ABSTRACT
Nucleoside analog inhibitors (NAIs) are an important class of antiviral agents. Although highly effective, some NAIs with activity against hepatitis C virus (HCV) can cause toxicity, presumably due to off-target inhibition of host mitochondrial RNA polymerase (POLRMT). The in vitro nucleotide substrate specificity of POLRMT was studied in order to explore structure-activity relationships that can facilitate the identification of nontoxic NAIs. These findings have important implications for the development of all anti-RNA virus NAIs.
KEYWORDS: antiviral agents, hepatitis C virus, toxicity
TEXT
Nucleoside analog inhibitors (NAIs) have been important components of antiviral therapy for decades. 2′-Deoxyribonucleoside analogs are regularly used for treatment of viral infections such as human immunodeficiency virus (HIV), hepatitis B virus (HBV), and herpes simplex virus (HSV) (reviewed in references 1 and 2). Upon cellular entry, NAIs must be phosphorylated into their active 5′-triphosphate (TP) forms by viral or cellular kinases. NAI-TPs are substrates for the viral polymerase, and their incorporation often results in chain termination of the growing genome and subsequent inhibition of virus replication. Recently, sofosbuvir became the first ribonucleoside analog inhibitor to receive approval for treatment of hepatitis C virus (HCV), an RNA virus (3). Sofosbuvir is administered as a phosphoramidate prodrug in order to target the liver, to improve cell permeability, and to bypass the first phosphorylation step by host cell kinases (4, 5). Despite the excellent safety profile of sofosbuvir, the development of several preceding anti-HCV ribonucleotide prodrugs was halted due to toxicity (reviewed in reference 6). For example, phase II clinical trials with BMS-986094 (formerly known as INX-189) were terminated after reports of severe adverse events and one death (7–9). Although the exact mechanism of toxicity has not yet been fully elucidated, it has been shown that treatment with NAIs can sometimes lead to off-target inhibition of host polymerases. For example, off-target inhibition of mitochondrial DNA polymerase γ has been well documented for zidovudine triphosphate (AZT-TP), the first anti-HIV 2′-deoxyribonucleoside analog to receive FDA approval (10–13). Importantly, it was recently shown that ribonucleoside triphosphate (rNTP) analogs could be substrates for recombinant human mitochondrial RNA polymerase (POLRMT) (14–16). Unlike the nuclear counterpart RNA polymerase II, POLRMT lacks exonuclease activity; therefore, incorporation of chain-terminating nucleotides was shown to be irreversible in vitro (14). Consistent with these observations, Arnold et al. reported that increased POLRMT incorporation of NAI-TPs, as well as high intracellular NAI-TP levels, correlated positively with reductions in mitochondrial RNA levels and cytotoxicity (14). Similarly, we recently showed that treatment of Huh-7 cells with BMS-986094, which generates high levels of 2′-C-methyl-GTP intracellularly, inhibited mitochondrial RNA transcription (17). Interestingly, treatment with a 2,6-diaminopurine nucleotide (DAPN) prodrug, an investigational anti-HCV agent that also generates 2′-C-methyl-GTP (18), did not have similar deleterious effects. Lower levels of intracellular NAI-TP accumulation were proposed to account for the distinct cytotoxicity profiles of these compounds (17).
Considering the importance of safety in developing novel ribonucleoside analogs as inhibitors of HCV and other RNA viruses, we explored the nucleotide substrate specificity of POLRMT. Here we report on the in vitro POLRMT incorporation profiles of over 40 rNTP analogs, in order to shed light on structure-activity relationships that may lead to the identification of nontoxic NAIs. NAI-TPs that were not substrates for this enzyme were further examined for antiviral activity. Knowledge gained from this study has important implications for the development of antiviral NAIs not only for HCV but for all RNA viruses.
Incorporation of nucleoside analogs by POLRMT.
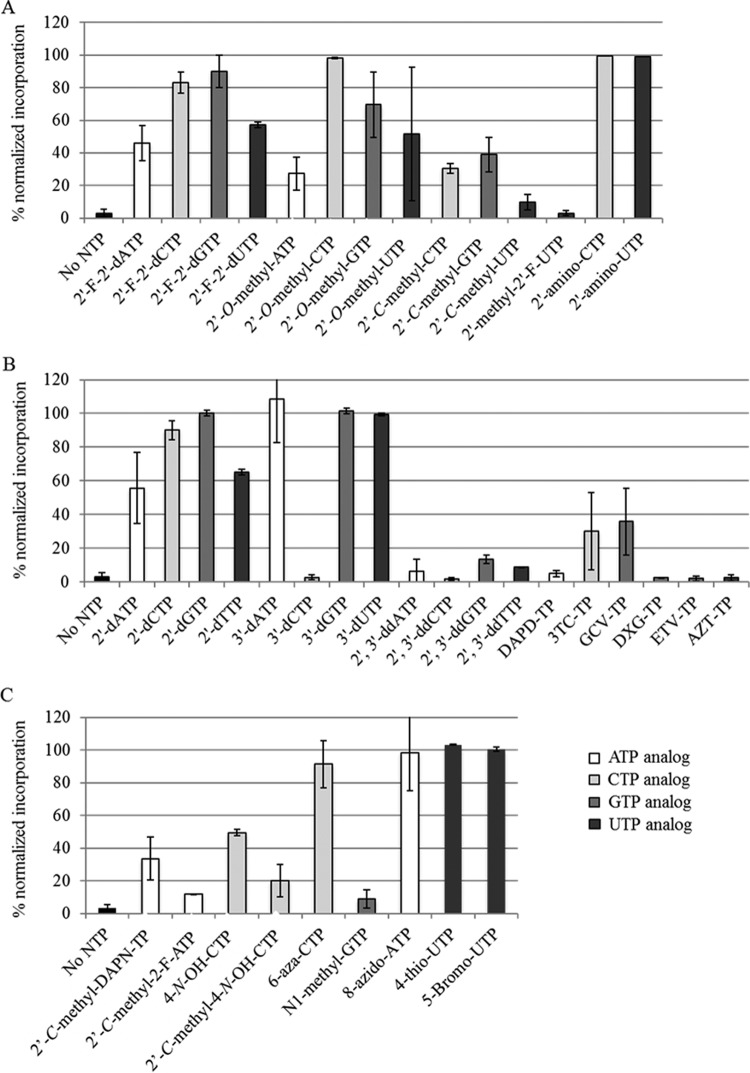
In order to examine the nucleoside substrate specificity of POLRMT, we evaluated the incorporation profiles of 41 nucleosides with various chemical modifications on the ribose or base moiety (the chemical structures are summarized in Fig. 1). As described previously (14, 17), 125 nM purified POLRMT enzyme was incubated with 500 nM 5′-radiolabeled RNA/DNA primer/template that allowed the incorporation of A, C, G, or U nucleotide analogs at position +1. Each nucleoside triphosphate (NTP) analog (100 μM) was incubated for 2 h at 30°C with the POLRMT-RNA/DNA complex containing the appropriate DNA template. The amount of NAI-TP incorporation was normalized to that observed with the corresponding natural nucleotide substrate (Fig. 2). We determined that most modifications on the ribose ring of nucleotide analogs did not completely neutralize incorporation by POLRMT. As depicted in Fig. 2A, the addition of 2′-O-methyl, 2′-fluoro, or 2′-amino modifications did not consistently affect POLRMT incorporation for all nucleotides tested. Consistent with previous findings (14, 15), the addition of a C-methyl group at the 2′ position generally led to notable reductions in incorporation for C, G, and U analogs. Nucleotides harboring this modification have been examined for their antiviral activities against HCV (19, 20) and dengue virus (21). Interestingly, the effect of the 2′-C-methyl modification was especially pronounced for 2′-C-methyl-UTP, whose incorporation was reduced to 10% of that of natural UTP, suggesting that U analogs may be particularly vulnerable to chemical modifications with regard to POLRMT incorporation. This is in agreement with the observation that 2′-C-methyl-2′-fluoro-UTP, the active metabolite of sofosbuvir, is an exceedingly poor substrate for POLRMT (Fig. 2A). Under the conditions tested, we found that 3′- or 2′-deoxynucleoside triphosphates (dNTPs) were generally good substrates for POLRMT (with the exception of 3′-dCTP) (Fig. 2B). However, the simultaneous removal of both hydroxyl groups (yielding 2′,3′-dideoxynucleoside triphosphates [ddNTPs]) completely abrogated incorporation, suggesting that the presence of at least one hydroxyl group is essential for NAI-TP incorporation. Our data are consistent with previous observations that little discrimination exists against sugar-modified nucleotides (22). The observed promiscuity of POLRMT has implications for the fidelity of this enzyme during mitochondrial RNA transcription.
FIG 1.
Chemical structures of NAI-TPs. (A) Structures of NAI-TPs with modifications on the ribose moiety. Compounds are grouped according to the base moiety. (B) Structures of dNAI-TPs with anti-HIV, anti-HBV, or anti-HSV activity. (C) Structures of rNTP analogs with modifications on the base moiety.
FIG 2.
NTP analog incorporation by POLRMT. (A) NTP analogs with modifications at the 2′ position of the ribose moiety were assessed for incorporation by POLRMT. Incorporation reactions were allowed to proceed for 2 h in the presence of 100 μM each substrate. Percentage incorporation of each NTP analog was normalized to that of the corresponding natural nucleotide substrate (ATP, CTP, GTP, or UTP). (B) POLRMT incorporation of 2′-deoxyribonucleoside analogs and NAI-TPs with anti-HBV, anti-HIV, or anti-HSV activity was assessed as described above. (C) POLRMT incorporation was assessed for rNTP analogs with modifications on the base moiety. Error bars represent standard deviations (SDs) for two or three separate experiments.
Considering that 2′-dNTP analogs were substrates for POLRMT, we next asked whether deoxynucleoside analog inhibitor triphosphates (dNAI-TPs) active against DNA viruses or retroviruses might also be substrates for POLRMT (see Fig. 1B for chemical structures). As expected, diaminopurine dioxolane triphosphate (DAPD-TP), guanosine dioxolane triphosphate (DXG-TP), AZT-TP, and entecavir triphosphate (ETV-TP) were not incorporated by POLRMT. Surprisingly, we found 30% and 36% incorporation for lamivudine triphosphate (3TC-TP) and ganciclovir triphosphate (GCV-TP), respectively, at 100 μM NTP (Fig. 2B). Although extensive literature on the impact of dNAI-TPs on mitochondrial DNA synthesis exists, it is worth noting that little information on the role of these compounds with regard to interference with mitochondrial RNA transcription in cells is available.
In our search for nucleoside analogs that are not substrates for POLRMT but are active against the viral RNA polymerase of HCV, we next investigated the incorporation profiles of NTP analogs harboring various base moiety modifications (Fig. 1C). Of the nine compounds tested, we found that the majority of modifications at positions 4, 5, 6, and 8 of purines and pyrimidines did not significantly reduce incorporation by POLRMT (Fig. 2C). Similarly, NTP analogs containing bulky modifications (such as 8-azido-ATP, 4-thio-UTP, and 5-bromo-UTP) were readily incorporated by POLRMT. The combined presence of 2′-C-methyl and 2-fluoro modifications resulted in poor POLRMT incorporation (12% of that of the natural ATP substrate). We reported previously on the anti-HCV activity of this compound (23); the phosphoramidate prodrug of 2′-C-methyl-2-fluoro-ATP was found to inhibit HCV replicons with submicromolar activity.
The addition of the NHOH chemical group at position 4 of the pyrimidine ring (yielding 4-N-OH-CTP) was observed to reduce incorporation by POLRMT to 50% of that of CTP (Fig. 2C). The addition of the 2′-C-methyl group further reduced incorporation, to 20% of that of CTP. Both 4-N-OH-CTP and 2′-C-methyl-4-N-OH-CTP demonstrated anti-HCV activity in cell culture (24, 25).
Incorporation of N1-methyl-GTP by POLRMT.
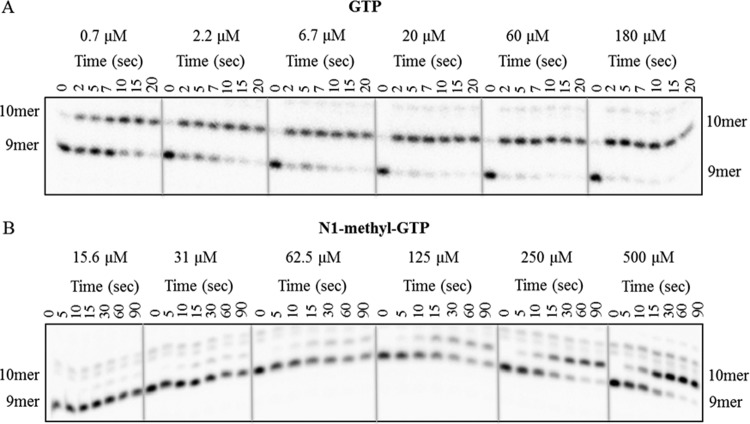
We identified N1-methyl-GTP as a NTP analog with minimal incorporation by POLRMT. Considering that the addition of the N1-methyl group on the base moiety reduced POLRMT incorporation to 10% of that of the natural GTP substrate (Fig. 2C), N1-methyl-GTP was selected for further analysis with regard to incorporation by viral nonstructural protein 5B (NS5B) RNA polymerase. NS5B incorporation assays were performed as described previously (17). NS5B-RNA/RNA complexes were incubated with increasing concentrations of GTP or N1-methyl-GTP. Single-nucleotide incorporation was observed over time and visualized on a denaturing polyacrylamide gel (Fig. 3), with the 5′-radiolabeled 9-mer primer extended to a 10-mer product. As expected, the natural GTP substrate was rapidly incorporated, with an apparent dissociation constant (Kd,app) of 4.1 ± 1.6 μM (Fig. 3A). Conversely, at least 125 μM N1-methyl-GTP was required for 50% incorporation by the HCV NS5B enzyme (Kd,app of >100 μM), suggesting compromised incorporation, compared to natural GTP (Fig. 3B).
FIG 3.
Incorporation profile for N1-methyl-GTP. (A) Increasing concentrations of GTP (0.7 μM to 180 μM) were incubated with a preformed NS5B-RNA/RNA complex, and nucleotide extension was measured over time. The extended 10-mer RNA product was visualized on a 20% denaturing acrylamide gel. Rates of incorporation at various nucleotide concentrations were plotted as described previously, in order to obtain an apparent dissociation constant (Kd,app) value of 4.1 ± 1.6 μM for GTP (17). (B) Increasing concentrations of N1-methyl-GTP (15.6 μM to 500 μM) were incubated with the preformed NS5B-RNA/RNA complex as described above. The Kd,app value for N1-methyl-GTP was estimated to be >100 μM.
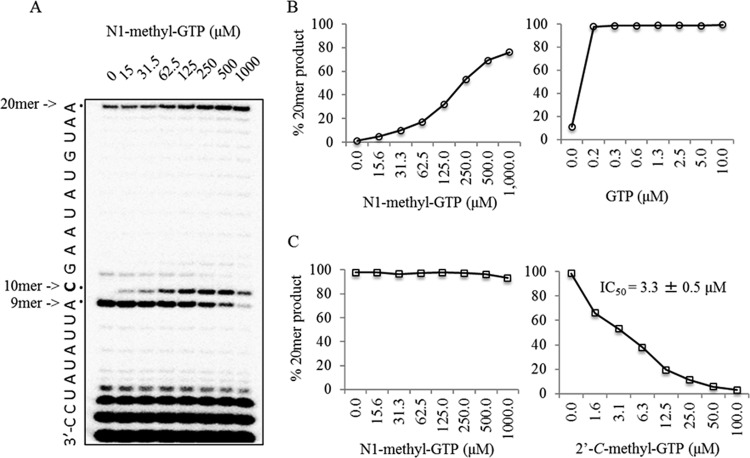
We next asked whether RNA extension could occur after N1-methyl-GTP incorporation by the NS5B enzyme. NAI-TP incorporation was assessed in the presence of 10 μM CTP, ATP, and UTP and increasing amounts of N1-methyl-GTP (Fig. 4A). We found that, as N1-methyl-GTP levels increased, increases in full-length 20-mer product levels were observed, suggesting that nucleotide extension can occur following N1-methyl-GTP incorporation (Fig. 4B, left). As expected, full-length RNA synthesis was more readily observed with low levels of GTP (Fig. 4B, right). Overall, these data suggested that, although N1-methyl-GTP was a poor substrate for the NS5B enzyme, its incorporation did not result in RNA chain termination. We next asked whether N1-methyl-GTP could inhibit RNA polymerization in the presence of competing GTP. We found that, when 1 μM GTP was present in addition to 10 μM CTP, ATP, and UTP, N1-methyl-GTP had no effect on full-length RNA synthesis (Fig. 4C). This is in contrast to the control compound 2′-C-methyl-GTP, which inhibited RNA synthesis with a 50% inhibitory concentration (IC50) of 3.3 ± 0.5 μM (Fig. 4C). Finally, we asked whether N1-methyl-GTP might inhibit viral genome synthesis in a manner that might not be detectable in our in vitro assay. To address this issue, we chemically synthesized a phosphoramidate prodrug of N1-methyl-GTP for cell culture testing (Fig. 5). We observed no inhibition when up to 200 μM N1-methyl-GTP phosphoramidate prodrug was incubated with HCV genotype 1b replicon cells (see reference 24 for the experimental methods). Further experiments suggested that this prodrug was not efficiently triphosphorylated intracellularly (data not shown). Together, these data suggest that, although N1-methyl-GTP may be a safe NAI-TP with regard to off-target incorporation by human POLRMT, it will likely not be effective as an anti-HCV agent. We also did not detect any antiviral activity against chikungunya virus, influenza A virus, or respiratory syncytial virus with testing at up to 100 μM. It remains to be determined whether this compound can be of interest as an antiviral agent for other RNA viruses.
FIG 4.
Effect of N1-methyl-GTP incorporation on NS5B-mediated RNA extension. (A) Increasing concentrations of N1-methyl-GTP were incubated with the NS5B enzyme, 5′-radiolabeled GG primer, and a 20-mer RNA template in the presence of 10 μM ATP, CTP, and UTP. RNA synthesis was allowed to proceed for 2 h at 30°C. In the absence of N1-methyl-GTP (lane 0), a strong pausing site was observed at position 9, while small amounts of full-length 20-mer RNA product accumulated as a result of nucleotide misincorporation. Increasing N1-methyl-GTP concentrations were correlated with the appearance of a 10-mer band (site of G incorporation) and the full-length 20-mer product. (B) Amounts of 20-mer product accumulation were plotted as a function of the N1-methyl-GTP concentration (left). Data from parallel experiments with increasing concentrations of GTP were also plotted (right). (C) RNA synthesis was monitored as described above, with the addition of 1 μM GTP in the presence of increasing concentrations of N1-methyl-GTP (left) or the control inhibitor 2′-C-methyl-GTP (right). No inhibition of RNA synthesis was observed with up to 1,000 μM N1-methyl-GTP, while 2′-C-methyl-GTP inhibited RNA synthesis with an IC50 of 3.3 ± 0.5 μM (average ± SD of two separate experiments).
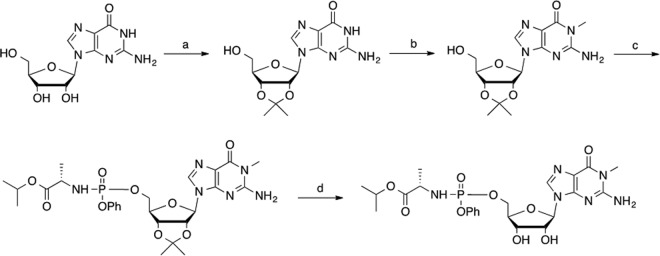
FIG 5.
Chemical synthesis of the N1-methyl-G phosphoramidate prodrug. Step a, p-toluenesulfonic acid and 2,2-dimethoxypropane in acetone, overnight at room temperature (yield, 93%); step b, NaH and MeI in dimethyl sulfoxide, overnight at room temperature (yield, 90%); step c, phosphorochloridate and N-methylimidazole in tetrahydrofuran-acetonitrile, 3 h at room temperature (yield, 91%); step d, 85% trifluoroacetic acid, 1 h at room temperature (yield, 86%).
In conclusion, in this study we examined the in vitro nucleotide substrate specificity of POLRMT. To our surprise, we found that the POLRMT active site was relatively tolerant in incorporating most of the NTP analogs tested. Several anti-HCV NAI-TPs were identified as poor substrates for POLRMT. In conclusion, the information on the NAI-TP incorporation profile of POLRMT presented herein sheds light on the biochemical properties of this enzyme active site and can inform future ribonucleotide analog drug design for all RNA viruses.
ACKNOWLEDGMENTS
This work was supported in part by NIH grant 5P30-AI-50409 (Center for AIDS Research) (to R.F.S.). M.E. is the recipient of a CIHR-NCRTP postdoctoral fellowship award and an American Liver Foundation postdoctoral fellowship award.
We thank Sijia Tao for performing the cellular pharmacology studies and Louise McCormick for conducting the influenza virus and respiratory syncytial virus assays.
REFERENCES
- 1.Menendez-Arias L, Alvarez M, Pacheco B. 2014. Nucleoside/nucleotide analog inhibitors of hepatitis B virus polymerase: mechanism of action and resistance. Curr Opin Virol 8:1–9. doi: 10.1016/j.coviro.2014.04.005. [DOI] [PubMed] [Google Scholar]
- 2.Das K, Arnold E. 2013. HIV-1 reverse transcriptase and antiviral drug resistance. Part 1. Curr Opin Virol 3:111–118. doi: 10.1016/j.coviro.2013.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Food and Drug Administration. 2013. Approval of Sovaldi (sofosbuvir) tablets for the treatment of chronic hepatitis C. Food and Drug Administration, Silver Spring, MD: http://www.fda.gov/forpatients/illness/hepatitisbc/ucm377920.htm. [Google Scholar]
- 4.Sofia MJ. 2013. Nucleotide prodrugs for the treatment of HCV infection. Adv Pharmacol 67:39–73. doi: 10.1016/B978-0-12-405880-4.00002-0. [DOI] [PubMed] [Google Scholar]
- 5.Pradere U, Garnier-Amblard EC, Coats SJ, Amblard F, Schinazi RF. 2014. Synthesis of nucleoside phosphate and phosphonate prodrugs. Chem Rev 114:9154–9218. doi: 10.1021/cr5002035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coats SJ, Garnier-Amblard EC, Amblard F, Ehteshami M, Amiralaei S, Zhang H, Zhou L, Boucle SR, Lu X, Bondada L, Shelton JR, Li H, Liu P, Li C, Cho JH, Chavre SN, Zhou S, Mathew J, Schinazi RF. 2014. Chutes and ladders in hepatitis C nucleoside drug development. Antiviral Res 102:119–147. doi: 10.1016/j.antiviral.2013.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pollack A, Gale J. 2012. BMS suspends study of nucleotide BMS094 formerly INX189. http://www.natap.org/2012/HCV/080312_01.htm.
- 8.Sheridan C. 2012. Calamitous HCV trial casts shadow over nucleoside drugs. Nat Biotechnol 30:1015–1016. doi: 10.1038/nbt1112-1015. [DOI] [PubMed] [Google Scholar]
- 9.Ahmad T, Yin P, Saffitz J, Pockros PJ, Lalezari J, Shiffman M, Freilich B, Zamparo J, Brown K, Dimitrova D, Kumar M, Manion D, Heath-Chiozzi M, Wolf R, Hughes E, Muir AJ, Hernandez AF. 2015. Cardiac dysfunction associated with a nucleotide polymerase inhibitor for treatment of hepatitis C. Hepatology 62:409–416. doi: 10.1002/hep.27488. [DOI] [PubMed] [Google Scholar]
- 10.Kohler JJ, Lewis W. 2007. A brief overview of mechanisms of mitochondrial toxicity from NRTIs. Environ Mol Mutagen 48:166–172. doi: 10.1002/em.20223. [DOI] [PubMed] [Google Scholar]
- 11.Dykens JA, Marroquin LD, Will Y. 2007. Strategies to reduce late-stage drug attrition due to mitochondrial toxicity. Expert Rev Mol Diagn 7:161–175. doi: 10.1586/14737159.7.2.161. [DOI] [PubMed] [Google Scholar]
- 12.Bailey CM, Anderson KS. 2010. A mechanistic view of human mitochondrial DNA polymerase γ: providing insight into drug toxicity and mitochondrial disease. Biochim Biophys Acta 1804:1213–1222. doi: 10.1016/j.bbapap.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sohl CD, Kasiviswanathan R, Kim J, Pradere U, Schinazi RF, Copeland WC, Mitsuya H, Baba M, Anderson KS. 2012. Balancing antiviral potency and host toxicity: identifying a nucleotide inhibitor with an optimal kinetic phenotype for HIV-1 reverse transcriptase. Mol Pharmacol 82:125–133. doi: 10.1124/mol.112.078758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arnold JJ, Sharma SD, Feng JY, Ray AS, Smidansky ED, Kireeva ML, Cho A, Perry J, Vela JE, Park Y, Xu Y, Tian Y, Babusis D, Barauskus O, Peterson BR, Gnatt A, Kashlev M, Zhong W, Cameron CE. 2012. Sensitivity of mitochondrial transcription and resistance of RNA polymerase II dependent nuclear transcription to antiviral ribonucleosides. PLoS Pathog 8:e1003030. doi: 10.1371/journal.ppat.1003030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feng JY, Xu Y, Barauskas O, Perry JK, Ahmadyar S, Stepan G, Yu H, Babusis D, Park Y, McCutcheon K, Perron M, Schultz BE, Sakowicz R, Ray AS. 2016. Role of the mitochondrial RNA polymerase in the toxicity of nucleotide inhibitors of the hepatitis C virus. Antimicrob Agents Chemother 60:806–817. doi: 10.1128/AAC.01922-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fenaux M, Lin X, Yokokawa F, Sweeney Z, Saunders O, Xie L, Lim SP, Uteng M, Uehara K, Warne R, Gang W, Jones C, Yendluri S, Gu H, Mansfield K, Boisclair J, Heimbach T, Catoire A, Bracken K, Weaver M, Moser H, Zhong W. 2016. Antiviral nucleotide incorporation by recombinant human mitochondrial RNA polymerase is predictive of increased in vivo mitochondrial toxicity risk. Antimicrob Agents Chemother 60:7077–7085. doi: 10.1128/AAC.01253-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ehteshami M, Tao S, Ozturk T, Zhou L, Cho JH, Zhang H, Amiralaei S, Shelton JR, Lu X, Khalil A, Domaoal RA, Stanton RA, Suesserman JE, Lin B, Lee SS, Amblard F, Whitaker T, Coats SJ, Schinazi RF. 2016. Biochemical characterization of the active anti-hepatitis C virus metabolites of 2,6-diaminopurine ribonucleoside prodrug compared to sofosbuvir and BMS-986094. Antimicrob Agents Chemother 60:4659–4669. doi: 10.1128/AAC.00318-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou L, Zhang HW, Tao S, Bassit L, Whitaker T, McBrayer TR, Ehteshami M, Amiralaei S, Pradere U, Cho JH, Amblard F, Bobeck D, Detorio M, Coats SJ, Schinazi RF. 2015. β-D-2′-C-Methyl-2,6-diaminopurine ribonucleoside phosphoramidates are potent and selective inhibitors of hepatitis C virus (HCV) and are bioconverted intracellularly to bioactive 2,6-diaminopurine and guanosine 5′-triphosphate forms. J Med Chem 58:3445–3458. doi: 10.1021/jm501874e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eldrup AB, Allerson CR, Bennett CF, Bera S, Bhat B, Bhat N, Bosserman MR, Brooks J, Burlein C, Carroll SS, Cook PD, Getty KL, MacCoss M, McMasters DR, Olsen DB, Prakash TP, Prhavc M, Song Q, Tomassini JE, Xia J. 2004. Structure-activity relationship of purine ribonucleosides for inhibition of hepatitis C virus RNA-dependent RNA polymerase. J Med Chem 47:2283–2295. doi: 10.1021/jm030424e. [DOI] [PubMed] [Google Scholar]
- 20.Migliaccio G, Tomassini JE, Carroll SS, Tomei L, Altamura S, Bhat B, Bartholomew L, Bosserman MR, Ceccacci A, Colwell LF, Cortese R, De Francesco R, Eldrup AB, Getty KL, Hou XS, LaFemina RL, Ludmerer SW, MacCoss M, McMasters DR, Stahlhut MW, Olsen DB, Hazuda DJ, Flores OA. 2003. Characterization of resistance to non-obligate chain-terminating ribonucleoside analogs that inhibit hepatitis C virus replication in vitro. J Biol Chem 278:49164–49170. doi: 10.1074/jbc.M305041200. [DOI] [PubMed] [Google Scholar]
- 21.Yin Z, Chen YL, Schul W, Wang QY, Gu F, Duraiswamy J, Kondreddi RR, Niyomrattanakit P, Lakshminarayana SB, Goh A, Xu HY, Liu W, Liu B, Lim JY, Ng CY, Qing M, Lim CC, Yip A, Wang G, Chan WL, Tan HP, Lin K, Zhang B, Zou G, Bernard KA, Garrett C, Beltz K, Dong M, Weaver M, He H, Pichota A, Dartois V, Keller TH, Shi PY. 2009. An adenosine nucleoside inhibitor of dengue virus. Proc Natl Acad Sci U S A 106:20435–20439. doi: 10.1073/pnas.0907010106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smidansky ED, Arnold JJ, Reynolds SL, Cameron CE. 2011. Human mitochondrial RNA polymerase: evaluation of the single-nucleotide-addition cycle on synthetic RNA/DNA scaffolds. Biochemistry 50:5016–5032. doi: 10.1021/bi200350d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou L, Zhang H, Tao S, Ehteshami M, Cho JH, McBrayer TR, Tharnish P, Whitaker T, Amblard F, Coats SJ, Schinazi RF. 2016. Synthesis and evaluation of 2,6-modified purine 2′-C-methyl ribonucleosides as inhibitors of HCV replication. ACS Med Chem Lett 7:17–22. doi: 10.1021/acsmedchemlett.5b00402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stuyver LJ, Whitaker T, McBrayer TR, Hernandez-Santiago BI, Lostia S, Tharnish PM, Ramesh M, Chu CK, Jordan R, Shi J, Rachakonda S, Watanabe KA, Otto MJ, Schinazi RF. 2003. Ribonucleoside analogue that blocks replication of bovine viral diarrhea and hepatitis C viruses in culture. Antimicrob Agents Chemother 47:244–254. doi: 10.1128/AAC.47.1.244-254.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ehteshami M, Tao S, Amiralaei S, Li H, Lu X, Ozturk T, Amblard F, McBrayer T, Whitaker T, Coats SJ, Schinazi RF. 2015. Biochemical characterization of potent inhibitors of hepatitis C virus polymerase generated by β-D-2′-C-methyl-4-N-hydroxycitidine nucleoside prodrug, p 21. Abstr 4th Can Symp HCV. http://www.canhepc.ca/sites/default/files/media/documents/4th_cshcv_abstract_book_2015.pdf.