ABSTRACT
Here we describe the spread of colistin resistance in clinical isolates of carbapenem-resistant Klebsiella pneumoniae in Medellín, Colombia. Among 32 isolates collected between 2012 and 2014, 24 showed genetic alterations in mgrB. Nineteen isolates belonged to sequence type 512 (ST512) (or its single locus variant [SLV]) and harbored an 8.1-kb hsdMSR insertion corresponding to ISKpn25, indicating a clonal expansion of the resistant strain. The insertion region showed 100% identity to several plasmids, suggesting that the colistin resistance is mediated by chromosomal integration of plasmid DNA.
KEYWORDS: Klebsiella pneumoniae, carbapenem resistance, colistin resistance, mgrB
TEXT
Resistance to colistin in Klebsiella pneumoniae is frequently associated with the inactivation of MgrB, a small protein that acts as a negative regulator of the PhoP/PhoQ two-component system. The upregulation of this pathway leads to lipopolysaccharide (LPS) modifications by the addition of phosphoethanolamine, 4-amino-4-arabinose or 2-hydroxymyristate to lipid A (1), resulting in the overall decrease in negative charge and the decrease in affinity of colistin to its target (2). Genotypic analysis of the mgrB gene regulator among colistin-resistant isolates revealed a high degree of plasticity and genetic disruptions (3, 4). Complementation experiments with a cloned wild-type mgrB gene restored susceptibility to the drug and reduced PhoQ and PmrK expression in resistant isolates to colistin-susceptible levels, in support of the role of mgrB in colistin resistance (2–4).
Here we describe the emergence and clonal expansion of a colistin- and carbapenem-resistant K. pneumoniae strain in Medellín, Colombia, where a unique disruption of the mgrB gene was caused by a chromosomal insertion of ISKpn25.
In a study conducted in four referral centers between 2012 and 2014 in Medellín, Colombia, we found 32 patients infected with carbapenem- and colistin-resistant K. pneumoniae, of which 27 belonged to sequence type 512 (ST512) or its single locus variant (SLV) (Table 1). Genotyping the mgrB gene among the 32 K. pneumoniae isolates showed that 8 isolates carried the wild-type gene, 1 isolate had a stop codon, another isolate had a frameshift, and 3 isolates had insertion sequence (IS) insertions; whereas 19 isolates were negative for the PCR amplification of mgrB (all belonged to ST512 or its SLV) (Table 1). Three mgrB-PCR negative ST512 (K. pneumoniae carbapenemase [KPC]-3 positive) isolates and one colistin-susceptible ST512 K. pneumoniae (KPC-3 positive) isolate were then selected for whole-genome sequencing analysis. Genome sequencing was done using the platform NextSeq (Illumina). The quality of the readings (150-bp insert size) was assessed using FastQC software (www.bioinformatics.babraham.ac.uk/projects/), and de novo assembly was done using SPAdes assembler (5). The plasmid replicons and resistance genes were mined using PlasmidFinder (6) and ResFinder (7), respectively. The sequence of mgrB and flanking regions reported in GenBank by Cannatelli (3) (GenBank accession number AVFC01000053, 155,512-155,655; 253 bp) was used to locate the gene in the de novo assembly. The whole-genome sequences (WGS) of four K. pneumoniae isolates have been deposited into GenBank under Bioproject PRJNA353361. In addition, two sets of primers (junction 1: a-F 5′-GCTCAGCCACATCCTCTTTC-3′ and c-R 5′-CTTGAAGTCACCCCACAGGT-3′, junction 2: d-F 5′-CCAGAAGCAGCTTTTTGTCC-3′ and b-R 5′-CCGCTCATTAATACGCCAAT-3′) targeting the junctions between chromosomal and plasmid regions identified in the whole-genome sequencing analysis were designed to investigate the presence of this element. The PCR products for both junctions were sequenced in both the sense and anti-sense directions. pKpQIL-like plasmids were screened by PCR (8), and blaKPC plasmid identification was confirmed by S1-pulsed-field gel electrophoresis (PFGE) and blaKPC-probe hybridization in one strain (KL027).
TABLE 1.
Characteristics of colistin- and carbapenem-resistant K. pneumoniae isolates from Medellín, Colombia, 2012–2014a
| Isolate | Hospital | Date of sampling (month/year) | ST | KPC variant | mgrB | Coresistance | Infection type |
|---|---|---|---|---|---|---|---|
| KL004 | C | 6/2012 | 512 | KPC-3 | ISKpn25 | Amk-Cip | Urinary tract |
| KL008 | C | 7/2012 | 512 | KPC-3 | ISKpn25 | Amk-Cip-Tig | Intraabdominal |
| KL011 | C | 8/2012 | 512 | KPC-3 | ISKpn25 | Amk-Cip-Tig | Urinary tract |
| KL015 | C | 11/2012 | 512 | KPC-3 | ISKpn25 | Amk-Cip-Tig | Urinary tract |
| KL026 | C | 3/2013 | 512 | KPC-3 | ISKpn25 | Amk-Cip-Tig | Urinary tract |
| KL027 | C | 3/2013 | 512 | KPC-3 | ISKpn25 | Amk-Gen-Cip-Tig | Surgical site |
| KL029 | C | 4/2013 | 512 | KPC-3 | ISKpn25 | Amk-Gen-Cip-Tig | Pneumonia |
| KL030 | C | 4/2013 | 512 | KPC-3 | ISKpn25 | Amk-Gen-Cip | Surgical site |
| KL038 | C | 7/2013 | 512 | KPC-3 | ISKpn25 | Amk-Cip-Tig | Bacteremia |
| KL041 | C | 8/2013 | 512 | KPC-3 | ISKpn25 | Amk-Cip-Tig | Bacteremia |
| KL043 | C | 10/2013 | 512 | KPC-3 | ISKpn25 | Amk-Gen-Cip-Tig | Intraabdominal |
| KL046 | C | 10/2013 | 512 | KPC-3 | ISKpn25 | Gen-Cip-Tig | Bacteremia |
| KL050 | C | 11/2013 | 512 | KPC-3 | ISKpn25 | Amk-Gen-Cip-Tig | Urinary tract |
| KL051 | C | 12/2013 | 512 | KPC-3 | ISKpn25 | Amk-Gen-Cip-Tig | Bacteremia |
| KL052 | C | 1/2014 | 512 | KPC-3 | ISKpn25 | Amk-Gen-Cip | Bacteremia |
| KL053 | C | 1/2014 | 512 | KPC-3 | ISKpn25 | Amk-Gen-Cip-Tig | Urinary tract |
| KL056 | C | 2/2014 | 512 | KPC-2 | ISKpn25 | Amk-Gen-Cip-Tig | Urinary tract |
| KL061 | C | 5/2014 | 512 | KPC-3 | ISKpn25 | Amk-Cip-Tig | Urinary tract |
| KL064 | C | 5/2014 | 868b | KPC-3 | ISKpn25 | Amk-Gen-Cip-Tig | Intraabdominal |
| KP018 | B | 11/2012 | 512 | KPC-3 | Frameshift | Amk-Gen-Cip-Tig | Osteomyelitis |
| KP032 | B | 4/2013 | 14 | KPC-2 | Stop codon | Gen-Cip-Tig | Intraabdominal |
| KL014 | C | 11/2012 | 512 | KPC-3 | IS5-like (+74 nt) | Amk-Cip-Tig | Urinary tract |
| KP001 | B | 6/2012 | 512 | KPC-3 | IS5-like (+74 nt) | Amk-Cip-Tig | Urinary tract |
| KP008 | B | 9/2012 | ND | Negative | IS903 (+83 nt) | Cip-Tig | Intraabdominal |
| KH036 | A | 1/2014 | 512 | KPC-3 | WT | Amk-Gen-Cip-Tig | Urinary tract |
| KL016 | C | 11/2012 | 512 | KPC-3 | WT | Amk-Cip-Tig | Urinary tract |
| KL039 | C | 7/2013 | 512 | KPC-3 | WT | Amk-Cip | Bacteremia |
| KL045 | C | 10/2013 | 512 | KPC-3 | WT | Amk-Gen-Cip-Tig | Intraabdominal |
| KP007 | B | 8/2012 | 1708 | KPC-2 | WT | Ciip | Intraabdominal |
| KP011 | B | 9/2012 | 636 | Negative | WT | Gen-Cip-Tig | Bacteremia |
| KP025 | B | 1/2013 | 1706 | KPC-2 | WT | Cip-Tig | Intraabdominal |
| KP028 | B | 12/2012 | 512 | KPC-3 | WT | Amk-Cip | Urinary tract |
ST, sequence type; WT, wild type; Amk, amikacin; Gen, gentamicin; Cip, ciprofloxacin; Tig, tigecycline.
Single-locus variant of ST512.
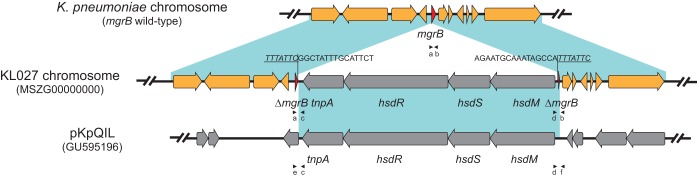
Whole-genome sequence analysis of 3 (KL027, GenBank accession number MSZG00000000; KL015, GenBank accession number MSZL00000000; and KL064, GenBank accession number MSZM00000000) of the 19 PCR-negative mgrB isolates identified an 8.1-kb fragment insertion into the mgrB gene. No additional mutations were detected in other genes associated with colistin resistance (phoPQ, pmrCAB, and crrAB) when they were compared to a colistin-susceptible strain (KL033, GenBank accession number MSZH00000000). Analyses of the inserted region identified four open reading frames (ORFs), corresponding to predicted proteins from a type I restriction-methylation system (hsdM, hsdS, hsdR) and one corresponding to a transposase (tnpA). A BLAST search of the inserted region in ISfinder (9) revealed that this element corresponded to ISKpn25. Further PCR and Sanger sequencing, targeting the chromosome insertion junction, showed that all 19 mgrB “negative” isolates carried the same ISKpn25 insertion. This insertion showed >99% identity and query coverage to a number of plasmids in GenBank, ranging in size from 48 to 307 kb, including the blaKPC-harboring plasmid pKpQIL which has been frequently identified in ST258 and ST512 isolates. In our study, pKpQIL-like plasmids were detected in 79% (15/19) of isolates harboring the ISKpn25 insertion. The presence of plasmid-borne ISKpn25 was further examined by two primers (junction 3: e-F: CCTACTGGATCCGTGTCGTT, c-R 5′-CTTGAAGTCACCCCACAGGT; junction 4: d-F 5′-CCAGAAGCAGCTTTTTGTCC-3′; f-R: TTTCCCCTGGCATAAACAAC) targeting the junctions between the 8.1-kb hsdMSR locus and the plasmid backbone (Fig. 1). The results showed that all 15 isolates carrying pKpQIL-like plasmids harbored a second copy of ISKpn25 on the plasmids. Moreover, S1-PFGE and blaKPC-hybridization to KL027 showed that blaKPC was present in a 113-kb plasmid, consistent with the size reported for pKpQIL. Plasmid analysis revealed sequences from incompatibility groups IncFII(K), IncFIB(K), IncFIB (pQIL), and IncX3 of plasmids pKpQIL, pKpn3, and pIncX. In addition to KPC genes, other resistant genes were detected, including those for TEM, SHV, and OXA beta-lactamases; aminoglycoside-, fluoroquinolone-, and phenicol-modifying enzymes; and genes conferring resistance to fosfomycin and quinolones. It is clinically significant to note that the 19 isolates were all the same ST (or its SLV) and were recovered from the same hospital between June 2012 and May 2014; also, only one patient had a history of using colistin previously. Additionally, PFGE examination showed that these isolates had indistinguishable profiles, in support of the clonal expansion of this strain.
FIG 1.
Schematic representation of the plasmid sequence (ISKpn25) inserted into mgrB in the KL027 K. pneumoniae chromosome and comparison of the inserted region to plasmid pKpQIL. Direct repeats are underlined and inverted repeats are in italic type. Primers amplifying mgrB (a b), the junctions between the chromosome and the 8.1-kb element (a c and d b), and the junctions between the plasmid backbone and the 8.1-kb element (e c and d f) are indicated by black arrowheads.
Insertional inactivation of mgrB gene has been shown to be a predominant cause of colistin resistance in K. pneumoniae. The role of IS elements in the inactivation of mgrB gene was described first by Cannatelli et al. (2) in one colistin resistant KPC-3 producing K. pneumoniae strain belonging to ST258 and selected in vivo after multiple rounds of antibiotic therapy, including colistin. The inactivation proved to be an insertion of an IS5-like element at nucleotide 75 in the mgrB gene, similar to two strains in our collection. Additional studies have confirmed the role of several IS sequences disrupting mgrB (IS5-like, IS1F and ISKpn14, ISKpn13 and IS10R) in colistin resistance (3, 4).
In this study, we described the largest insertional inactivation of mgrB reported, which includes an 8.1-kb element corresponding to ISKpn25. Complementation experiments with the wild-type mgrB gene are still needed to confirm that this insertion and the disruption of mgrB is the cause of colistin resistance. However, both the current literature regarding IS-element disruptions of the mgrB gene among colistin-resistant strains and whole-genome sequencing failing to find any mutations in the numerous two-component regulatory genes associated with colistin resistance give support to our hypothesis that colistin resistance is likely due to the ISKpn25 insertion in the mgrB gene.
The ISKpn25 region had an hsdMSR locus that encodes a type I restriction-modification (R-M) system. Most R-M systems are present in the bacterial accessory genome, and they are also frequently found associated with mobile elements, such as plasmids, phages, conjugative elements, transposons, and integrons (10). Our results showed that the ISKpn25 is present in several plasmids, including blaKPC-harboring plasmids, such as pKpQIL. The presence of two copies of ISKpn25 on the chromosome and the plasmid suggested that the chromosomal insertion of the hsdMSR locus in mgrB was likely acquired from the plasmid through transposition or homologous recombination. Similarly, it has been suggested ISKpn25 is involved in DNA rearrangements among pKpQIL-like and possibly other plasmids (11). An additional study showed that a mobile element containing the carbapenemase OXA-181 disrupted the mgrB gene in a carbapenem- and colistin-resistant K. pneumoniae isolate from a patient from the United Arab Emirates (12). In the same isolate, a copy of a second element harboring CTX-M-15 (ISEcp1-blaCTX-M-15) was inserted into ompK35, inactivating the gene. These findings underscore the potential role of mobile elements associated with carbapenemase genes in the evolution of colistin-resistant clones.
In addition, the presence of the same insertion element in a cluster of 19 isolates from the same clonal group isolated from the same hospital support the hypothesis that this spread was the result of clonal expansion and that the inactivation of the mgrB gene was a unique event. Consistent with the clonal spread reported in this study, large outbreaks of colistin-resistant, KPC-3-harboring K. pneumoniae from ST258 and ST512 (SLV of ST258) were reported in Italy, Greece, and the United States (13, 14), and at least one of the outbreaks was caused by the clonal expansion of a mutant with a deletion of 11 bp in the mgrB coding sequence (15).
The repeated observation that the mgrB gene is a hot spot for insertions and that its loss is associated with colistin resistance, plus the recent finding that mcr-1 genes that confer colistin resistance are plasmid encoded (16), raises significant clinical and public health issues in a background where the pipeline for new and novel antibiotics is limited. As a consequence, close monitoring of colistin- and carbapenem- resistant strains should be maintained to control their spread.
Accession number(s).
Whole-genome sequence analysis of 3 PCR-negative mgrB isolates, KL027, KL015, and KL064, have been deposited at GenBank under the respective accession numbers MSZG00000000, MSZL00000000, and MSZM00000000. A colistin-susceptible strain, KL033, has been deposited in GenBank under the accession number MSZH00000000.
ACKNOWLEDGMENTS
This work was supported by Colciencias project 111565741641 (to J.N.J.). This work was also supported by the National Institute of Allergy and Infectious Diseases (grants R01AI090155 [to B.N.K.] and R21AI117338 [to L.C.]).
We declare no competing interests.
REFERENCES
- 1.Kidd TJ, Mills G, Sá-Pessoa J, Dumigan A, Frank CG, Insua JL, Ingram R, Hobley L, Bengoechea JA. 2017. A Klebsiella pneumoniae antibiotic resistance mechanism that subdues host defences and promotes virulence. EMBO Mol Med 9:430–447. doi: 10.15252/emmm.201607336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cannatelli A, D'Andrea MM, Giani T, Di Pilato V, Arena F, Ambretti S, Gaibani P, Rossolini GM. 2013. In vivo emergence of colistin resistance in Klebsiella pneumoniae producing KPC-type carbapenemases mediated by insertional inactivation of the PhoQ/PhoP mgrB regulator. Antimicrob Agents Chemother 57:5521–5526. doi: 10.1128/AAC.01480-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cannatelli A, Giani T, D'Andrea MM, Pilato Di V, Arena F, Conte V, Tryfinopoulou K, Vatopoulos A, Rossolini GM. 2014. MgrB inactivation is a common mechanism of colistin resistance in KPC-producing Klebsiella pneumoniae of clinical origin. Antimicrob Agents Chemother 58:5696–5703. doi: 10.1128/AAC.03110-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Poirel L, Jayol A, Bontron S, Villegas MV, Ozdamar M, Turkoglu S, Nordmann P. 2015. The mgrB gene as a key target for acquired resistance to colistin in Klebsiella pneumoniae. J Antimicrob Chemother 70:75–80. doi: 10.1093/jac/dku323. [DOI] [PubMed] [Google Scholar]
- 5.Nurk S, Bankevich A, Antipov D, Gurevich AA, Korobeynikov A, Lapidus A, Prjibelski AD, Pyshkin A, Sirotkin A, Sirotkin Y, Stepanauskas R, Clingenpeel SR, Woyke T, McLean JS, Lasken R, Tesler G, Alekseyev MA, Pevzner PA. 2013. Assembling single-cell genomes and mini-metagenomes from chimeric MDA products. J Comput Biol 20:714–737. doi: 10.1089/cmb.2013.0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carattoli A, Zankari E, Garciá-Fernández A, Larsen MV, Lund O, Villa L, Aarestrup FM, Hasman H. 2014. In silico detection and typing of plasmids using Plasmid Finder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58:3895–3903. doi: 10.1128/AAC.02412-14.24777092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen L, Chavda KD, Melano RG, Jacobs MR, Koll B, Hong T, Rojtman AD, Levi MH, Bonomo RA, Kreiswirtha BN. 2014. Comparative genomic analysis of KPC-encoding pKpQIL-like plasmids and their distribution in New Jersey and New York hospitals. Antimicrob Agents Chemother 58:2871–2877. doi: 10.1128/AAC.00120-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M. 2006. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res 2006:D32–D36. doi: 10.1093/nar/gkj014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Furuta Y, Kobayashi I. 2013. Restriction-modification systems as mobile epigenetic elements, p 1–19. In Roberts AP, Mullany P (ed), Bacterial integrative mobile genetic elements. Landes Bioscience, Austin, TX. [Google Scholar]
- 11.He S, Chandler M, Varani AM, Hickman AB, Dekker JP, Dyda F. 2016. Mechanisms of evolution in high-consequence drug resistance. MBio 2016:e01987-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zowawi HM, Forde BM, Alfaresi M, Alzarouni A, Farahat Y, Chong T-M, Yin W-F, Chan K-G, Li J, Schembri MA, Beatson SA, Paterson DL. 2015. Stepwise evolution of pandrug-resistance in Klebsiella pneumoniae. Sci Rep 5:15082. doi: 10.1038/srep15082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marchaim D, Chopra T, Pogue JM, Perez F, Hujer AM, Rudin S, Endimiani A, Navon-Venezia S, Hothi J, Slim J, Blunden C, Shango M, Lephart PR, Salimnia H, Reid D, Moshos J, Hafeez W, Bheemreddy S, Chen TY, Dhar S, Bonomo RA, Kaye KS. 2011. Outbreak of colistin-resistant, carbapenem-resistant Klebsiella pneumoniae in metropolitan Detroit, Michigan. Antimicrob Agents Chemother 55:593–599. doi: 10.1128/AAC.01020-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mezzatesta ML, Gona F, Caio C, Petrolito V, Sciortino D, Sciacca A, Santangelo C, Stefani S. 2011. Outbreak of KPC-3-producing, and colistin-resistant, Klebsiella pneumoniae infections in two Sicilian hospitals. Clin Microbiol Infect 17:1444–1447. doi: 10.1111/j.1469-0691.2011.03572.x. [DOI] [PubMed] [Google Scholar]
- 15.Giani T, Arena F, Vaggelli G, Conte V, Chiarelli A, De Angelis LH, Fornaini R, Grazzini M, Niccolini F, Pecile P, Rossolini GM. 2015. Large nosocomial outbreak of colistin-resistant, carbapenemase-producing Klebsiella pneumoniae traced to clonal expansion of an mgrb deletion mutant. J Clin Microbiol 53:3341–3344. doi: 10.1128/JCM.01017-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Y, Wang Y, Walsh TR, Yi L, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Park H. 2016. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]