Abstract
CD133, also known as Prominin-1, is expressed on stem cells present in many tissues and tumors. In this work, we have identified and characterized a single chain variable fragment (scFv) for efficient and specific recognition of CD133. Phage display was used to develop the scFv from a previously reported anti-CD133 hybridoma clone 7, which was capable of recognizing both glycosylated and non-glycosylated forms of human CD133. The scFv immunostained CD133+ Caco-2 cells but not CD133-/low U87 cells. Significantly, it immunostained CD133- cells transiently transfected with the mouse CD133 gene as well as CD133+ mouse cells. Co-immunostaining studies in mouse bone marrow cells, using anti-CD133 scFv-FITC and anti-mouse CD133-PE (clone 13A4) commercial antibody, indicated that the epitopes recognized by these reagents partially overlap. Taken together, these results suggest that the scFv can recognize mouse CD133 protein in addition to recognizing human CD133. This new scFv is expected to be valuable both as a molecular diagnostic reagent for identifying CD133+ cells and as a ligand for targeting therapeutics to CD133+ tumor initiating cells.
Keywords: CD133, ScFv, cancer stem cells, prominin-1
Introduction
Human CD133 is a penta-span transmembrane glycoprotein. It is a marker of neural, endothelial and hematopoietic stem cells [1–4]. Its expression is also seen in cancer initiating cells in many tumors including that of brain, prostrate, colon and breast [5–7] and is, therefore, used widely as a molecular marker to isolate and characterize these cell phenotypes. Targeting CD133 may also enable enhanced delivery of cytotoxic therapies to cancer initiating cells [8, 9]. Further development of CD133 as a diagnostic marker and/or therapeutic target, requires highly sensitive reagents that exclusively recognize CD133. Currently, AC133 and 293C3 antibodies are widely used to study CD133 [10]. These reagents identify glycosylated epitopes of CD133 and not the native protein. Hence the reliability of these reagents for accurate detection of CD133 protein levels is a major concern [11, 10, 12]. To overcome this limitation, we have earlier identified a hybridoma (Clone 7) that specifically recognizes CD133 in a range of cell-based assays [13]. Clone 7 recognized both glycosylated and non-glycosylated epitopes of CD133 protein, and hence could be used to investigate CD133 expression with greater reliability. For applications such as targeted drug delivery, single chain variable fragments (scFv) are more convenient and more effective than larger antibodies. In this work, we employed phage-display to derive a scFv targeting CD133. Our studies show that the new scFv is capable of recognizing both human and mouse CD133 protein and can be potentially used as a diagnostic tool to characterize CD133 expression.
Materials and Methods
1. Generation of ScFv library from CD133 hybridoma
The scFv was generated from anti-CD133 hybridoma Clone 7 cells [13]. Total RNA was isolated from these cells by using RNeasy micro kit (Qiagen) and first strand cDNA synthesis was carried out using oligo (dT)20 primer and Superscript II reverse transcriptase (Invitrogen). Following this, a unique two-step PCR protocol was used to amplify the scFv-encoding gene repertoire. DNA sequences encoding the variable region of light-chain (VL) and heavy-chain (VH) fragments were amplified independently, fused by an overlap-extension PCR, and cloned directionally into pCDisplay-4 vector using the rare cutter SfiI enzyme. The digested vector and PCR amplicons were ligated using T4 ligase, precipitated by sodium acetate-ethanol mixture along with glycogen and yeast tRNA carriers, and then transformed into electro-competent E.coli XLI-BLUE. After overnight culture at 37° C, the amplified recombinant phagemid was extracted from bacteria using Qiagen kit and stored at −20° C. When required, E.Coli transformed with the library was infected with VCSM13 helper phage to package the RNA into phage particles.
2. Library panning against human CD133
96-well ELISA plates were coated with 2 μg of human CD133 protein in 50 μL of 0.1M NaHCO3 (pH 8.6) and allowed to incubate overnight at 4° C. The next day, the protein solution was removed and the wells were blocked with a blocking buffer containing 1% BSA in TBS. The plates were sealed and incubated overnight at 4° C. The blocking solution was then removed and replaced with 50 μl of freshly prepared scFv phage library and incubated for 1–2 hrs at 37° C. After multiple washes, the bound phage particles were eluted, with a low pH buffer containing 0.1 M glycine-HCl pH 2.2, and amplified in E.coli for next round of panning. After 3 rounds of panning, 30 randomly selected clones were checked for their reactivity against CD133 by phage ELISA. In this assay, phage clones were diluted in 5% milk solution and incubated with CD133 protein coated wells for 2 hrs at 37° C. Wells were washed five times before incubation with 1:3000 diluted HRP-conjugated anti-M13 antibody (GE Healthcare) for 1 hour at 37° C. The secondary antibody was then removed, washed five times and incubated with HRP-substrate TMB (50 μl / well) for 30 min at RT. The color development was monitored by measuring absorbance at 450 nm. Plasmid DNA from clones that did bind to the CD133 protein was extracted, and sequenced using primers: P1: 5′aagacagctatcgcgattgcag3′, P2: 5′gcccccttattagcgtttgccatc3′. The DNA sequences were then analyzed using lasergene software (DNASTAR Inc, WI, USA)
3. Subcloning of CD133 scFv
The CD133 scFv cDNA was cloned into the pET28c bacterial expression vector as previously described [9]. The sequence of the resulting clone was verified by DNA sequencing analysis performed at the University of Minnesota biomedical genomics center.
4. Expression, purification and characterization
Purification of the CD133 scFv was performed as described previously [9]. Briefly, the protein was expressed and purified from inclusion bodies using a Novagen pET expression system (Novagen, Madison WI), followed by a 2-step purification process consisting of ion exchange fast protein liquid chromatography (Q sepharose Fast Flow, Sigma) and size exclusion chromatography (Hiload Superdex 200, Pharmacia). About 3 μg of the purified CD133 scFv protein was resolved by 4–15% SDS PAGE gel (Biorad) and then stained by E-Zinc Reversible stain kit (Thermo scientific) to determine purity.
5. Cell culture
All established cell lines used in this work except ONC94M2 were purchased from the American Type Culture Collection. ONC94M2 is a primary glioma cell line derived from a C57BL/6 mouse where the primary tumor was induced by oncogene transfer using NRAS and SV40LgT as described in [14]. Caco-2 (a human colorectal adenocarcinoma) and U87 cells (a human glioblastoma) were grown in Minimum Essential Medium (MEM) containing 20% FBS, 1% non-essential amino acids (Sigma), 1% sodium pyruvate (Sigma) and 1% penicillin-streptomycin (Invitrogen). 4T1, a mouse mammary tumor cell line, was cultured in RPMI medium supplemented with 10% FBS and 1% penicillin-streptomycin. NIH3T3 (a mouse embryo fibroblast cell line) was grown in Dulbecco’s Modified Eagle’s Medium (DMEM) containing 10% FBS and 1% penicillin-streptomycin. All cells were maintained at 37°C / 5% CO2 in a humidified incubator.
6. DNA transfection
NIH 3T3 cells were seeded in 6-well plates. Following attachment, cells were transfected with 1.5 μg mCD133 construct (ATCC Catalog Number MGC-25280, Image ID: 4502359) or empty vector using lipofectamine reagent. After 72 hrs, cells were trypsinized, immunostained, and then analyzed by flow cytometry.
7. Isolation of bone marrow cells
Bone marrow was obtained from femur of CD-1 mice. A single cell suspension of the bone marrow was obtained by dilution in cold PBS, vigorous pipetting, followed by passage through a 70-μm cell strainer. Collected cells were centrifuged at 1000 rpm for 7 min, washed once with cold PBS, and counted using a hemocytometer.
8. FACS staining protocol
One million cells were suspended in 100 μl FACS buffer (1× PBS with 1% BSA and 0.1% sodium azide) in a flow cytometry tube. The cell suspensions were incubated with 2.5 μg FITC-labeled anti-CD133 scFv for 30 min on ice. For the competitive binding assay, 1 million CD-1 mouse bone marrow cells suspended in FACS buffer were pre-incubated with 1 μg Fc Block (BD Pharmingen) on ice for 10 min followed by FITC-labeled anti-CD133 scFv as described above and then with 1 μg anti-CD133 Ab-PE (clone 13A4-PE, eBioscience) for another 30 min on ice. Another group received the two reagents in the reverse order. In addition, one group of cells was incubated with both the reagents simultaneously. Finally, cells were washed twice with FACS buffer and resuspended in 500 μl of FACS buffer for flow cytometric analysis. For immunostaining 4T1 cells with anti-CD133 polyclonal antibody (Abnova), cells were fixed with 2% formaldehyde for 10 min and then permeabilized with ice-cold 90% methanol for 30 min. Cells were then incubated with FACS buffer containing 5 μg antibody for 30 min on ice followed by incubation with FITC-labeled anti-rabbit secondary antibody (BD Pharmingen) for 30 min. Cells were then washed twice with FACS buffer before analysis by flow cytometry. The acquired data was analyzed by FLOWJO software (Tree Star Inc., OR, USA)
9. Western blot
ONC94M2 and 4T1 cells were pelleted and lysed using RIPA buffer (Pierce). Protein levels were estimated using BCA kit (Pierce). Western blotting was done as described in [15]. Anti-human CD133 polyclonal antibody (Abnova) was used as the primary antibody. Anti-rabbit IgG-HRP (Cell Signal) was used as the secondary antibody.
10. scFv ELISA
E.Coli TOP10F′ transformed with the scFv clone-11(HA tagged, in pCDisplay-4 vector background) was induced with IPTG (1 mM, final concentration) and then cultured overnight at 30° C. Next day, the bacterial culture was pelleted by centrifugation at 15,000 rpm for 30 min at 4° C and the soluble scFv in the culture supernatant was analysed by ELISA. Briefly, human CD133 protein coated wells were incubated with either scFv, parental monoclonal antibody or blocking solution (5% non-fat milk in phosphate-buffered saline) for 1 hour at 37° C. The treatments were removed and the wells were washed five times with PBS before incubation with HRP-conjugated anti-HA (obtained from GenScript USA; 1:2000 diluted in 5% milk solution) for 1 hour at 37° C. The wells were washed with PBS and then incubated with the HRP substrate TMB. The color development was analysed by measuring the absorbance at 450 nm using a plate-reader.
Results
1. Identification of a novel anti-CD133 scFv
Recombinant anti-CD133 scFv was constructed using VL and VH fragments of the clone 7 anti-CD133 antibody. The VL and VH fragments were amplified by RT-PCR. The scFv was constructed using these fragments and a DNA linker that encodes for Ser7Gly10Arg by performing an overlap extension PCR. The resulting PCR products were analyzed by gel electrophoresis. The individual VL and VH components were about 400 bp in size and the assembled scFv was found to be 800 bp in size, which was in agreement with the theoretical predictions (Figure 1a). The scFv was then cloned into a pCDisplay-4 vector, transformed into E.coli XL-1 BLUE and were subsequently infected with VCSM13 helper phage in-order to release the complete phage particles that displayed a library of scFv on their coat surface. The phage library was then screened for reactivity to CD133 by a standard panning process which involved incubation of the phage particles with CD133 protein coated plates. Only a fraction of the phage library was found to bind to the plates. These binders were then retrieved, amplified in bacteria and screened by subsequent panning. After 3 such rounds, a 394-fold (6.3 *10−5 / 1.6 * 10−7) enrichment of phage binders was achieved (Figure 1b). At this stage, 30 phage clones were randomly picked to confirm their reactivity against human CD133 by phage ELISA (Figure 1c). In this assay, 9 phage clones were found to have high reactivity with CD133. Plasmid DNA of these clones were extracted and then sequenced. Five of them were found to have identical DNA sequence (data not shown). Phage clone 11 showed a high reactivity. Hence this clone was selected for subsequent analysis; the plasmid DNA, encoding the scFv as a HA-tagged recombinant protein, was extracted. This DNA construct has an amber stop codon between the scFv sequence and the vector backbone. So a non-suppressor E.Coli TOP10F’ bacterial strain was used for the scFv protein expression. The scFv was predominantly secreted into bacterial culture media (data not shown). The bacterial culture supernatant containing the secreted scFv protein was diluted and analyzed by ELISA to investigate the specificity of the clone 11 scFv to the human CD133 protein substrate (Figure 1d). In this assay, the parent monoclonal antibody was used as a positive control and it showed a higher absorbance (1.4 absorbance units, AU) than the scFv protein (0.9 AU). The background absorbance of the blank treatment was found to be 0.2 AU. These results indicate that the CD133 scFv could recognize and bind to human CD133 protein, however with a lower binding affinity than its parental monoclonal antibody. Further experiments reported in this study were done using anti-CD133- scFv protein without HA epitope tag. To obtain this, ORF of the scFv was sub-cloned into a PET-28C expression system, bacterially expressed, purified and analyzed by SDS-PAGE gel (Figure 2a). The molecular weight of the anti-CD133 scFv protein was found to be 26 kDa, and was in agreement with the theoretical predictions and with our own published results [9]. Amino acid sequence of the recombinant anti-CD133 scFv is shown in Figure 2b.
Fig. 1. Cloning of anti-CD133 scFv.
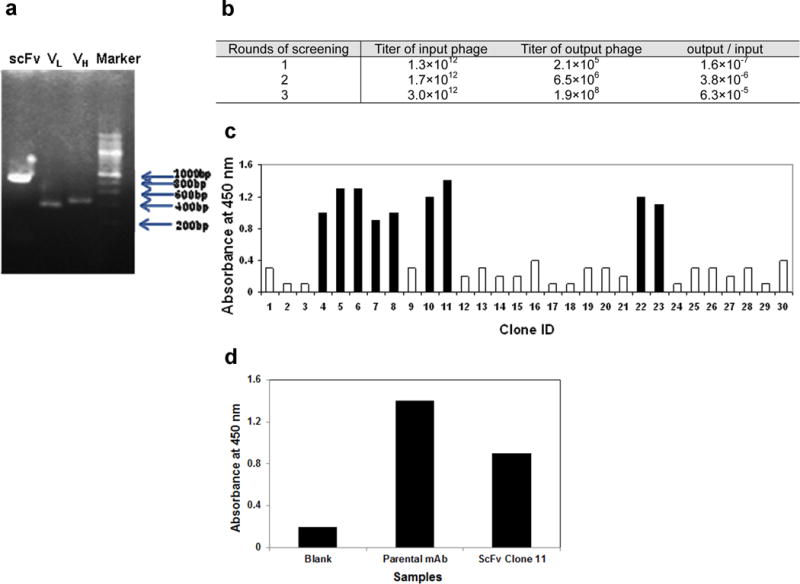
Panel a- Gel electrophoresis of PCR products. Lane 1- recombinant scFv constructed using VL and VH fragments and a DNA linker encoding Ser7Gly10Arg; Lane 2- amplification of VL fragment alone; Lane 3- amplification of VH fragment alone; Lane 4- molecular weight ladder.
Panel b- The recombinant scFv is cloned into a phagemid, which were then packaged into M13 phage. They were screened by a biopanning assay by incubating phage particles with CD133 coated plates. Three rounds of panning were done. Titers of phage particles in each round are shown. At the end of three rounds, phage particles bound to CD133-coated wells were significantly enriched.
Panel c- Phage ELISA of 30 randomly picked phage clones. 9 positive clones that showed A450 >1 are indicated in black. Clone 11 was chosen for further characterization.
Panel d- Clone 11 plasmid DNA encodes a HA-tagged anti-CD133 scFv recombinant protein. It was bacterially expressed and the bacterial culture supernatant was analyzed by ELISA. Human CD133 protein coated wells were incubated with either diluted bacterial supernatant, parental monoclonal antibody (positive control) or blocking solution (blank). The treatments were removed and incubated with HRP conjugated anti-HA secondary antibody. The HRP reaction was then analyzed by measuring the absorbance at 450 nm after addition of TMB substrate.
Fig. 2. Expression and specificity of anti-CD133 scFv.
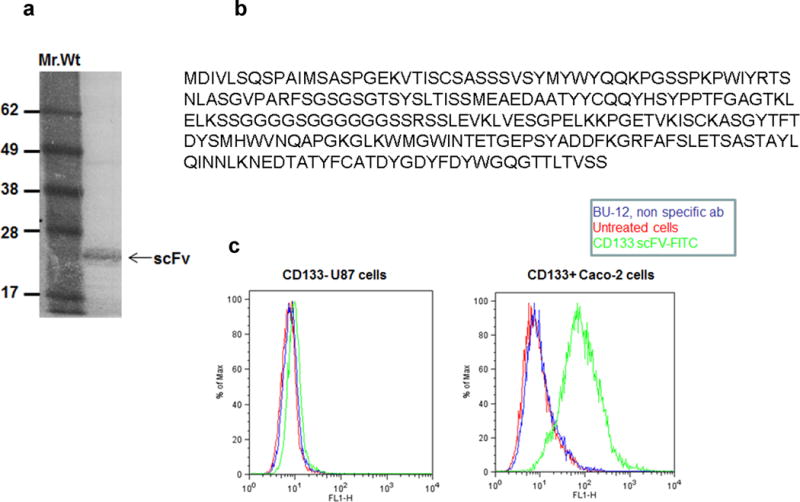
Panel a- anti-CD133 scFv clone 11 was subcloned into a PET-28c expression cassette, bacterially expressed. This resulted in the expression of HA-tagless anti-CD133 scFv. SDS-PAGE gel electrophoresis of the scFv under non-reducing conditions.
Panel b- Sequence of recombinant anti CD133 scFV (clone 11).
Panel c- Immunostaining of Caco-2 and U87 cells. Cells were stained with anti-CD133 scFv (blue). Cells stained with BU-12-FITC (red) and unstained cells (black) served as controls. Anti-CD133 scFv stained only Caco-2 cells but not U87 cells.
2. Specificity of anti-CD133 scFv
Specificity of the anti-CD133 scFv was investigated by immunostaining. The scFv was initially conjugated to FITC to enable direct staining of cells. Anti-CD133 scFv-FITC specifically stained human colon carcinoma Caco-2 cells that express high levels of CD133 [16, 9] but not CD133low/− U87 cells (Figure 2c). Unstained cells and cells stained with a non-specific antibody (BU-12 FITC) were used as controls. These results established the specificity of anti-CD133 scFv for human CD133 protein.
3. Cross-reactivity of anti-CD133 scFv to mouse CD133
A plasmid encoding the mCD133 gene was transfected into NIH3T3 cells. The cells were then immunostained using anti-CD133 scFv-FITC (Figure 3). About 13 % of the transfected cells were stained by anti-CD133 scFv-FITC. Unstained cells, transfected cells stained with a non-specific RFB4 (anti-B-cells)-FITC served as controls. Cells transfected with vector backbone served as additional control. The results obtained with this study establish the cross-reactivity of the anti-CD133 scFv to mouse CD133 protein. When the transfection experiment was repeated, about 20% of the transfected cells were found to be immunoreactive with CD133 scFv (data not shown). The low number of cells stained by the scFv may be due to weak expression of the transfected plasmid and/or due to cell-cycle associated variations in CD133 surface mobilization affecting its detection by commercial antibodies [17].
Fig. 3. Cross-reactivity of anti-CD133 scFv with mouse CD133.
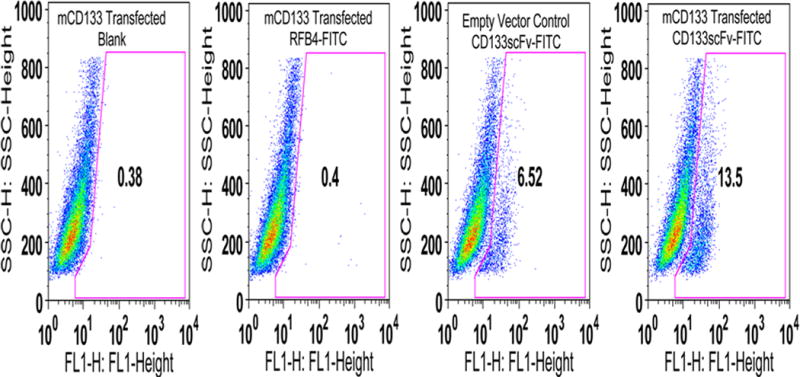
Immunostaining of mCD133 transfected NIH 3T3 cells. Transfected cells were stained with anti-CD133 scFv-FITC. Untransfected cells (FITC Blank), cells transfected with vector alone or transfected cells stained with RFB4-FITC antibody served as controls.
To further study the cross-reactivity of anti-CD133 scFv to mouse CD133 protein, immunostaining experiments were done in mouse tumor cells and bone marrow cells. Anti-CD133 scFv-FITC immunostained ONC49M2- mouse glioblastoma cells (Figure 4a) as well as 4T1 breast carcinoma cells (Figure 4b). CD133 expression in these cell lines was confirmed by Western blotting using a polyclonal anti-mouse CD133 antibody (Figure 4c). CD133 expression in 4T1 cells was further confirmed by indirect immunostaining done with anti-mouse CD133 polyclonal antibody (Figure 4C). This anti-CD133 polyclonal antibody was raised against c-terminal region of the CD133 protein, which is localized within the cytosol. Hence, 4T1 cells were fixed and permeabilized before immunostaining.
Fig. 4. Immunostaining of mouse cells with anti-CD133 scFv.
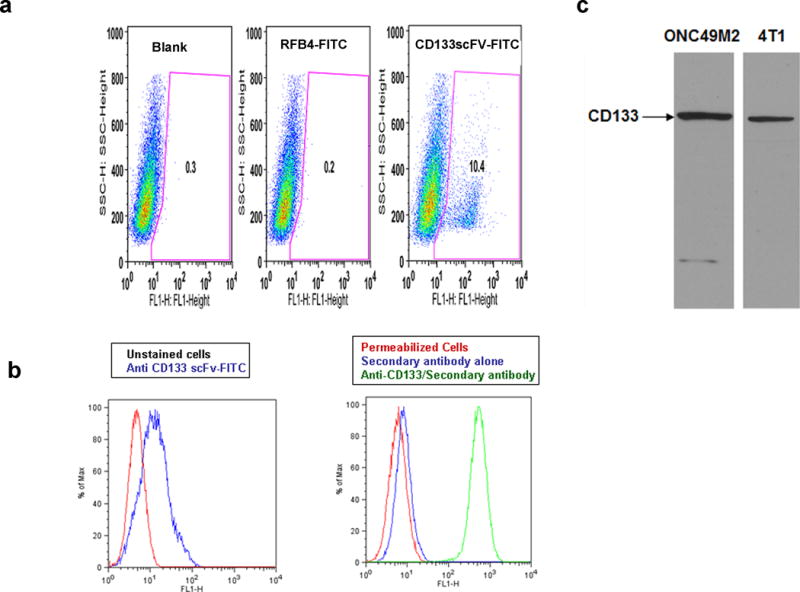
Panel a- Immunostaining of ONC49M2 mouse glioblastoma cells. Unstained cells or cells stained with RFB4-FITC antibody served as controls.
Panel b- Immunostaining of 4T1 cells. (Left): Staining with anti-CD133 scFv-FITC; (Right): Staining with polyclonal anti-CD133 antibody. 4T1 cells were PFA fixed and methanol permeabilized before immunostaining. Permeabilized cells with or without anti-rabbit secondary antibody staining served as controls.
Panel c- Western blotting to demonstrate CD133 expression in ONC49M2 and 4T1 cells
Progenitor cells in bone marrow are reported to express CD133 [18]. Anti-CD133 scFv-FITC immunostained 4% of the mouse bone marrow cells (data not shown). In order to confirm that anti-CD133 scFv binds to mouse CD133 protein, we co-stained mouse bone marrow cells with a commercially available PE-labeled- monoclonal antibody (Figure 5). The order of addition of scFv and 13A4 clone resulted in distinct immunostaining profiles. When clone 13A4 was added first, around 40% of CD133 scFv FITC stained fraction was co-stained by 13A4. However, when anti-CD133 scFv-FITC was added first or when added along with 13A4-PE clone, this fraction decreased to 25%. This indicates that epitopes recognized by 13A4 and anti-CD133 scFv partially overlap. As clone 13A4 is known to bind to mouse CD133 [19], the data from these results confirms that anti-CD133 scFv binds to the mouse CD133 protein.
Fig. 5. Immunostaining of mouse bone marrow cells using anti-CD133 scFv.
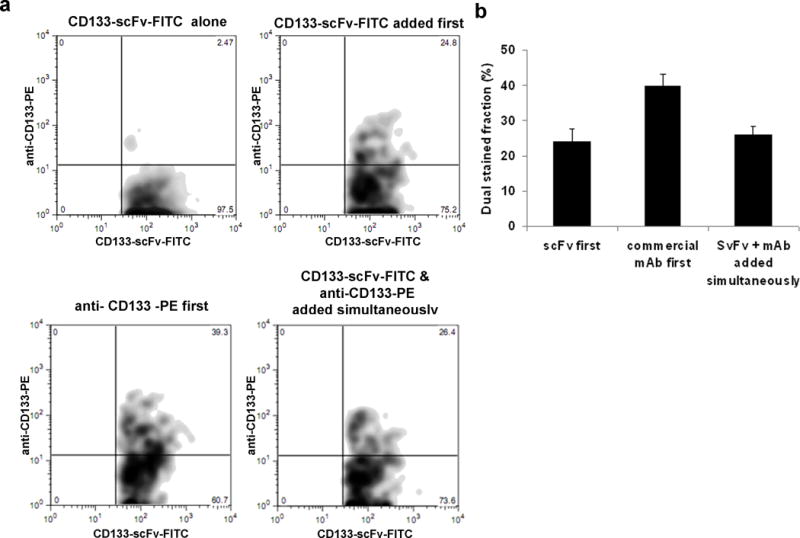
Panel a- Competitive binding assay of anti-CD133 (clone13A4-PE) and anti-CD133 scFv-FITC. One million mouse bone marrow cells suspended in FACS buffer were incubated with anti-CD133 scFv, anti-CD133 scFv followed by anti-CD133 Ab-PE, anti-CD133 Ab-PE followed by anti-CD133 scFv, or both anti-CD133 scFv and anti-CD133 Ab-PE added simultaneously. Cells were washed twice with FACS buffer and subjected to flow cytometric analysis. The scFv stained fraction was gated and then double stained population within them was analyzed.
Panel b- Bar graph of the results obtained in the above study. Data Shown is mean± S.D. (n=3).
Discussion
Human CD133 is a five trans-membrane domain protein composed of 865 amino acids with a molecular weight of 120 kDa [20]. Detailed molecular analysis indicate that CD133 expression is regulated and it involves transcription from five alternative promoters, promoter hypermethylation, tissue-specific alternative splicing [20–22] and lipid-raft associated sorting of resulting protein to apical membrane [23]. The CD133 protein undergoes extensive post-translational glycosylation modification at 8-N linked glycosylation sites found in its extracellular loops [24]. Recently a set of genes that may contribute to the CD133 glycosylation has been reported [25].
Emerging studies indicate that cellular conditions like stem cell differentiation [11], cell cycle status [17] and differential glycosylation processing [25] can selectively modify CD133 glycosylation levels without affecting the native CD133 protein levels. Hence reactivity to AC133 and AC141 reagents that bind to the glycosylation epitopes may not accurately reflect CD133 protein levels present within cell. To circumvent this problem, we have earlier identified a monoclonal antibody (clone7) against CD133. Although clone 7 was raised against a non-glycosylated recombinant CD133 protein, it recognized CD133 protein from both tunicamycin treated and untreated cell lysates by western-blot assay [13]. This indicated that clone 7 can recognize glycosylated CD133 protein and non-glycosylated CD133 protein. Since the anti-CD133 scFv described in this study is derived from the clone 7 antibody, it is expected to recognize and bind to both glycosylated and non-glycosylated CD133 protein. Also, it will be interesting to determine whether clone 7 and the scFv identify the different isoforms of CD133 as well as the mature and premature forms of the protein.
An important feature of the anti-CD133 scFv is its ability to cross-react with mouse CD133 protein. This observation is not totally surprising, given the fact that human and mouse orthologs of CD133 share a 60% sequence identity. It is worthwhile to note here that the parental hybridoma, clone 7, was raised against a recombinant protein consisting of immunogenic amino acids of the human CD133 protein. This stretch of amino acids shares a significant sequence similarity with its mouse counterpart (Figure 6).
Fig. 6. CD133 protein sequence alignment.
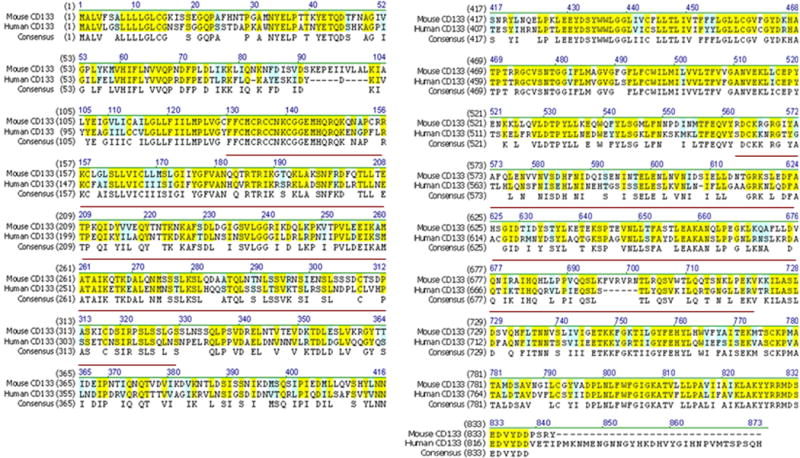
Amino acid sequences of mouse (accession number: AAH28286.1) and human CD133 (accession number: AAH12089.1) were used as input sequences. The consensus sequences are marked with yellow (high similarity) and blue (weak similarity). Red line indicates amino acids of human CD133 used in the generation of parental hybridoma (clone 7).
The scFvs retain target specificity but are smaller in size compared to their parental monoclonal antibodies and hence, are often used for targeted drug delivery. Because the CD133 protein is a known marker for cancer stem cells, we expect the newly developed scFv to be of significant value in developing therapies that selectively target and eliminate cancer stem cells. We are currently investigating the use of anti-CD133 scFv for constructing a targeted immunotoxin, dCD133KDEL, by fusing the anti-CD133 scFv with pseudomonas exotoxin A-PE38 and a c-terminal Lys-Asp-Glu-Leu (KDEL) peptide. This toxin was found to be very potent in selectively targeting and killing CD133+ cancer stem cells in metastatic breast carcinoma [8] and head neck carcinoma xenografts in mouse [9]. Expression of CD133 is not restricted to tumor-initiating cells. So, it is possible that this targeted therapy may have side-effects on other CD133 expressing cells like stem cells present in a normal tissue. In our initial studies with dCD133KDEL, we did not observe any cytotoxicity to normal cells. However, for clinical realization of this immunotoxin, an elaborate evaluation of its effect on normal cells must be carried out. As the anti-CD133 scFv reagent can react with both mouse and human CD133 protein, it is uniquely suited for evaluating the safety of CD133-targeted therapy in mouse xenograft tumor models.
Acknowledgments
We thank Dong Chen at Creative Dynamics Inc. for contributing reagents and expertise used in generating the CD133-reactive antibodies.
References
- 1.Yin AH, Miraglia S, Zanjani ED, Almeida-Porada G, Ogawa M, Leary AG, et al. AC133, a novel marker for human hematopoietic stem and progenitor cells. Blood. 1997;90(12):5002–12. [PubMed] [Google Scholar]
- 2.Uchida N, Buck DW, He D, Reitsma MJ, Masek M, Phan TV, et al. Direct isolation of human central nervous system stem cells. Proc Natl Acad Sci U S A. 2000;97(26):14720–5. doi: 10.1073/pnas.97.26.14720. doi:10.1073/pnas.97.26.14720 97/26/14720 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peichev M, Naiyer AJ, Pereira D, Zhu Z, Lane WJ, Williams M, et al. Expression of VEGFR-2 and AC133 by circulating human CD34(+) cells identifies a population of functional endothelial precursors. Blood. 2000;95(3):952–8. [PubMed] [Google Scholar]
- 4.Weigmann A, Corbeil D, Hellwig A, Huttner WB. Prominin, a novel microvilli-specific polytopic membrane protein of the apical surface of epithelial cells, is targeted to plasmalemmal protrusions of non-epithelial cells. Proc Natl Acad Sci U S A. 1997;94(23):12425–30. doi: 10.1073/pnas.94.23.12425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fabrizi E, di Martino S, Pelacchi F, Ricci-Vitiani L. Therapeutic implications of colon cancer stem cells. World J Gastroenterol. 2010;16(31):3871–7. doi: 10.3748/wjg.v16.i31.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harper LJ, Piper K, Common J, Fortune F, Mackenzie IC. Stem cell patterns in cell lines derived from head and neck squamous cell carcinoma. J Oral Pathol Med. 2007;36(10):594–603. doi: 10.1111/j.1600-0714.2007.00617.x. doi:JOP617 [pii] 10.1111/j.1600-0714.2007.00617.x. [DOI] [PubMed] [Google Scholar]
- 7.Du Z, Qin R, Wei C, Wang M, Shi C, Tian R, et al. Pancreatic cancer cells resistant to chemoradiotherapy rich in “stem-cell-like” tumor cells. Dig Dis Sci. 2010;56(3):741–50. doi: 10.1007/s10620-010-1340-0. [DOI] [PubMed] [Google Scholar]
- 8.Ohlfest JR, Zellmer D, Panyam J, Swaminathan SK, Oh S, Waldron N, et al. Immunotoxin targeting CD133+ breast carcinoma cells. Drug Deliv and Transl Res. 2012 doi: 10.1007/s13346-012-0066-2. In press. [DOI] [PubMed] [Google Scholar]
- 9.Waldron NN, Kaufman DS, Oh S, Inde Z, Hexum MK, Ohlfest JR, et al. Targeting Tumor-Initiating Cancer Cells with dCD133KDEL Shows Impressive Tumor Reductions in a Xenotransplant Model of Human Head and Neck Cancer. Mol Cancer Ther. 2011;10(10):1829–38. doi: 10.1158/1535-7163.MCT-11-0206. doi:1535-7163.MCT-11-0206 [pii] 10.1158/1535-7163.MCT-11-0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bidlingmaier S, Zhu X, Liu B. The utility and limitations of glycosylated human CD133 epitopes in defining cancer stem cells. J Mol Med. 2008;86(9):1025–32. doi: 10.1007/s00109-008-0357-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kemper K, Sprick MR, de Bree M, Scopelliti A, Vermeulen L, Hoek M, et al. The AC133 epitope, but not the CD133 protein, is lost upon cancer stem cell differentiation. Cancer Res. 2010;70(2):719–29. doi: 10.1158/0008-5472.CAN-09-1820. doi:0008-5472.CAN-09-1820 [pii] 10.1158/0008-5472.CAN-09-1820. [DOI] [PubMed] [Google Scholar]
- 12.Florek M, Haase M, Marzesco AM, Freund D, Ehninger G, Huttner WB, et al. Prominin-1/CD133, a neural and hematopoietic stem cell marker, is expressed in adult human differentiated cells and certain types of kidney cancer. Cell Tissue Res. 2005;319(1):15–26. doi: 10.1007/s00441-004-1018-z. [DOI] [PubMed] [Google Scholar]
- 13.Swaminathan SK, Olin MR, Forster CL, Cruz KSS, Panyam J, Ohlfest JR. Identification of a novel monoclonal antibody recognizing CD133. J Immunol Methods. 2010;361(1–2):110–5. doi: 10.1016/j.jim.2010.07.007. [DOI] [PubMed] [Google Scholar]
- 14.Wiesner SM, Decker SA, Larson JD, Ericson K, Forster C, Gallardo JL, et al. De novo induction of genetically engineered brain tumors in mice using plasmid DNA. Cancer Res. 2009;69(2):431–9. doi: 10.1158/0008-5472.CAN-08-1800. doi:69/2/431 [pii] 10.1158/0008-5472.CAN-08-1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Swaminathan SK, Olin MR, Forster CL, Cruz KS, Panyam J, Ohlfest JR. Identification of a novel monoclonal antibody recognizing CD133. J Immunol Methods. 2010;361(1–2):110–5. doi: 10.1016/j.jim.2010.07.007. doi:S0022-1759(10)00203-6 [pii] 10.1016/j.jim.2010.07.007. [DOI] [PubMed] [Google Scholar]
- 16.Vander Griend DJ, Karthaus WL, Dalrymple S, Meeker A, DeMarzo AM, Isaacs JT. The role of CD133 in normal human prostate stem cells and malignant cancer-initiating cells. Cancer Res. 2008;68(23):9703–11. doi: 10.1158/0008-5472.CAN-08-3084. doi:68/23/9703 [pii] 10.1158/0008-5472.CAN-08-3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jaksch M, Munera J, Bajpai R, Terskikh A, Oshima RG. Cell cycle-dependent variation of a CD133 epitope in human embryonic stem cell, colon cancer, and melanoma cell lines. Cancer Res. 2008;68(19):7882–6. doi: 10.1158/0008-5472.CAN-08-0723. doi:68/19/7882 [pii] 10.1158/0008-5472.CAN-08-0723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Quirici N, Soligo D, Caneva L, Servida F, Bossolasco P, Deliliers GL. Differentiation and expansion of endothelial cells from human bone marrow CD133(+) cells. Br J Haematol. 2001;115(1):186–94. doi: 10.1046/j.1365-2141.2001.03077.x. doi:3077 [pii] [DOI] [PubMed] [Google Scholar]
- 19.Sugiyama T, Rodriguez RT, McLean GW, Kim SK. Conserved markers of fetal pancreatic epithelium permit prospective isolation of islet progenitor cells by FACS. Proc Natl Acad Sci U S A. 2007;104(1):175–80. doi: 10.1073/pnas.0609490104. doi:0609490104 [pii] 10.1073/pnas.0609490104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shmelkov SV, St Clair R, Lyden D, Rafii S. AC133/CD133/Prominin-1. Int J Biochem Cell Biol. 2005;37(4):715–9. doi: 10.1016/j.biocel.2004.08.010. doi:S1357-2725(04)00350-4 [pii] 10.1016/j.biocel.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 21.Shmelkov SV, Jun L, St Clair R, McGarrigle D, Derderian CA, Usenko JK, et al. Alternative promoters regulate transcription of the gene that encodes stem cell surface protein AC133. Blood. 2004;103(6):2055–61. doi: 10.1182/blood-2003-06-1881. [DOI] [PubMed] [Google Scholar]
- 22.Pellacani D, Packer RJ, Frame FM, Oldridge EE, Berry PA, Labarthe M-C, et al. Regulation of the stem cell marker CD133 is independent of promoter hypermethylation in human epithelial differentiation and cancer. Mol Cancer. 10:94. doi: 10.1186/1476-4598-10-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roper K, Corbeil D, Huttner WB. Retention of prominin in microvilli reveals distinct cholesterol-based lipid micro-domains in the apical plasma membrane. Nat Cell Biol. 2000;2(9):582–92. doi: 10.1038/35023524. [DOI] [PubMed] [Google Scholar]
- 24.Miraglia S, Godfrey W, Yin AH, Atkins K, Warnke R, Holden JT, et al. A novel five-transmembrane hematopoietic stem cell antigen: isolation, characterization, and molecular cloning. Blood. 1997;90(12):5013–21. [PubMed] [Google Scholar]
- 25.Mak AB, Blakely KM, Williams RA, Penttila P-A, Shukalyuk AI, Osman KT, et al. CD133 protein N-glycosylation processing contributes to cell surface recognition of the primitive cell marker AC133 epitope. J Biol Chem. 2011;286(47):41046–56. doi: 10.1074/jbc.M111.261545. [DOI] [PMC free article] [PubMed] [Google Scholar]


