Abstract
Background
Cetuximab is an epidermal growth factor receptor (EGFR)-blocking antibody that has been approved for the treatment of patients with head and neck squamous cell carcinoma (HNSCC) and metastatic colorectal cancer, but no predictive biomarkers of activity have been yet identified. Establishment of such biomarkers will help identify a subset of patients that will benefit from cetuximab therapy.
Methods
In this paper, we report on a patient with HNSCC who had a complete tumour regression following treatment with cetuximab given as a single agent after initial surgery and radiation therapy. The EGFR protein expression level, the EGFR gene copy number and the EGFR gene sequence were assessed from both normal and tumour tissues.
Results
Besides protein overexpression and gene amplification in the tumour tissue, sequencing of the EGFR gene from the patient revealed the presence of two somatic mutations, one in the kinase domain (R705G) and the other in the ligand binding domain (P546S). Cells that stably express these EGFR mutants were treated with cetuximab and their sensitivity to the drug was compared to cells expressing the wildtype gene. While P546S mutation sensitised NIH-3T3 cells to cetuximab, R705G had a marginal effect. The double mutant (P546S/R705G) behaved like the P546S mutant, indicating that the mutation in the kinase domain does not contribute to the increased sensitivity to cetuximab. No mutations were found in K-RAS or B-RAF genes and no HPV protein or DNA was detected in the tumour. This is the first report of a somatic mutation in the EGFR ligand binding domain that may contribute to increased sensitivity to cetuximab.
Conclusions
Our results support a role for the P546S mutation in cetuximab sensitivity. Other factors including EGFR protein high copy number and protein overexpression may have also contributed to the observed response. The severity of a skin rash developed by this patient and its correlation with the antitumour activity does not exclude the involvement of the immune system (i.e. complement-mediated immune response) as well. The occurrence of the P546S mutation needs to be evaluated in HNSCC, as a well as a prospective evaluation of cetuximab anti-tumour activity in patients with tumours harbouring the mutation.
Keywords: EGFR ligand-binding, Cetuximab, Head and neck squamous, cell carcinoma, Somatic mutation
1. Introduction
Head and neck squamous cell carcinoma (HNSCC) contributes to approximately 5% of all cancers in males and 2% in females in the Western world.1 Over 95% of HNSCCs express elevated epidermal growth factor receptor (EGFR) levels compared with levels in normal mucosa from patients without cancer,2 and elevated EGFR expression levels in HNSCC serve as an independent indicator of poor prognosis and decreased overall survival.3,4
EGFR is a ubiquitously expressed receptor tyrosine kinase (RTK) that is activated in response to ligand stimulation.5,6 It is a key mediator of proliferation and progression in many human tumours, and strategies to inhibit its signalling have emerged as highly promising cancer therapy approaches.7 Monoclonal antibodies (mAbs) have been developed to target the extracellular domain of EGFR and block its natural ligand binding.8–10 Cetuximab (Erbitux), an Immunoglobulin G1 monoclonal antibody directed against the extracellular domain of EGFR, prevents receptor activation and dimerisation and ultimately induces receptor internalisation and downregulation.11
Increasing evidence suggests that patients who initially respond to EGFR inhibitors may subsequently become refractory.12 The identification of catalytic domain EGFR mutations that predict response to EGFR-Tyrosine Kinase Inhibitors (TKIs) in selected lung cancer patients represents a landmark development in the EGFR field.13 Mutation in exon 21 of the EGFR-Tyrosine Kinase Domain (TKD), L858R, may predict increased sensitivity to TKIs, whereas the T790M mutation in exon 20 is associated with acquired resistance to TKI therapy.14 These recent findings suggest that patient selection may be critical for successful therapies using EGFR-TKIs.15 By contrast, no predictive biomarkers of cetuximab antitumour activity have been clearly identified.16,17
In this paper we report on a 33 year old patient with an HNSCC who achieved complete remission following treatment with cetuximab as a single agent. Analysis showed that there was EGFR gene amplification and EGFR overexpression in the tumour, and that the EGFR gene also had a somatic mutation in the EGFR-TKD (R705G) and in the ligand-binding domain (P546S). The P546S mutation seems to be involved in cell sensitivity to cetuximab in cultured cells.
2. Methods
2.1. DNA extraction and mutation analyses
We assessed the mutation status of the region corresponding to the catalytic domain of EGFR (i.e. exons 18–21) and the ligand-binding domains (i.e. exons 5 and 6 and exons 13–15). DNA was extracted from paraffin-embedded normal and tumoural tissues from haematoxylin and eosin stained sections that were analysed for detailed morphology by a pathologist. Written informed consent was obtained from the patient. Regions of tumour tissue were marked, and this tissue was extracted. After extraction, DNA was purified with Qiagen PCR purification kit (Qiagen). Exon-specific primers for EGFR were either as previously described18 for the catalytic domain of EGFR, or designed for the EGFR ligand-binding domain (exons 5 and 6 and exons 13–15) by use of Primer3 software and synthesised by IDTDNA (Iowa, United States of America (USA)) (Table 1). We also assessed the mutation status of two EGFR intracellular effectors (KRAS, B-RAF) by analysing exons where mutations occur with the highest frequencies in cancers (K-RAS exon 2, B-RAF exon 15). The nucleotide sequence corresponding to every exon was amplified from tumour-extracted genomic DNA using previously described primers,18 cloned in a TA-cloning vector (Invitrogen, CA) and sequenced using the M-13 universal primer. Conditions for the amplification of exon-specific regions from tumour genomic DNA by PCR and for the identification of mutations have been described.19
Table 1.
A list of primers used to amplify the exons that encode the ligand-binding domain of epidermal growth factor receptor (EGFR).
| Primer | Sequence |
|---|---|
| Exon5-F | 5′-CATCATTCACTGAGATATGC-3′ |
| Exon5-R | 5′-CAATCACCTAAGCAAGTGAAG-3′ |
| Exon6-F | 5′-CCTACCCTCACTCTTCAGCTC-3′ |
| Exon6-R | 5′-CACAGGAAGTCTTCTGTCCTG-3′ |
| Exon13-F | 5′-GCTCTGTCACTGACTGCTGTG-3′ |
| Exon13-R | 5′-CTATAACAACAACCTGGAGCC-3′ |
| Exon14-F | 5′-GTTCCTGCAATAATGTCTCAG-3′ |
| Exon14-R | 5′-CTGTTCGGCTTCTGTGAAGGC-3′ |
| Exon15-F | 5′-GCTTTCCCCACTCACACACAC-3′ |
| Exon15-R | 5′-CCTCGGCAATTTGTTGCCGGA-3′ |
2.2. EGFR copy number determination and protein expression
For gene copy number determination, genomic DNA was extracted from both normal and tumoural tissue samples using ZR Genomic DNA II kit (Cat. #D3024, Zymo Research). TaqMan copy number assay was performed according to the protocol from Applied Biosystems. Briefly, 20 ng of genomic DNA was transferred into a 384-well MicroAmp Optical Reaction Plate (Applied Biosystems). TaqMan Genotyping Master Mix (Product #4371355, Applied Biosystems), TaqMan Copy Number Assay for EGFR (Cat. #4400291, Applied Biosystems) and for RNaseP (Cat. #4401631, Applied Biosystems) were mixed and pipetted into the same well. Real time PCR was performed using the 7900HT Sequence Detection System (Applied Biosystems) at 95 °C for 10 min followed by 50 cycles at 95 °C for 15 s and 60 °C for 1 min. Three technical replicates were run for the normal and tumoural tissues from three different slides sectioned from the same tumour.
For the detection of EGFR protein expression in tumoural (versus normal) tissues, immunohistochemical stains were performed as part of a routine processing in the Department of Pathology at the University of Cincinnati using the standard streptavidin–biotin-peroxidase immunostaining procedure. The antibody used is a mouse monoclonal anti-EGFR antibody (Cat. #790-2988, clone 31G7; Ventana Medical Systems, Inc., Tucson, AZ).
2.3. HPV in situ hybridisation
Slides were deparaffinised and subjected to in situ hybridisation using standard DNA probes provided by Ventana Medical Systems that detect several high risk HPV types including HPV 16. Sections were incubated with a fluorescein-tagged DNA probe and counterstained using the automated Ventana BenchMark instrument (Ventana Medical System).
2.4. Genomic DNA PCR amplification of HPV 16
Total genomic DNA was isolated from Caski and SiHa which are both HPV 16 positive (integrated) cervical cancer cell lines and HeLa which is HPV 18 positive (integrated) cervical cancer cell line by phenol/chloroform extraction. PCR was subsequently performed for 35 cycles at an annealing temperature of 45 °C by using the following primers: for GAPDH, 5′-GGG AGC CAA AAG GGT CAT CA-3′ and 5′-TTT CTA GAC GGC AGG TCA GGT-3′; for HPV consensus primers, 5′-CGT CCM ARR GGA WAC TGA TC-3′ and 5′-GCM CAG GGW CAT AAY AAT GG-3′.
2.5. EGFR mutant construction and retrovirus production
Wild-type EGFR cDNA in pBabe retroviral vector was purchased from Addgene (Cambridge, MA). The point mutations were created in the wild-type EGFR gene by using QuickChange II Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA). The primers used to create the mutations are: 5′-CCT TCT GGA GGG TGA GTC AAG GGA GTT TGT GGA GAA C-3′ and 5′-GTT CTC CAC AAA CTC CCT TGA CTC ACC CTC CAG AAG G-3′ for P546S mutation and 5′-CCC AAC CAA GCT CTC TTG GGG ATC TTG AAG GAA ACT G-3′ and 5′-CAG TTT CCT TCA AGA TCC CCA AGA GAG CTT GGT TGG G-3 for R705G mutation. To create the double mutant (P546S and R705G) the latter primers were used to introduce the R705G mutation in EGFR cDNA already containing the P546S mutation. The following primers 5′-CAA AAA GAT CAA AGT GCT GAG CTC CGG TGC GTT CGG CAC-3′ and 5′-GTG CCG AAC GCA CCG GAG CTC AGC ACT TTG ATC TTT TTG-3′ were used to introduce the G719S mutation. All wildtype and mutant EGFR constructs were verified by sequencing of the full length gene.
Replication incompetent retroviruses were produced from pBabe vectors by transfection into the Phoenix 293T packaging cell line (Orbigen, San Diego, CA) using Lipofectamine 2000 (Invitrogen).
2.6. Cell culture and cetuximab cytotoxicity assay
NIH-3T3 cells recently tested by ATCC were cultured in HyClone DMEM/HIGH Glucose medium (Thermo Scientific), supplemented with 10% FBS, 100 μg/ml penicillin, 100 μg/ml streptomycin and 2 mM glutamine, and were infected with the retroviruses in the presence of polybrene. Two days after infection, puromycin (0.2 μg/ml) was added and stable clones were selected over a period of 8–12 days. Several clones were expanded from each group and the EGFR protein expression was tested in each clone by Western blot and immunocytochemistry.
For cetuximab cell cytotoxicity assay, 6.103 cells/well were plated in 96-well flat-bottom plates. The viability of the NIH-3T3 cells was determined by using the trypan blue dye exclusion assay and the number of cells was counted using a hemocytometer. Twenty-four hours after plating, cell culture medium was replaced with DMEM containing 0.5% FBS with or without cetuximab (Bristol-Myers Squibb Co., Princeton, NJ). A serial dilution of cetuximab from 1.25 μg/ml up to 80 μg/ml was added to the cells and the cells were incubated for another 48 h. Growth inhibition was measured using the CellTiter 96 Aqueous One Solution (Promega, Madison, WI). The MTS solution was prepared according to the manufacturer’s instructions. The measurement was performed at 490 nm with a 96-well plate reader (Wallac, 1420 Victor 2, Perkin Elmer Life Science). Each experiment was conducted using two different single clones expressing comparable amounts of EGFR. The data from all experiments were analysed by using GraphPad Prism version 3.00 for Windows (GraphPad Software, San Diego, CA).
2.7. Immunoblotting, immunocytochemistry and immunohistochemistry
Cells were lysed in a buffer containing 50 mM Tris–HCl (pH 7.4), 150 mM NaCl, 2.5 mM EDTA and 1% Triton X-100. Protease and phosphatase inhibitors (Sigma) were added prior to use. Lysates were boiled in sample buffer, separated by SDS–PAGE on 10% polyacrylamide gels, transferred to nitrocellulose, and incubated with anti-EGFR antibody (#2232, Cell Signaling Technologies), anti-p-EGFR antibody (#2234, Cell Signaling Technologies), anti-AKT antibody (#4691, Cell Signaling Technologies), anti-p-AKT antibody (#4060, Cell Signaling Technologies), anti-ERK1/2 antibody (#4695, Cell Signaling Technologies), anti-pERK1/2 antibody (#4370, Cell Signaling Technologies) or anti-β-actin monoclonal antibody (#2228, Sigma) as loading control. Bound antibodies were probed using anti-mouse HRP conjugate (1:1000 dilution) (Pierce Chemical Co.) in PBS containing 5% milk powder and 0.05% Tween 20, and detected via chemiluminescence with ECL (Amersham Pharmacia Biotech).
For detection of EGFR protein by immunofluorescence, cells were cultured on coverslips in DMEM with 10% FBS, washed with PBS, fixed with 4% paraformaldehyde and permeabilised for 10 min at room temperature with 0.2% Triton X-100 in PBS. After washing, coverslips were blocked with 10% horse serum, 1% bovine serum albumin and 0.02% NaN3 in 1% PBS. Cells were incubated with antibody against EGFR (#2232, Cell Signaling Technologies) or tubulin (#T5161, Sigma) for 1 h at 37 °C. Coverslips were washed extensively with PBS and further incubated with 13.2 nM of anti-mouse secondary antibody conjugated to Alexafluor568 phalloidin (Invitrogen, Eugene, OR, USA) in PBS for 30 min at room temperature, washed three times with PBS and mounted with Gelmount (Fisher Scientific, Pittsburgh, PA, USA). Cells were visualised with the use of an LSM 510 laser scanning confocal microscope (Zeiss, Oberkochen, Germany). For immunohistochemistry, tissue sections were deparaffinised, rehydrated, subjected to antigen retrieval and blocked for 20 min. Sections were incubated with γH2AX (Clone JBW301, Millipore), anti-p-EGFR antibody (#2234, Cell Signaling Technologies), anti-p-AKT antibody (#4060, Cell Signaling Technologies) and anti-p-S6K (#9208, Cell Signaling Technologies). Sections were washed in phosphate buffered saline (PBS) and incubated in biotinylated secondary antibody for 30 min. Sections were washed in PBS and incubated in Vectastain Elite ABC reagent (Vector Laboratories) for 30 min, washed again in PBS and incubated with 3,3′-diaminobenzidine. Sections were then washed in water and counterstained in nuclear fast red prior to mounting. Pictures were captured at 20×.
3. Results
3.1. Cetuximab treatment of a patient with HNSCC tumour led to complete tumour regression
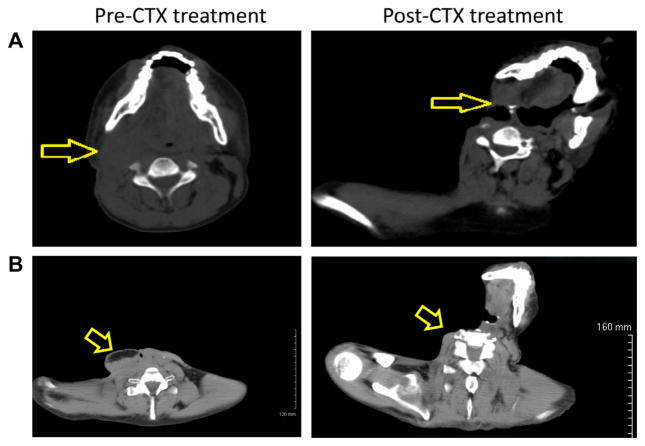
A 33 year-old male from Southern India without any known risk-factors (tobacco and alcohol use) and without a significant past medical or family history presented in July 2008 with hoarseness, sore throat and odynophagia. An endoscopic exam revealed an irregular mass of the left vocal fold. Pathology revealed an infiltrating squamous cell carcinoma (SCC) involving the anterior commissure stage I (T1N0M0). The patient was treated with definitive laryngeal radiation. He continued to be symptomatic after this initial therapy, when a local recurrence was diagnosed in December 2008. At that time, a total laryngectomy was performed. He again had a local neck recurrence for which he underwent selective right lymph node dissection on May 2009 followed by adjuvant radiotherapy completed in August 2009. He then developed a carotid oesophageal fistula and underwent a neck exploration with ligation of the right internal carotid artery. On August 2009, his wound (right neck) was biopsied and continued to demonstrate active well differentiated SCC. In December 2009, after 4 months of rehabilitation, the patient was severely debilitated, emaciated, vomiting daily, had multiple areas of drainage from a complex neck wound and nodular densities on palpation of the right neck (Fig. 1A). Positron emission/computed tomography showed a 3.8 × 2.8 cm round right retromandibular soft tissue mass and extensive F-18 FDG concentration along the right neck fistulous tract with maximum standardised uptake value of 14. The patient was started on a loading dose of cetuximab (400 mg/m2) then a maintenance dose (250 mg/m2) weekly for an additional 12 weeks. Weeks 7 and 8 were held secondary to skin toxicity (Fig. 3B). Five weeks after the initial loading dose of cetuximab, the patient’s pain was under better control, he no longer had palpable tumour on physical exam (Fig. 1B) and a follow up positron emission/computed tomography scan showed post-surgical changes with no areas of F-18 FDG uptake to indicate locoregional or distant metastatic disease.
Fig. 1.
Tumour regression following cetuximab treatment. (A) Computed tomography (CT) of the head and neck before and after treatment with cetuximab (same vertebral level). (B) A CT scan that better shows the initial tumour but at different vertebral levels.
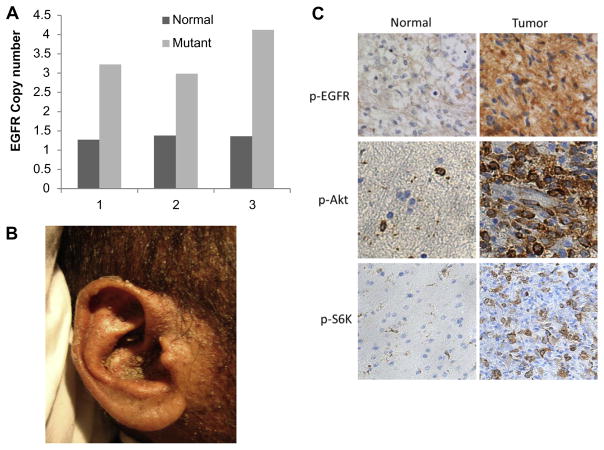
Fig. 3.
Epidermal growth factor receptor (EGFR) gene amplification and protein overexpression in the head and neck squamous cell carcinoma (HNSCC) patient’s tumour tissue. (A) EGFR gene copy number in the normal and tumoural tissues determined by qPCR from three different slides sectioned from the same tumour. (B) A picture showing the excessive skin rash that the patient had during treatment with cetuximab. (C) IHC showing p-EGFR in the tumour tissue and to a lesser extent in the normal skin tissue. Downstream targets of EGFR, namely AKT and S6K are also highly activated in the tumour compared to the normal tissue.
In March 2010, 1 week after the completion of maintenance cetuximab, the patient presented to the emergency room with altered mental status. He was admitted to the medical intensive care unit with septic shock secondary to multi-drug resistant Escherichia coli pneumonia and a biopsy proven cetuximab skin reaction with secondary MRSA infection. Ultrasound-guided fine needle aspiration of a 3 × 5 cm right neck mass was positive for cancer recurrence. The patient subsequently entered hospice and expired 5 months after initiation of cetuximab.
3.2. The HNSCC patient is free of oncogenic HPV infection
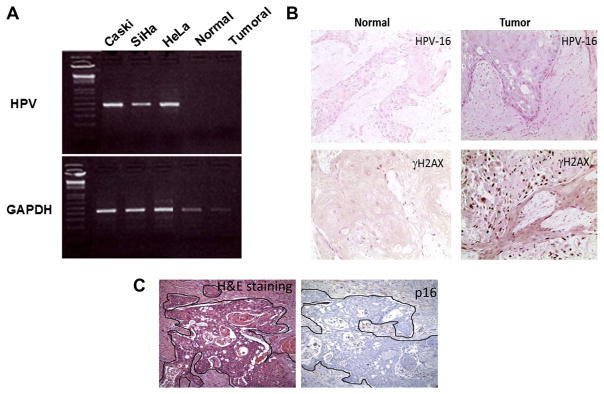
HNSCC tumours with HPV have been associated with a more favourable clinical outcome than HPV negative tumours.20 To test the tumour HPV status, we used two independent approaches; a PCR amplification of genomic DNA for HPV 16 and HPV 18 and HPV in situ hybridisation. The data in Fig. 2A show that several known HPV-infected cervical cancer cell lines, such as Caski and SiHa which have integrated HPV 16, and HeLa which harbours HPV18 display the correct size band when assayed for HPV DNA by PCR. In contrast, neither normal nor tumour tissues from the patient show a PCR-derived diagnostic band, indicating that within the limits of detection the patient’s tumours lack HPV DNA sequences. To confirm this result, we showed that in situ hybridisation also failed to detect HPV sequences (Fig. 2B). Fig. 2B also shows that tumour cells stained positive for γH2AX, an indicator of genomic instability. The tumour is also negative for p16 protein expression (Fig. 3C).
Fig. 2.
Absence of HPV detection in tumour biopsy. (A) PCR amplification of the genomic DNA for HPV 16 and HPV 18 shows that both normal and tumoural tissues from the patient are negative for HPV, while cell lines with integrated HPV 16 or HPV 18 show the correct size band for HPV. (B) In situ hybridisation confirms the HPV negative status of the patient. γH2AX, a marker for genomic instability, was used as a positive control. (C) A staining for p16 confirms the absence of the protein in both normal and tumour tissues.
3.3. EGFR gene amplification and protein overexpression in the patient’s tumour tissue
Overexpression of EGFR is a common hallmark in many cancers including HNSCC. Deregulated expression of EGFR ultimately leads to the activation of Ras/MAPK and PI-3K/Akt signalling cascades promoting cellular proliferation and cell survival. Cetuximab would block such activation by interfering with ligand binding, leading to tumour regression. In colorectal-cancer cell lines, the concentration of cetuximab that completely inhibited proliferation of cells with amplified EGFR copy number did not affect proliferation of cells with unamplified EGFR, indicating that the response to anti-EGFR treatment has a genetic basis and suggesting that patients might be selected for treatment on the basis of EGFR copy number.19,21
We therefore investigated EGFR protein expression status by immunohistochemistry as well as gene copy number by q-PCR. As seen in Fig. 3A, there is an almost threefold EGFR gene amplification in the tumour [ΔΔCt (EGFR-RNaseP) = 3.18] versus normal tissue [ΔΔCt (EGFR-RNaseP) = 1.35]. Consistent with EGFR increased copy number, the level of EGFR protein expression and the activated forms of its downstream targets AKT and S6K are elevated in the tumour tissue compared with normal skin tissue (Fig. 3C).
3.4. The catalytic and ligand-binding domains contain EGFR somatic mutations
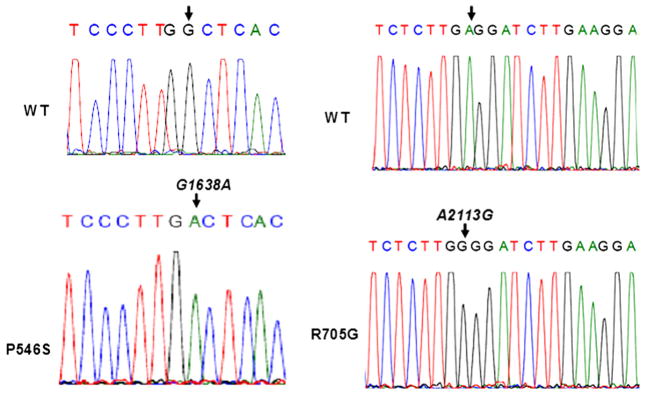
We tested whether the tumour DNA contained EGFR somatic mutations that may associate with the observed increased sensitivity to cetuximab. We therefore cloned and sequenced the EGFR exons that encode both the ligand-binding domain (exons 5 and 6 and exons 13 through 15) and the kinase domain (exons 18 through 21). Sequencing results revealed the presence of two somatic mutations that are not present in the normal tissue (Fig. 4). One of these mutations is in the ligand-binding domain, leading to substitution of a proline residue in position 546 for a serine (P546S). The second somatic mutation is in the kinase domain and leads to substitution of an arginine for a glycine (R705G). Both mutations may affect the increased sensitivity of EGFR to cetuximab. Indeed, the mutation in the ligand-binding domain may alter the local structure of the receptor leading to greater affinity of EGFR to cetuximab than to its natural ligands.
Fig. 4.
Detection of two somatic mutations in the epidermal growth factor receptor (EGFR) gene of the HNSCC patient. Sequencing of the EGFR exons 5 through 6; 13 through 15 and 18 through 21 from both normal and tumoural tissues, revealed the existence of two somatic mutations, one in the ligand binding domain and the other in the catalytic domain. Sequences of the forward strand from single clones are shown.
3.5. Differential effect of P546S and R705G mutations on EGFR sensitivity to cetuximab
To assess the oncogenic potential of the EGFR ligand binding domain (P546S) and kinase domain (R705G) mutants, the tumour-derived mutations were introduced into wildtype human EGFR cDNA by site-directed mutagenesis either singly or in combination. The previously described mutation in exon 18 of the kinase domain (G719S) was also introduced and used as a control. The resulting wildtype and mutant cDNAs in the pBabe-Puro retroviral vector were transferred into NIH-3T3 cells by retroviral infection. Clones that stably expressed EGFR protein were obtained following Puromycin selection. Clones expressing comparable amounts of EGFR protein were used to determine the cetuximab half-maximal-inhibitory concentration (IC50) values of wildtype and mutant EGFR-NIH-3T3 expressing cells.
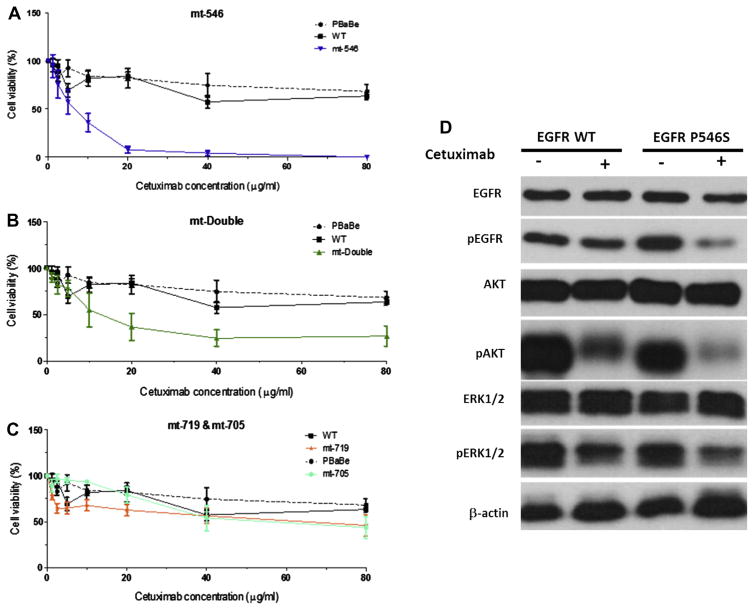
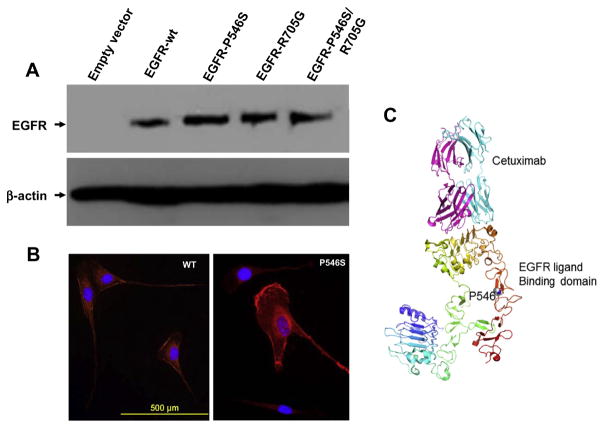
Two clones from each group were treated with increasing concentrations of cetuximab and incubated for 24 h after which cell growth inhibition was assayed using the MTS assay. The data in Fig. 5A show that increased concentrations of cetuximab led to a very quick and efficient killing of cells that express the P546S mutation (IC50 = 7.5 μg/ml) while wildtype expressing cells and cells transfected with the pBabe vector alone failed to reach 50% cell death even at the highest concentration of cetuximab (80 μg/ml). Comparison of the double mutant P546S/R705G with wildtype and empty vector expressing cells showed that the presence of the additional mutation in the kinase domain did not affect the overall sensitivity of P546S mutant to cetuximab, suggesting that the R705G mutation does not contribute to the response to cetuximab (Fig. 5B). Cells expressing EGFR protein with the R705G mutation behaved very similarly to cells expressing the wildtype protein (Fig. 5C), further supporting the neutrality of this mutation to cetuximab sensitivity. Consistent with previous observations,17 cells that express the EGFR kinase activating mutation G719S showed a modest response to cetuximab treatment (Fig. 5C). Cetuximab seems to have a stronger effect on the EGFR signalling pathway in cells expressing the EGFR-P546S mutant than in cells expressing the wildtype EGFR as indicated by a lower expression of the activated forms of EGFR, AKT and ERK1/2 following treatment (Fig. 5D). The P546S mutation does not seem to affect protein stability, as indicated by comparable protein expression patterns in NIH-3T3 cells stably expressing either the wild-type or mutant proteins by Western blotting (Fig. 6A), nor does it affect protein recycling to the membrane as indicated by similar EGFR protein staining intensities at the membrane by immunocytochemistry (Fig. 6B). Whether the mutation increases cetuximab affinity to the receptor is under investigation. Preliminary modelling of the effect of the mutation on EGFR ligand binding shows that the mutation is likely to disturb local interactions despite the fact that the mutation is remote from the core binding residues (Fig. 6C).
Fig. 5.
The P546S mutation affects NIH-3T3 cells sensitivity to cetuximab. (A) NIH-3T3 cells that stably express the P546S mutation show a dramatic increase in sensitivity to cetuximab both singly or when combined with R705G mutation (B), compared to wild type or empty vector only transfected cells. The R705G and G719S mutations do not significantly affect sensitivity to cetuximab (C). (D) Cetuximab treatment has a more profound effect on the epidermal growth factor receptor (EGFR) signalling pathway in cells expressing mutant EGFR-P546S than it did on cells expressing the wild type protein.
Fig. 6.
The P546S does not affect protein stability or epidermal growth factor receptor (EGFR) localisation to the membrane. (A) Western blotting for EGFR protein shows comparable expression levels of wild type and mutant EGFR proteins. (B) IF of EGFR wild type and P546S mutant showing similar amounts of EGFR protein at the membrane. (C) A modelling of the EGFR ligand-binding domain showing the core interaction site with cetuximab and the location of the P546S mutation.
4. Discussion
Overexpression of EGFR is a hallmark of many epithelial tumours, particularly HNSCC,22,23 and is associated with tumour progression, early metastatic spread and poor prognosis.23,24
In this report we show that a patient with HNSCC, who responded positively to treatment with cetuximab had amplified EGFR gene, overexpressed EGFR protein and more importantly carries two EGFR somatic mutations. Further analysis showed that while the mutation in the catalytic domain conferred a very marginal sensitivity to cetuximab in a cell viability assay, the mutation in the ligand-binding domain led to a significant increase in sensitivity to the drug. To our knowledge this is the first tumour-associated mutation in the ligand-binding domain of EGFR that modulates its sensitivity to cetuximab in a cell based assay. Whether this mutation alone was responsible for the positive response to cetuximab is unlikely since this patient’s tumour also overexpressed EGFR protein and the patient developed an adverse side effect that correlated with the antitumour activity indicating that the immune response may also have played a major role. The finding that the protein is overexpressed in the tumour tissue by IHC further indicates that the mutation does not affect protein internalisation and subsequent ER-mediated degradation. However, the protein overexpression may also be the result of gene amplification that we have observed in the tumour tissue. An increase in cetuximab affinity to EGFR is the most plausible explanation for the observed increase in sensitivity. This possibility is supported by our preliminary modelling of the effect of the mutation on EGFR ligand binding.
Robust biomarkers to predict clinical activity of cetuximab have not yet been established. Their identification could enhance the risk-to-benefit ratio of this class of compounds and also reduce the spiraling cost of cancer care.25 The lack of available predictive biomarkers has been recently reported in the BMS009 study when cetuximab is combined with chemotherapy in NSCLC.26 No significant correlations were found between mutations in EGFR exons 18–21 in this retrospective study, and interestingly the mutational status of the other exons has not been investigated. In addition, EGFR gene copy number and protein expression level were not correlated with drug activity. However, van Cutsem et al. have reported a powerful predictive value of KRAS mutation status for the efficacy of cetuximab when combined with chemotherapy (FOLFIRI) in metastatic colorectal cancers.27
Montagut et al. have recently identified a mutation (S492R) in the extracellular EGFR domain that confers resistance, both in vivo and in humans, to cetuximab.28 This mutation may affect cetuximab binding to the EGFR ectodomain, but not EGFR ligand activation. Wang et al. have isolated an EGFR variant in which exon-4 deleted (del4 EGFR). This variant is associated with a lack of cetuximab activity in human glioma xenografts.29 In contrast; an EGFR R521K polymorphism is associated with a favourable outcome in colorectal patients treated with cetuximab.30 R521K affects affinity of ligand biding to EGFR and reduces activity of its downstream target genes.
A direct correlation of the clinical observations with the effects of this mutation on the activity of EGFR will require solving the structure of the mutant protein. Although several questions remain to be addressed in order to better understand how this mutation led to such a clinical outcome, mapping of this mutation represents an excellent opportunity for patients with HNSCC and possibly other cancers to benefit from cetuximab and other ligand-blocker therapy. It still remains to be determined how common this mutation is in HNSCC as this observation represents an unusual presentation for a 33 year-old male who was originally from India with no definite aetiologic factor for laryngeal cancer (tobacco or Bidi smoking, tobacco chewing, HPV infection or alcohol31). Based on the frequency of the mutation, we will prospectively evaluate the clinical activity of single-agent cetuximab in patients with tumours harbouring the P546S mutation.
Acknowledgments
We would like to thank the Cincinnati Children’s Hospital, Division of Pathology for the ventana staining and Grace Hallenbeck and Julianne Qualtieri for technical assistance. We also thank Dr. Rhett Kovall for the structure modelling of the EGFR mutation. We also thank Dr. Susan Robbins for editing the manuscript and Dr. George Atweh, Director of the University of Cincinnati Cancer Institute for the financial support of this work. Funding sources had no influence on the design and implementation of the experiments or on the interpretation of the results of this study. The corresponding author had full access to the data and had final responsibility for the decision to submit for publication.
Footnotes
Conflict of interest statement
None declared.
References
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.Grandis JR, Tweardy DJ. Elevated levels of transforming growth factor a and epidermal growth factor receptor messenger RNA are earlymarkers of carcinogenesis in head and neck cancer. Cancer Res. 1993;53:3579–84. [PubMed] [Google Scholar]
- 3.Grandis JR, Melhem MF, Gooding WE, et al. Levels of TGF-a and EGFR protein in head and neck squamous cell carcinoma and patient survival. J Natl Cancer Inst. 1998;90:824–32. doi: 10.1093/jnci/90.11.824. [DOI] [PubMed] [Google Scholar]
- 4.Ang KK, Berkey BA, Tu X, et al. Impact of epidermal growth factor receptor expression on survival and pattern of relapse in patients with advanced head and neck carcinoma. Cancer Res. 2002;62:7350–6. [PubMed] [Google Scholar]
- 5.Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2000;103:211–25. doi: 10.1016/s0092-8674(00)00114-8. [DOI] [PubMed] [Google Scholar]
- 6.Jorissen RN, Walker F, Pouliot N, Garrett TP, Ward CW, Burgess AW. Epidermal growth factor receptor: mechanisms of activation and signalling. Exp Cell Res. 2003;284:31–53. doi: 10.1016/s0014-4827(02)00098-8. [DOI] [PubMed] [Google Scholar]
- 7.Yarden Y. The EGFR family and its ligands in human cancer. Signalling mechanisms and therapeutic opportunities. Eur J Cancer. 2001;37:S3–8. doi: 10.1016/s0959-8049(01)00230-1. [DOI] [PubMed] [Google Scholar]
- 8.Mendelsohn J, Baselga J. Status of epidermal growth factor receptor antagonists in the biology and treatment of cancer. J Clin Oncol. 2003;21:2787–99. doi: 10.1200/JCO.2003.01.504. [DOI] [PubMed] [Google Scholar]
- 9.Mendelsohn J, Baselga J. Epidermal growth factor receptor targeting in cancer. Semin Oncol. 2006;33:369–85. doi: 10.1053/j.seminoncol.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 10.Ng M, Cunningham D. Cetuximab (Erbitux)-an emerging targeted therapy for epidermal growth factor receptor-expressing tumours. Int J Clin Pract. 2004;58:970–6. doi: 10.1111/j.1368-5031.2004.00369.x. [DOI] [PubMed] [Google Scholar]
- 11.Sunada H, Magun BE, Mendelsohn J, MacLeod CL. Monoclonal antibody against epidermal growth factor receptor is internalized without stimulating receptor phosphorylation. Proc Natl Acad Sci USA. 1986;83:3825–9. doi: 10.1073/pnas.83.11.3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pao W, Miller VA, Politi KA, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2:e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sequist LV, Lynch TJ. EGFR tyrosine kinase inhibitors in lung cancer: an evolving story. Annu Rev Med. 2008;59:429–42. doi: 10.1146/annurev.med.59.090506.202405. [DOI] [PubMed] [Google Scholar]
- 14.Riely GJ, Pao W. Combining EGFR targeted therapy with chemotherapy in pancreatic cancer: is timing important? Cancer Biol Ther. 2005;4:1096–7. doi: 10.4161/cbt.4.10.2102. [DOI] [PubMed] [Google Scholar]
- 15.Arteaga CL, Baselga J. Tyrosine kinase inhibitors: why does the current process of clinical development not apply to them? Cancer Cell. 2004;5:525–31. doi: 10.1016/j.ccr.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 16.Mukohara T, Engelman JA, Hanna NH, et al. Differential effects of gefitinib and cetuximab on non-small-cell lung cancers bearing epidermal growth factor receptor mutations. J Natl Cancer Inst. 2005;97:1185–94. doi: 10.1093/jnci/dji238. [DOI] [PubMed] [Google Scholar]
- 17.Doody JF, Wang Y, Patel SN, et al. Inhibitory activity of cetuximab on epidermal growth factor receptor mutations in non-small cell lung cancers. Mol Cancer Ther. 2007;6:2642–51. doi: 10.1158/1535-7163.MCT-06-0506. [DOI] [PubMed] [Google Scholar]
- 18.Moroni M, Sartore-Bianchi A, Benvenuti S, Artale S, Bardelli A, Siena S. Somatic mutation of EGFR catalytic domain and treatment with gefitinib in colorectal cancer. Ann Oncol. 2005;16:1848–9. doi: 10.1093/annonc/mdi356. [DOI] [PubMed] [Google Scholar]
- 19.Moroni M, Veronese S, Benvenuti S, et al. Gene copy number for epidermal growth factor receptor (EGFR) and clinical response to anti EGFR treatment in colorectal cancer: a cohort study. Lancet Oncol. 2005;6:279–86. doi: 10.1016/S1470-2045(05)70102-9. [DOI] [PubMed] [Google Scholar]
- 20.Gillison ML, Koch WM, Capone RB, et al. Evidence for a causal association between human papillomavirus and a subset of head and neck cancers. J Natl Cancer Inst. 2000;92:709–20. doi: 10.1093/jnci/92.9.709. [DOI] [PubMed] [Google Scholar]
- 21.Bardelli A, Parsons DW, Silliman N, et al. Mutational analysis of the tyrosine kinome in colorectal cancers. Science. 2003;300:949. doi: 10.1126/science.1082596. [DOI] [PubMed] [Google Scholar]
- 22.Kamata N, Chida K, Rikimaru K, et al. Growth-inhibitory effects of epidermal growth factor and overexpression of its receptors on human squamous cell carcinomas in culture. Cancer Res. 1986;46:1648–53. [PubMed] [Google Scholar]
- 23.Mandic R, Eikelkamp N, Peldszus R, et al. Variations of EGF-R surface expression in squamous cell carcinomas of the head and neck region. Anticancer Res. 2001;21:3413–8. [PubMed] [Google Scholar]
- 24.Chung CH, Parker JS, Karaca G, et al. Molecular classification of head and neck squamous cell carcinomas using patterns of gene expression. Cancer Cell. 2004;5:489–500. doi: 10.1016/s1535-6108(04)00112-6. [DOI] [PubMed] [Google Scholar]
- 25.Fojo T, Grady C. How much is life worth: cetuximab, non-small cell lung cancer, and the $440 billion question. J Natl Cancer Inst. 2009;101:1044–8. doi: 10.1093/jnci/djp177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khambata-Ford S, Harbison CT, Hart LL, et al. Analysis of potential predictive markers of cetuximab benefit in BMS099, a phase III study of cetuximab and first-line taxane/carboplatin in advanced non-small-cell lung cancer. J Clin Oncol. 2010;28:918–27. doi: 10.1200/JCO.2009.25.2890. [DOI] [PubMed] [Google Scholar]
- 27.Van Cutsem E, Köhne CH, Láng I, et al. Cetuximab plus irinotecan, fluorouracil, and leucovorin as first-line treatment for metastatic colorectal cancer: updated analysis of overall survival according to tumor KRAS and BRAF mutation status. J Clin Oncol. 2011;29:2011–9. doi: 10.1200/JCO.2010.33.5091. [DOI] [PubMed] [Google Scholar]
- 28.Montagut C, Dalmases A, Bellosillo B, et al. Identification of a mutation in the extracellular domain of the Epidermal Growth Factor Receptor conferring cetuximab resistance in colorectal cancer. Nat Med. 2012;18:221–3. doi: 10.1038/nm.2609. [DOI] [PubMed] [Google Scholar]
- 29.Wang H, Shi B, Zhang Q, et al. Growth and metastasis suppression of glioma xenografts expressing exon 4-deletion variant of epidermal growth factor receptor by monoclonal antibody CH12-mediated receptor degradation. FASEB J. 2012;26:73–80. doi: 10.1096/fj.11-191064. [DOI] [PubMed] [Google Scholar]
- 30.Hsieh YY, Tzeng CH, Chen MH, Chen PM, Wang WS. Epidermal growth factor receptor R521K polymorphism shows favorable outcomes in KRAS wild-type colorectal cancer patients treated with cetuximab-based chemotherapy. Cancer Sci. 2012;103:791–6. doi: 10.1111/j.1349-7006.2012.02225.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Addala L, Pentapati CK, Reddy Thavanati PK, et al. Risk factor profiles of head and neck cancer patients of Andhra Pradesh, India. Indian J Cancer. 2012;49(2):215–9. doi: 10.4103/0019-509X.102865. [DOI] [PubMed] [Google Scholar]