (Becton Dickinson, Heidelberg, Germany). S. aureus strains containing plasmids with a temperature-sensitive origin of replication were incubated at 34°C in TSB or BHI medium supplemented with 20 μg/ml of chloramphenicol, 25 μg/ml of erythromycin, or 150 μg/ml of spectinomycin.
Growth kinetic measurements and antibiotic resistance testing.
For growth experiments, an overnight culture was diluted to an optical density at 600 nm (OD600) of 0.1 in TSB, BHI, or MH medium supplemented with selective concentrations of 4 μg/ml of erythromycin or 64 μg/ml of spectinomycin. The growth curves were measured over a period of 12 h at 37°C in a microplate reader (Sunrise; Tecan, Crailsheim, Germany) at intervals of 12 min, and the microtiter plate was shaken for two periods of 3 min within each interval. Data were analyzed by Magellan data analysis software (Tecan, Crailsheim, Germany).
Some growth experiments with the S. aureus HG001 guaA::IS256-ermB insertion mutant were performed in modified RPMI 1640 medium R7509 (Sigma-Aldrich, Taufkirchen, Germany) (53). Before each experiment, 500 ml of RPMI medium was supplemented with glutamine (end concentration, 2 mM) and 10 ml of a sterile solution of Wolfe's mineral salts [500 mg/liter of Titriplex I, 180 mg/liter of ZnSO4·7 H2O, 50 mg/liter of CuSO4·5 H2O, 170 mg/liter of CoCl2·6 H2O, 130 mg/liter of CaCl2·2 H2O, 11 mg/liter of NaMoO4·H2O, 26 mg/liter of Al2(SO4)3, 10 mg/liter of H3BO3, 5 ml/liter of 1 M NaOH]. In order to show the guanine auxotrophy of the S. aureus HG001 guaA::IS256-ermB insertion mutant, the main cultures were inoculated with 2 or 3 colonies from a fresh TSA plate resuspended in RPMI medium (OD600, 0.05 to 0.1) to prevent contamination with trace amounts of guanine present in the TSB overnight culture. Guanine was added to the RPMI medium at different concentrations between 0.005 mM and 0.5 mM. To demonstrate the effect of the guaA insertion on vancomycin resistance, the strains were incubated in the presence of different concentrations of vancomycin in BHI growth medium in the microplate reader for 12 h exactly as described above; also, MICs were determined in BHI.
Fosfomycin MICs were determined by Etest (bioMérieux, Mercy-l'Étoile, France). For the MIC determinations for spectinomycin and aminoglycosides, fresh colonies of S. aureus HG001 G2, S. aureus HG001 hemY::IS256-spc, and the revertant S. aureus HG001 R1, which had lost the insertion in hemY, were grown on TSA agar plates (150 μg/ml of spectinomycin for the strains containing IS256-spc). The colonies were resuspended in MH broth (no antibiotic) and diluted to 106 cells per ml. A total of 100 μl of this suspension was mixed with 100 μl of the antibiotics dissolved in MH broth in the wells of a microtiter plate. The MICs were read after 24 and 48 h of incubation at 37°C.
Mapping of all IS256 insertions in three ST247 strains by 454 sequencing.
The genomic sequences of the ST247 strains S. aureus SA1450/94, SA137/93A, and SA137/93G were determined by next-generation sequencing (NGS) using 454 sequencing technology (54). In the first de novo assembly of the NGS reads with the GS De Novo Assembler (Roche Applied Science GmbH, Mannheim, Germany), the sequences of the IS elements IS256, IS431mec, and IS1181 were recognized as repetitive sequences and were summarized as three single contigs. Only the first or last bases of the IS elements were present in the neighboring contigs. To identify all insertion sites of IS256 and other insertion sequences and to close the gaps between the neighboring contigs, the nucleotide sequences of IS256, IS431mec, and IS1181 were compared via a BLAST search against all reads of strain SA137/93G using Geneious 6.1.8 (Biomatters Limited, Auckland, New Zealand). The resulting hits were assembled de novo and generated contigs, which then were analyzed via BLAST against the reference strain S. aureus COL (GenBank accession no. NC_002951.2), identifying the precise insertion sites. In BLAST analyses of SA137/93A and SA1450/94, the contigs, which were joined by an IS256 sequence in vivo, were identified by the 8-bp direct repeat that frames every IS256 insertion. All insertion sites of the IS elements were confirmed for all three strains by PCR and Sanger sequencing and integrated into the draft genome sequences.
Mapping of single IS256, IS256-spc, and IS256-ermB insertions.
The localizations of IS256 and recombinant IS256 elements in different insertion mutants of S. aureus HG001 were determined by inverse PCR as previously described (12) (Table 4). In brief, the genomic DNA was digested with XmnI (New England BioLabs, Frankfurt am Main, Germany) or AluI (Roche Applied Science GmbH, Mannheim, Germany). After circularization of the fragments using the T4 ligase (Roche Applied Science GmbH, Mannheim, Germany), the insertion sites were detected by PCR analysis using the outgoing primers IS91rev and IS141for, which anneal within the 5′ end of IS256 (Table 4). Afterwards, the insertion site of the insertion element was detected by Sanger sequencing (SEQLAB Sequence Laboratories Göttingen GmbH, Göttingen, Germany).
TABLE 4.
Oligonucleotides used in this study
| Oligonucleotide | Sequence (5′–3′)a |
|---|---|
| ISIRLfor | GTA GAA TTC GGA TAA AGT CCG TAT AAT TGT GT |
| ISIRRrev | TGT TCT AGA TGC TAT ACA ATG TTT TTA CCA TTT C |
| IS91rev | CTT TTT ACT TTT ACA CAA TTA TAC GG |
| IS141for | CCC AGG AGG ACT TTT ACA TGA C |
| Spc-for_HindIII | ACG AAG CTT AGT CAA GTC CAG ACT CCT GTG TAA AAT CGT CCA ATC TAG GGT A |
| Spc-rev_XbaI | AAG TCT AGA TGA ATT TCA CAA GAG GAC |
| guaAfor | CAT GTA TCT TTG TAG ACC ATG |
| guaArev | AGG ATC TGA AAC ACC TTT TAA T |
| guaA_forMH | TTC GTG AAA TGG GCG TTT AT |
| guaA_revMH | TAG ACT ACG CGG TTG ACG TG |
| hemY_forMH | GTG ACT AAA TCA GTG GCT ATT |
| hemY_revMH | TGC GAT TAC TTC TTC AGC |
| hemLBDCA_for | GTT GAT CCG ATT TGC ACA C |
| hemLBDCA_int1 | CCT AAT GGT GCT ATA TGA TCC |
| hemLBDCA_int2 | CGT ACT GGA CTA TTT ACA CC |
| hemLBDCA_int3 | GTT GCC TGT TGA ATA ACA CC |
| hemLBDCA_int4 | GCG CTG TCT TAC TTC CTA TC |
| hemLBDCA_int5 | CAA TAA TAC TGC CTT CTG GC |
| hemLBDCA_int6 | AAC TTA ACA CAG CTA GAC CG |
| hemLBDCA_int7 | ATC CAC TCA TTG TGT GCA TG |
| hemLBDCA_rev | GGA TGT TCC AGG GTA TGG AT |
| hemQ_for | ATT GTA TAG AGT AGC GAC TGT |
| hemQ_rev | GAA CCT TAG TCT CAT CAC CT |
| hemN_for | TGT CAA TGC TGA TGA TAC ATC |
| hemN_rev | AGG ACA TAT TAC ATC TGG CT |
| hemYHE_for | CAT CTG TCA TCT CAT TCT CTC C |
| hemYHE_int1 | ATG CCA ATA CAA CAG TTG C |
| hemYHE_int2 | ATT CAG GCC CTA GTT CAA TC |
| hemYHE_int3 | ATT GCT TCA GTA ATG CCA TC |
| hemYHE_rev | GTT GAT GGG TCG GGT TAT TG |
| hemYHE_int2.5.1 | GGT ACG GAA CTC GTT ATT TC |
| hemYHE_int2.5.2 | CAC AGG AGT CTG GAC TTG AC |
| SA2050_for | TAT CGC TTT ATG TTA TTT CG |
| SA2050_int1 | TCC TAT ACC TGC TGA AAC AG |
| SA2050_int2 | TAC CGC TTC TTT CCC TTT AC |
| SA2050_int3 | GCG TGC TTG GTC TTT ATA TG |
| SA2050_rev | CCT CAA CAG TAC AAT GAA ATA G |
| 377504 F | TGT CAC GAA CCG CTA CAA CT |
| 378517R | TGC GAT ACC GGA AGC TTC AA |
| 1,112,585 F | GTT CAT CCA TCA CCA GGG CA |
| 1,116,051 R | TAC CAT CAC CAG CAT TCG CA |
| 1113415_int1 | CCA GAG AGA CGT CAA AGA C |
| 1114264_int2 | CTT GTA GCA ACT TGT CAC AC |
| 1114821_int3 | CCT GGA GAT ATG AAA GGT TAC |
Restriction sites used for cloning are underlined.
Passaging with increasing vancomycin concentrations.
In order to select vancomycin-resistant IS256 insertion mutants, different strains (see Results) were passaged in the presence of increasing vancomycin concentrations. Each strain was inoculated in 5 ml of BHI medium with three different vancomycin concentrations and was shaken at 170 rpm and 34°C. After 24 h, the culture that had yielded visible bacterial growth and contained the highest vancomycin concentration was used to inoculate three new BHI cultures supplemented with the same vancomycin concentration as the preculture and two higher vancomycin concentrations (preculture plus 0.5 μg/ml and preculture plus 1 μg/ml). The passaging was performed for at least 10 days. In these tests, the appropriate antibiotics were used to maintain the different plasmids and several inoculum volumes were tested.
Selection of an IS256-mediated SCV in the presence of spectinomycin.
The spc gene employed in this study encodes the spectinomycin adenyl transferase ANT (9)-Ia, which confers resistance to spectinomycin but not to streptomycin (28). In order to select IS256-spc insertion mutants, the strain S. aureus HG001 containing the temperature-sensitive plasmid pA6 (7) was shaken overnight in TSB medium supplemented with 20 μg/ml of chloramphenicol at 30°C. Then the culture was diluted 1:100 in 20 ml TSB medium without chloramphenicol and was shaken for 6 h at 30°C. Aliquots of the culture were diluted and plated onto TSA containing 150 μg/ml of spectinomycin. After incubation of the plates for 48 h at 45°C, a nonpermissive temperature for plasmid replication, a yellow small-colony variant, which exhibited spectinomycin resistance and chloramphenicol susceptibility (indicating the presence of the IS element and loss of the plasmid), was selected from an agar plate containing less than 200 CFU.
Phage transduction.
Phage transduction was used to transfer the guaA::IS256-ermB insertion of the S. aureus HG001 insertion mutant (which had been isolated after passaging for 24 days in the presence of vancomycin) to the wild-type strain S. aureus HG001 employing phage 85 as previously described (55). The transductants were selected on LB agar containing 25 μg/ml of erythromycin. The chromosomal guaA::IS256-ermB insertion in wild-type S. aureus HG001 was then verified by PCR using primers guaAfor and guaArev and sequencing.
Determination of revertants of the SCV phenotype.
To analyze the appearance of revertants of the SCV phenotype, a single SCV colony (yielding 2 × 105 to 6 ×105 CFU/ml as determined by plating immediately after inoculation) from a fresh agar plate was resuspended in 5 ml of TSB or BHI medium and was shaken at 37°C. In some experiments the medium was also supplemented with 4 μg/ml of erythromycin (S. aureus HG001 guaA::IS256-ermB) or 150 μg/ml of spectinomycin (S. aureus HG001 hemY::IS256-spc). Every 24 h, the culture was diluted to an OD600 of 0.05 with fresh medium and an aliquot was plated in dilution on TSA or BHI agar. After incubation at 37°C for 24 to 48 h, the small and large colonies (phenotypic revertants) were counted on agar plates containing less than 200 CFU. All experiments were performed at least in triplicate, and mean values are presented. After 4 days, phenotypic revertants were isolated and analyzed by PCR to monitor the loss of the IS256 insertion in guaA and hemY. The primer pairs guaA_forMH/guaA_revMH and hemY_forMH/hemY_revMH (Table 4) yielded a large band (>3.5 kb) if the recombinant IS256 was inserted into the gene and a small band (about 1.5 kb) after loss of the recombinant IS256. Colonies used for the inoculum of the culture were also tested for the correct IS256 insertions for both insertion mutants and only the large PCR band was visible, indicating that the IS256 element was still present in hemY or guaA, respectively.
Competition experiments with S. aureus HG001 guaA::IS256-ermB and S. aureus HG001 pMAD were performed at 30°C in the presence of 25 μg/ml of erythromycin. For the inoculum of the mixed culture, successive 10-fold dilutions were prepared from an S. aureus HG001 pMAD preculture. Then 0.1 ml of each dilution was plated on TSA and another 0.1-ml aliquot was used as an inoculum for a separate main culture that already contained one resuspended colony of S. aureus HG001 guaA::IS256-ermB. The CFU of S. aureus HG001 pMAD were counted after 24 h, and only the main culture that had been inoculated with approximately 1 CFU/ml was incubated further for the competition experiment.
Identification of the number of IS256 elements in sequenced genomes in NCBI.
The numbers of IS256 elements in sequenced complete genomes were identified in December 2016 by NCBI Nucleotide BLAST search employing the IS256 sequence (accession no. AF535088.1 [56]) and to this end, all hits were counted, i.e., the insertion that was formed by the recombination of two IS256 elements after head-to-head insertion was counted as two elements. In Fig. 1B, this counts as one insertion site, however, since both IS256s were inserted at the same site. Multilocus sequence types of S. aureus were determined from the published sequences using MLST 1.8 (https://cge.cbs.dtu.dk//services/MLST/) (57).
Accession number(s).
The sequences have been deposited in GenBank with the following accession numbers: for S. aureus SA137/93A, JWMH00000000 (JWMH01000001 to JWMH01000036); for S. aureus SA137/93G, JWMG00000000 (JWMG01000001 to JWMG01000041); and for S. aureus SA1450/94, JWMI00000000 (JWMI01000001 to JWMI01000029) (54).
Supplementary Material
ACKNOWLEDGMENTS
F.K. and this work were supported by grant Bi 504/10-1 from the German Research Foundation (DFG) to G.B. and a Maria von Linden fellowship of the University of Bonn to F.K. Additional support was provided by the BONFOR program of the Medical Faculty, University of Bonn.
We thank S. Herbert and F. Götz for kindly providing strain S. aureus HG001 and C. Gurgui and J. Piel (Zürich) for the donation of the plasmid pBPsy1Prom containing the spectinomycin resistance cassette.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.00144-17.
REFERENCES
- 1.Lyon BR, Gillespie MT, Skurray RA. 1987. Detection and characterization of IS256, an insertion sequence in Staphylococcus aureus. J Gen Microbiol 133:3031–3038. [DOI] [PubMed] [Google Scholar]
- 2.Byrne ME, Rouch DA, Skurray RA. 1989. Nucleotide sequence analysis of IS256 from the Staphylococcus aureus gentamicin-tobramycin-kanamycin-resistance transposon Tn4001. Gene 81:361–367. doi: 10.1016/0378-1119(89)90197-2. [DOI] [PubMed] [Google Scholar]
- 3.Hennig S, Ziebuhr W. 2008. A transposase-independent mechanism gives rise to precise excision of IS256 from insertion sites in Staphylococcus epidermidis. J Bacteriol 190:1488–1490. doi: 10.1128/JB.01290-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ziebuhr W, Krimmer V, Rachid S, Lössner I, Götz F, Hacker J. 1999. A novel mechanism of phase variation of virulence in Staphylococcus epidermidis: evidence for control of the polysaccharide intercellular adhesin synthesis by alternating insertion and excision of the insertion sequence element IS256. Mol Microbiol 32:345–356. doi: 10.1046/j.1365-2958.1999.01353.x. [DOI] [PubMed] [Google Scholar]
- 5.Loessner I, Dietrich K, Dittrich D, Hacker J, Ziebuhr W. 2002. Transposase-dependent formation of circular IS256 derivatives in Staphylococcus epidermidis and Staphylococcus aureus. J Bacteriol 184:4709–4714. doi: 10.1128/JB.184.17.4709-4714.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kiem S, Oh WS, Peck KR, Lee NY, Lee JY, Song JH, Hwang ES, Kim EC, Cha CY, Choe KW. 2004. Phase variation of biofilm formation in Staphylococcus aureus by IS256 insertion and its impact on the capacity adhering to polyurethane surface. J Korean Med Sci 19:779–782. doi: 10.3346/jkms.2004.19.6.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schreiber F, Szekat C, Josten M, Sahl HG, Bierbaum G. 2013. Antibiotic-induced auto-activation of IS256 in Staphylococcus aureus. Antimicrob Agents Chemother 57:6381–6384. doi: 10.1128/AAC.01585-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conlon KM, Humphreys H, O'Gara JP. 2004. Inactivations of rsbU and sarA by IS256 represent novel mechanisms of biofilm phenotypic variation in Staphylococcus epidermidis. J Bacteriol 186:6208–6219. doi: 10.1128/JB.186.18.6208-6219.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Valle J, Vergara-Irigaray M, Merino N, Penades JR, Lasa I. 2007. sigmaB regulates IS256-mediated Staphylococcus aureus biofilm phenotypic variation. J Bacteriol 189:2886–2896. doi: 10.1128/JB.01767-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maki H, McCallum N, Bischoff M, Wada A, Berger-Bächi B. 2004. tcaA inactivation increases glycopeptide resistance in Staphylococcus aureus. Antimicrob Agents Chemother 48:1953–1959. doi: 10.1128/AAC.48.6.1953-1959.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maki H, Murakami K. 1997. Formation of potent hybrid promoters of the mutant llm gene by IS256 transposition in methicillin-resistant Staphylococcus aureus. J Bacteriol 179:6944–6948. doi: 10.1128/jb.179.22.6944-6948.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jansen A, Türck M, Szekat C, Nagel M, Clever I, Bierbaum G. 2007. Role of insertion elements and yycFG in the development of decreased susceptibility to vancomycin in Staphylococcus aureus. Int J Med Microbiol 297:205–215. doi: 10.1016/j.ijmm.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 13.McEvoy CR, Tsuji B, Gao W, Seemann T, Porter JL, Doig K, Ngo D, Howden BP, Stinear TP. 2013. Decreased vancomycin susceptibility in Staphylococcus aureus caused by IS256 tempering of WalKR expression. Antimicrob Agents Chemother 57:3240–3249. doi: 10.1128/AAC.00279-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Benson MA, Ohneck EA, Ryan C, Alonzo F III, Smith H, Narechania A, Kolokotronis SO, Satola SW, Uhlemann AC, Sebra R, Deikus G, Shopsin B, Planet PJ, Torres VJ. 2014. Evolution of hypervirulence by a MRSA clone through acquisition of a transposable element. Mol Microbiol 93:664–681. doi: 10.1111/mmi.12682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gill SR, Fouts DE, Archer GL, Mongodin EF, Deboy RT, Ravel J, Paulsen IT, Kolonay JF, Brinkac L, Beanan M, Dodson RJ, Daugherty SC, Madupu R, Angiuoli SV, Durkin AS, Haft DH, Vamathevan J, Khouri H, Utterback T, Lee C, Dimitrov G, Jiang L, Qin H, Weidman J, Tran K, Kang K, Hance IR, Nelson KE, Fraser CM. 2005. Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J Bacteriol 187:2426–2438. doi: 10.1128/JB.187.7.2426-2438.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Monecke S, Coombs G, Shore AC, Coleman DC, Akpaka P, Borg M, Chow H, Ip M, Jatzwauk L, Jonas D, Kadlec K, Kearns A, Laurent F, O'Brien FG, Pearson J, Ruppelt A, Schwarz S, Scicluna E, Slickers P, Tan HL, Weber S, Ehricht R. 2011. A field guide to pandemic, epidemic and sporadic clones of methicillin-resistant Staphylococcus aureus. PLoS One 6:e17936. doi: 10.1371/journal.pone.0017936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bierbaum G, Fuchs K, Lenz W, Szekat C, Sahl HG. 1999. Presence of Staphylococcus aureus with reduced susceptibility to vancomycin in Germany. Eur J Clin Microbiol Infect Dis 18:691–696. doi: 10.1007/s100960050380. [DOI] [PubMed] [Google Scholar]
- 18.Reipert A, Ehlert K, Kast T, Bierbaum G. 2003. Morphological and genetic differences in two isogenic Staphylococcus aureus strains with decreased susceptibilities to vancomycin. Antimicrob Agents Chemother 47:568–576. doi: 10.1128/AAC.47.2.568-576.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tuchscherr L, Bischoff M, Lattar SM, Noto Llana M, Pförtner H, Niemann S, Geraci J, van de Vyver H, Fraunholz MJ, Cheung AL, Herrmann M, Völker U, Sordelli DO, Peters G, Löffler B. 2015. Sigma factor SigB is crucial to mediate Staphylococcus aureus adaptation during chronic infections. PLoS Pathog 11:e1004870. doi: 10.1371/journal.ppat.1004870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kahl BC, Becker K, Löffler B. 2016. Clinical significance and pathogenesis of staphylococcal small colony variants in persistent infections. Clin Microbiol Rev 29:401–427. doi: 10.1128/CMR.00069-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.von Eiff C, Heilmann C, Proctor RA, Woltz C, Peters G, Götz F. 1997. A site-directed Staphylococcus aureus hemB mutant is a small-colony variant which persists intracellularly. J Bacteriol 179:4706–4712. doi: 10.1128/jb.179.15.4706-4712.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.von Eiff C, Peters G, Becker K. 2006. The small colony variant (SCV) concept—the role of staphylococcal SCVs in persistent infections. Injury 37(Suppl 2):S26–S33. doi: 10.1016/j.injury.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 23.Schaaff F, Bierbaum G, Baumert N, Bartmann P, Sahl HG. 2003. Mutations are involved in emergence of aminoglycoside-induced small colony variants of Staphylococcus aureus. Int J Med Microbiol 293:427–435. doi: 10.1078/1438-4221-00282. [DOI] [PubMed] [Google Scholar]
- 24.Tuchscherr L, Medina E, Hussain M, Völker W, Heitmann V, Niemann S, Holzinger D, Roth J, Proctor RA, Becker K, Peters G, Löffler B. 2011. Staphylococcus aureus phenotype switching: an effective bacterial strategy to escape host immune response and establish a chronic infection. EMBO Mol Med 3:129–141. doi: 10.1002/emmm.201000115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jansen A, Szekat C, Schröder W, Wolz C, Goerke C, Lee JC, Türck M, Bierbaum G. 2013. Production of capsular polysaccharide does not influence Staphylococcus aureus vancomycin susceptibility. BMC Microbiol 13:65. doi: 10.1186/1471-2180-13-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nagel M, Reuter T, Jansen A, Szekat C, Bierbaum G. 2011. Influence of ciprofloxacin and vancomycin on mutation rate and transposition of IS256 in Staphylococcus aureus. Int J Med Microbiol 301:229–236. doi: 10.1016/j.ijmm.2010.08.021. [DOI] [PubMed] [Google Scholar]
- 27.Ramirez MS, Tolmasky ME. 2010. Aminoglycoside modifying enzymes. Drug Resist Updat 13:151–171. doi: 10.1016/j.drup.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murphy E. 1985. Nucleotide sequence of a spectinomycin adenyltransferase AAD(9) determinant from Staphylococcus aureus and its relationship to AAD(3″) (9). Mol Gen Genet 200:33–39. doi: 10.1007/BF00383309. [DOI] [PubMed] [Google Scholar]
- 29.Proctor RA, Balwit JM, Vesga O. 1994. Variant subpopulations of Staphylococcus aureus as cause of persistent and recurrent infections. Infect Agents Dis 3:302–312. [PubMed] [Google Scholar]
- 30.Xu T, Han J, Zhang J, Chen J, Wu N, Zhang W, Zhang Y. 2016. Absence of protoheme IX farnesyltransferase CtaB causes virulence attenuation but enhances pigment production and persister survival in MRSA. Front Microbiol 7:1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jensen KF, Dandanell G, Hove-Jensen B, WillemoËs M. 2008. Nucleotides, nucleosides, and nucleobases. EcoSal Plus 3:10. doi: 10.1128/ecosalplus.3.6.2. [DOI] [PubMed] [Google Scholar]
- 32.Mulhbacher J, Brouillette E, Allard M, Fortier LC, Malouin F, Lafontaine DA. 2010. Novel riboswitch ligand analogs as selective inhibitors of guanine-related metabolic pathways. PLoS Pathog 6:e1000865. doi: 10.1371/journal.ppat.1000865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moran NA, Plague GR. 2004. Genomic changes following host restriction in bacteria. Curr Opin Genet Dev 14:627–633. doi: 10.1016/j.gde.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 34.Cerveau N, Leclercq S, Leroy E, Bouchon D, Cordaux R. 2011. Short- and long-term evolutionary dynamics of bacterial insertion sequences: insights from Wolbachia endosymbionts. Genome Biol Evol 3:1175–1186. doi: 10.1093/gbe/evr096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wan TW, Khokhlova OE, Iwao Y, Higuchi W, Hung WC, Reva IV, Singur OA, Gostev VV, Sidorenko SV, Peryanova OV, Salmina AB, Reva GV, Teng LJ, Yamamoto T. 2016. Complete circular genome sequence of successful ST8/SCCmecIV community-associated methicillin-resistant Staphylococcus aureus (OC8) in Russia: one-megabase genomic inversion, IS256's spread, and evolution of Russia ST8-IV. PLoS One 11:e0164168. doi: 10.1371/journal.pone.0164168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Noto MJ, Fox PM, Archer GL. 2008. Spontaneous deletion of the methicillin resistance determinant, mecA, partially compensates for the fitness cost associated with high-level vancomycin resistance in Staphylococcus aureus. Antimicrob Agents Chemother 52:1221–1229. doi: 10.1128/AAC.01164-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Villafane R, Bechhofer DH, Narayanan CS, Dubnau D. 1987. Replication control genes of plasmid pE194. J Bacteriol 169:4822–4829. doi: 10.1128/jb.169.10.4822-4829.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dordel J, Kim C, Chung M, Pardos de la Gandara M, Holden MT, Parkhill J, de Lencastre H, Bentley SD, Tomasz A. 2014. Novel determinants of antibiotic resistance: identification of mutated loci in highly methicillin-resistant subpopulations of methicillin-resistant Staphylococcus aureus. mBio 5:e01000-13. doi: 10.1128/mBio.01000-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abranches J, Martinez AR, Kajfasz JK, Chavez V, Garsin DA, Lemos JA. 2009. The molecular alarmone (p)ppGpp mediates stress responses, vancomycin tolerance, and virulence in Enterococcus faecalis. J Bacteriol 191:2248–2256. doi: 10.1128/JB.01726-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Berscheid A, Francois P, Strittmatter A, Gottschalk G, Schrenzel J, Sass P, Bierbaum G. 2014. Generation of a vancomycin-intermediate Staphylococcus aureus (VISA) strain by two amino acid exchanges in VraS. J Antimicrob Chemother 69:3190–3198. doi: 10.1093/jac/dku297. [DOI] [PubMed] [Google Scholar]
- 41.Geiger T, Kästle B, Gratani FL, Goerke C, Wolz C. 2014. Two small (p)ppGpp synthases in Staphylococcus aureus mediate tolerance against cell envelope stress conditions. J Bacteriol 196:894–902. doi: 10.1128/JB.01201-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gao W, Chua K, Davies JK, Newton HJ, Seemann T, Harrison PF, Holmes NE, Rhee HW, Hong JI, Hartland EL, Stinear TP, Howden BP. 2010. Two novel point mutations in clinical Staphylococcus aureus reduce linezolid susceptibility and switch on the stringent response to promote persistent infection. PLoS Pathog 6:e1000944. doi: 10.1371/journal.ppat.1000944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kofoed EM, Yan D, Katakam AK, Reichelt M, Lin B, Kim J, Park S, Date SV, Monk IR, Xu M, Austin CD, Maurer T, Tan MW. 2016. De novo guanine biosynthesis but not the riboswitch-regulated purine salvage pathway is required for Staphylococcus aureus infection in vivo. J Bacteriol 198:2001–2015. doi: 10.1128/JB.00051-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lobo SA, Scott A, Videira MA, Winpenny D, Gardner M, Palmer MJ, Schroeder S, Lawrence AD, Parkinson T, Warren MJ, Saraiva LM. 2015. Staphylococcus aureus haem biosynthesis: characterisation of the enzymes involved in final steps of the pathway. Mol Microbiol 97:472–487. doi: 10.1111/mmi.13041. [DOI] [PubMed] [Google Scholar]
- 45.Balwit JM, van Langevelde P, Vann JM, Proctor RA. 1994. Gentamicin-resistant menadione and hemin auxotrophic Staphylococcus aureus persist within cultured endothelial cells. J Infect Dis 170:1033–1037. doi: 10.1093/infdis/170.4.1033. [DOI] [PubMed] [Google Scholar]
- 46.Köser CU, Holden MT, Ellington MJ, Cartwright EJ, Brown NM, Ogilvy-Stuart AL, Hsu LY, Chewapreecha C, Croucher NJ, Harris SR, Sanders M, Enright MC, Dougan G, Bentley SD, Parkhill J, Fraser LJ, Betley JR, Schulz-Trieglaff OB, Smith GP, Peacock SJ. 2012. Rapid whole-genome sequencing for investigation of a neonatal MRSA outbreak. N Engl J Med 366:2267–2275. doi: 10.1056/NEJMoa1109910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bzymek M, Lovett ST. 2001. Evidence for two mechanisms of palindrome-stimulated deletion in Escherichia coli: single-strand annealing and replication slipped mispairing. Genetics 158:527–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peeters BP, de Boer JH, Bron S, Venema G. 1988. Structural plasmid instability in Bacillus subtilis: effect of direct and inverted repeats. Mol Gen Genet 212:450–458. doi: 10.1007/BF00330849. [DOI] [PubMed] [Google Scholar]
- 49.Kriegeskorte A, Lore NI, Bragonzi A, Riva C, Kelkenberg M, Becker K, Proctor RA, Peters G, Kahl BC. 2015. Thymidine-dependent Staphylococcus aureus small-colony variants are induced by trimethoprim-sulfamethoxazole (SXT) and have increased fitness during SXT challenge. Antimicrob Agents Chemother 59:7265–7272. doi: 10.1128/AAC.00742-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Iqbal S, Parker G, Davidson H, Moslehi-Rahmani E, Robson RL. 2004. Reversible phase variation in the phnE gene, which is required for phosphonate metabolism in Escherichia coli K-12. J Bacteriol 186:6118–6123. doi: 10.1128/JB.186.18.6118-6123.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brooks JL, Jefferson KK. 2014. Phase variation of poly-N-acetylglucosamine expression in Staphylococcus aureus. PLoS Pathog 10:e1004292. doi: 10.1371/journal.ppat.1004292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bertani G. 1951. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J Bacteriol 62:293–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moore GE, Gerner RE, Franklin HA. 1967. Culture of normal human leukocytes. JAMA 199:519–524. [PubMed] [Google Scholar]
- 54.Kleinert F, Kallies R, Zweynert A, Bierbaum G. 2016. Draft genome sequences of three Northern German epidemic Staphylococcus aureus (ST247) strains containing multiple copies of IS256. Genome Announc 4:e00936-16. doi: 10.1128/genomeA.00936-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Berger-Bächi B, Kohler M. 1983. A novel site on the chromosome of Staphylococcus aureus influencing the level of methicillin resistance: genetic mapping. FEMS Microbiol Lett 20:305–309. [Google Scholar]
- 56.Alam MM, Kobayashi N, Uehara N, Watanabe N. 2003. Analysis on distribution and genomic diversity of high-level antiseptic resistance genes qacA and qacB in human clinical isolates of Staphylococcus aureus. Microb Drug Resist 9:109–121. doi: 10.1089/107662903765826697. [DOI] [PubMed] [Google Scholar]
- 57.Larsen MV, Cosentino S, Rasmussen S, Friis C, Hasman H, Marvig RL, Jelsbak L, Sicheritz-Ponten T, Ussery DW, Aarestrup FM, Lund O. 2012. Multilocus sequence typing of total-genome-sequenced bacteria. J Clin Microbiol 50:1355–1361. doi: 10.1128/JCM.06094-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Watanabe Y, Cui L, Katayama Y, Kozue K, Hiramatsu K. 2011. Impact of rpoB mutations on reduced vancomycin susceptibility in Staphylococcus aureus. J Clin Microbiol 49:2680–2684. doi: 10.1128/JCM.02144-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Herbert S, Ziebandt AK, Ohlsen K, Schäfer T, Hecker M, Albrecht D, Novick R, Götz F. 2010. Repair of global regulators in Staphylococcus aureus 8325 and comparative analysis with other clinical isolates. Infect Immun 78:2877–2889. doi: 10.1128/IAI.00088-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Arnaud M, Chastanet A, Debarbouille M. 2004. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria. Appl Environ Microbiol 70:6887–6891. doi: 10.1128/AEM.70.11.6887-6891.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.