ABSTRACT
A novel HIV-1 integrase mutation pattern, L74F V75I, which conferred resistance to first-generation integrase strand transfer inhibitors (INSTIs), was identified in a clinical case with virological failure under a raltegravir-based regimen. Addition of L74F V75I to N155H or G140S Q148H increased resistance levels to the second-generation INSTIs dolutegravir (>385- and 100-fold, respectively) and cabotegravir (153- and 197-fold, respectively). These findings are important for the development of an accurate system for interpretation of INSTI resistance and the rational design of next-generation INSTIs.
KEYWORDS: dolutegravir, drug resistance mechanisms, human immunodeficiency virus, integrase, integrase strand transfer inhibitor
TEXT
Integrase strand transfer inhibitors (INSTIs) constitute the latest class of available antiretroviral agents exhibiting potent antiretroviral effects in vitro and vivo (1–8). The first-generation INSTIs raltegravir (RAL) and elvitegravir (EVG) display broad cross-resistance, whereas second-generation INSTIs of carbamoyl pyridine analogues, dolutegravir (DTG) and cabotegravir (CAB), demonstrate superior activity against first-generation INSTI-resistant HIV-1 variants (4, 9–13). Although a DTG resistance mutation, R263K, has been reported, and its resistance mechanism has been well studied (10, 12, 14, 15), other potential DTG resistance mutations and their mechanisms are not fully understood.
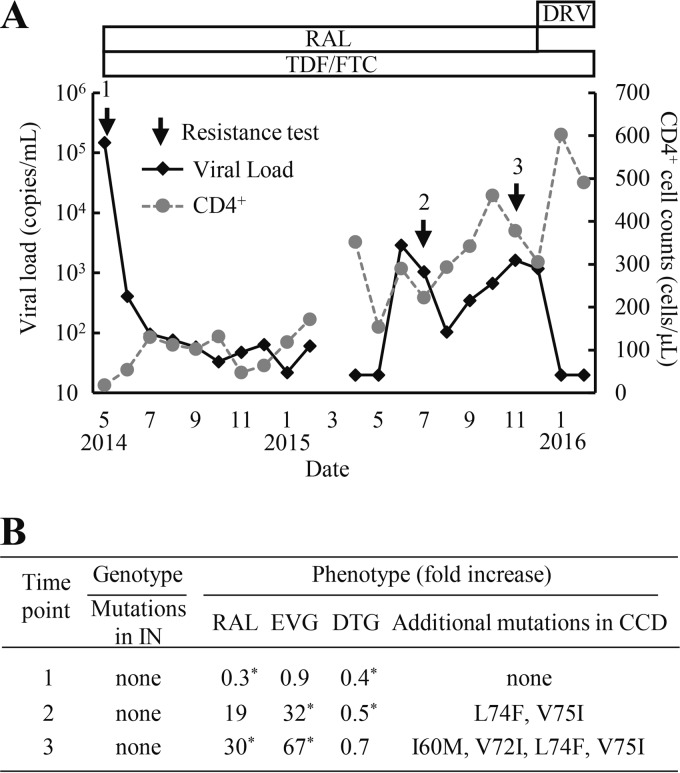
In a clinical case (Fig. 1A), the plasma HIV-1 RNA level rebounded to 2,900 copies/ml 1 year after starting antiretroviral therapy (ART) that included tenofovir disoproxil fumarate (TDF), emtricitabine (FTC), and RAL. However, no known INSTI resistance mutations were identified based on major drug resistance mutation lists (IAS-USA drug resistance mutations list [16] and the HIV Drug Resistance Database at Stanford University [HIVDB]) at time points 2 and 3 (Fig. 1B). Clinical samples were obtained from the fresh plasma of a patient attending the outpatient clinic of the National Hospital Organization Nagoya Medical Center. The Institutional Review Board approved this study (2010-310), and written informed consent was obtained from this patient. To identify novel mutations associated with RAL resistance in the clinical isolates, we constructed infectious HIV-1 clones with cDNA fragments of the integrase (IN)-coding region derived from the clinical isolates and performed phenotypic resistance assays using TZM-bl cells as previously described (8, 17). Briefly, viral RNA was extracted from plasma and subjected to reverse transcription-PCR (RT-PCR) and nested PCR using the Superscript III one-step RT-PCR system (Thermo Fisher Scientific, Waltham, MA) and PrimeSTAR GXL (TaKaRa Bio, Otsu, Japan), respectively. The DNA fragments amplified from the clinical samples were cloned into the XbaI-NdeI region (891 bp) of pSLINwt, which encodes nucleotides 4232 to 5122 of pNL101. Next, the XbaI-NdeI cassettes were inserted into pBNΔIN, which encodes nucleotides 5122 (NdeI) to 5785 (SalI) of pNL101. Finally, the Xba-SalI region (1,544 bp) was inserted back into pNL101. Each HIV-1 proviral molecular clone was transfected into human embryonic kidney 293T cells using FuGENE HD (Promega, WI). Viral infectivity was determined by serially diluting each stock of virus and by applying it to the TZM-bl cell assay (104 cells per well). Luciferase marker gene expression was measured using the Bright-Glo luciferase assay system (Promega, WI) after 48 h. For the INSTI susceptibility assay, RAL, EVG, and DTG were purchased commercially from Selleck Chemicals (Houston, TX). TZM-bl cells (104 cells per well) were infected with diluted virus stock at 100,000 relative light units (RLU) in the presence of increasing concentrations of each INSTI. The 50% effective concentration (EC50) was calculated as the concentration that is required to reduce RLU by 50%. Recombinant virus at time point 1 exhibited susceptibility to RAL, EVG, and DTG, whereas the recombinant viruses at time point 2 had significantly reduced RAL and EVG susceptibilities (19- and 32-fold, respectively). Mutations L74F and V75I first appeared during the periods of virological failure, followed by two additional mutations, I60M and V72I.
FIG 1.
Clinical course and drug resistance profiles of a patient on an RAL-based ART regimen. (A) The treatment history and clinical course. Arrows indicate the time points for drug resistance assays. (B) HIV-1 genotypic and phenotypic resistance assay results. Genotypic results were analyzed according to the major drug resistance mutation lists (HIV Drug Resistance Database at Stanford University [March 2015] and IAS-USA drug resistance mutations list [16]). Resistance levels to INSTIs were calculated as the fold increase in the EC50 of the HIV-1 variants relative to that of WT. The data shown were obtained from at least three independent experiments. Statistical significance was calculated for difference between the WT and recombinant virus derived from a clinical isolate using a Student t test with a statistical cutoff of P < 0.02 (*). Additional mutations in the catalytic core domain (CCD) were observed in recombinant clones of time points 2 and 3 compared to time point 1 (GenBank accession no. LC201871 to LC201873).
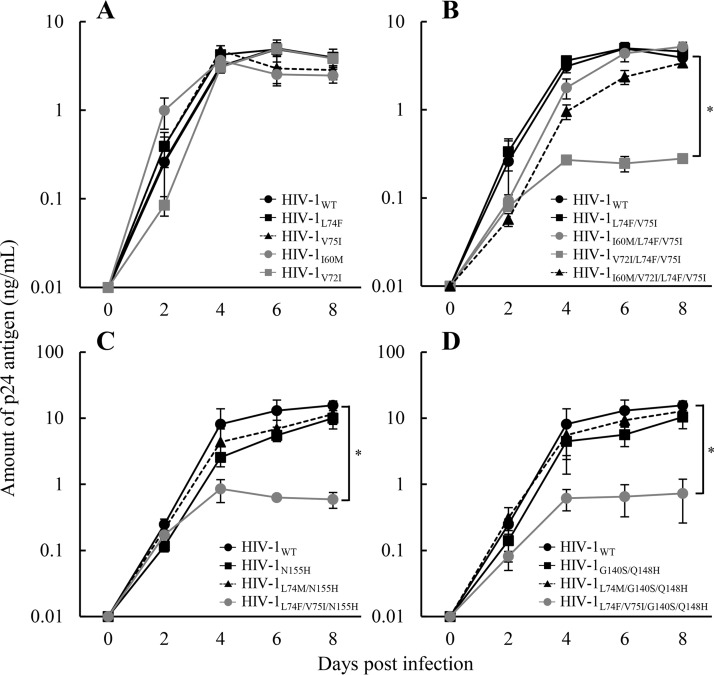
To evaluate the impact of the four novel mutations observed in the clinical isolates on INSTI resistance levels, the IN mutant variants of HIV-1 were prepared by transfection with pNL101-based plasmids (8) and were applied to the INSTI susceptibility assay using TZM-bl cells (17) (Table 1). Single mutation L74F or V75I had no effect on INSTI resistance, while the combination L74F V75I increased both RAL and EVG resistance levels (5.5- and 11-fold, respectively). I60M caused no change in the RAL and EVG resistance of HIV-1L74F V75I, although V72I marginally increased the resistance level. There are minor discrepancies in the RAL and EVG susceptibilities between the clinical isolate (19- and 32-fold, respectively) and laboratory virus (5.5 and 11-fold, respectively). It is possible that strain-specific polymorphisms present in our clinical isolate may also affect drug susceptibility, leading to minor discrepancies with the results from laboratory virus. To investigate the effect of I60M and/or V72I on HIV-1L74F V75I replication, the replication kinetics of the corresponding recombinant viruses were examined. Briefly, MT-2 cells (1 × 106 cells/flask) were infected with each diluted virus (750,000 relative fluorescence units [RFU]) for 4 h. The infected cells were then washed and cultured in a final volume of 2.5 ml (18). Culture supernatants were collected at various times postinfection, and p24 concentrations were measured using AlphaLISA (PerkinElmer, Waltham, MA, USA). The replication capacities of HIV-1 strains carrying I60M, V72I, or L74F V75I were found to be comparable to that of the wild type (WT) (Fig. 2A). The kinetics of HIV-1V72I L74F V75I replication were significantly lower than the HIV-1L74F V75I (P = 0.0041), whereas the additional I60M mutation improved the impaired replication capacity of HIV-1V72I L74F V75I (Fig. 2B).
TABLE 1.
Susceptibility of recombinant HIV-1 strains to integrase strand transfer inhibitors
| HIV-1 strain typea | EC50, nM (fold increase)b |
||
|---|---|---|---|
| RAL | EVG | DTG | |
| WT | 12 ± 1.2 | 2.6 ± 0.5 | 2.5 ± 0.1 |
| Mutant | |||
| I60M | 8.4 ± 0.8 (0.7) | 2.2 ± 0.4 (0.9) | 2.4 ± 0.1 (1.0) |
| V72I | 16 ± 2.9 (1.4) | 2.8 ± 0.2 (1.1) | 2.5 ± 0.3 (1.0) |
| L74F | 18 ± 2.1 (1.5) | 5.5 ± 0.4 (2.3) | 1.1 ± 0.1 (0.4)c |
| V75I | 7.5 ± 0.3 (0.7) | 3.5 ± 0.2 (1.2) | 1.3 ± 0.2 (0.5)c |
| L74F V75I | 66 ± 1.2 (5.5)c,d | 27 ± 3.6 (11)c,d | 0.8 ± 0.1 (0.3)c |
| L74I V75I | 12 ± 2.1 (1.0) | 5.3 ± 0.3 (1.9)c | 1.6 ± 0.1 (0.6)c |
| L74M V75I | 21 ± 3.9 (1.8) | 8.0 ± 0.5 (3.1)c | 1.2 ± 0.1 (0.5)c |
| I60M L74F V75I | 44 ± 8.2 (3.7) | 32 ± 8.8 (12) | 1.2 ± 0.1 (0.5)c |
| V72I L74F V75I | 137 ± 17 (11)c | 45 ± 11 (17) | 1.3 ± 0.2 (0.5)c |
| I60M V72I L74F V75I | 47 ± 1.2 (3.9)c | 38 ± 5.9 (15)c | 0.7 ± 0.1 (0.3)c |
Select L74 mutations in the double and triple mutants have been highlighted for emphasis. In the V75I variants, underlining indicates the L74M or L74I mutation did not have an impact on susceptibility, whereas boldface indicates the L74F mutation increased INSTI resistance.
The data shown are means ± standard deviations from three independent experiments. Values in parentheses are fold increases of the EC50 relative to that in the HIV-1 WT.
Statistical significance was calculated for difference between the WT and mutant using a Student t test with a statistical cutoff of P < 0.02.
Statistical significance was calculated for difference between single (L74F or V75I) and corresponding double mutants using a Student t test with a statistical cutoff of P < 0.02.
FIG 2.
Replication kinetics of HIV-1L74F V75I variants. Multicycle replication kinetics in MT-2 cells were assessed by measuring the p24 levels in culture supernatants. (A) Effect of each single mutation on HIV-1 replication. The data shown compare the replication kinetics of HIV-1WT (solid line with circles), HIV-1L74F (solid line with squares), HIV-1V75I (dotted line with triangles), HIV-1I60M (solid gray line with circles), and HIV-1V72I (solid gray line with squares). (B) Effect of I60M or V72I on HIV-1L74F V75I replication. The data shown compare the replication kinetics of HIV-1WT (solid line with circles), HIV-1L74F V75I (solid line with squares), HIV-1I60M L74F V75I (solid gray line with circles), HIV-1V72I L74F V75I (solid gray line with squares), and HIV-1I60M V72I L74F V75I (dotted line with triangles). (C) Effect of N155H on HIV-1L74F V75I replication. The data shown compare the replication kinetics of HIV-1WT (solid line with circles), HIV-1N155H (solid line with squares), HIV-1L74M N155H (dotted line with triangles), and HIV-1L74F V75I N155H (solid gray line with circles). (D) Effect of G140S Q148H on HIV-1L74F V75I replication. The data shown compare the replication kinetics of HIV-1WT (solid line with circles), HIV-1G140S Q148H (solid line with squares), HIV-1L74M G140S Q148H (dotted line with triangles), and HIV-1L74F V75I G140S Q148H (solid gray line with circles). Error bars represent standard deviations from at least three independent experiments. Statistical significance was calculated for difference between the p24 levels of HIV-1L74F V75I and HIV-1V72I L74F V75I (B) and the WT and mutant (C and D) at day 8 using a Student t test with a statistical cutoff of P < 0.02 (*).
L74M and L74I were frequently observed in the INSTI-experienced individual as a secondary mutation (4.9 or 4.5% [subtype B, n = 957] by HIVDB [https://hivdb.stanford.edu/]), whereas L74F was rare (less than 0.1%). Therefore, to examine whether L74M or L74I has any impact on INSTI susceptibility in the presence of V75I, HIV-1 strains carrying L74M V75I or L74I V75I were tested for susceptibility (Table 1). L74M or L74I did not change susceptibility of the V75I variants, whereas L74F increased INSTI resistance. These results suggest that the genetic pathway to acquire INSTI resistance of the L74F mutant differs from that of mutants carrying the secondary mutation, L74M or -I.
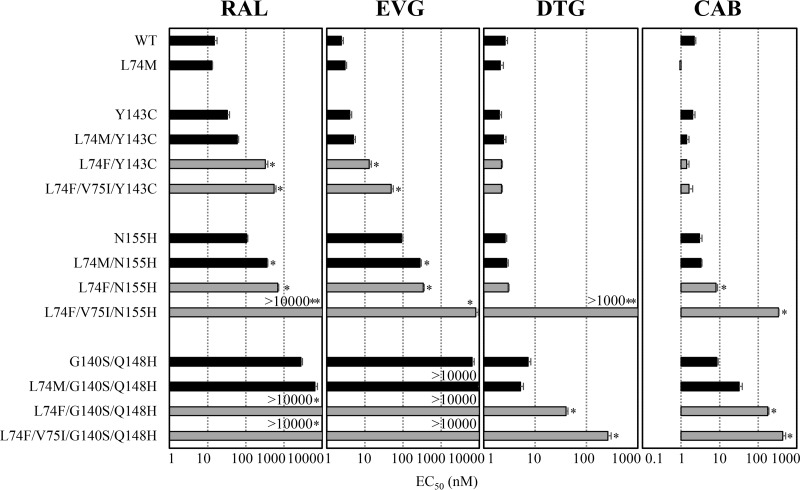
To date, clinical studies demonstrated that three major resistance pathways, Y143C/H/R, Q148H/K/R, and N155H were frequently observed in patients with virological failure under a RAL-based regimen (19–22). The N155H mutation was usually accompanied by the secondary mutation L74M, E92Q, T97A, G136R, or V151I (23–25). Although secondary mutations do not individually confer INSTI resistance, their combinations with N155H increase resistance and restore viral replication capacity (26, 27). To assess whether mutations at L74 alone or with V75I act as secondary mutations, we evaluated resistance level (Fig. 3) and viral replication capacity (Fig. 2C and D) in the presence of major mutations. The results demonstrated that either L74M or L74F enhanced RAL and EVG resistance induced by Y143C or N155H. L74M with the three major mutations showed no effect on DTG resistance level, although L74F with G140S Q148H increased the levels of DTG and CAB resistance (15- and 80-fold, respectively). Notably, L74F V75I exhibited high-level cross-resistance to both DTG and CAB when combined with N155H (>385- and 153-fold, respectively) or G140S Q148H (100- and 197-fold, respectively) mutations. The combination L74F V75I further impaired the replication capacity of HIV-1N155H and HIV-1G140S/Q148H (Fig. 2C and D). These results suggest that L74F and V75I are not simply involved in INSTI resistance as secondary mutations as previously reported. This is supported by our findings that L74F and V75I enhanced the resistance level of major mutations to second-generation INSTIs, while they decreased the viral replication capacity.
FIG 3.
Effect of L74F V75I on susceptibility of INSTI-resistant variants to RAL, EVG, DTG, or CAB. The antiviral activities of RAL, EVG, DTG, or CAB in HIV-1 isolates carrying major INSTI mutations Y143C, N155H, or G140S Q148H with no additional mutations (black bars) or L74M (black bars), L74F (gray bars), or L74F V75I (gray bars) were determined by TZM-bl cell-based assay. Each EC50 is represented as a bar graph. Error bars represent standard deviations from at least three independent experiments. Statistical significance was calculated for difference between strains with a major mutation solely and a major mutation with L74M, L74F, or L74F V75I using a Student t test with a statistical cutoff of P < 0.02 (*) or P < 0.0001 (**).
INSTIs such as DTG efficiently chelate two divalent metal ions that are coordinated by the catalytic carboxylates of HIV-1 IN (DDE motif D64-D116-E152) (4, 28, 29). Molecular modeling studies have shown that the N155H mutation deforms the α4 helix and widens the bottom of the catalytic pocket between residues D64 and E152. Given the proximity of N155 to D64 in the wild-type HIV-1 IN, it is possible that this mutation may also affect the position of the active site divalent metals (28). Binding of DTG to the prototype foamy virus (PFV) N224H mutant (equivalent to HIV-1 N155H) was reported to affect electrostatic interactions between H224 and the phosphate of the 3′ nucleotide (29, 30). Additionally, the metal-chelating oxygen atoms of the DTG were found to shift in response to changes in the DDE motif to accommodate the histidine residue (29, 30). In contrast to the first-generation INSTIs, DTG seems to adapt its position and conformation to the altered geometry of the active site of N224H mutant. Such structural changes in the active site and minor alteration of the catalytic metal coordination may contribute to the mechanism of DTG resistance. This notion is supported by a previous biochemical study demonstrating that the N155H mutation increases dissociation (koff) of the three clinically available INSTIs (4). The recently reported cryo-electron microscopy structure of the HIV-1 intasome (31) shows that L74 and V75, located in the β2 strand, are juxtaposed to the β1 strand containing D64 (Fig. 4A), and a similar motif is shown at the active site of PFV IN (Fig. 4B) (29). Additionally, a V75I mutation may alter the positioning of N155 and the α4 helix, thus also affecting the positions of key residues at the IN active site, which in turn may also affect divalent metal and DNA substrate binding, leading to HIV-1 resistance to the second-generation INSTIs. Such changes are consistent with the significant decrease in viral replication capacity that the L74F V75I mutations impart on V72I, N155H, and G140S Q148H (Fig. 2B, C, and D).
FIG 4.
Locations of L74 and V75 near the IN active site. The structures of the HIV-1 IN active site (A, gray [PDB ID no. 5U1C]) and the PFV IN active site (B, yellow [PDB ID no. 3S3M]) in complex with DTG (cyan sticks) are shown. Both panels are shown in cross-eyed stereo view with divalent metal ions shown in blue and viral DNA strands in orange. Panel A contains only one Mg2+ ion, as the structure was solved using HIV-1 IN with an E152Q active site mutation. Residue numbers in parentheses in panel B represent equivalent residue numbers in HIV-1 IN. Figures were created using PyMOL.
In this study, we identified a novel INSTI resistance mutation pattern, L74F V75I, in a clinical case with virological failure under a RAL-based regimen. L74F and V75I were associated with high-level resistance to second-generation INSTIs when combined with frequently observed major INSTI mutations. However, L74F and V75I could not compensate for the impaired replication capacity of the viruses with major mutations, suggesting their involvement as resistance mutations, not just secondary mutations. Although the emergence of L74F V75I with major mutations has not been reported in clinical isolates, emergence of such mutations should be monitored under long-term INSTI-based regimens. These findings may help future design of therapeutic strategies that can further improve clinical outcomes. Accumulation of information on INSTI resistance may provide insights into improvement of INSTI affinity and lead to the rational design of next-generation INSTIs.
Accession number(s).
Direct sequence data reported in this paper have been deposited in GenBank under accession no. LC201871 to LC201873.
ACKNOWLEDGMENTS
We thank Eiichi Kodama for the generous gift of the plasmids used in this study and the CARES staff and Masaaki Nakashima for dedicated assistance. We are indebted to the AIDS Research Reference and Reagent Program, NIAID and NIH, for the TZM-bl cells.
This work was supported in part by NIH grants AI076119 and AI120860 to S.G.S. and by a grant for the promotion of AIDS Research from the Ministry of Health, Labor and Welfare (A.H., Y.Y., and Y.I.). This publication is based on work supported by award no. OISE-17-62937-1 of the U.S. Civilian Research and Development Foundation (CRDF Global), by the National Science Foundation under Cooperative Agreement no. OISE-9531011 (K.K.), and by the U.S.-Japan Cooperative Medical Science Program (A.H.).
REFERENCES
- 1.Goethals O, Clayton R, Van Ginderen M, Vereycken I, Wagemans E, Geluykens P, Dockx K, Strijbos R, Smits V, Vos A, Meersseman G, Jochmans D, Vermeire K, Schols D, Hallenberger S, Hertogs K. 2008. Resistance mutations in human immunodeficiency virus type 1 integrase selected with elvitegravir confer reduced susceptibility to a wide range of integrase inhibitors. J Virol 82:10366–10374. doi: 10.1128/JVI.00470-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grinsztejn B, Nguyen BY, Katlama C, Gatell JM, Lazzarin A, Vittecoq D, Gonzalez CJ, Chen J, Harvey CM, Isaacs RD, Protocol 005 Team. 2007. Safety and efficacy of the HIV-1 integrase inhibitor raltegravir (MK-0518) in treatment-experienced patients with multidrug-resistant virus: a phase II randomised controlled trial. Lancet 369:1261–1269. doi: 10.1016/S0140-6736(07)60597-2. [DOI] [PubMed] [Google Scholar]
- 3.Hazuda DJ, Felock P, Witmer M, Wolfe A, Stillmock K, Grobler JA, Espeseth A, Gabryelski L, Schleif W, Blau C, Miller MD. 2000. Inhibitors of strand transfer that prevent integration and inhibit HIV-1 replication in cells. Science 287:646–650. doi: 10.1126/science.287.5453.646. [DOI] [PubMed] [Google Scholar]
- 4.Hightower KE, Wang R, Deanda F, Johns BA, Weaver K, Shen Y, Tomberlin GH, Carter HL III, Broderick T, Sigethy S, Seki T, Kobayashi M, Underwood MR. 2011. Dolutegravir (S/GSK1349572) exhibits significantly slower dissociation than raltegravir and elvitegravir from wild-type and integrase inhibitor-resistant HIV-1 integrase-DNA complexes. Antimicrob Agents Chemother 55:4552–4559. doi: 10.1128/AAC.00157-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lennox JL, DeJesus E, Lazzarin A, Pollard RB, Madruga JV, Berger DS, Zhao J, Xu X, Williams-Diaz A, Rodgers AJ, Barnard RJ, Miller MD, DiNubile MJ, Nguyen BY, Leavitt R, Sklar P, STARTMRK Investigators . 2009. Safety and efficacy of raltegravir-based versus efavirenz-based combination therapy in treatment-naive patients with HIV-1 infection: a multicentre, double-blind randomised controlled trial. Lancet 374:796–806. doi: 10.1016/S0140-6736(09)60918-1. [DOI] [PubMed] [Google Scholar]
- 6.Markowitz M, Morales-Ramirez JO, Nguyen BY, Kovacs CM, Steigbigel RT, Cooper DA, Liporace R, Schwartz R, Isaacs R, Gilde LR, Wenning L, Zhao J, Teppler H. 2006. Antiretroviral activity, pharmacokinetics, and tolerability of MK-0518, a novel inhibitor of HIV-1 integrase, dosed as monotherapy for 10 days in treatment-naive HIV-1-infected individuals. J Acquir Immune Defic Syndr 43:509–515. doi: 10.1097/QAI.0b013e31802b4956. [DOI] [PubMed] [Google Scholar]
- 7.Raffi F, Rachlis A, Brinson C, Arasteh K, Gorgolas M, Brennan C, Pappa K, Almond S, Granier C, Nichols WG, Cuffe RL, Eron J Jr, Walmsley S. 2015. Dolutegravir efficacy at 48 weeks in key subgroups of treatment-naive HIV-infected individuals in three randomized trials. AIDS 29:167–174. doi: 10.1097/QAD.0000000000000519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shimura K, Kodama E, Sakagami Y, Matsuzaki Y, Watanabe W, Yamataka K, Watanabe Y, Ohata Y, Doi S, Sato M, Kano M, Ikeda S, Matsuoka M. 2008. Broad antiretroviral activity and resistance profile of the novel human immunodeficiency virus integrase inhibitor elvitegravir (JTK-303/GS-9137). J Virol 82:764–774. doi: 10.1128/JVI.01534-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kobayashi M, Yoshinaga T, Seki T, Wakasa-Morimoto C, Brown KW, Ferris R, Foster SA, Hazen RJ, Miki S, Suyama-Kagitani A, Kawauchi-Miki S, Taishi T, Kawasuji T, Johns BA, Underwood MR, Garvey EP, Sato A, Fujiwara T. 2011. In vitro antiretroviral properties of S/GSK1349572, a next-generation HIV integrase inhibitor. Antimicrob Agents Chemother 55:813–821. doi: 10.1128/AAC.01209-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Quashie PK, Mesplede T, Han YS, Oliveira M, Singhroy DN, Fujiwara T, Underwood MR, Wainberg MA. 2012. Characterization of the R263K mutation in HIV-1 integrase that confers low-level resistance to the second-generation integrase strand transfer inhibitor dolutegravir. J Virol 86:2696–2705. doi: 10.1128/JVI.06591-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seki T, Suyama-Kagitani A, Kawauchi-Miki S, Miki S, Wakasa-Morimoto C, Akihisa E, Nakahara K, Kobayashi M, Underwood MR, Sato A, Fujiwara T, Yoshinaga T. 2015. Effects of raltegravir or elvitegravir resistance signature mutations on the barrier to dolutegravir resistance in vitro. Antimicrob Agents Chemother 59:2596–2606. doi: 10.1128/AAC.04844-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wares M, Mesplede T, Quashie PK, Osman N, Han Y, Wainberg MA. 2014. The M50I polymorphic substitution in association with the R263K mutation in HIV-1 subtype B integrase increases drug resistance but does not restore viral replicative fitness. Retrovirology 11:7. doi: 10.1186/1742-4690-11-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yoshinaga T, Kobayashi M, Seki T, Miki S, Wakasa-Morimoto C, Suyama-Kagitani A, Kawauchi-Miki S, Taishi T, Kawasuji T, Johns BA, Underwood MR, Garvey EP, Sato A, Fujiwara T. 2015. Antiviral characteristics of GSK1265744, an HIV integrase inhibitor dosed orally or by long-acting injection. Antimicrob Agents Chemother 59:397–406. doi: 10.1128/AAC.03909-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anstett K, Fusco R, Cutillas V, Mesplede T, Wainberg MA. 2015. Dolutegravir-selected HIV-1 containing the N155H and R263K resistance substitutions does not acquire additional compensatory mutations under drug pressure that lead to higher-level resistance and increased replicative capacity. J Virol 89:10482–10488. doi: 10.1128/JVI.01725-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cahn P, Pozniak AL, Mingrone H, Shuldyakov A, Brites C, Andrade-Villanueva JF, Richmond G, Buendia CB, Fourie J, Ramgopal M, Hagins D, Felizarta F, Madruga J, Reuter T, Newman T, Small CB, Lombaard J, Grinsztejn B, Dorey D, Underwood M, Griffith S, Min S, SAILING Study Team . 2013. Dolutegravir versus raltegravir in antiretroviral-experienced, integrase-inhibitor-naive adults with HIV: week 48 results from the randomised, double-blind, non-inferiority SAILING study. Lancet 382:700–708. doi: 10.1016/S0140-6736(13)61221-0. [DOI] [PubMed] [Google Scholar]
- 16.Wensing AM, Calvez V, Gunthard HF, Johnson VA, Paredes R, Pillay D, Shafer RW, Richman DD. 2017. 2017 update of the drug resistance mutations in HIV-1. Top Antivir Med 24:132–133. [PMC free article] [PubMed] [Google Scholar]
- 17.Hachiya A, Reeve AB, Marchand B, Michailidis E, Ong YT, Kirby KA, Leslie MD, Oka S, Kodama EN, Rohan LC, Mitsuya H, Parniak MA, Sarafianos SG. 2013. Evaluation of combinations of 4′-ethynyl-2-fluoro-2′-deoxyadenosine with clinically used antiretroviral drugs. Antimicrob Agents Chemother 57:4554–4558. doi: 10.1128/AAC.00283-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hachiya A, Ode H, Matsuda M, Kito Y, Shigemi U, Matsuoka K, Imamura J, Yokomaku Y, Iwatani Y, Sugiura W. 2015. Natural polymorphism S119R of HIV-1 integrase enhances primary INSTI resistance. Antiviral Res 119:84–88. doi: 10.1016/j.antiviral.2015.04.014. [DOI] [PubMed] [Google Scholar]
- 19.Blanco JL, Varghese V, Rhee SY, Gatell JM, Shafer RW. 2011. HIV-1 integrase inhibitor resistance and its clinical implications. J Infect Dis 203:1204–1214. doi: 10.1093/infdis/jir025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cooper DA, Steigbigel RT, Gatell JM, Rockstroh JK, Katlama C, Yeni P, Lazzarin A, Clotet B, Kumar PN, Eron JE, Schechter M, Markowitz M, Loutfy MR, Lennox JL, Zhao J, Chen J, Ryan DM, Rhodes RR, Killar JA, Gilde LR, Strohmaier KM, Meibohm AR, Miller MD, Hazuda DJ, Nessly ML, DiNubile MJ, Isaacs RD, Teppler H, Nguyen BY, BENCHMRK Study Teams . 2008. Subgroup and resistance analyses of raltegravir for resistant HIV-1 infection. N Engl J Med 359:355–365. doi: 10.1056/NEJMoa0708978. [DOI] [PubMed] [Google Scholar]
- 21.Fransen S, Gupta S, Frantzell A, Petropoulos CJ, Huang W. 2012. Substitutions at amino acid positions 143, 148, and 155 of HIV-1 integrase define distinct genetic barriers to raltegravir resistance in vivo. J Virol 86:7249–7255. doi: 10.1128/JVI.06618-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hurt CB, Sebastian J, Hicks CB, Eron JJ. 2014. Resistance to HIV integrase strand transfer inhibitors among clinical specimens in the United States, 2009–2012. Clin Infect Dis 58:423–431. doi: 10.1093/cid/cit697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fransen S, Gupta S, Danovich R, Hazuda D, Miller M, Witmer M, Petropoulos CJ, Huang W. 2009. Loss of raltegravir susceptibility by human immunodeficiency virus type 1 is conferred via multiple nonoverlapping genetic pathways. J Virol 83:11440–11446. doi: 10.1128/JVI.01168-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malet I, Delelis O, Valantin MA, Montes B, Soulie C, Wirden M, Tchertanov L, Peytavin G, Reynes J, Mouscadet JF, Katlama C, Calvez V, Marcelin AG. 2008. Mutations associated with failure of raltegravir treatment affect integrase sensitivity to the inhibitor in vitro. Antimicrob Agents Chemother 52:1351–1358. doi: 10.1128/AAC.01228-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Menendez-Arias L. 2013. Molecular basis of human immunodeficiency virus type 1 drug resistance: overview and recent developments. Antiviral Res 98:93–120. doi: 10.1016/j.antiviral.2013.01.007. [DOI] [PubMed] [Google Scholar]
- 26.Goethals O, Vos A, Van Ginderen M, Geluykens P, Smits V, Schols D, Hertogs K, Clayton R. 2010. Primary mutations selected in vitro with raltegravir confer large fold changes in susceptibility to first-generation integrase inhibitors, but minor fold changes to inhibitors with second-generation resistance profiles. Virology 402:338–346. doi: 10.1016/j.virol.2010.03.034. [DOI] [PubMed] [Google Scholar]
- 27.Jones GS, Yu F, Zeynalzadegan A, Hesselgesser J, Chen X, Chen J, Jin H, Kim CU, Wright M, Geleziunas R, Tsiang M. 2009. Preclinical evaluation of GS-9160, a novel inhibitor of human immunodeficiency virus type 1 integrase. Antimicrob Agents Chemother 53:1194–1203. doi: 10.1128/AAC.00984-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.DeAnda F, Hightower KE, Nolte RT, Hattori K, Yoshinaga T, Kawasuji T, Underwood MR. 2013. Dolutegravir interactions with HIV-1 integrase-DNA: structural rationale for drug resistance and dissociation kinetics. PLoS One 8:e77448. doi: 10.1371/journal.pone.0077448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hare S, Smith SJ, Metifiot M, Jaxa-Chamiec A, Pommier Y, Hughes SH, Cherepanov P. 2011. Structural and functional analyses of the second-generation integrase strand transfer inhibitor dolutegravir (S/GSK1349572). Mol Pharmacol 80:565–572. doi: 10.1124/mol.111.073189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hare S, Vos AM, Clayton RF, Thuring JW, Cummings MD, Cherepanov P. 2010. Molecular mechanisms of retroviral integrase inhibition and the evolution of viral resistance. Proc Natl Acad Sci U S A 107:20057–20062. doi: 10.1073/pnas.1010246107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Passos DO, Li M, Yang R, Rebensburg SV, Ghirlando R, Jeon Y, Shkriabai N, Kvaratskhelia M, Craigie R, Lyumkis D. 2017. Cryo-EM structures and atomic model of the HIV-1 strand transfer complex intasome. Science 355:89–92. doi: 10.1126/science.aah5163. [DOI] [PMC free article] [PubMed] [Google Scholar]