ABSTRACT
The H30 subclone of Escherichia coli sequence type 131 (ST131-H30) has become the leading antimicrobial resistance E. coli lineage in the United States and often exhibits resistance to one or both of the two key antimicrobial classes for treating Gram-negative infections, extended-spectrum cephalosporins (ESCs) and fluoroquinolones (FQs). However, the timing of and reasons for its recent emergence are inadequately defined. Accordingly, from E. coli clinical isolates collected systematically across the United States by the SENTRY Antimicrobial Surveillance Program in 2000, 2003, 2006, and 2009, 234 isolates were selected randomly, stratified by year, within three resistance categories: (i) ESC-reduced susceptibility, regardless of FQ phenotype (ESC-RS); (ii) FQ resistance, ESC susceptible (FQ-R); and (iii) FQ susceptible, ESC susceptible (FQ-S). Susceptibility profiles, phylogroup, ST, ST131 subclone, and virulence genotypes were determined, and temporal trends and between-variable associations were assessed statistically. From 2000 to 2006, concurrently with the emergence of ESC-RS and FQ-R strains, the prevalence of (virulence-associated) phylogroup B2 among such strains also rose dramatically, due entirely to rapid emergence of ST131, especially H30. By 2009, H30 was the dominant E. coli lineage overall (22%), accounting for a median of 43% of all single-agent and multidrug resistance (68% for ciprofloxacin). H30's emergence increased the net virulence gene content of resistant (especially FQ-R) isolates, giving stable overall virulence gene scores despite an approximately 4-fold expansion of the historically less virulent resistant population. These findings define more precisely the timing and tempo of H30's emergence in the United States, identify possible reasons for it, and suggest potential consequences, including more frequent and/or aggressive antimicrobial-resistant infections.
KEYWORDS: Escherichia coli ST131, antimicrobial resistance, clonality, epidemic emergence, extended-spectrum beta-lactamases, fluoroquinolone resistance, molecular epidemiology
INTRODUCTION
Extraintestinal Escherichia coli infections are an important source of morbidity, mortality, and increased health care costs (1). Over the past decade, they have become increasingly frequent (2) and challenging to treat (3), with treatment challenges occurring from the rising prevalence of resistance to most first-line antimicrobial agents.
Sequence type 131 (ST131), a recently emerged globally disseminated clonal group from virulence-associated E. coli phylogenetic group B2, was first reported in 2008 as an important contributor to the resistance pandemic in E. coli (4, 5). Multiple studies have documented ST131 as the most prevalent clonal group among resistant E. coli isolates (6). ST131 isolates often exhibit resistance to one or both of two key antimicrobial classes for treating Gram-negative infection, fluoroquinolones (FQs) and extended-spectrum cephalosporins (ESCs), which severely limits the available treatment options (6).
Recent studies have shown ST131 to be clonally complex (7–10). Its H30 subset (so named for carrying allele 30 of the type-1 fimbrial adhesin gene fimH) accounts for nearly all FQ resistance and most extended-spectrum cephalosporin (ESC) resistance in ST131, and it is by far the most expanded ST131 subset, although some ancestral members remain antimicrobial susceptible. H30 in turn comprises two important subclones, H30Rx and H30R1, the members of which are uniformly FQ resistant (FQ-R). Whereas H30Rx isolates are often also ESC resistant due to the production of the CTX-M-15 extended-spectrum beta-lactamase (ESBL), H30R1 isolates are usually ESC susceptible, although some, especially those in Asia, are ESC resistant due to the production of different ESBLs, e.g., CTX-M-14 or CTX-M-27 (10–13).
Multiple cross-sectional and convenience sample-based studies have suggested that ST131, and specifically H30, expanded during the first decade of this millennium (6). However, very few studies have provided longitudinal evidence for this, and those that did so were limited with respect to geography, resistance phenotypes, phylogenetic and clonal typing, and virulence gene analysis (11, 13–19). Accordingly, due to the unprecedented nature of the H30 expansion and its tremendous importance to clinicians, patients, and microbiologists, we undertook the present study, using E. coli isolates from across the United States collected prospectively by an established national surveillance program. Specifically, we sought to define the timing and tempo of the H30 expansion in the United States, which E. coli types H30 displaced, and the consequences of the H30 expansion for resistance prevalence, prevalence of extraintestinal pathogenic E. coli (ExPEC), and virulence gene content.
RESULTS
Phylogroups.
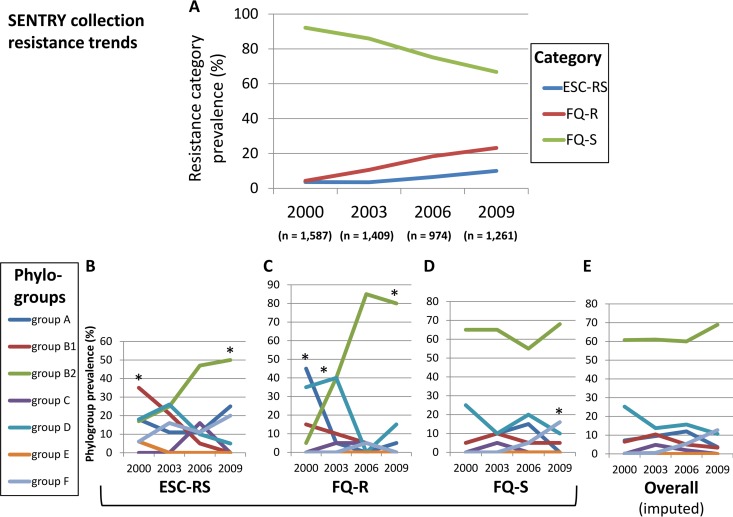
Over the 9-year study period (2000 to 2009), among clinical E. coli isolates from across the United States, the relative prevalence of the three mutually exclusive resistance categories (ESC-reduced susceptibility, irrespective of fluoroquinolone phenotype [here, ESC-RS]; fluoroquinolone resistant, ESC susceptible [FQ-R]; and fluoroquinolone susceptible, ESC susceptible [FQ-S]) shifted steadily toward increasing resistance (Fig. 1A). Concurrently, the relative prevalences of the seven E. coli phylogroups shifted significantly within each of the three resistance categories in a category-specific manner (Fig. 1B to E).
FIG 1.
Temporal trends for resistance category prevalence and phylogroup distribution. Data are for 2000, 2003, 2006, and 2009; intervening points are interpolated. Resistance categories: ESC-RS, extended-spectrum cephalosporin-reduced susceptibility; FQ-R, fluoroquinolone resistant, ESC susceptible; FQ-S, fluoroquinolone susceptible, ESC susceptible. *, P < 0.05 for linear trend. (A) Relative prevalence of the three resistance categories among all SENTRY collection Escherichia coli isolates. (B) Measured relative prevalence of seven E. coli phylogroups (A, B1, B2, C, D, E, and F) among the 75 selected ESC-RS study isolates (17 to 20 per year). (C) Measured relative prevalences of seven E. coli phylogroups among the 80 selected FQ-R study isolates (20 per year). (D) Measured relative prevalences of seven E. coli phylogroups among the 79 selected FQ-S study isolates (19 or 20 per year). (E) Imputed relative prevalences of seven E. coli phylogroups within the overall SENTRY source population.
Specifically, among ESC-RS isolates, group B2 increased in relative prevalence from 6% to 50% (P < 0.001), thereby assuming the dominant position held initially by group B1, which declined correspondingly from 35% to 0% (P < 0.001) (Fig. 1B). Among FQ-R isolates, group B2 expanded even more dramatically (5% to 80%; P < 0.001), accompanied by significant declines for groups A (45% to 5%; P < 0.001) and D (35% to 15%; P = 0.02), and a similar trend for group B1 (15% to 0%; P = 0.06) (Fig. 1C). In contrast, among FQ-S isolates, group B2 dominated throughout, with a median prevalence of 62.5%; no other phylogroup reached even 30% prevalence (Fig. 1D). The only statistically significant temporal prevalence shift among FQ-S isolates was an increase for group F (0% to 16%; P = 0.02).
Adjustment of these resistance category-specific prevalence values for each category's annual fractional contribution to the total SENTRY population allowed estimates of overall trends for phylogroup distribution (Fig. 1E). Group B2 predominated throughout, and increasingly so over time. In contrast, other phylogroups had a stable or declining prevalence, except for group F, which in 2009 appeared to supplant group D as the second-most-prevalent phylogroup. (Statistical testing was not possible since these values were imputed.)
Clonal groups.
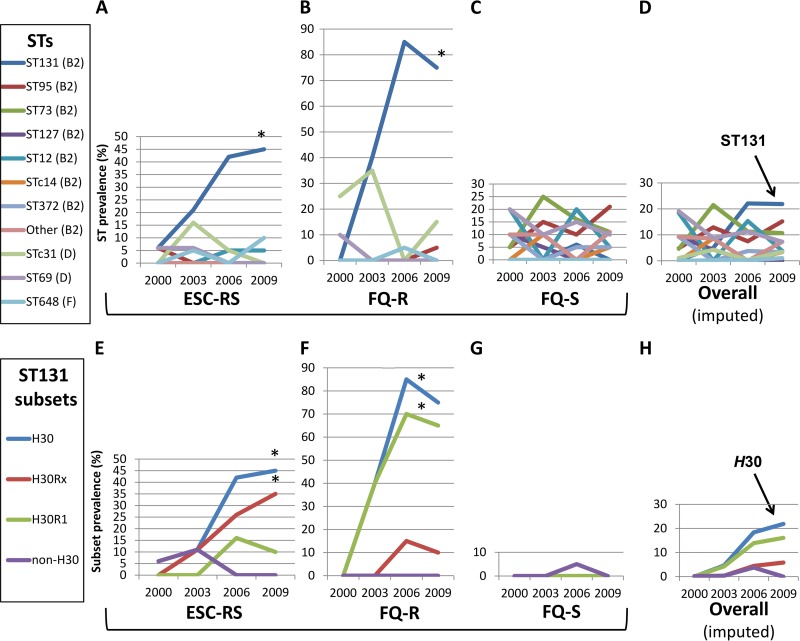
A similar analysis focusing on individual STs showed that within a given resistance category, the only ST that shifted significantly over time in relative prevalence was ST131, which accounted for nearly all group B2 isolates among the ESC-RS and FQ-R isolates (Fig. 2A and B). There, ST131's prevalence increased from 0% (2000, both ESC-RS and FQ-R isolates) to peaks of 45% (2009, ESC-RS isolates) and 85% (2006, FQ-R isolates). In contrast, among the FQ-S isolates, no ST shifted significantly in prevalence over time, and ST131 was scarcely detected, with group B2 being represented instead mainly by ST73 and ST95 (Fig. 2C). In the population-adjusted analysis, the main temporal shift in relative ST prevalence was the dramatic rise in ST131, which by 2006 had become the dominant ST overall (22.8% total prevalence), supplanting the previously dominant STs 69, 73, and 95 (Fig. 2D).
FIG 2.
Temporal trends for sequence type (ST) and ST131 clonal subset prevalence. Data are for 2000, 2003, 2006, and 2009; intervening points are interpolated. Resistance categories: ESC-RS, extended-spectrum cephalosporin-reduced susceptibility; FQ-R, fluoroquinolone resistant, ESC susceptible; FQ-S, fluoroquinolone susceptible, ESC susceptible. The key identifies STs by phylogroup. Only STs that accounted for ≥10% of isolates in ≥1 resistance subset during ≥1 study year are shown. Other (B2), combined group B2 isolates from STs other than the STs studied. Within ST131 (E to H), values for H30Rx and H30R1 sum to those for H30, and values for H30 and non-H30 sum to those for ST131. *, P < 0.05 for linear trend. (A) Measured relative prevalence of STs among the 75 selected ESC-RS study isolates (17 to 20 per year). (B) Measured relative prevalences of STs among the 80 selected FQ-R study isolates (20 per year). (C) Measured relative prevalences of STs among the 79 selected FQ-S study isolates (19 or 20 per year). (D) Imputed relative prevalences of STs within the overall SENTRY source population. (E) Measured relative prevalences of ST131 and four clonal subsets among the 75 selected ESC-RS study isolates (17 to 20 per year). (F) Measured relative prevalences of ST131 and four clonal subsets among the 80 selected FQ-R study isolates (20 per year). (G) Measured relative prevalences of ST131 and four clonal subsets among the 79 selected FQ-S study isolates (19 or 20 per year). (H) Imputed relative prevalences of ST131 and four clonal subsets within the overall SENTRY source population.
ST131 subsets.
A similar analysis focusing on clonal subsets within ST131 showed that the ST131 emergence was due entirely to the H30 subclone (Fig. 2E to H). Both of the main H30 subsets, i.e., H30Rx and H30R1, contributed importantly to this expansion. However, their relative prevalences varied in relation to resistance category, i.e., H30Rx strains predominated among ESC-RS isolates (Fig. 2E) and H30R1 strains among FQ-R isolates (Fig. 2F).
In contrast, non-H30 ST131 strains were quite rare. Although in 2000 they were the only ST131 strains identified, and in 2003, they were as prevalent within the ESC-RS category as H30 isolates (11%) (Fig. 2E), in 2006, they were detected only as a single FQ-S isolate (Fig. 2G) and in 2009 were not detected at all.
The corresponding population-adjusted analysis identified H30R1 as the main overall driver of the H30 expansion, with H30Rx making a much smaller, albeit proportionately consistent, contribution (Fig. 2H). In contrast, non-H30 strains registered minimally and in 2006 only.
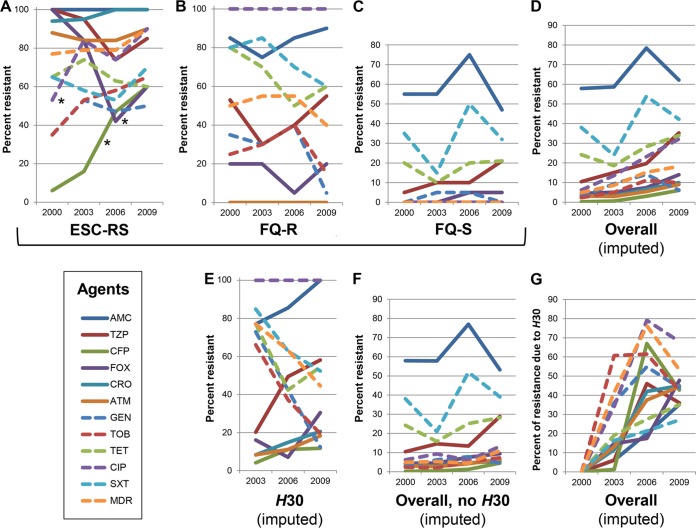
Antimicrobial resistance.
Resistance to imipenem (n = 1) and amikacin (n = 2) was rare, so it was not analyzed statistically. For the remaining 11 agents, the prevalence of resistance to individual agents and to ≥3 antimicrobial classes (i.e., multidrug resistance [MDR] status) varied considerably by resistance category, agent, and year (Fig. 3A to D). In general, resistance prevalence was highest among ESC-RS isolates (Fig. 3A), intermediate among FQ-R isolates (except for ciprofloxacin resistance, which was 100% prevalent, by definition) (Fig. 3B), and lowest among FQ-S isolates (Fig. 3C). Although most of the observed temporal variation appeared to be nondirectional, resistance increased significantly among ESC-RS isolates for cefepime and ciprofloxacin and decreased significantly for cefoxitin (Fig. 3A).
FIG 3.
Temporal trends for individual antimicrobial agent and multidrug resistance. Data are for 2000, 2003, 2006, and 2009; intervening points are interpolated. Resistance categories: ESC-RS, extended-spectrum cephalosporin-reduced susceptibility; FQ-R, fluoroquinolone resistant, ESC susceptible; FQ-S, fluoroquinolone susceptible, ESC susceptible. AMC, amoxicillin-clavulanate; TZP, piperacillin-tazobactam; CFP, cefepime; FOX, cefoxitin; CRO, ceftriaxone; ATM, aztreonam; GEN, gentamicin; TOB, tobramycin; TET, tetracycline; CIP, ciprofloxacin; SXT, sulfamethoxazole-trimethoprim; MDR, resistant to ≥3 antimicrobial classes. *, P < 0.05 for linear trend. (A) Measured prevalences of antimicrobial resistance among the 75 selected ESC-RS study isolates (17 to 20 per year). (B) Measured prevalences of antimicrobial resistance among the 80 selected FQ-R study isolates (20 per year). (C) Measured prevalences of antimicrobial resistance among the 79 selected FQ-S study isolates (19 or 20 per year). (D) Imputed prevalences of resistance within the overall SENTRY source population. (E) Imputed prevalences of resistance among ST131-H30 isolates within the overall SENTRY source population. (F) Imputed prevalences of resistance among non-H30 isolates within the overall SENTRY source population. (G) Imputed fractions of resistance attributable to H30 within the overall SENTRY source population.
In the overall population-adjusted analyses, although resistance prevalence generally increased over time, most values remained at <20% (Fig. 3D). In contrast, a similar analysis limited to H30 isolates showed much higher resistance prevalence values, mostly >20% (Fig. 3E). Considerable year-to-year variation was evident, some directional (either up or down) and some nondirectional. Conversely, an analysis limited to non-H30 isolates yielded time-trend curves that, compared with the overall population curves, were downshifted (most values were <10%) and flattened, indicating slower temporal increases (Fig. 3F). An analysis of the by-year proportional contribution of H30 to each resistance phenotype within the total population showed a striking net rise across the study period, from 0% in 2000 to a median of 43% (range, 25% to 68%) in 2009 (Fig. 3G).
Resistance and virulence gene scores.
To gain insights into the basis for and the potential clinical implications of H30's emergence, we assessed resistance scores and virulence gene scores for temporal trends and differences between H30 isolates and all non-H30 isolates (whether ST131 or non-ST131), both overall and stratified by resistance category. Within a given resistance category, resistance scores neither changed significantly over time nor differed significantly between H30 and non-H30 isolates (data not shown). In contrast, virulence gene scores, although temporally stable among FQ-S isolates, increased significantly over time among FQ-R isolates (annual means, 5 [2000] to 8.7 [2009]; P < 0.001) and numerically but nonsignificantly among ESC-RS isolates (annual means, 7.4 [2000] to 8.1 [2009]; P = 0.78), whereas they were stable among FQ-S isolates (not shown). Correspondingly, when virulence gene scores were aggregated across study years, they were significantly higher for H30 isolates than for all non-H30 isolates, among both the ESC-RS isolates (means, 10.4 [H30] versus 6.7 [non-H30]; P < 0.001) and the FQ-R isolates (means, 9.3 [H30] versus 5.5 [non-H30]; P < 0.001). In a population-adjusted analysis that included all three resistance categories and the H30 and non-H30 isolates, despite the overall temporal increase in resistance prevalence (Fig. 1A and 3D), the population's mean virulence gene scores remained stable (mean scores, 9.9 [2000], 10.3 [2003], 9.7 [2006], and 10.0 [2009]).
DISCUSSION
In this study, we analyzed temporal trends (2000 to 2009) in the clonal distribution, antimicrobial resistance profiles, and virulence gene contents of a nationally representative U.S.-based collection of E. coli clinical isolates and then compared clonal background with resistance profiles and virulence genes. Our findings support three main conclusions. First, the study period captured a dramatic rise in the relative prevalence of phylogroup B2 among ESC-RS and FQ-R isolates that was due entirely to the rapid emergence of ST131, and specifically its H30 subclone (7), which by the end of the study period was the dominant E. coli lineage overall. Second, because of H30's expansion and extensive antimicrobial resistance, by the study end, it accounted overall for a median of 43% of antimicrobial resistance, including up to 68% for ciprofloxacin. Third, due to H30's extensive virulence gene content, the H30 emergence led to increased virulence gene content among resistant isolates, especially the FQ-R subset, resulting in stable overall virulence gene scores despite expansion of the resistant population, which historically has had a relative deficit of virulence genes (20). These findings underscore the uniqueness and importance of H30, thereby emphasizing the critical need for effective interventions against this remarkable multidrug-resistant pathogen.
This study is one of few that provide a systematic longitudinal analysis of the clonal distribution of E. coli clinical isolates at a national level (19). Consequently, it distinctively documents in the United States the emergence of ST131 and its various clonal subsets, including H30, H30R1, H30Rx, and non-H30 strains, and defines over time the relative contribution of H30 to antimicrobial resistance within E. coli. It also is one of only a few studies to document temporal trends in virulence gene content in relation to clonal background and resistance. As such, it supersedes previous longitudinal studies based on smaller geographical regions (11, 13–18, 21), more narrowly selected bacterial populations (17, 18, 21), and convenience samples (8–10, 12, 22), as well as those that provided broadly inclusive national-level surveillance but only cross-sectionally (23).
Regarding our first main finding, we found that H30's expansion in the United States began only after 2000 yet was nearly complete by 2006, a remarkably rapid rise. This fits with previous estimates based on less reliably representative strain sets (8, 24). Although much attention has been given to H30Rx, due largely to its clinically important association with the CTX-M-15 ESBL (8), which confers ESC resistance, our findings indicate that in the United States, H30Rx was only a minor contributor to the H30 expansion, which instead was driven mainly by H30R1. It is conceivable that although, on average, H30Rx is more extensively resistant than H30R1, it is less fit in some way that has limited its relative prevalence among clinical isolates. Alternatively, these two H30 subclones may be equally fit but were subject to a founder effect, whereby H30R1 emerged first in the United States and disseminated slightly before H30Rx, thereby occupying relevant niches and excluding the soon-to-follow H30Rx. A more granular analysis of isolates from 2000, 2001, 2002, and 2003 could be used to hone in on the comparative timing of the first appearances of H30R and H30Rx. Notably, in a longitudinal Calgary region-based study, H30R1 had a similar prevalence advantage over H30Rx early on but was overtaken by H30Rx in the late 2000s (17), suggesting regional differences in clonal dynamics (16).
Regarding our second main finding, our results provide novel insights into the shifts over time in H30's contribution to total antimicrobial resistance in E. coli. H30's share of total resistance clearly varied both by agent, being greatest for ciprofloxacin, tobramycin, and multidrug resistance, and by year, increasing over time for nearly all agents. These results confirm H30 as the single main contributor to the rising prevalence of resistance among SENTRY collection E. coli isolates during the study period (23). However, it likewise establishes that resistance increased also among non-H30 E. coli isolates, which indicates that a focus on H30 alone would only partially address the E. coli resistance problem. Similar analyses of other lineages could determine which may warrant special attention with respect to surveillance, diagnostics, and therapeutics, analogous (albeit secondary) to H30 (25, 26).
Regarding our third main finding, the more extensive virulence gene content of H30 strains, compared with that of other resistant strains, may both underlie H30's expansion and create an added threat from it. That is, to the extent that virulence gene content predicts actual virulence potential (27), strains with more extensive virulence gene profiles may be more likely to cause infections and/or to cause more severe infections. As such, the H30 expansion may have both enlarged the resistant population and made it more aggressive, thereby resulting in the double threat of more severe infections that also are harder to treat. Although available epidemiological data do not consistently support the idea that infections caused by H30 are more aggressive than those caused by other E. coli types (17, 28, 29), H30 has not been compared adequately with similarly resistant strains, over which it may have a virulence or aggressiveness advantage by virtue of its greater virulence gene content. This hypothesis awaits specific testing.
The study has limitations. First, clinical and epidemiological data were unavailable. Second, generalizability beyond the participating SENTRY sites is uncertain. Third, in some instances, small subgroups created unstable prevalence estimates and limited statistical power. Fourth, mechanisms for reduced ESC susceptibility (e.g., ESBL production) were not assessed phenotypically or genotypically. The study also has important strengths. First, the isolates were collected prospectively and systematically from across the United States, making them uniquely suitable for the proposed analyses. Second, the study period was selected to correspond with the presumed time of emergence in the United States of H30. Third, the molecular typing was extensive and state-of-the-art. Fourth, extensive standardized antimicrobial susceptibility data were available, allowing analyses of resistance in relation to clonal background.
In summary, by analyzing E. coli isolates selected systematically from the nationally representative SENTRY collection, we documented the rapid emergence of ST131-H30 in the United States between 2000 and 2009. This emergence resulted in marked increases in the overall prevalence of antimicrobial resistance in E. coli and, among resistant isolates, significant shifts toward phylogenetic group B2 and greater virulence gene content. These findings clarify the timing and tempo of H30's emergence in the United States, identify possible reasons for its occurrence, and suggest the likely consequences of it, including more frequent and, potentially, more aggressive antimicrobial-resistant infections.
MATERIALS AND METHODS
Isolates.
The SENTRY Antimicrobial Surveillance Program systematically collected 1,587 (in 2000), 1,409 (2003), 974 (2006), and 1,261 (2009) nonduplicate E. coli clinical isolates from patients hospitalized in multiple widely distributed U.S. medical centers, without regard for susceptibility testing results, and prioritizing blood isolates. The contributing centers represented multiple cities and states within all nine U.S. geographical regions.
The isolates initially underwent standardized antimicrobial susceptibility testing at JMI Laboratories (see below). Those that exhibited reduced broth microdilution susceptibility to ESCs (30), regardless of FQ phenotype, were classified as ESC-RS, which suggests ESBL production, although alternative resistance mechanisms (e.g., AmpC production and porin mutations) are possible. Based on ciprofloxacin MICs, the remaining (fully ESC-susceptible) isolates were classified as being either FQ-R or FQ-S. This stratification yielded three resistance categories, i.e., ESC-RS, FQ-R, and FQ-S.
To obtain representative subsamples for molecular analysis, 20 isolates per resistance category per year were selected randomly for 2000, 2003, 2006, and 2009, giving 60 study isolates per year, or 240 total isolates. Of these, 6 isolates were excluded later for failure to confirm identity and/or resistance category, leaving 234 confirmed study isolates. Due in part to the SENTRY program's criteria for isolate submission, 84% of the study isolates were from blood, 8% were from urine, and 8% were from miscellaneous other sources. Clinical and epidemiological data regarding the source patients were unavailable.
Susceptibility testing.
Susceptibility to 13 antimicrobial agents was assessed by broth microdilution according to procedures and interpretive criteria specified by the Clinical and Laboratory Standards Institute (30, 31). Intermediate results were analyzed as resistant. Multidrug-resistant isolates were those resistant to at least 1 representative of ≥3 antimicrobial classes, i.e., penicillins (amoxicillin-clavulanate and piperacillin-tazobactam), cephalosporins and monobactams (cefoxitin, ceftriaxone, cefepime, and aztreonam), carbapenems (imipenem), aminoglycosides (gentamicin, tobramycin, and amikacin), fluoroquinolones (ciprofloxacin), tetracyclines (tetracycline), and antifolate agents (trimethoprim-sulfamethoxazole) (16). The resistance score was the number of antimicrobial classes to which resistance was detected (16).
Genotyping.
Molecular typing was done using established multiplex PCR-based assays, with duplicate boiled lysates used as the template DNA and inclusion of relevant positive and negative controls. Phylogroups A, B1, B2, C, D, E, and F were resolved per Clermont et al. (32). ST131, H30, and H30Rx, plus 12 other STs, including 8 from group B2 (ST12, ST14, ST73, ST95, ST127, ST141, ST144, and ST372), 3 from group D (ST31 complex, ST69, and ST405), and 1 from group F (ST648), were detected by clade-specific PCR assays, as described previously (25, 33–40). Fluoroquinolone-resistant H30 isolates were classified as H30R, and among these, the H30Rx-negative isolates were classified as H30R1. Extended virulence genotypes for 51 extraintestinal virulence-associated genes and variants thereof, including those for diverse adhesins, toxins, siderophores, protectins, invasins, and miscellaneous factors, were detected as described previously (27). An isolate's virulence gene score, which corresponds with virulence both epidemiologically and experimentally (41, 42), was the number of virulence gene operons detected. Isolates were classified presumptively as ExPEC if positive for >2 of papAH and/or papC (P fimbriae, counted as one), sfa or focDE (S and F1C fimbriae), afa or draBC (Dr-binding adhesins), kpsM II (group 2 capsule), and iutA (aerobactin receptor) (43).
Statistical methods.
To allow prevalence values derived from the artificially selected study subsets to be scaled proportionately to the SENTRY source population, resistance category-specific results were adjusted for the relative prevalence of the corresponding resistance category within the source population during a particular study year, with additional adjustment (as needed) for the relative prevalence of specific clonal subsets within each resistance category. The resulting fractional prevalence values for a given year for individual resistance categories were summed to yield authentic annual prevalence values applicable to the total source population.
Statistical testing was done using SPSS version 16 (IBM Analytics). Comparisons involving dichotomous variables were tested by Fisher's exact test (two-tailed); those involving continuous variables were tested by analysis of variance (ANOVA) or a two-tailed t test. The significance threshold was a P value of <0.05.
Ethics approval.
The Minneapolis VA Health Care System institutional review board reviewed and approved the study.
ACKNOWLEDGMENTS
This work was supported by merit review award numbers I01 CX000192 01 and I01 CX000920 from the U.S. Department of Veterans Affairs Clinical Sciences R&D (CSRD) Service (both to J.R.J.). The sponsor had no involvement in the conduct of the study, data analysis, manuscript preparation, or decision to publish.
J. R. Johnson has received contracts, grants, or consultancies from Actavis, Jannsen, Merck, and Tetraphase and has patent applications pertaining to tests for specific E. coli strains. M. Castanheira is an employee of JMI Laboratories, which was contracted to perform services in 2016 for Achaogen, Actelion, Allecra, Allergan, Ampliphi, API, Astellas, AstraZeneca, Basilea, Bayer, BD, Biomodels, Cardeas, CEM-102 Pharma, Cempra, Cidara, Cormedix, CSA Biotech, Cubist, Debiopharm, Dipexium, Duke, Durata, Entasis, Fortress, Fox Chase Chemical, GSK, Medpace, Melinta, Merck, Micurx, Motif, N8 Medical, Nabriva, Nexcida, Novartis, Paratek, Pfizer, Polyphor, Rempex, Scynexis, Shionogi, Spero Therapeutics, Symbal Therapeutics, Synolgoic, TGV Therapeutics, The Medicines Company, Theravance, Thermo Fisher, VenatoRx, Wockhardt, and Zavante. Additionally, some JMI employees are advisors/consultants for Allergan, Astellas, Cubist, Pfizer, Cempra, and Theravance. The other authors report no conflicts of interest.
REFERENCES
- 1.Russo TA, Johnson JR. 2003. Medical and economic impact of extraintestinal infections due to Escherichia coli: an overlooked epidemic. Microbes Infect 5:449–456. doi: 10.1016/S1286-4579(03)00049-2. [DOI] [PubMed] [Google Scholar]
- 2.Thelwall S, Nsonwu O, Wasti S, Elmi M, Gerver S, Davies J, Hope R. 2016. Annual epidemiological commentary: mandatory MRSA, MSSA and E. coli bacteraemia and C. difficile infection data 2015/16, p 33–42. Public Health England, London, United Kingdom: https://www.researchgate.net/publication/305392637_Annual_Epidemiological_Commentary_Mandatory_MRSA_MSSA_and_E_coli_bacteraemia_and_C_difficile_infection_data_201516. [Google Scholar]
- 3.Walker EB, Lyman A, Gupta K, Mahoney MV, Snyder GM, Hirsch EB. 2016. Clinical management of an increasing threat: outpatient urinary tract infections due to multidrug-resistant uropathogens. Clin Infect Dis 63:960–965. doi: 10.1093/cid/ciw396. [DOI] [PubMed] [Google Scholar]
- 4.Nicolas-Chanoine M-H, Blanco J, Leflon-Guibout V, Demarty R, Alonso MP, Caniça MM, Park Y-J, Lavigne J-P, Pitout J, Johnson JR. 2008. Intercontinental emergence of Escherichia coli clone O25:H4-ST131 producing CTX-M-15. J Antimicrob Chemother 61:273–281. doi: 10.1093/jac/dkm464. [DOI] [PubMed] [Google Scholar]
- 5.Coque TM, Novais A, Carattoli A, Poirel L, Pitout J, Peixe L, Baquero F, Cantón R, Nordmann P. 2008. Dissemination of clonally related Escherichia coli strains expressing extended-spectrum β-lactamase CTX-M-15. Emerg Infect Dis 14:195–200. doi: 10.3201/eid1402.070350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nicolas-Chanoine M, Bertrand X, Madec J-Y. 2014. Escherichia coli ST131, an intriguing clonal group. Clin Microbiol Rev 27:543–574. doi: 10.1128/CMR.00125-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson JR, Tchesnokova V, Johnston B, Clabots C, Roberts PL, Billig M, Riddel K, Rogers P, Qin X, Butler-Wu S, Price LB, Aziz M, Nicolas-Chanoine M, Debroy C, Robicsek A, Hansen G, Urban C, Platell JL, Trott DJ, Zhanel G, Weissman SJ, Cookson B, Fang F, Limaye AP, Scholes D, Chattopadhyay S, Hooper DC, Sokurenko E. 2013. Abrupt emergence of a single dominant multi-drug-resistant strain of Escherichia coli. J Infect Dis 207:919–928. doi: 10.1093/infdis/jis933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Price LB, Johnson JR, Aziz M, Clabots C, Johnston B, Tchesnokova V, Nordstrom L, Billig M, Chattopadhyay S, Stegger M, Andersen PS, Pearson T, Riddell K, Rogers P, Scholes D, Kahl B, Keim P, Sokurenko EV. 2013. The epidemic of ESBL-producing Escherichia coli ST131 is driven by a single highly virulent subclone, H30-Rx. mBio 4(6):e00377-13. doi: 10.1128/mBio.00377-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Petty NK, Ben Zakour N, Stanton-Cook M, Skippington E, Totsika M, Forde B, Phan M-D, Moriela DG, Peters K, Davies M, Rogers BA, Dougand G, Rodriguez-Baño J, Pascual A, Pitout J, Upton M, Paterson DL, Walsh TR, Schembri MA, Beatson SA. 2014. Global dissemination of a multidrug resistant Escherichia coli clone. Proc Natl Acad Sci U S A 111:5694–5699. doi: 10.1073/pnas.1322678111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stoesser N, Sheppard AE, Pankhurst L, de Maio N, Moore C, Sebra R, Turner P, Anson L, Kasarkis A, Batty E, Kos V, Wilson D, Phetsouvanh R, Wyllie D, Sokurenko E, Manges A, Johnson TJ, Price LB, Peto T, Johnson J, Didelot X, Walker AS, Crook D, Modernizing Medical Microbiology Informatics Group (MMMIG). 2016. Evolutionary history of the global emergence of the Escherichia coli epidemic clone ST131. mBio 7(2):e02162-15. doi: 10.1128/mBio.02162-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matsumura Y, Pitout J, Gommi R, Matsuda T, Noguchi T, Yamamoto M, Peirano G, DeVinney R, Bradfor PA, Motyl M, Tanaka M, Nagao M, Takakura S, Ichiyama S. 2016. Global Escherichia coli sequence type 131 clade with blaCTX-M-27 gene. Emerg Infect Dis 22:1900–1907. doi: 10.3201/eid2211.160519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ben Zakour N, Alsheich-Hussain A, Ashcroft M, Nhu N, Roberts L, Stanton-Cook M, Schembri MA, Beatson SA. 2016. Sequential acquisition of virulence and fluoroquinolone resistance has shaped the evolution of Escherichia coli ST131. mBio 7:e00347-16. doi: 10.1128/mBio.00347-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matsumura Y, Johnson JR, Yamamoto M, Nagao M, Tanaka K, Takakura S, Ichiyama S, Kyoto-Shiga Clinical Microbiology Study Group. 2015. CTX-M-27- and CTX-M-14-producing, ciprofloxacin-resistant Escherichia coli of the H30 subclonal group within ST131 drive a Japanese regional ESBL epidemic. J Antimicrob Chemother 70:1639–1649. [DOI] [PubMed] [Google Scholar]
- 14.Peirano G, van der Bij A, Gregson DB, Pitout JDD. 2012. Molecular epidemiology over an 11-year period (2000 to 2010) of extended-spectrum β-lactamase-producing Escherichia coli causing bacteremia in a centralized Canadian region. J Clin Microbiol 50:294–299. doi: 10.1128/JCM.06025-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karfunkel D, Carmeli Y, Chmelnitsky I, Kotlovsky T, Navon-Venezia S. 2013. The emergence and dissemination of CTX-M-producing Escherichia coli sequence type 131 causing community-onset bacteremia in Israel. Eur J Clin Microbiol Infect Dis 32:513–521. doi: 10.1007/s10096-012-1765-9. [DOI] [PubMed] [Google Scholar]
- 16.Johnson JR, Johnston B, Thuras P, Launer B, Sokurenko EV, Miller L. 2016. Escherichia coli sequence type 131 H30 is the main driver of emerging extended-spectrum-β-lactamase-producing E. coli at a tertiary care center. mSphere 1(6):e00314-16. doi: 10.1128/mSphere.00314-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peirano G, Pitout JDD. 2014. Fluoroquinolone-resistant Escherichia coli sequence type 131 isolates causing bloodstream infections in a Canadian region with a centralized laboratory system: rapid emergence of the H30-Rx sublineage. Antimicrob Agents Chemother 58:2600–2703. doi: 10.1128/AAC.00119-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Olesen B, Hansen DS, Nilsson F, Frimodt-Møller J, Leihof RF, Struve C, Scheutz F, Johnston B, Krogfelt KA, Johnson JR. 2013. Prevalence and characteristics of the epidemic multiresistant Escherichia coli ST131 clonal group among extended-spectrum β-lactamase (ESBL)-producing E. coli in Copenhagen, Denmark. J Clin Microbiol 51:1779–1785. doi: 10.1128/JCM.00346-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Day MJ, Doumith M, Abernethy J, Hope R, Reynolds R, Wain J, Livermore DM, Woodford N. 2016. Population structure of Escherichia coli causing bacteraemia in the UK and Ireland between 2001 and 2010. J Antimicrob Chemother 71:2139–2142. doi: 10.1093/jac/dkw145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vila J, Simon K, Ruiz J, Horcajada JP, Velasco M, Barranco M, Moreno A, Mensa J. 2002. Are quinolone-resistant uropathogenic Escherichia coli less virulent? J Infect Dis 186:1039–1042. doi: 10.1086/342955. [DOI] [PubMed] [Google Scholar]
- 21.Peirano G, van der Bij AK, Freeman JL, Poirel L, Nordmann P, Costello M, Tchesnokova VL, Pitout JDD. 2014. Characteristics of Escherichia coli sequence type 131 isolates that produce extended-spectrum beta-lactamases: global distribution of the H30-Rx sublineage. Antimicrob Agents Chemother 58:3762–3767. doi: 10.1128/AAC.02428-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McNally A, Oren Y, Kelly D, Pascoe B, Dunn S, Sreecharan T, Vehkala M, Välimäki N, Prentice M, Ashour A, Avram O, Pupko T, Dobrindt U, Literak I, Guenther S, Schaufler K, Wieler LH, Zhiyong Z, Sheppard SK, McInerney JO, Corander J. 2016. Combined analysis of variation in core, accessory and regulatory genome regions provides a super-resolution view into the evolution of bacterial populations. PLoS Genet 12:e1006280. doi: 10.1371/journal.pgen.1006280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson JR, Johnston B, Clabots C, Kuskowski MA, Castanheira M. 2010. Escherichia coli sequence type ST131 as the major cause of serious multidrug-resistant E. coli infections in the United States (2007). Clin Infect Dis 51:286–294. doi: 10.1086/653932. [DOI] [PubMed] [Google Scholar]
- 24.Johnson JR, Nicolas-Chanoine M, Debroy C, Castanheira M, Robiscek A, Hansen G, Weissman SJ, Urban C, Platell JL, Trott DJ, Zhanel GG, Clabots C, Johnston BD, Kuskowski MA, the MASTER Investigators. 2012. Comparison of Escherichia coli sequence type ST131 pulsotypes by epidemiologic traits, 1967–2009. Emerg Infect Dis 18:598–607. doi: 10.3201/eid1804.111627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson JR, Johnston BD, Gordon DM. 2017. Rapid and specific detection of the Escherichia coli sequence type 648 complex (STc648) within phylogroup F. J Clin Microbiol 55:1116–1121. doi: 10.1128/JCM.01949-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Riley LW. 2014. Pandemic lineages of extraintestinal pathogenic Escherichia coli. Clin Microbiol Infect 20:380–390. doi: 10.1111/1469-0691.12646. [DOI] [PubMed] [Google Scholar]
- 27.Johnson JR, Porter S, Johnston B, Kuskowski MA, Spurbeck RR, Mobley HLT, Williamson DA. 2015. Host characteristics and bacterial traits predict experimental virulence for Escherichia coli bloodstream isolates from patients with urosepsis. Open Forum Infect Dis 2:ofv083. doi: 10.1093/ofid/ofv083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson J, Thuras P, Johnston B, Weissman SJ, Limaye AP, Riddell K, Scholes D, Tchesnokova V, Sokurenko E. 2016. The pandemic H30 subclone of Escherichia coli sequence type 131 is associated with persistent infections and adverse outcomes independent from its multidrug resistance and associations with compromised hosts. Clin Infect Dis 62:1529–1536. doi: 10.1093/cid/ciw193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.López-Cerero L, Navarro MD, M B, Martín-Peña A, Vinas L, Cisneros JM, Gómez-Langley SL, Sánchez-Monteseirín H, Morales I, Pascual A, Rodríguez-Baño J. 2014. Escherichia coli belonging to the worldwide emerging epidemic clonal group O25b/ST131: risk factors and clinical implications. J Antimicrob Chemother 69:809–814. doi: 10.1093/jac/dkt405. [DOI] [PubMed] [Google Scholar]
- 30.Clinical and Laboratory Standards Institute. 2013. Performance standards for antimicrobial susceptibility testing; 23rd informational supplement CLSI document M100-S23. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 31.Clinical and Laboratory Standards Institute. 2012. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, 9th ed CLSI document M07-A9. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 32.Clermont O, Christenson JK, Denamur E, Gordon DM. 2012. The Clermont Escherichia coli phylo-typing method revisited: improvement of specificity and detection of new phylo-groups. Environ Microbiol Rep 5:58–65. doi: 10.1111/1758-2229.12019. [DOI] [PubMed] [Google Scholar]
- 33.Johnson JR, Menard M, Johnston B, Kuskowski MA, Nichol K, Zhanel GG. 2009. Epidemic clonal groups of Escherichia coli as a cause of antimicrobial-resistant urinary tract infections in Canada, 2002–2004. Antimicrob Agents Chemother 53:2733–2739. doi: 10.1128/AAC.00297-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Colpan A, Johnston B, Porter S, Clabots C, Anway R, Thao L, Kuskowski MA, Tchesnokova V, Sokurenko EV, Johnson JR, VICTORY (Veterans Influence of Clonal Types on Resistance: Year 2011) Investigators. 2013. Escherichia coli sequence type 131 (ST131) as an emergent multidrug-resistant pathogen among U.S. veterans. Clin Infect Dis 57:1256–1265. doi: 10.1093/cid/cit503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Banerjee R, Strahilevitz J, Johnson J, Nagwekar P, Schora D, Shevrin I, Du H, Peterson L, Robicsek A. 2013. Predictors and molecular epidemiology of community-onset extended-spectrum beta-lactamase-producing Escherichia coli infection in a midwestern community. Infect Control Hosp Epidemiol 34:947–953. doi: 10.1086/671725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clermont O, Christenson JK, Daubie A, Gordon DM, Denamur E. 2014. Development of an allele-specific PCR for Escherichia coli B2 sub-typing, a rapid and easy to perform substitute of multilocus sequence typing. J Microbiol Methods 101:24–27. doi: 10.1016/j.mimet.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 37.Johnson JR, Owens K, O'Bryan TT, Sabate M, Prats G. 2004. Rapid and specific detection of the O15:K52:H1 clonal group of Escherichia coli by gene-specific PCR. J Clin Microbiol 42:3841–3843. doi: 10.1128/JCM.42.8.3841-3843.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johnson JR, Owens K, Manges AR, Riley LW. 2004. Rapid and specific detection of Escherichia coli clonal group A by gene-specific PCR. J Clin Microbiol 42:2618–2622. doi: 10.1128/JCM.42.6.2618-2622.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matsumura Y, Yamamoto M, Nagao M, Hotta G, Matsushima A, Ito Y, Takakura S, Ichiyama S, Kyoto-Shiga Clinical Microbiology Study Group. 2012. Emergence and spread of B2-ST131-O25b, B2-ST131-O16 and D-ST405 clonal groups among extended-spectrum-β-lactamase-producing Escherichia coli in Japan. J Antimicrob Chemother 67:2612–2620. doi: 10.1093/jac/dks278. [DOI] [PubMed] [Google Scholar]
- 40.Johnson JR, Clermont O, Johnston B, Clabots C, Tchesnokova V, Sokurenko E, Junka AF, Maczynska B, Denamur E. 2014. Rapid and specific detection, molecular epidemiology, and experimental virulence of the O16 subgroup within Escherichia coli sequence type 131. J Clin Microbiol 52:1358–1365. doi: 10.1128/JCM.03502-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johnson JR, Owens K, Gajewski A, Kuskowski MA. 2005. Bacterial characteristics in relation to clinical source among Escherichia coli isolates from women with acute cystitis or pyelonephritis and uninfected women. J Clin Microbiol 43:6064–6072. doi: 10.1128/JCM.43.12.6064-6072.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Johnson JR, Porter SB, Zhanel G, Kuskowski MA, Denamur E. 2012. Virulence of Escherichia coli clinical isolates in a murine sepsis model in relation to sequence type ST131 status, fluoroquinolone resistance, and virulence genotype. Infect Immun 80:1554–1562. doi: 10.1128/IAI.06388-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johnson JR, Murray AC, Gajewski A, Sullivan M, Snippes P, Kuskowski MA, Smith KE. 2003. Isolation and molecular characterization of nalidixic acid-resistant extraintestinal pathogenic Escherichia coli from retail chicken products. Antimicrob Agents Chemother 47:2161–2168. doi: 10.1128/AAC.47.7.2161-2168.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]