ABSTRACT
Antibiotics excreted into the intestinal tract may disrupt the microbiota that provide colonization resistance against enteric pathogens and alter normal metabolic functions of the microbiota. Many of the bacterial metabolites produced in the intestinal tract are absorbed systemically and excreted in urine. Here, we used a mouse model to test the hypothesis that alterations in levels of targeted bacterial metabolites in urine specimens could provide useful biomarkers indicating disrupted or intact colonization resistance. To assess in vivo colonization resistance, mice were challenged with Clostridium difficile spores orally 3, 6, and 11 days after the completion of 2 days of treatment with piperacillin-tazobactam, aztreonam, or saline. For concurrent groups of antibiotic-treated mice, urine samples were analyzed by using liquid chromatography-tandem mass spectrometry (LC-MS/MS) to quantify the concentrations of 11 compounds targeted as potential biomarkers of colonization resistance. Aztreonam did not affect colonization resistance, whereas piperacillin-tazobactam disrupted colonization resistance 3 days after piperacillin-tazobactam treatment, with complete recovery by 11 days after treatment. Three of the 11 compounds exhibited a statistically significant and >10-fold increase (the tryptophan metabolite N-acetyltryptophan) or decrease (the plant polyphenyl derivatives cinnamoylglycine and enterodiol) in concentrations in urine 3 days after piperacillin-tazobactam treatment, followed by recovery to baseline that coincided with the restoration of in vivo colonization resistance. These urinary metabolites could provide useful and easily accessible biomarkers indicating intact or disrupted colonization resistance during and after antibiotic treatment.
KEYWORDS: intestinal microbiota
INTRODUCTION
The gastrointestinal tract of adult mammals is inhabited by a complex microbial community that includes hundreds of bacterial species (1). These organisms complement host physiology by providing an array of metabolic functions that benefit the host (e.g., digestion of complex polysaccharides and proteins) (1–4). The indigenous microbiota of the colon also provide an important host defense, termed colonization resistance, by inhibiting the growth of potentially pathogenic microorganisms such as Clostridium difficile (5, 6). Antibiotics that are excreted into the intestinal tract may suppress the microbiota and disrupt bacterial metabolic functions and colonization resistance (5–9). We demonstrated previously that clindamycin or piperacillin-tazobactam treatment of mice resulted in the disruption of colonization resistance and marked changes in bacterial metabolites present in fecal specimens based on nontargeted metabolic profiling (9). Of 484 compounds analyzed, 146 (30%) exhibited a significant increase or decrease in concentrations during antibiotic treatment, followed by recovery to baseline that coincided with the restoration of in vivo colonization resistance. Identified as potential biomarkers of colonization resistance, these compounds included intermediates in carbohydrate or protein metabolism that showed increased (pentitols, gamma-glutamyl amino acids, and inositol metabolites) or decreased (pentoses and dipeptides) levels with antibiotic treatment.
Although measurements of concentrations of bacterial metabolites in stool could provide useful biomarkers of colonization resistance, the collection and processing of stool specimens can be challenging. In health care settings, urine or blood specimens are much more frequently used for diagnostic testing due to ease of collection and the ability to collect specimens in a timely manner. Because many metabolites produced by intestinal bacteria are absorbed with subsequent excretion in urine (10–14), we hypothesized that urine metabolites might provide useful indicators of intact or disrupted colonization resistance. To test this hypothesis, we used a mouse model to determine if the timing of recovery of in vivo colonization resistance to C. difficile after antibiotic treatment correlated with changes in concentrations of selected metabolites of intestinal bacteria in urine.
RESULTS
Restoration of in vivo colonization resistance after antibiotic treatment.
Figure 1 shows the results of the assessment of in vivo colonization resistance to C. difficile. Aztreonam-treated mice maintained intact colonization resistance with no increase in the density of C. difficile colonization in comparison to saline controls (P > 0.92). Piperacillin-tazobactam-treated mice had altered colonization resistance when challenged with C. difficile 3 days after the final antibiotic dose, with high-density colonization in comparison to the controls and aztreonam-treated mice (P < 0.001); 6 days after the final antibiotic dose, there was a nonsignificant trend toward an increased density of colonization in the piperacillin-tazobactam-treated versus control and aztreonam-treated mice (P = 0.07). At day 11 after the final dose, piperacillin-tazobactam-treated mice demonstrated intact colonization resistance.
FIG 1.
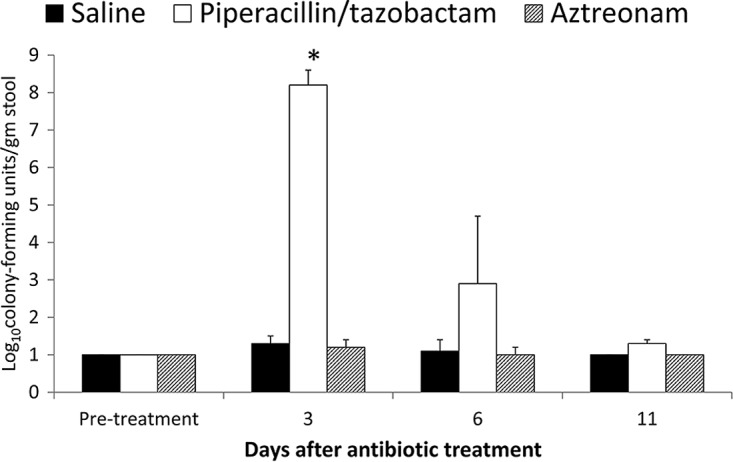
Effect of antibiotic treatment on in vivo colonization resistance to Clostridium difficile. Mice received subcutaneous antibiotic treatment for 2 days, and subgroups (8 per group) were challenged with 10,000 CFU of C. difficile spores before treatment or on days 3, 6, and 11 after antibiotic treatment by gastric gavage. *, P < 0.001.
Identification of urinary biomarkers of colonization resistance.
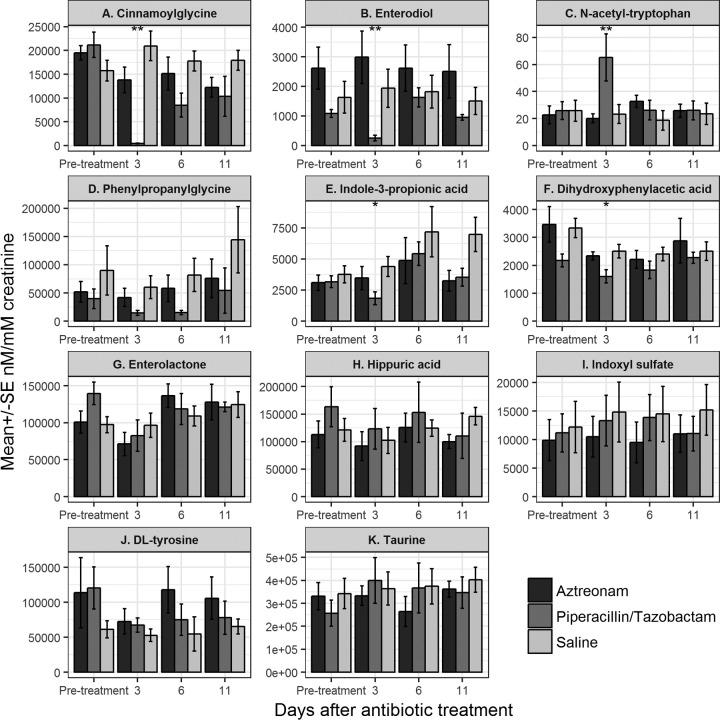
Figure 2 shows the effects of antibiotic treatment on the concentrations of the 11 urinary compounds studied. Concentrations of 5 of the 11 (45%) compounds differed significantly between the piperacillin-tazobactam-treated and control or aztreonam-treated mice, including N-acetyltryptophan, indole-3 propionic acid, cinnamoylglycine, enterodiol, and 3,4-dihydroxyphenylacetic acid. Three compounds, including N-acetyltryptophan, cinnamoylglycine, and enterodiol, were identified as potential biomarkers of colonization resistance because they exhibited a 10-fold difference in concentrations between piperacillin-tazobactam- and saline- or aztreonam-treated mice on day 3 after treatment with normalization by day 11 after treatment, in conjunction with the recovery of colonization resistance. The mean concentration of N-acetyltryptophan was >10-fold higher in urine of piperacillin-tazobactam-treated mice than in urine of saline controls or aztreonam-treated mice on day 3 after antibiotic treatment; 7 of 8 piperacillin-tazobactam-treated mice had increased levels of N-acetyltryptophan in comparison to the pretreatment levels. The concentrations of cinnamoylglycine and enterodiol were >100-fold lower in urine of all of the piperacillin-tazobactam-treated mice than in urine of the saline controls or aztreonam-treated mice on day 3 after antibiotic treatment.
FIG 2.
Effect of antibiotic treatment on the concentrations of the 11 urinary compounds. Compounds whose concentrations differed significantly between the piperacillin-tazobactam-treated and control mice included N-acetyltryptophan, indole-3 propionic acid, cinnamoylglycine, enterodiol, and 3,4-dihydroxyphenylacetic acid. N-Acetyltryptophan, cinnamoylglycine, and enterodiol were identified as potential biomarkers of colonization resistance because they exhibited a 10-fold difference in concentrations between piperacillin-tazobactam- and saline-treated mice on day 3 after treatment with normalization by day 11, in conjunction with the recovery of colonization resistance. SE, standard error. *, P < 0.05; **, P < 0.001.
DISCUSSION
Our findings are consistent with data from previous studies in demonstrating that piperacillin-tazobactam treatment causes a disruption of colonization resistance, whereas aztreonam, an agent lacking activity against anaerobes, does not (7–9). The rapid restoration of colonization resistance against C. difficile by 6 to 11 days after treatment highlights the resilience of the microbiota. By correlating the timing of the functional recovery of colonization resistance with changes in concentrations of selected urine metabolites, we identified 3 potential urinary biomarkers of colonization resistance (N-acetyltryptophan, cinnamoylglycine, and enterodiol).
The biomarkers identified are biologically plausible, and our findings for these compounds are consistent with data from previous studies with germfree mice and antibiotic-treated mice or humans (4, 9, 15–20). Cinnamoylglycine is a glycine conjugate of cinnamic acid that was previously shown to be abundant in the serum of conventional mice but present in minimal concentrations in the serum of germfree mice (4). Cinnamic acid is present in a variety of foods or can be produced from phenylalanine metabolism, and cinnamoylglycine has been shown to be excreted in the urine of mice and humans (16–17). Dietary tryptophan is metabolized to indole in the colon by tryptophanase produced by enteric bacteria, with subsequent conversion by other bacterial enzymes to indole-3-propionic acid (4). Our finding of increased N-acetyltryptophan and decreased indole-3-propionic acid concentrations in urine of piperacillin-tazobactam-treated mice is consistent with previously reported data for stool of antibiotic-treated mice and for serum of germfree mice (4, 9, 15). Finally, dietary lignans are plant-derived polyphenols that are metabolized by anaerobic intestinal bacteria to produce the enterolignans enterodiol and enterolactone (18–19). Oral antibiotic therapy has been associated with reduced serum enterolactone concentrations in humans (20). It is not clear why enterodiol, but not enterolactone, was significantly suppressed by piperacillin-tazobactam treatment in our study.
Our findings have important clinical implications. There is a need for biomarkers that can be used to monitor whether colonization resistance is intact. The finding that urinary biomarkers may potentially be useful to monitor the functional status of the gut microbiota is significant because collection of urine samples is relatively easy compared to collection of stool specimens, and samples may be collected in a timely manner. Although we identified only 3 candidate biomarkers of colonization resistance, we anticipate that it may be feasible to identify many additional urinary biomarkers if larger-scale nontargeted and targeted studies are conducted. Previous studies demonstrated antibiotic-induced changes in the concentrations of several other urinary metabolites (e.g., urobilin, short-chain fatty acids, and trimethylamine-N-oxide) (10, 11, 13, 15). The availability of urine biomarkers of colonization resistance could be useful for a wide range of studies examining the impact of antibiotic treatment on the intestinal microbiota.
Our study has several limitations. First, we did not examine changes in the intestinal microbiota that correlated with the recovery of colonization resistance. However, we previously demonstrated that the recovery of colonization resistance after piperacillin-tazobactam treatment in mice correlates with the recovery of bacteria from the families Lachnospiraceae and Ruminococcaceae (phylum Firmicutes, order Clostridiales) in stool specimens (9). Organisms of the phylum Firmicutes have been associated with colonization resistance against C. difficile (21–22). Second, additional studies are needed to determine which bacterial species are responsible for the production of specific metabolites. Given the degree of functional redundancy of the intestinal microbiota, it is possible that multiple families of bacteria may be able to carry out the metabolic conversions required to produce the metabolites identified here. Third, findings in mice may differ from findings in humans given differences in diet and microbiota. Finally, we cannot exclude the possibility that metabolites whose concentrations differed significantly between piperacillin-tazobactam- and saline-treated mice on day 3 after treatment (i.e., indole-3 propionic acid and 3,4-dihydroxyphenylacetic acid) without achieving a 10-fold difference could be useful as biomarkers. However, these compounds exhibited substantial day-to-day variation in concentrations in the saline control group, which would make them potentially less valuable as biomarkers of colonization resistance.
In summary, using a targeted approach, we identified N-acetyltryptophan, cinnamoylglycine, and enterodiol as potential urinary biomarkers of colonization resistance. Such compounds could provide useful and easily accessible biomarkers indicating intact or disrupted colonization resistance during and after antibiotic treatment. Studies are needed to determine if antibiotic treatment in humans results in alterations of the concentrations of these urinary metabolites. Ultimately, studies will be needed to identify or design compounds metabolized by the intestinal microbiota that provide colonization resistance that would provide a unique urine or serum indicator of colonization resistance.
MATERIALS AND METHODS
C. difficile strain.
VA17 is an epidemic North American pulsed-field gel electrophoresis type 1 (NAP1) C. difficile strain. For VA17, the MICs of piperacillin-tazobactam and aztreonam are 2 μg/ml and >256 μg/ml, respectively (8). C. difficile spores were prepared as previously described (23).
In vivo mouse model of colonization resistance.
We used a mouse model described previously to evaluate the recovery of colonization resistance after antibacterial treatment (9). Female CF-1 mice weighing 25 to 30 g (Harlan Sprague-Dawley, Indianapolis, IN) were housed in individual microisolator cages and fed sterilized Teklad Global 18% protein-extruded rodent diet (Harlan Teklad, Madison, WI). Mice (32 per group) received daily subcutaneous injections (0.1-ml total volume) of saline, piperacillin-tazobactam (8 mg/day), or aztreonam (3 mg/day) for 2 days. The antibiotic dose was equal to the usual human doses administered over a 24-h period (milligrams of antibiotic per gram of body weight).
To assess in vivo colonization resistance, subgroups of the antibiotic-treated mice were challenged with 104 CFU of C. difficile VA17 spores by orogastric gavage either before or 3, 6, or 11 days after the completion of antibiotic treatment (8 mice from each treatment group were challenged at each time point). Fresh stool specimens were collected 2 days after gavage, and the concentration of C. difficile was measured by plating serially diluted samples onto selective agar as previously described (9). Colonization resistance was deemed intact if there was no significant increase in concentrations of C. difficile in the stool of antibiotic-treated mice in comparison to the control mice.
Urine specimen collection and processing.
For the group of mice challenged with C. difficile on day 11 (8 mice), fresh urine specimens were collected in sterile Eppendorf tubes before antibiotic treatment and on days 3, 6, and 11 after antibiotic treatment. For urine collection, individual mice were moved into clean cages with no bedding for 1-h periods and observed continuously for the production of urine or stool. Urine specimens were collected with a pipette tip immediately after they were produced. Urine was collected for analysis only if there was no contact with stool pellets. Mice were moved into new clean cages as needed to avoid stool contamination of urine. Urine specimens were centrifuged at 13,000 × g for 15 min at 8°C. To reduce glucuronide and sulfate concentrations in urine, which can interfere with analyses, urine supernatants were digested with beta-glucuronidase and aryl sulfatase for 24 h according to the manufacturer's recommendations (Roche Diagnostics GmbH, Mannheim, Germany). Digested urine supernatants were frozen at −80°C prior to analysis at the Cleveland Clinic Small Molecule Mass Spectrometry Core Facility.
Liquid chromatography-mass spectrometry analysis.
Targeted metabolic profiling by liquid chromatography-tandem mass spectrometry (LC-MS/MS) was used to quantify the concentrations of 11 urinary metabolites that were hypothesized to be associated with disrupted versus intact colonization resistance. The 11 metabolites included tryptophan metabolites (N-acetyltryptophan, indole-3 propionic acid, and indoxyl sulfate), plant polyphenyl derivatives (cinnamoylglycine, hippuric acid, enterolactone, enterodiol, 3,4-dihydroxyphenylacetic acid, and phenylpropionylglycine), dl-tyrosine, and taurine. These metabolites were chosen because they were identified as potential biomarkers of colonization resistance in our recent nontargeted study of mouse fecal specimens or in previous studies by others (4, 9–11, 13, 15).
Twenty-microliter aliquots of the urine supernatant were injected onto a Waters 2690 high-performance liquid chromatography (HPLC) system (Quattro Ultima Micromass, Beverly, MA) and the metabolites were separated by using a C18 column (Phenomenex, Rancho Palos Verdes, CA) under gradient conditions at a flow rate of 0.2 ml/min. A gradient was formed by mixing mobile phases A (water containing 5 mM ammonium acetate) and B (acetonitrile containing 0.2% acetic acid and 5 ammonium acetate) as follows: isocratic elution with 100% mobile phase A from 0 to 2 min with an increase to 100% mobile phase B from 2 to 8 minutes, which was kept at 100% mobile phase B for 10 min and then equilibrated with 100% mobile phase A for 10 min.
The HPLC column effluent was introduced onto an electrospray ionization triple quadrupole mass spectrometer (Waters Corporation, Milford, MA) and analyzed by using negative electrospray ionization in the multiple-reaction-monitoring mode. The multiple-reaction-monitoring precursor ion and product ion transitions are shown in Table S1 in the supplemental material. Internal standard calibration curves were used to quantify the concentrations of the metabolites in urine. Urine metabolite concentrations were normalized to urine creatinine concentrations and expressed as nanomolar per millimolar creatinine. Deuterated internal standards (100 ng each) detected in the negative mode were used for hippuric acid-d5 (Toronto Research Chemicals, North York, Canada) and indoxyl sulfate-d4 (Sigma-Aldrich).
Data analysis.
Analysis of variance (ANOVA) was used to compare concentrations of C. difficile recovered from stool samples for control versus antibiotic-treated mice. For the LC-MS/MS analyses, data processing was done by using MassLynx software v4.1 from Waters. For metabolite analysis, two-way ANOVA was performed to assess differences in log-transformed concentrations across days and antibiotic treatments to identify compounds whose concentrations differed significantly between control and antibiotic-treated groups at specific points in time. Contrast tests we performed to compare antibiotic groups on specific days. Significance was assessed at an alpha value of 0.05. As these steps were hypothesis generating, no adjustments were made for multiple comparisons. Compounds were assessed as possible biomarkers of colonization resistance if there was a significant difference in concentrations between the piperacillin-tazobactam-treated and control mice on day 3 after piperacillin-tazobactam treatment but not at baseline or day 11; this choice was based on the finding that colonization resistance was disrupted on day 3 after piperacillin-tazobactam treatment but not at baseline or on day 11 after treatment. For the compounds whose concentrations differed significantly from those in the saline controls, those that exhibited a 10-fold difference between the piperacillin-tazobactam-treated and control mice on day 3 but not at baseline or on day 11 were identified as potential biomarkers of colonization resistance; a 10-fold difference was chosen in order to identify potential biomarkers with marked differences that were robust to log transformation. Concentrations of the metabolites were analyzed for aztreonam-treated mice to determine if there were differences from those in saline-treated mice, but metabolites with differences among these groups were not considered potential biomarkers of colonization resistance because aztreonam did not disrupt colonization resistance.
Supplementary Material
ACKNOWLEDGMENTS
We thank Belinda Willard, Stanley Hazen, and Bruce Levinson of the Cleveland Clinic for helpful discussion. We thank Vincent Monnier and David Sell for providing several small molecular compounds and sugars used for targeting many of these analytes.
We declare that no competing interests exist.
This work was supported by a grant from the Case Western Reserve University Clinical and Translational Science Collaborative and by the Department of Veterans Affairs.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.00477-17.
REFERENCES
- 1.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. 2005. Diversity of the human intestinal microbial flora. Science 308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blaut M, Clavel T. 2007. Metabolic diversity of the intestinal microbiota: implications for health and disease. J Nutr 137:751S–755S. [DOI] [PubMed] [Google Scholar]
- 3.Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, Pettersson S. 2012. Host-gut microbiota metabolic interactions. Science 336:1262–1267. doi: 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- 4.Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, Siuzdak G. 2009. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci U S A 106:3698–3703. doi: 10.1073/pnas.0812874106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Donskey CJ. 2004. The role of the intestinal tract as a reservoir and source for transmission of nosocomial pathogens. Clin Infect Dis 39:219–226. doi: 10.1086/422002. [DOI] [PubMed] [Google Scholar]
- 6.Buffie CG, Pamer EG. 2013. Microbiota-mediated colonization resistance against intestinal pathogens. Nat Rev Immunol 13:790–801. doi: 10.1038/nri3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stiefel U, Pultz NJ, Helfand MS, Donskey CJ. 2004. Increased susceptibility to vancomycin-resistant Enterococcus intestinal colonization persists after completion of anti-anaerobic antibiotic treatment in mice. Infect Control Hosp Epidemiol 25:373–379. doi: 10.1086/502408. [DOI] [PubMed] [Google Scholar]
- 8.Pultz NJ, Donskey CJ. 2005. Effect of antibiotic treatment on growth of and toxin production by Clostridium difficile in the cecal contents of mice. Antimicrob Agents Chemother 49:3529–3532. doi: 10.1128/AAC.49.8.3529-3532.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jump RL, Polinkovsky A, Hurless K, Sitzlar B, Eckart K, Tomas M, Deshpande A, Nerandzic MM, Donskey CJ. 2014. Metabolomics analysis identifies intestinal microbiota-derived biomarkers of colonization resistance in clindamycin-treated mice. PLoS One 9:e101267. doi: 10.1371/journal.pone.0101267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yap IK, Li JV, Saric J, Martin FP, Davies H, Wang Y, Wilson ID, Nicholson JK, Utzinger J, Marchesi JR, Holmes E. 2008. Metabonomic and microbiological analysis of the dynamic effect of vancomycin-induced gut microbiota modification in the mouse. J Proteome Res 7:3718–3728. doi: 10.1021/pr700864x. [DOI] [PubMed] [Google Scholar]
- 11.Romick-Rosendale LE, Goodpaster AM, Hanwright PJ, Patel NB, Wheeler ET, Chona DL, Kennedy MA. 2009. NMR-based metabonomics analysis of mouse urine and fecal extracts following oral treatment with the broad-spectrum antibiotic enrofloxacin (Baytril). Magn Reson Chem 47(Suppl 1):S36–S46. doi: 10.1002/mrc.2511. [DOI] [PubMed] [Google Scholar]
- 12.Bouatra S, Aziat F, Mandal R, Guo AC, Wilson MR, Knox C, Bjorndahl TC, Krishnamurthy R, Saleem F, Liu P, Dame ZT, Poelzer J, Huynh J, Yallou FS, Psychogios N, Dong E, Bogumil R, Roehring C, Wishart DS. 2013. The human urine metabolome. PLoS One 8:e73076. doi: 10.1371/journal.pone.0073076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Swann JR, Tuohy KM, Lindfors P, Brown DT, Gibson GR, Wilson ID, Sidaway J, Nicholson JK, Holmes E. 2011. Variation in antibiotic-induced microbial recolonization impacts on the host metabolic phenotypes of rats. J Proteome Res 10:3590–3603. doi: 10.1021/pr200243t. [DOI] [PubMed] [Google Scholar]
- 14.Auger C, Mullen W, Hara Y, Crozier A. 2008. Bioavailability of polyphenon E flavan-3-ols in humans with an ileostomy. J Nutr 138:1535S–1542S. [DOI] [PubMed] [Google Scholar]
- 15.Zheng X, Xie G, Zhao A, Zhao L, Yao C, Chiu NH, Zhou Z, Bao Y, Jia W, Nicholson JK, Jia W. 2011. The footprints of gut microbial-mammalian co-metabolism. J Proteome Res 10:5512–5522. doi: 10.1021/pr2007945. [DOI] [PubMed] [Google Scholar]
- 16.Lustgarten MS, Price LL, Chalé A, Fielding RA. 2014. Metabolites related to gut bacterial metabolism, peroxisome proliferator-activated receptor-alpha activation, and insulin sensitivity are associated with physical function in functionally-limited older adults. Aging Cell 13:918–925. doi: 10.1111/acel.12251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang J, Sun R, Chen Y, Tan K, Wei H, Yin L, Pu Y. 2014. Small molecule metabolite biomarker candidates in urine from mice exposed to formaldehyde. Int J Mol Sci 15:16458–16468. doi: 10.3390/ijms150916458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clavel T, Henderson G, Engst W, Doré J, Blaut M. 2006. Phylogeny of human intestinal bacteria that activate the dietary lignan secoisolariciresinol diglucoside. FEMS Microbiol Ecol 55:471–478. doi: 10.1111/j.1574-6941.2005.00057.x. [DOI] [PubMed] [Google Scholar]
- 19.Setchell KD, Brown NM, Zimmer-Nechemias L, Wolfe B, Jha P, Heubi JE. 2014. Metabolism of secoisolariciresinol-diglycoside the dietary precursor to the intestinally derived lignan enterolactone in humans. Food Funct 5:491–501. doi: 10.1039/c3fo60402k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kilkkinen A, Pietinen P, Klaukka T, Virtamo J, Korhonen P, Adlercreutz H. 2002. Use of oral antimicrobials decreases serum enterolactone concentration. Am J Epidemiol 155:472–477. doi: 10.1093/aje/155.5.472. [DOI] [PubMed] [Google Scholar]
- 21.Vincent C, Stephens DA, Loo VG, Edens TJ, Behr MA, Dewar K, Manges AR. 2013. Reductions in intestinal Clostridiales precede the development of nosocomial Clostridium difficile infection. Microbiome 1:18. doi: 10.1186/2049-2618-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Antharam VC, Li EC, Ishmael A, Sharma A, Mai V, Rand KH, Wang GP. 2013. Intestinal dysbiosis and depletion of butyrogenic bacteria in Clostridium difficile infection and nosocomial diarrhea. J Clin Microbiol 51:2884–2892. doi: 10.1128/JCM.00845-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nerandzic MM, Donskey CJ. 2009. Effective and reduced-cost modified selective medium for isolation of Clostridium difficile. J Clin Microbiol 47:397–400. doi: 10.1128/JCM.01591-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.