LETTER
RmtB is a 16S rRNA methyltransferase that confers a high level of resistance to both old and new aminoglycoside agents (1). rmtB was first reported in a Serratia marcescens clinical isolate from Japan in 2002 (2). Since then, it has been widely reported in isolates recovered from environmental and hospital settings. Usually, IncFII plasmids harboring rmtB also carry carbapenemase-encoding genes that make its spread easier (3, 4). The rmtB genetic background is frequently related to blaTEM-1, which is usually located upstream of rmtB and is recognized as a Tn3 transposon passenger gene (tnpA, tnpR, and blaTEM-1) (2–6). However, this β-lactamase-encoding gene has been encountered in diverse transposons, like Tn1, Tn2, and Tn3, which were the first transposons characterized as related to antimicrobial resistance. Although Tn2 and Tn3 show high similarity, sharing the same mechanisms of transposition, they possess distinct passenger genes, like blaTEM-1a in Tn3 and blaTEM-1b in Tn2 (7). In addition, other nucleotide differences can be found in transposase- and resolvase-encoding genes and at the resolution site (res). The main mutations are usually found in a region located between nucleotides 94 and 150 at the res site (7). In this manner, the four Tn2 sequences regions (tnpA, res site, tnpR, and blaTEM) contained genetic characteristics capable of differentiating the other related transposons. Based on that, we analyzed the sequences of 19 whole-plasmid sequences carrying rmtB. We also studied 15 partial sequences of rmtB-associated transposons, which were deposited in GenBank to investigate the rmtB genetic background and its association with the blaTEM-1 variants.
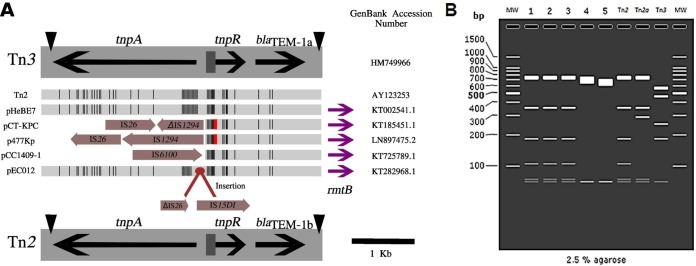
Three conserved rmtB genetic backgrounds (tnpA, tnpR, blaTEM-1b, and rmtB or tnpA, ΔtnpR, blaTEM-1b, and rmtB or ΔtnpA, tnpR, blaTEM-1b, and rmtB) were observed in all 34 analyzed sequences (Fig. 1A). In all structures, blaTEM-1b was detected. In this manner, we conclude that rmtB is in fact related to Tn2 and not Tn3, as suggested by many authors (2, 3, 5, 6). blaTEM-1a and blaTEM-1b show three nucleotide differences (18C→T, 228C→T, 396G→T) but encode identical TEM-1 enzymes. Although silent, these mutations may be helpful in identifying and suggesting the evolution of mobile genetic elements like Tn2 and Tn3. According to Partridge and Hall (7), the similarity between Tn2 and Tn3 has corroborated the misannotation of Tn2 as Tn3. In fact, a Tn2 or ΔTn2 transposon located upstream of rmtB has been recognized by only a few authors (8–12). Two factors have contributed to the misannotation of Tn2 as Tn3: the high similarity found between these two transposons and an annotation error in the databases. When a Tn2 nucleotide BLAST analysis is performed using the ISfinder, Tn3 is the first obtained hit (E value, 0.0; score, 8,387; 97% identity). UniProt and NCBI BLAST analyses also indicate the Tn3 transposon as the main result, with rare entries for Tn2. To make easier the proper automatic annotation, the Tn2 sequence was submitted to the ISfinder database. We also confirmed the presence of Tn2 by in silico MslI digestion using SnapGene version 3.3 (8). This analysis was able to differentiate Tn2 from its correlated variant Tn2a (Fig. 1B). Tn2a was described in 2011 (8) and differs from Tn2 by 9 nucleotides located in the resolution site.
FIG 1.
(A) Comparison of the main transposon sequence regions (tnpA, res, tnpR, and blaTEM) with Tn3. Genes are shown by labeled arrows, inverted repeats of Tn3 are shown as black triangles, and the resolution site is a dark-gray box. Inverted repeats are located in the same positions of Tn2. Vertical black lines indicate differences in the nucleotide sequences of Tn3 (reference sequence) (GenBank accession number HM749966) from those of the other analyzed transposons. The red box in pCT-KPC and the p477Kp transposons indicate the presence of truncated tnpR. Insertion sequences were identified by ISfinder and are demonstrated by light-purple arrows. The pEC012 plasmid shows an insertion of a partial IS26 (ΔIS26) sequence and an IS15DI sequence that show 99% similarity with IS26, according to ISfinder. Purple arrows indicate the presence of rmtB downstream of Tn2 transposons in pHeBE7 (Tn2), pCT-KPC (ΔTn2), p477Kp (ΔTn2), pCC1409-1 (ΔTn2), and pEC012 (Tn2). This figure was compiled from alignments created using SeaView version 4.6.1. (B) In silico MslI restriction analysis of Tn2 and its variants proposed by Bailey and colleagues (8). Lane 1, Tn2 of pHeBE7; lane 2, Tn2 of pIP1206; lane 3, Tn2 of pMC-NDM; lane 4, ΔTn2 (tnpR and blaTEM-1b) of pC06114; lane 5, ΔTn2 of p397Kp (ΔtnpR and blaTEM-1b). In both ΔTn2 sequences analyzed, tnpA was missing. MslI restriction confirmed the presence of Tn2 in isolates with the complete transposon present (tnpA, tnpR, and blaTEM-1b). However, this methodology was shown to not be accurate in identifying the ΔTn2 sequence, since the digested fragment comprises the end of tnpA (absent in ΔTn2) and the beginning of blaTEM-1b. MW, molecular weight markers.
The origin of the rmtB and Tn2 association remains unclear, since the right and left inverted repeats (IRR and IRL, respectively) of Tn2 have been located downstream of tnpA and upstream of blaTEM-1b (Fig. 1A), respectively. The absence of these sequences upstream of rmtB suggests the nonmobilization of rmtB by Tn2. For this mobilization to occur, the IRL upstream blaTEM-1b gene would have been inactivated by a new passenger gene (in this case, rmtB) and a more distant “surrogate” IR sequence recruited (13). This kind of mobilization had occurred with Tn4401, a derivate transposon of Tn3 that carries blaKPC-2 (14). Despite this genetic evidence, transposition in vitro experiments are necessary to confirm this hypothesis.
In conclusion, we reinforce the need for a proper annotation of the rmtB genetic structure through manual checking of automatic annotation to avoid a misperception about an epidemiological change. To date, rmtB has always been associated with Tn2 and never with Tn3.
Accession number(s).
The following GenBank accession numbers have been assigned: AM886293.1, KR078259.1, JF927996.1, KP893385.1, NC_016839.1, KT185451.1, KT002541.1, KT725788.1, KT725789.1, KT282968.1, KJ020575.1, KR259132.1, JN232517.1, LN897475.2, LN897474.2, NC_025106.1, NC_020278.2, NZ_CP015725.1, NZ_CP016035.1, EU213261.1, JN315966.1, FJ556899.1, EU491958.1, FJ167861.1, FJ410927.1, KM598665.1, HQ174461.1, KX064436.1, FJ556900.1, FJ539137.1, FJ183463.1, JQ941741.1, FJ744121.1, and HQ665010.1.
REFERENCES
- 1.Wachino J, Arakawa Y. 2012. Exogenously acquired 16S rRNA methyltransferases found in aminoglycoside-resistant pathogenic Gram-negative bacteria: an update. Drug Resist Updat 15:133–148. doi: 10.1016/j.drup.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 2.Doi Y, Yokoyama K, Yamane K, Wachino J, Shibata N, Yagi T, Shibayama K, Kato H, Arakawa Y. 2004. Plasmid-mediated 16S rRNA methylase in Serratia marcescens conferring high-level resistance to aminoglycosides. Antimicrob Agents Chemother 48:491–496. doi: 10.1128/AAC.48.2.491-496.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li JJ, Sheng ZK, Deng M, Bi S, Hu FS, Miao HF, Ji ZK, Sheng JF, Li LJ. 2012. Epidemic of Klebsiella pneumoniae ST11 clone coproducing KPC-2 and 16S rRNA methylase RmtB in a Chinese university hospital. BMC Infect Dis 12:373. doi: 10.1186/1471-2334-12-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pitart C, Solé M, Roca I, Román A, Moreno A, Vila J, Marco F. 2015. Molecular characterization of blaNDM-5 carried on an IncFII plasmid in an Escherichia coli isolate from a nontraveler patient in Spain. Antimicrob Agents Chemother 59:659–662. doi: 10.1128/AAC.04040-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu FY, Yao D, Pan JY, Chen C, Qin ZQ, Parsons C, Yang LH, Li QQ, Zhang XQ, Qu D, Wang LX. 2010. High prevalence of plasmid-mediated 16S rRNA methylase gene rmtB among Escherichia coli clinical isolates from a Chinese teaching hospital. BMC Infect Dis 23:184. doi: 10.1186/1471-2334-10-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deng YT, Zeng ZL, Tian W, Yang T, Liu JH. 2013. Prevalence and characteristics of rmtB and qepA in Escherichia coli isolated from diseased animals in China. Front Microbiol 4:198. doi: 10.3389/fmicb.2013.00198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Partridge SR, Hall RM. 2005. Evolution of transposons containing blaTEM genes. Antimicrob Agents Chemother 49:1267–1268. doi: 10.1128/AAC.49.3.1267-1268.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bailey JK, Pinyon JL, Anantham S, Hall RM. 2011. Distribution of the blaTEM gene and blaTEM-containing transposons in commensal Escherichia coli. J Antimicrob Chemother 66:745–751. doi: 10.1093/jac/dkq529. [DOI] [PubMed] [Google Scholar]
- 9.He L, Partridge SR, Yang X, Hou J, Deng Y, Yao Q, Zeng Z, Chen Z, Liu JH. 2013. Complete nucleotide sequence of pHN7A8, an F33:A-:B- type epidemic plasmid carrying blaCTX-M-65, fosA3 and rmtB from China. J Antimicrob Chemother 68:46–50. doi: 10.1093/jac/dks369. [DOI] [PubMed] [Google Scholar]
- 10.Chen X, He L, Li Y, Zeng Z, Deng Y, Liu Y, Liu JH. 2014. Complete sequence of a F2:A-:B- plasmid pHN3A11 carrying rmtB and qepA, and its dissemination in China. Vet Microbiol 174:267–271. doi: 10.1016/j.vetmic.2014.08.023. [DOI] [PubMed] [Google Scholar]
- 11.Pan YS, Zong ZY, Yuan L, Du XD, Huang H, Zhong XH, Hu GZ. 2016. Complete sequence of pEC012, a multidrug-resistant IncI1 ST71 plasmid carrying blaCTX-M-65, rmtB, fosA3, floR, and oqxAB in an avian Escherichia coli ST117 strain. Front Microbiol 7:1117. doi: 10.3389/fmicb.2016.01117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fu T, Du XD, Cheng PP, Li XR, Zhao XF, Pan YS. 2016. Characterization of an rmtB-carrying IncI1 ST136 plasmid in avian Escherichia coli isolates from chickens. J Med Microbiol 65:387–391. doi: 10.1099/jmm.0.000240. [DOI] [PubMed] [Google Scholar]
- 13.Nicolas E, Lambin M, Dandoy D, Galloy C, Nguyen N, Oger CA, Hallet B. 2014. The Tn3-family of replicative transposons. Microbiol Spectr 3:MDNA3-0060. doi: 10.1128/microbiolspec.MDNA3-0060-2014. [DOI] [PubMed] [Google Scholar]
- 14.Naas T, Cuzon G, Villegas MV, Lartigue MF, Quinn JP, Nordmann P. 2008. Genetic structures at the origin of acquisition of the beta-lactamase blaKPC gene. Antimicrob Agents Chemother 52:1257–1263. doi: 10.1128/AAC.01451-07. [DOI] [PMC free article] [PubMed] [Google Scholar]