ABSTRACT
Escherichia coli sequence type 131 (ST131) is a pandemic clonal lineage that is responsible for the global increase in fluoroquinolone resistance and extended-spectrum-β-lactamase (ESBL) producers. The members of ST131 clade C, especially subclades C2 and C1-M27, are associated with ESBLs. We developed a multiplex conventional PCR assay with the ability to detect all ST131 clades (A, B, and C), as well as C subclades (C1-M27, C1-nM27 [C1-non-M27], and C2). To validate the assay, we used 80 ST131 global isolates that had been fully sequenced. We then used the assay to define the prevalence of each clade in two Japanese collections consisting of 460 ESBL-producing E. coli ST131 (2001-12) and 329 E. coli isolates from extraintestinal sites (ExPEC) (2014). The assay correctly identified the different clades in all 80 global isolates: clades A (n = 12), B (n = 12), and C, including subclades C1-M27 (n = 16), C1-nM27 (n = 20), C2 (n = 17), and other C (n = 3). The assay also detected all 565 ST131 isolates in both collections without any false positives. Isolates from clades A (n = 54), B (n = 23), and C (n = 483) corresponded to the O serotypes and the fimH types of O16-H41, O25b-H22, and O25b-H30, respectively. Of the 483 clade C isolates, C1-M27 was the most common subclade (36%), followed by C1-nM27 (32%) and C2 (15%). The C1-M27 subclade with blaCTX-M-27 became especially prominent after 2009. Our novel multiplex PCR assay revealed the predominance of the C1-M27 subclade in recent Japanese ESBL-producing E. coli isolates and is a promising tool for epidemiological studies of ST131.
KEYWORDS: Escherichia coli, assay development, beta-lactamases, clonality, whole-genome sequencing
INTRODUCTION
The global increase in resistance to third-generation cephalosporins and fluoroquinolones among extraintestinal pathogenic Escherichia coli (ExPEC) isolates is a public health emergency due to the importance of these drugs in the treatment of serious infections (1). The increase in antimicrobial resistance among ExPEC isolates is mainly due to the recent expansion of a pandemic clonal group known as E. coli ST131, which is usually fluoroquinolone resistant and often associated with extended-spectrum β-lactamases (ESBLs) responsible for third-generation cephalosporin resistance (2, 3).
Recent studies using whole-genome sequencing (WGS) analysis revealed that ST131 consists of different lineages or clades: A/H41, B/H22, and C/H30 (4–6). The H numbers indicate the fimH allele, which mostly corresponds to each clade (4–6). Since the 2000s, clade C has become the most dominant lineage among ST131 isolates (currently, up to 80% of global ST131 isolates belong to clade C) (7). Clade C mainly consists of the subclades C1 and C2 that are fluoroquinolone resistant with characteristic quinolone resistance-determining region (QRDR) mutations (4–6). Interestingly, C2 (also known as H30Rx) is associated with blaCTX-M-15, while C1 is often negative for ESBLs. We recently described a novel global C1 subclade named C1-M27, which is associated with blaCTX-M-27 and is present among Japanese ESBL-producing ST131 isolates (8). The other clade C subclade, C0, is considered an ancestor of subclades C1 and C2, and the clade B subclade B0 is considered an ancestor of subclade C0 (6). These C0 and B0 subclades were regarded as belonging to an “intermediate” clade because they had intermediate characteristics between those of clades B and C (6).
The detection of ST131 and its clades is important for epidemiological studies. WGS- or sequence-based strain typing using multilocus sequence typing is considered the gold standard for identifying ST131; however, this approach is expensive and time-consuming. Simple conventional PCR assays to detect ST131, O16-H41, H30, or H30Rx (9–13) are helpful and are often used because they are rapid and cheap, but they cannot detect other clades or subclades. Reliance on the fimH alleles for classification of ST131 clades has a risk due to the presence of alternative fimH alleles (6, 14). Therefore, a more convenient and reliable multiplex method with the ability to detect all of the major ST131 clades, including the C subclades, is required for the optimal detection of ST131 clades for global surveillance studies. Here, we designed a novel multiplex PCR assay to detect ST131 clades in a single reaction. We used this method to identify ST131 clades in two different Japanese E. coli collections.
RESULTS AND DISCUSSION
Definition of clades and subclades of the 390 ST131 genomes.
A total of 390 ST131 genomes were used for the identification of the genomic regions and single nucleotide polymorphisms (SNPs) specific to each clade and subclade. A recombination-free maximum-likelihood phylogeny made from 6,667 core SNP sites represented three major clades, A, B, and C, including four subclades within C (C0, C1, C1-M27, and C2) in agreement with previous studies (6, 8) (see Fig. S1 in the supplemental material). The 390 genomes included clades A (n = 46), B (n = 77), and C (n = 267) and subclades C0 (n = 7), C1-nM27 (n = 85), C1-M27 (n = 29), and C2 (n = 146) (see Data Set S1 in the supplemental material).
The C1-M27 subclade-unique M27PP1 region was found exclusively in a cluster of 29 isolates within the C1 clade. This cluster included all 19 C1-M27 subclade isolates from the original C1-M27 study (8), which had included isolates with blaCTX-M-27 from the Petty et al. study (5), eight isolates from the Stoesser et al. study (14), one isolate sequenced in this study, and one isolate from the Price et al. study (4). Based on the phylogeny and the M27PP1 result, these 10 isolates not derived from the original C1-M27 study (8) were defined to belong to the C1-M27 subclade. They are from Laos (n = 4; 2007 to 2009), Thailand (n = 2; 2009 to 2011), Cambodia (n = 1; 2010), Japan (n = 1; 2014), Germany (n = 1; 2010), and the United States (n = 1; 2010). The original C1-M27 study focused on isolates with ESBLs and the C1-M27 subclade that uniformly had blaCTX-M-27 (8). Among these ten new isolates, six had blaCTX-M-27, one had blaCTX-M-24, and three lacked any ESBL genes. The discovery of C1-M27 in European (Germany) and Asian countries (Laos and Cambodia) further supports its global distribution.
Identification of genomic regions and SNPs specific to each clade and subclade.
The pangenome analyses found four clade A-specific regions (see Table S1 in the supplemental material), but no other regions specific to clade or subclade B, C, C1, C1-M27, or C2 were found. We designed a primer that targeted region 4 because this region was absent in non-ST131 E. coli genomes (Table 1; see also Table S1 in the supplemental material).
TABLE 1.
Primers used for the detection of E. coli ST131 clades
| Assay and primer | Nucleotide sequence (5′-3′) | Primer concn (μM) | Target | PCR product size (bp) | Reference or source |
|---|---|---|---|---|---|
| ST131 clade assay | |||||
| CladeAspe4-YF5 | TGACGGGACGTGAGCAAATTA | 0.15 | Clade A specific (region 4) | 707 | This study (Table S1) |
| CladeAspe4-YR5 | AGTCAGACCTAGCCACCCTT | 0.15 | |||
| ST131_R19-YF1 | AGCAACGATATTTGCCCATT | 0.15 | ST131 specific (region 19) | 580 | 11; this study |
| ST131_R19-YR1 | GGCGATAACAGTACGCCATT | 0.15 | |||
| prfC-1615spe0-YF1 | CAACGTTGAAGCAGTGTATGAG | 0.08 | prfC SNPs specific for the clade B | 442 | This study (Table S2) |
| prfC-d2034-YR1 | TGACAATCGACGGCTTTAGA | 0.08 | |||
| C1-578spe-YF1 | GGCCCCACAAATTGCTT | 0.1 | Clade C1 | 337 | This study (Table S2) |
| C1-898-YR1 | CGCACCTCCGATACCAAA | 0.1 | |||
| M27PP1C-YF1 | TGAATCAAAGGTCCGAGCTG | 0.08 | M27PP1 region specific for the C1-M27 subclade | 232 | 8; LC209430a |
| M27PP1C-YR1 | TATGGCTGGCAGATGCTTTA | 0.08 | |||
| nrdI-534spe2-YF1 | ACGGATTCAGGTAGACGATT | 0.25 | nrdI SNP specific for the C2 clade | 164 | This study (Table S2) |
| nrdI-678R | CCTCACCAAAGTTGCGATTAC | 0.25 | |||
| C-SNP1-700spe-YF1 | CGCTGGCCAGTTATCTGAAAT | 0.2 | mgtA SNPs specific for the C clade | 103 | This study (Table S2) |
| C-SNP1-762spe-YR2 | CCTTTCACCAACTGGGTTACT | 0.2 | |||
| C1-M27 subclade assay | |||||
| ST131_R19-YF1 | AGCAACGATATTTGCCCATT | 0.15 | ST131 specific (region 19) | 580 | 11; this study |
| ST131_R19-YR1 | GGCGATAACAGTACGCCATT | 0.15 | |||
| M27aer-spe-YF1 | GCCGATGGGCTTTCCT | 0.15 | aer SNP specific for the C1-M27 subclade | 140 | This study (Table S2) |
| M27aer-YR2 | GTCACCGCGTCTTCCAGT | 0.15 |
GenBank/ENA/DDBJ accession number.
Analysis of SNPs specific for clades and subclades B, C, C1, C1-M27, and C2 found 4, 16, 1, 5, and 1 SNP sites, respectively (see Table S2 in the supplemental material). We used some of these SNPs to design primers for the ST131 clade and the C1-M27 subclade assays (Table 1; see also Table S2 in the supplemental material). The only C2-specific SNP found in this study was situated in nrdI, although the PCR assay that has often been used for the detection of the C2 clade utilized ybbW SNP (9). One C2 genome sequenced in this study (BRG145; marked with an arrow in Fig. S1 in the supplemental material) lacked the ybbW SNP.
Validation of the ST131 clade and C1-M27 subclade assays.
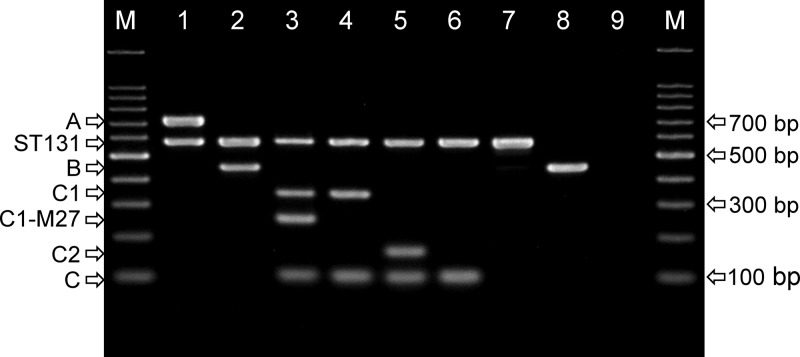
We validated two PCR assays using the WGS collection that consisted of 80 isolates. Both assays produced the expected amplicons (Fig. 1 and Table 2; see also Fig. S2 in the supplemental material) for all isolates. Thus, the ST131 assay correctly identified clades and subclades A, B, C1-M27, C1-nM27, and C2. The three clade C0 isolates were classified to clade C other than subclades C1 or C2, as expected. The C1-M27 assay correctly identified the C1-M27 subclade.
FIG 1.
Agarose gel electrophoresis of the ST131 clade PCR amplicons. Lanes: M, 100-bp DNA ladder; 1, strain SNEC15, clade A; 2, strain KFEC6, clade B; 3, strain KFEC8, subclade C1-M27; 4, strain SNEC5, subclade C1-nM27 (C1-non-M27); 5, strain ONEC14, clade C2; 6, strain KSEC7, subclade C0; 7, strain BRG210, subclade I1; 8, strain BRG28, non-ST131; 9, no-template control. All strains underwent WGS except for BRG28.
TABLE 2.
Identification of ST131 clades and subclades using the ST131 clade assay
| ST131 clade/subclade | Amplicons of the ST131 clade assay |
||||||
|---|---|---|---|---|---|---|---|
| A specific | ST131 specific | B specific | C1 specific | C1-M27 specific | C2 specific | C specific | |
| A | + | + | – | – | – | – | – |
| B | – | + | + | – | – | – | – |
| C | |||||||
| C1 | |||||||
| C1-M27 | – | + | – | + | + | – | + |
| C1-nM27 (non-C1-M27) | – | + | – | + | – | – | + |
| C2 | – | + | – | – | – | + | + |
| C other than C1 or C2a | – | + | – | – | – | – | + |
| Unclassifiedb | – | + | – | – | – | – | – |
| Non-ST131 | –/+ | – | –/+ | –/+ | –/+ | –/+ | –/+ |
Isolates found in this study belonged to subclade C0 or C3.
Isolates found in this study belonged to subclade I1, belonging to the intermediate clade between clades B and C.
Application of the ST131 clade assay for the ESBL and ExPEC collections.
Table 3 shows that the ST131 clade assay correctly detected the ST131 status of the ESBL and ExPEC collection isolates (n = 460 and 105, respectively). In agreement with previous ST131 studies (5, 14), the O serotypes and fimH types of O16-H41, O25b-H22, and O25b (or O nontypeable) H30 perfectly corresponded to clades A (n = 54), B (n = 23), and C (n = 483), respectively. The other five ST131 isolates that contained O25b-H54 were unclassified by the ST131 clade assay and further investigated (detailed below). Among the clade C isolates, the H30Rx status defined by the ybbW SNP-based PCR assay corresponded to subclade C2, except for 1 subclade C2 isolate in the ExPEC collection (BRG145, as mentioned above). These 86 C2 isolates were associated with blaCTX-M-15 (94.2%; whose presence was defined in previous studies [15, 16]), confirming their relationship (4). The 202 C1-M27 and 180 C1-nM27 subclade isolates were associated with blaCTX-M-27 (98.0%) and blaCTX-M-14 (61.1%), as expected from our previous studies (8, 15, 16). The other 15 clade C isolates other than C1 or C2 were considered to belong to subclade C0 according to the current understanding of the clade C lineages (4–6). To confirm this expectation, we further investigated these isolates (detailed below).
TABLE 3.
Results of the ST131 clade PCR assay
| Strain collection and characteristicsa | Total | ST131 |
Non-ST131 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| A | B | C |
Unclassified | ||||||
| C1 |
C2 | C other than C1, C2 | |||||||
| C1-M27 | C1-nM27 | ||||||||
| ESBL (n = 460) | |||||||||
| ST131 | 460 | 45 | 13 | 178 | 140 | 66 | 15 | 3 | |
| fimH41 group,b O16 | 45 | 45 | |||||||
| fimH22 group,c O25b | 13 | 13 | |||||||
| fimH30, O25b/ONT,d H30Rx negative | 333 | 178d | 140d | 15 | |||||
| fimH30, O25b, H30Rx | 66 | 66 | |||||||
| fimH54, O25b | 3 | 3 | |||||||
| blaCTX-M-27 | 179 | 2 | 176 | 1 | |||||
| blaCTX-M-14 | 157 | 31 | 4 | 1 | 104e | 3f | 11 | 3 | |
| blaCTX-M-15 | 72 | 3 | 5 | 64f | |||||
| Other ESBLs | 56 | 9 | 9 | 1 | 33 | 4 | |||
| ExPEC (n = 329) | |||||||||
| Non-ST131 | 224 | 224 | |||||||
| ST131 | 105 | 9 | 10 | 24 | 40 | 20 | 2 | ||
| fimH41 group, O16/O12g | 9 | 9 | |||||||
| fimH22 group,h O25b | 10 | 10 | |||||||
| fimH30 group,i O25b, H30Rx negative | 65 | 24 | 40i | 1j | |||||
| fimH30, O25b/ONT,k H30Rx | 19 | 19 | |||||||
| fimH54, O25b | 2 | 2 | |||||||
| blaCTX-M-27 | 25 | 22 | 1 | 2 | |||||
| blaCTX-M-14 | 17 | 2 | 1 | 6 | 1f | 7 | |||
| blaCTX-M-15 | 20 | 1 | 17f | 2 | |||||
| Other ESBLs | 10 | 3 | 7 | ||||||
| Non-ESBL | 258 | 7 | 9 | 2 | 29 | 3 | 2 | 206 | |
H30Rx status was defined by PCR detection of ybbW SNP.
One isolate had fimH89 (1 SNP to fimH41) and was included in the WGS collection (clade A).
Two isolates had fimH376 (1 SNP to fimH22) and were included in the WGS collection (clade B).
Three C1-M27 and three C1-nM27 isolates were O nontypeable (ONT).
Two isolates had both blaCTX-M-14 and blaCTX-M-3. One isolate had both blaCTX-M-14 and blaCTX-M-2.
One isolate had both blaCTX-M-14 and blaCTX-M-15.
Two isolates had fimH488 (1 SNP to fimH41), one of which was included in the WGS collection (clade A). One isolate with fimH41 was O12.
Two isolates had fimH338 (1 SNP to fimH22), one of which was included in the WGS collection (clade B).
One isolate had fimH497 (2 SNPs to fimH30) and was included in the WGS collection (subclade C1-nM27).
This isolate (BRG145) included in the WGS collection belonged to the C2 subclade and lacked H30Rx-specific ybbW SNP.
One isolate was ONT.
C1-M27 subclade detection by the ST131 clade and the C1-M27 subclade assays.
The results from the ST131 clade and the C1-M27 subclade assays for the detection of the C1-M27 subclade in the ESBL and ExPEC collections were completely in agreement, which supports the presence of the C1-M27 subclade in the 188 isolates (of 202 isolates found) that did not undergo WGS. Thus, we concluded that a single ST131 clade assay is sufficient for the detection of the C1-M27 subclade.
Blinded assay validation.
A blinded validation study in the Shiga laboratory using the WGS and ExPEC collections yielded the same results except for one isolate in the ExPEC collections. The isolates did not produce any bands and were initially regarded as non-ST131. A repetition of the experiment produced the expected clade B amplicons, so we suspected that pipetting errors of the DNA template were responsible for the anomalous results.
Prevalence of the ST131 clades and subclade in Japanese ESBL-producing E. coli.
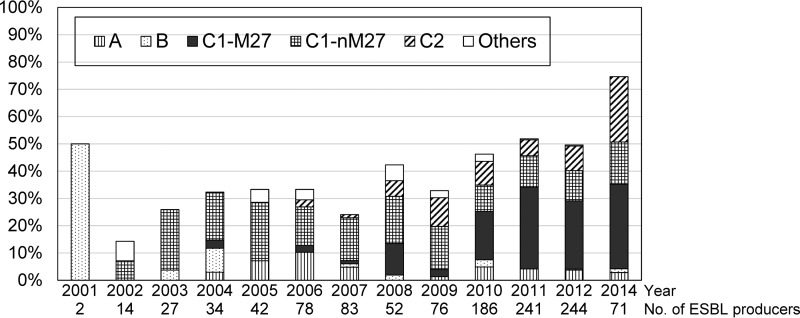
Figure 2 shows the rates of ST131 clades and subclades between 2001 and 2014 calculated from the results of the ST131 clade assay in the ESBL and ExPEC collections with ESBL genes. The C1-M27, C1-nM27, and C2 subclades contributed to the recent expansion of ESBL-producing E. coli in Japan. The C1-M27 subclade was the most prevalent ST131 subclade after 2009. These results clarified that the main contributor of the recent ESBL-producing E. coli in Japan is the C1-M27 subclade, which was regarded as the CTX-M-27-producing H30R group in our previous study (15).
FIG 2.
Yearly rates of ST131 clades and subclades in Japanese ESBL-producing E. coli. Rates of ST131 and subclades C1-M27 and C2 increased yearly (P < 0.001 each). In 2014, the rates of clades A and B and subclades C1-M27, C1-non-M27, and C2 were 3, 1, 31, 15, and 24%, respectively.
New subclades I1 and C3 were found by genome analysis of isolates unclassified by the ST131 clade assay.
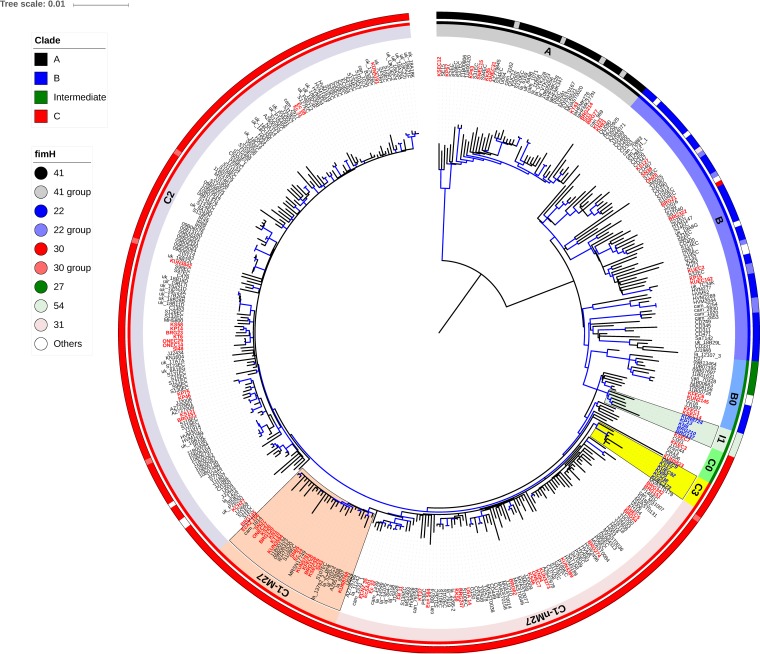
All of the 5 O25b-H54 isolates unclassified by the ST131 clade assay underwent WGS (see Data Set S2 in the supplemental material). The phylogenetic tree analysis indicated these isolates belonged to a new lineage between B0 and C0 in the intermediate clade (Fig. 3) and were presumptively placed in a new subclade designated subclade I1 (Fig. 3). The genome analysis indicated that the subclade I1 isolates lacked all of the clade B or C characteristic SNPs, including the sites used for the ST131 clade assay (n = 4 and n = 16, respectively; see Table S2 in the supplemental material). The subclade I1 isolates contained the prophages Phi2 and Phi4 and the parC-1a genotype characteristic of subclades B0 and C0 (6). Among the genomic markers for clade C, including subclade C0 (6), the prophage Phi1 and the ST131 genomic island GI-pheV were present, whereas GI-leuX and ISEc55 insertions in fimB were absent. These findings suggest that subclade I1 shared an ancestor with subclade C0 and underwent subsequent recombination events in the core genome, including fimH. The five subclade I1 isolates were susceptible to ciprofloxacin, had O25b-H54 had emerged in 2010, and were collected from five Japanese hospitals. Three isolates had blaCTX-M-14.
FIG 3.
Recombination-free core SNP-based phylogeny of 401 ST131 genomes. This maximum-likelihood phylogenetic tree is rooted by using the clade A isolates. A total of 6,881 core SNP sites were used after excluding the 18,693 core SNP sites that were located within the 1,955,761-bp recombination region. Branches that had >90% bootstrap support from 100 replicates are highlighted in blue. The bootstrap values for the root of subclades I1 and C3 were 100 and 99%, respectively. Genomes of the WGS collection isolates are marked in red. Eleven genomes (marked in blue) were sequenced to investigate their phylogeny in addition to the 390 genomes (see Fig. S1 in the supplemental material).
Of fifteen clade C isolates other than C1 or C2 indicated by the ST131 clade assay, three (KSEC7, KKEC3, and KUN5823) had been sequenced and defined as subclade C0 (see Fig. S1 in the supplemental material) without the C1/C2-characteristic QRDR mutations of gyrA-1AB and parC-1aAB (6). Six of the remaining twelve isolates underwent WGS (see Data Set S2 in the supplemental material). The genome analysis confirmed that these isolates lacked the subclade C1- and C2-specific SNPs (see Table S2 in the supplemental material). The phylogenetic tree analysis indicated that these isolates belonged to a new lineage in clade C and they were presumptively named subclade C3 (Fig. 3). Subclade C3 diverged from subclade C0 and had contained gyrA-1AB and parC-1aAB genotypes, similar to subclades C1 and C2. Among the 401 ST131 genomes in Fig. 3, the average number of SNPs between subclades C3 and C1 (n = 54) or C2 (n = 55) were similar to that between subclades C1 and C2 (n = 50) and was larger than that among C1 (n = 40), C2 (n = 38), and C3 (n = 26), which supports the hypothesis that subclade C3 is independent of C1 or C2. The other six isolates that did not undergo WGS also contained the gyrA-1AB and parC-1aAB genotypes, suggesting subclade C3 status. The subclade C3 isolates were not susceptible to ciprofloxacin, had O25b-H30 and ESBLs (blaCTX-M-14, n = 10; blaCTX-M-2, n = 2), had emerged in 2005, and were found in 2.6% of the ESBL-producing E. coli collection from five Japanese hospitals. Further analysis of both subclades I1 and C3 is needed to define their characteristics and their importance in evolutional history and global epidemiology.
The study limitations include the lack of extensive validation studies in other laboratories. The strengths include the use of large numbers of well-characterized ESBL-producing isolates and ExPEC isolates without any selection bias, which encompass the study population with which the assay will likely be used.
Conclusions.
To our knowledge, this is the first study to develop a PCR assay (the ST131 clade assay) to detect ST131 clades and subclades in a single reaction. This assay will elucidate the global molecular epidemiology of ST131 and provide potential knowledge to establish infection control policies for ESBL-producing E. coli. Application of this assay to Japanese isolates and subsequent genomic analysis identified two novel ST131 lineages that may be important to understand the evolutional history of ST131. We also showed that the C1-M27 subclade with blaCTX-M-27 is responsible for the expansion of ESBL-producing E. coli in Japan, as well as the underlying global importance of this clade. We urgently need well-designed epidemiological and molecular studies to understand the dynamics of the transmission, risk factors, and reservoirs for C1-M27.
MATERIALS AND METHODS
Bacterial isolates.
We included three collections of clinical E. coli isolates for the PCR assays: the WGS collection for the validation of the method, as well as the ESBL and ExPEC collections for application. The WGS collection included 80 isolates, whose clades were defined using WGS-based analysis and phylogeny: A (n = 12), B (n = 12), C0 (n = 3), C1-M27 (n = 16), C1-nM27 (C1-non-M27; n = 20), and C2 (n = 17). These clades were derived from our previous study (8) (n = 53) and the present study (n = 27). Part of the WGS collection of isolates was selected from the ESBL (n = 55) and ExPEC collections (n = 15).
We assessed the prevalence of the ST131 clades and subclades using the ESBL and ExPEC collections of E. coli clinical isolates. The ESBL collections comprised 460 ESBL-producing ST131 isolates that were collected in 10 acute-care hospitals in the Kyoto and Shiga prefectures in Japan from 2001 to 2012 (15). The ExPEC collection comprised 329 isolates that were collected in ten acute-care hospitals in five prefectures in Japan during December 2014 (all clinical isolates during the collection period) and included seven major ST complexes containing ST131 (32%), ST95 (13%), ST73 (8%), ST357/1876 (5%), ST69 (4%), ST38 (3%), and ST1193 (2%) (16). These isolates from the two collections were previously defined for ciprofloxacin susceptibility, the presence of ESBL genes, ST131 status using PCR detection of specific SNPs in mdh and gyrB (12) or fumC-fimH typing (17), O typing (rfb alleles) (18, 19), fimH alleles using sequencing or H30-specific PCR (10), and H30Rx status using PCR detection of ybbW SNP (9). The clade C isolates that were negative for the C1 or C2 subclades according to the developed ST131 clade assay underwent Sanger sequencing to define the gyrA and parC genotypes (3).
Genome analysis.
We performed WGS using Illumina MiSeq as previously described (8) for the 27 isolates from the WGS collection and 11 isolates from the ESBL and ExPEC collections whose clades or subclades had not been classified by the ST131 clade assay we developed. In silico multilocus sequence typing, fimH typing, gyrA and parC typing, O serotyping, and C2-specific ybbW SNP typing were performed using the assembled draft genomes as previously described (8).
To design the PCR assay, we used data on 390 ST131 genomes from the three major ST131 studies reported to date (Petty et al. [5], n = 95; Stoesser et al. [14], n = 119; and Ben Zakour et al. [6]—originally by Price et al. [4], n = 90), 6 ST131 draft or complete genomes, and our WGS data set (see Data Set S1 in the supplemental material). The reads were assembled using SPAdes v3.10.1 (20). First, we created a recombination-free, core SNP-based phylogenetic tree to define the clades and subclades. We identified the SNPs by mapping the reads or aligning the genomes against a subclade C2 reference genome of EC958 (21) as previously described (8). Recombination sites identified according to Gubbins (22) were excluded and recombination-free core SNPs, sites that are present in all genomes, were included to create a maximum-likelihood tree using RAxML with the GTR GAMMA substitution model (23). The tree was visualized using iTOL v3 (24). We defined the C1-M27 subclade based on the phylogenetic tree and the presence of the C1-M27 subclade-specific prophage-like region (M27PP1) based on the original study (8). Second, to identify a genomic region specific to each clade or subclade, we performed a pangenome analysis using Panseq (25). The unique core SNPs of each clade or subclade were determined using an in-house Perl script. The genomes of 11 isolates mentioned above from unclassified clades or subclades were analyzed to determine their phylogeny, by following the same steps as above, for specific SNPs and the ST131 genomic islands (6).
ST131 clade PCR assay.
Bacterial DNA was isolated using a QIAamp DNA minikit (Qiagen, Hilden, Germany). The assay utilized seven primer sets designed in the present study (Table 1). Table 2 shows the expected amplicons for each clade. Amplification was performed with 1× PCR buffer, 1.5 mM MgCl2, 0.2 mM deoxynucleoside triphosphates, each primer, 0.5 U of TaKaRa Ex Taq DNA polymerase (TaKaRa Bio, Inc., Otsu, Japan), and 1 μl of purified DNA in a total volume of 20 μl using a TaKaRa PCR Dice Touch thermal cycler (TaKaRa Bio). The cycling protocol was as follows: 30 cycles of 98°C for 10 s, 57°C for 20 s, and 72°C for 40 s. The PCR products were loaded on a 2.5% agarose gel with GelRed (Biotium, Hayward, CA) and photographed under UV light.
C1-M27 subclade PCR assay.
We developed another assay to detect only the C1-M27 subclade by using a primer set designed for the C1-M27 subclade-specific SNP in its core genome, which is different from the accessory genes target (M27PP1) used in the ST131 clade assay (Table 1), in order to support the presence of the C1-M27 subclade in the ESBL and ExPEC collections.
Blinded assay validation.
All of the PCR experiments were performed in the Kyoto laboratory. The ST131 clade assay was validated in another laboratory in Shiga using the WGS and ExPEC collections in a blinded manner.
Statistical analysis.
The yearly prevalence of isolates was analyzed using a chi-square test for linear trends. A P value of <0.05 was considered statistically significant. We conducted our statistical analysis using R version 3.3.2 (R Foundation for Statistical Computing [www.r-project.org]).
Accession number(s).
The sequences determined in this study have been deposited in the GenBank/ENA/DDBJ Sequence Read Archive database (DRA005691 and DRA005809).
Supplementary Material
ACKNOWLEDGMENTS
We thank the Kyoto–Shiga Clinical Microbiology Study Group and the 89th JAID BRG (89th Annual Meeting of the Japanese Association for Infectious Diseases Drug-Resistant Bacteria Research Group) for providing their E. coli collections.
This study was supported by the John Mung Program from Kyoto University, Kyoto, Japan (Y.M.), and by a research grant from the Calgary Laboratory Services (10006465; J.D.D.P.). The funders had no role in the study design, data collection and interpretation, or the decision to submit the work for publication. J.D.D.P. had previously received research funds from Merck and AstraZeneca.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.00179-17.
REFERENCES
- 1.Pitout JD, Laupland KB. 2008. Extended-spectrum beta-lactamase-producing Enterobacteriaceae: an emerging public-health concern. Lancet Infect Dis 8:159–166. doi: 10.1016/S1473-3099(08)70041-0. [DOI] [PubMed] [Google Scholar]
- 2.Nicolas-Chanoine MH, Bertrand X, Madec JY. 2014. Escherichia coli ST131, an intriguing clonal group. Clin Microbiol Rev 27:543–574. doi: 10.1128/CMR.00125-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matsumura Y, Yamamoto M, Nagao M, Ito Y, Takakura S, Ichiyama S. 2013. Association of fluoroquinolone resistance, virulence genes, and IncF plasmids with extended-spectrum-β-lactamase-producing Escherichia coli sequence type 131 (ST131) and ST405 clonal groups. Antimicrob Agents Chemother 57:4736–4742. doi: 10.1128/AAC.00641-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Price LB, Johnson JR, Aziz M, Clabots C, Johnston B, Tchesnokova V, Nordstrom L, Billig M, Chattopadhyay S, Stegger M, Andersen PS, Pearson T, Riddell K, Rogers P, Scholes D, Kahl B, Keim P, Sokurenko EV. 2013. The epidemic of extended-spectrum-β-lactamase-producing Escherichia coli ST131 is driven by a single highly pathogenic subclone, H30-Rx. mBio 4:e00377-13. doi: 10.1128/mBio.00388-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Petty NK, Ben Zakour NL, Stanton-Cook M, Skippington E, Totsika M, Forde BM, Phan MD, Gomes Moriel D, Peters KM, Davies M, Rogers BA, Dougan G, Rodriguez-Baño J, Pascual A, Pitout JD, Upton M, Paterson DL, Walsh TR, Schembri MA, Beatson SA. 2014. Global dissemination of a multidrug resistant Escherichia coli clone. Proc Natl Acad Sci U S A 111:5694–5699. doi: 10.1073/pnas.1322678111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ben Zakour NL, Alsheikh-Hussain AS, Ashcroft MM, Khanh Nhu NT, Roberts LW, Stanton-Cook M, Schembri MA, Beatson SA. 2016. Sequential acquisition of virulence and fluoroquinolone resistance has shaped the evolution of Escherichia coli ST131. mBio 7:e00347-16. doi: 10.1128/mBio.00958-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mathers AJ, Peirano G, Pitout JD. 2015. The role of epidemic resistance plasmids and international high-risk clones in the spread of multidrug-resistant Enterobacteriaceae. Clin Microbiol Rev 28:565–591. doi: 10.1128/CMR.00116-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matsumura Y, Pitout JD, Gomi R, Matsuda T, Noguchi T, Yamamoto M, Peirano G, DeVinney R, Bradford PA, Motyl MR, Tanaka M, Nagao M, Takakura S, Ichiyama S. 2016. Global Escherichia coli sequence type 131 clade with blaCTX-M-27 gene. Emerg Infect Dis 22:1900–1907. doi: 10.3201/eid2211.160519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Banerjee R, Robicsek A, Kuskowski MA, Porter S, Johnston BD, Sokurenko E, Tchesnokova V, Price LB, Johnson JR. 2013. Molecular epidemiology of Escherichia coli sequence type 131 and its H30 and H30-Rx subclones among extended-spectrum-β-lactamase-positive and -negative E. coli clinical isolates from the Chicago region, 2007 to 2010. Antimicrob Agents Chemother 57:6385–6388. doi: 10.1128/AAC.01604-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Colpan A, Johnston B, Porter S, Clabots C, Anway R, Thao L, Kuskowski MA, Tchesnokova V, Sokurenko EV, Johnson JR. 2013. Escherichia coli sequence type 131 (ST131) subclone H30 as an emergent multidrug-resistant pathogen among US Veterans. Clin Infect Dis 57:1256–1265. doi: 10.1093/cid/cit503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doumith M, Day M, Ciesielczuk H, Hope R, Underwood A, Reynolds R, Wain J, Livermore DM, Woodford N. 2015. Rapid identification of major Escherichia coli sequence types causing urinary tract and bloodstream infections. J Clin Microbiol 53:160–166. doi: 10.1128/JCM.02562-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson JR, Menard M, Johnston B, Kuskowski MA, Nichol K, Zhanel GG. 2009. Epidemic clonal groups of Escherichia coli as a cause of antimicrobial-resistant urinary tract infections in Canada, 2002 to 2004. Antimicrob Agents Chemother 53:2733–2739. doi: 10.1128/AAC.00297-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson JR, Clermont O, Johnston B, Clabots C, Tchesnokova V, Sokurenko E, Junka AF, Maczynska B, Denamur E. 2014. Rapid and specific detection, molecular epidemiology, and experimental virulence of the O16 subgroup within Escherichia coli sequence type 131. J Clin Microbiol 52:1358–1365. doi: 10.1128/JCM.03502-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stoesser N, Sheppard AE, Pankhurst L, De Maio N, Moore CE, Sebra R, Turner P, Anson LW, Kasarskis A, Batty EM, Kos V, Wilson DJ, Phetsouvanh R, Wyllie D, Sokurenko E, Manges AR, Johnson TJ, Price LB, Peto TE, Johnson JR, Didelot X, Walker AS, Crook DW. 2016. Evolutionary history of the global emergence of the Escherichia coli epidemic clone ST131. mBio 7:e02162-15. doi: 10.1128/mBio.02162-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matsumura Y, Johnson JR, Yamamoto M, Nagao M, Tanaka M, Takakura S, Ichiyama S. 2015. CTX-M-27- and CTX-M-14-producing, ciprofloxacin-resistant Escherichia coli of the H30 subclonal group within ST131 drive a Japanese regional ESBL epidemic. J Antimicrob Chemother 70:1639–1649. doi: 10.1093/jac/dkv017. [DOI] [PubMed] [Google Scholar]
- 16.Matsumura Y, Noguchi T, Tanaka M, Kanahashi T, Yamamoto M, Nagao M, Takakura S, Ichiyama S. 2017. Population structure of Japanese extraintestinal pathogenic Escherichia coli and its relationship with antimicrobial resistance. J Antimicrob Chemother 72:1040–1049. doi: 10.1093/jac/dkw530. [DOI] [PubMed] [Google Scholar]
- 17.Weissman SJ, Johnson JR, Tchesnokova V, Billig M, Dykhuizen D, Riddell K, Rogers P, Qin X, Butler-Wu S, Cookson BT, Fang FC, Scholes D, Chattopadhyay S, Sokurenko E. 2012. High-resolution two-locus clonal typing of extraintestinal pathogenic Escherichia coli. Appl Environ Microbiol 78:1353–1360. doi: 10.1128/AEM.06663-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clermont O, Johnson JR, Menard M, Denamur E. 2007. Determination of Escherichia coli O types by allele-specific polymerase chain reaction: application to the O types involved in human septicemia. Diagn Microbiol Infect Dis 57:129–136. doi: 10.1016/j.diagmicrobio.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 19.Clermont O, Lavollay M, Vimont S, Deschamps C, Forestier C, Branger C, Denamur E, Arlet G. 2008. The CTX-M-15-producing Escherichia coli diffusing clone belongs to a highly virulent B2 phylogenetic subgroup. J Antimicrob Chemother 61:1024–1028. doi: 10.1093/jac/dkn084. [DOI] [PubMed] [Google Scholar]
- 20.Nurk S, Bankevich A, Antipov D, Gurevich AA, Korobeynikov A, Lapidus A, Prjibelski AD, Pyshkin A, Sirotkin A, Sirotkin Y, Stepanauskas R, Clingenpeel SR, Woyke T, McLean JS, Lasken R, Tesler G, Alekseyev MA, Pevzner PA. 2013. Assembling single-cell genomes and mini-metagenomes from chimeric MDA products. J Comput Biol 20:714–737. doi: 10.1089/cmb.2013.0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Forde BM, Ben Zakour NL, Stanton-Cook M, Phan MD, Totsika M, Peters KM, Chan KG, Schembri MA, Upton M, Beatson SA. 2014. The complete genome sequence of Escherichia coli EC958: a high quality reference sequence for the globally disseminated multidrug-resistant E. coli O25b:H4-ST131 clone. PLoS One 9:e104400. doi: 10.1371/journal.pone.0104400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Croucher NJ, Page AJ, Connor TR, Delaney AJ, Keane JA, Bentley SD, Parkhill J, Harris SR. 2015. Rapid phylogenetic analysis of large samples of recombinant bacterial whole-genome sequences using Gubbins. Nucleic Acids Res 43:e15. doi: 10.1093/nar/gku1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Letunic I, Bork P. 2016. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res 44:W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laing C, Villegas A, Taboada EN, Kropinski A, Thomas JE, Gannon VP. 2011. Identification of Salmonella enterica species- and subgroup-specific genomic regions using Panseq 2.0. Infect Genet Evol 11:2151–2161. doi: 10.1016/j.meegid.2011.09.021. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.