Abstract
Rationale
Polycomb repressive complex 2 (PRC2) is a major epigenetic repressor that deposits methylation on histone H3 on lysine 27 (H3K27me) and controls differentiation and function of many cells, including cardiac myocytes. EZH1 and EZH2 are two alternative catalytic subunits with partial functional redundancy. The relative roles of EZH1 and EZH2 in heart development and regeneration are unknown.
Objective
We compared the roles of EZH1 versus EZH2 in heart development and neonatal heart regeneration.
Methods and Results
Heart development was normal in Ezh1−/− (E1KO) and Ezh2f/f::cTNT−Cre (E2KO) embryos. Ablation of both genes in Ezh1−/−::Ezh2f/f::cTNT−Cre (DKO) embryos caused lethal heart malformations, including hypertrabeculation, compact myocardial hypoplasia, and ventricular septal defect. Epigenome and transcriptome profiling showed that de-repressed genes were upregulated in a manner consistent with total “EZH” dose. In neonatal heart regeneration, Ezh1 was required but Ezh2 was dispensible. This finding was further supported by rescue experiments: cardiac myocyte-restricted re-expression of EZH1 but not EZH2 restored neonatal heart regeneration in E1KO. In MI performed outside of the neonatal regenerative window, EZH1 but not EZH2 likewise improved heart function and stimulated CM proliferation. Mechanistically, EZH1 occupied and activated genes related to cardiac growth.
Conclusions
Our work unravels divergent mechanisms of EZH1 in heart development and regeneration, which will empower efforts to overcome epigenetic barriers to heart regeneration.
Keywords: Heart development, heart regeneration, epigenetics, gene regulation, cardiomyopathy
Subject Terms: Developmental Biology, Myocardial Regeneration, Epigenetics, Gene Expression and Regulation
INTRODUCTION
Adult mammalian cardiac myocytes (CMs) have limited capacity to proliferate, and as a result the adult human heart has little capacity to regenerate from heart injury1, 2. Murine heart regeneration potential remains robust into the first postnatal week of life, as hearts injured by apical resection or myocardial infarction on postnatal day 1 (P1) recover with little scar through proliferation of pre-existing CMs3. This regenerative capacity is lost rapidly, as similar injury at P7 induces scar rather than myocardial regeneration. The mechanisms responsible for CM cell cycle exit remain incompletely understood4. Reactivation of some transcriptional regulatory networks essential for heart development, such as those controlled by YAP and TBX20, have been shown to promote adult CM cell cycle activity4–7. However, clinically meaningful strategies are still lacking. The barriers likely reside in the intrinsically distinct chromatin states and epigenetic pathways of adult compared to fetal and neonatal cardiac myocytes, which repress critical regulatory genomic loci required for effective regeneration8.
Polycomb repressive complex 2 (PRC2) is major epigenetic repressive complex, the core of which is composed of EED, SUZ12 and a catalytic subunit, either EZH1 or EZH2. PRC2 trimethylates histone H3 on lysine 27 (H3K27me3), an epigenetic mark associated with transcriptional repression9. Recently, EZH1 was also found to form a distinct complex containing SUZ12 but not EED that activates genes in association with increased H3K27me1 and H3K4me310, 11. In postnatal CMs, Ezh1/2 markedly decreases coincident with loss of regenerative potential and reduced CM cell cycle activity, leading us to hypothesize that Ezh1/2 is required for heart regeneration.
In this work, we studied the relative contribution of Ezh1 versus Ezh2 to heart development and regeneration. We show that Ezh1 and Ezh2 function redundantly to silence ectopic gene transcription in fetal CMs by depositing H3K27me3 in an “EZH” dose-dependent manner. Surprisingly, Ezh1 but not Ezh2 was required for neonatal heart regeneration. Moreover, over-expression of Ezh1 but not Ezh2 improved myocardial function after MI at postnatal stages with poor innate regenerative capacity. Thus our results indicate that heart regeneration requires a distinct set of epigenetic regulators from heart development.
METHODS
A detailed Materials and Methods section is available in the Online Data Supplement.
RESULTS
Cardiac Ezh1/2 DKO disrupted heart development and reduced CM proliferation in an “EZH” dose dependent manner
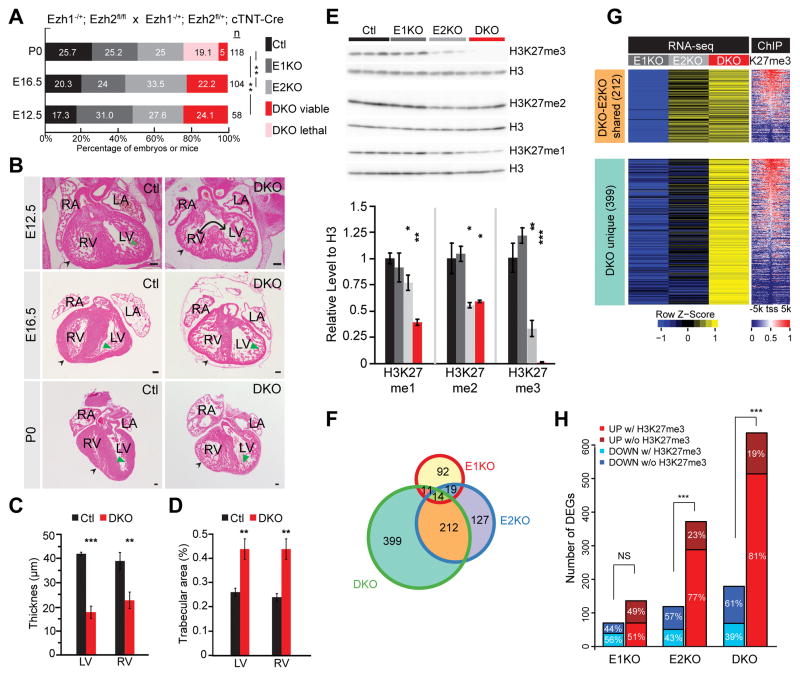
Consistent with previous studies12, constitutive Ezh1 knockout mice (E1KO) were phenotypically normal, viable, and fertile and showed no abnormalities in postnatal growth and heart function (Figure 1A and Online Figure IA). To further investigate the effect of PRC2 catalytic subunit EZH1 and EZH2 in early heart development, we inactivated an Ezh2 floxed allele (Ezh2fl/fl) in cardiac myocytes using cTNTCre (E2KO)13, which is active in differentiated cardiac myocytes by E8.514. By crossing Ezh1+/−; Ezh2fl/fl; cTNTCre+ and Ezh1−/−; Ezh2fl/fl mice, we generated compound double knockout embryos (Ezh1−/−; Ezh2fl/fl; cTNTCre+; abbreviated DKO). By embryonic day E12.5, DKO cardiac myocytes exhibited markedly reduced Ezh1 and Ezh2 expression (Online Figure IB). E12.5 and E16.5 DKO embryos were present at the expected Mendelian ratio, but exhibited abnormal heart development (Figure 1A and Online Figure IC). Unlike E1KO and E2KO, a majority of DKO mice died perinatally (Figure 1A). By E12.5 DKO mutant hearts exhibited hypoplasia of the compact myocardium of both left and right ventricles (Fig. 1B–D). Both ventricles also had excessive trabeculation.
Figure 1. Derangement of heart development and transcriptional regulation in cardiac Ezh1 and Ezh2 double knockout.
A. Survival analysis of mutants at indicated developmental stages. A majority of DKO died perinatally. Numbers next to bars indicating sample size. P, postnatal. E, embryonic. Pearson’s Chi-squared test was used for P-value calculation. B. Cardiac abnormalities of E12.5, E16.5 and P0 embryos. Representative HE stained transverse sections revealing thinning of compact myocardium (black arrowheads), excessive myocardial trabeculation (green arrowheads), and ventricular septal defect (double-headed arrow). Scale bar = 100 μm.C and D. Quantification showing decreased compact myocardial thickness (C) and increased myocardial trabecular area (D). E. Western blotting and quantification showing downregulation of bulk H3K27me1/2/3 in DKO. Heart apex of embryos at E16.5 was used for protein extraction. F. Venn diagram showing differentially expressed genes in E1KO, E2KO and DKO compared to Ctl (Ezh1−/+ or Ezh2fl/+; cTNT-Cre). Differentially expressed genes identified by Cuffdiff (P<0.05 and log2(fold change)>0.5 or <−0.5). G. Heat map showing RNA-seq and H3K27me3 at ± 5 kb of TSS of genes in F. Genes are ordered by decreasing promoter H3K27me3 signal. H. Bar graph showing association of differentially expressed genes with H3K27me3 enrichment at ± 5 kb of TSS. RV, right ventricle; LV: left ventricle; RA, right atrium; LA, left atrium; CM, cardiac myocyte. *, P<0.05; **, P<0.01; ***, P<0.001 compared to control.
Hypoplasia of compact myocardium suggested reduced CM proliferation. We stained histological sections of E13.5 DKO and control hearts for phosphorylated histone H3 (PH3), a cell cycle M-phase marker. Dual inactivation of Ezh1 and Ezh2 caused ~2.5-fold decrease in PH3 positive cardiac myocytes (P<0.05; Online Figure 1D,E). In line with decreased proliferation, the expression of cell cycle inhibitors Ink4a and Ink4b (Mouse Genome informatics: Cdkn2a/b), which we previously showed were repressed by PRC2 in fetal cardiac myocytes13, were dramatically upregulated in E12.5 DKO ventricles (Online Figure 1F), while the extent of upregulation was less in E2KO, coinciding with its normal heart development.
Next, we investigated the molecular regulation of gene expression in DKO hearts. By immunostaining, we found markedly reduced H3K27me3 in E13.5 DKO CMs (Online Figure 1G). Western blotting also showed the global reduction in H3K27me3 in E16.5 DKO heart (Figure 1E). Interestingly, H3K27me3 was also significantly reduced in E2KO, but not to the same degree as DKO, whereas it was normal in E1KO. These data suggest that Ezh2 is the main catalytic subunit of PRC2 in fetal CMs, but Ezh1 provides partial functional compensation after Ezh2 ablation. This level of compensation is sufficient to support normal heart development. We also found that H3K27me1 and H3K27me2, also deposited by PRC2, were reduced (Figure 1E) in DKO. Interestingly H3K27me2 was not different between E2KO and DKO, suggesting that Ezh1 does not compensate for its global level, and that global H3K27me2 level is not critically linked to normal heart development.
We globally measured gene expression by RNA-seq in E1KO, E2KO and DKO. A large number of genes (399) were uniquely upregulated in DKO and 212 genes were shared between DKO and E2KO (Figure 1F). Quantitative analysis of gene expression levels showed that the extent of gene dysregulation was much greater in DKO compared to E1KO or E2KO (Figure 1G). The promoters (TSS ± 5 kb) of the majority of these differentially expressed genes were occupied by H3K27me3 in controls (Figure 1G). Further analysis of the direction of differential expression (up or down regulated) and the presence of H3K27me3 at the promoter indicated that upregulated genes were significantly enriched for H3K27me3 (Figure 1H). These results suggest that most genes upregulated in DKO are normally directly repressed by PRC2. Gene ontology term analysis of upregulated genes showed that these upregulated genes are enriched for functional terms related to development, cell proliferation, and non-cardiac gene expression programs (Online Fig. II). Together, these results show that EZH1 and EZH2 function redundantly and regulate heart development by depositing H3K27me3 and repressing non-cardiac gene programs.
EZH1 but not EZH2 was essential for heart regeneration
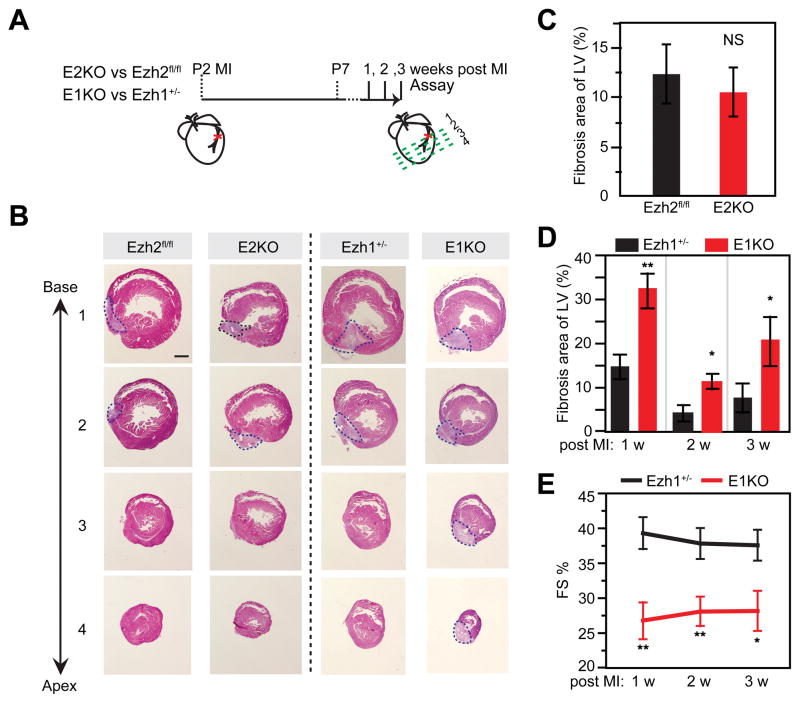
Reactivation of PRC2 has been linked to proliferation of tumor cells, and PRC2 is downregulated in postnatal cardiac myocytes coincident with loss of cardiac regenerative capacity. These observations led us to hypothesize that Ezh1 or Ezh2 are required for neonatal heart regeneration. To test this hypothesis, we studied the effect of Ezh1 or Ezh2 depletion on the neonatal heart’s capacity to regenerate after injury by induction of myocardial infarction (MI) by ligation of the left anterior descending coronary artery at postnatal day 2 (P2; Figure 2A). At 3 weeks after MI, both control (Ezh2fl/fl) and E2KO hearts had little fibrotic scar (Figure 2B,C), indicating that Ezh2 is dispensable for neonatal heart regeneration (data not shown). In contrast, E1KO mice had markedly increased fibrotic scar from 1–3 weeks after MI (Figure 2B,D). Consistent with impaired myocardial regeneration, E1KO mice had severely depressed heart function, as measured by echocardiography (Figure 2E). CM proliferation, measured by EdU incorporation and staining of transverse tissue sections, was clearly reduced in the border zone of E2KO (Online Figure III). These results indicate that Ezh1 but not Ezh2 is essential for cardiac regeneration following neonatal MI.
Figure 2. Ezh1 was required for neonatal heart regeneration after myocardial infarction.
A. Experimental design and timeline. B–D. Representative Masson’s trichrome-stained cross-sections from E2KO, Ezh2fl/fl, Ezh1+/−, and E1KO hearts one week after MI (B) and quantification of fibrotic area of LV (C–D). E. Quantification of heart function by echocardiography. FS%, Fractional shortening. *, P<0.05; **P<0.01; n=7 per group. Scale bar = 500 μm.
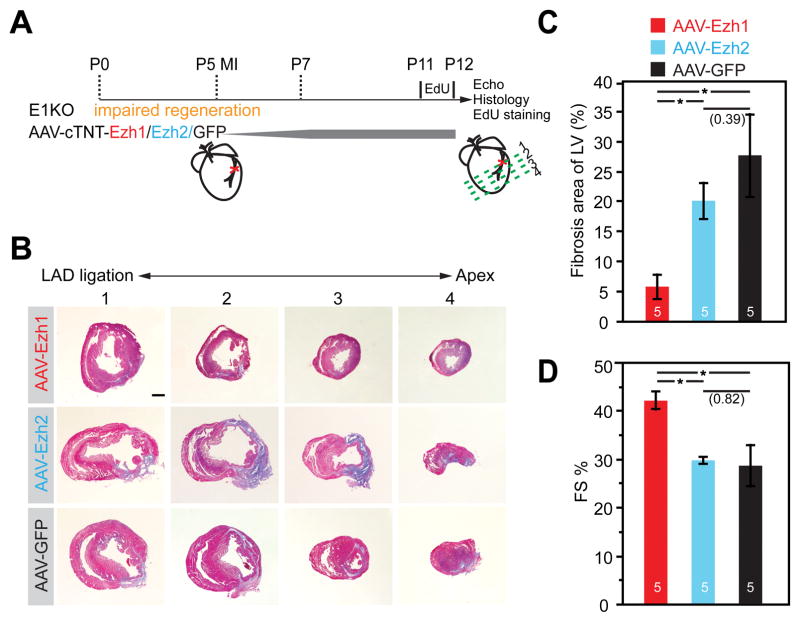
To confirm the differential requirement of Ezh1 and Ezh2 in this context, we performed a rescue experiment in which AAV9-cTNT-Ezh1, -Ezh2 or -GFP (control) were delivered to E1KO mice at P5, when MI was also induced (Figure 3A, and Online Figure IVA). We validated over-expression of EZH1, EZH2, and GFP proteins in CMs (Online Figure IVB–D). CM-specific re-expression of Ezh1, but not Ezh2 or GFP, dramatically reduced fibrosis area of injured E1KO hearts (Figure 3B, C) and improved heart function (Figure 3D). We also examined CM proliferation in the rescue experiment. Ezh1 over-expression increased the incidence of EdU+ CMs by nearly 7-fold relative to Ezh2 or GFP over-expression (Online Figure III). Unlike EZH1, overexpression of EZH2 in E1KO did not rescue fibrosis or heart function (Figure 3B–D). These results confirm the key role of Ezh1 in heart regeneration, and show that Ezh2 cannot functionally replace Ezh1 in cardiac regeneration. Furthermore, since AAV9 with Tnnt2 promoter selectively drives gene expression in cardiac myocytes15, these data demonstrate that CMs are the cell type that requires Ezh1 for effective cardiac regeneration. Collectively, these data support the crucial role of EZH1 in neonatal heart regeneration through modulating cardiac myocyte proliferation.
Figure 3. Re-expression of Ezh1 but not Ezh2 restored cardiac function and regeneration in E1KO hearts.
A. Schematic of rescue experimental design. AAV-cTNT-Ezh1/Ezh2/GFP were injected 24 hours subcutaneously before MI, and EdU was injected 24 hours before harvest. B–C. Representative Masson’s trichrome staining of cross-sections from E1KO mice treated with indicated AAV (B) and quantification of the percentage of the LV occupied by fibrotic scar (C). D. Echocardiographic measurement of heart function. FS%, fractional shortening. Scale bar = 500 μm; *, P<0.05. Numbers in parentheses indicate non-significant p-values. Numbers in bars indicate group size.
EZH1 over-expression prolongs the heart regeneration window
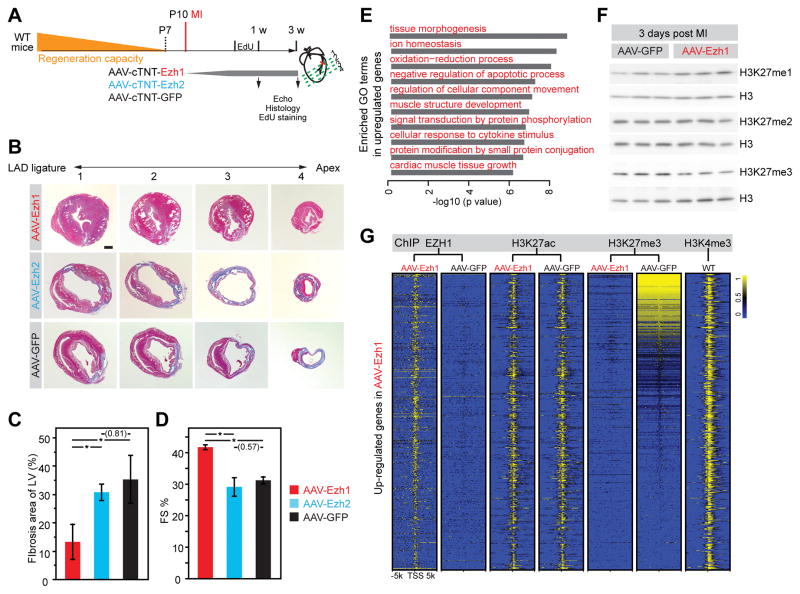
Because Ezh1 played a crucial role in both heart morphogenesis and regeneration, we next tested the hypothesis that Ezh1 might be sufficient to prolong the window during which the neonatal murine heart remains competent for regeneration. By P7, murine hearts have lost regenerative capacity, and as a result myocardium lost from myocardial infarction is replaced by fibrotic scar. We studied the effect of over-expression of AAV9-expressed Ezh1, Ezh2, or GFP on heart regeneration following MI induced at P10, well outside the innate regenerative window (Figure 4A). Histological evaluation 3 weeks after MI showed that AAV-Ezh1 markedly and significantly reduced fibrotic scar area compared to AAV-GFP, whereas AAV-Ezh2 had no significant effect (Figure 4B–C). Echocardiography demonstrated that AAV-Ezh1 significantly improved heart function compared to AAV-GFP, whereas AAV-Ezh2 was ineffective (Figure 4D).
Figure 4. Over-expression of Ezh1 but not Ezh2 promoted heart regeneration by upregulating cardiac muscle growth genes.
A. Schematic of rescue experimental design. AAV-cTNT-Ezh1, -Ezh2, or -GFP were injected subcutaneously 3 days before MI at P10, and EdU was injected 24 hours before harvest. Separate cohorts were analyzed at 1 week or 3 weeks post MI. B–C. Masson’s trichrome staining of cross-sections from hearts treated with AAV expressing indicated transgenes. B, representative sections. C, quantitative analysis. D. Echocardiographic measurement of heart function at 3 weeks post MI. FS%, fractional shortening. E. GO terms in which genes up-regulated in AAV-Ezh1-treated post MI hearts were enriched. F. Western blotting showing H3K27me1/2/3 bulk levels in hearts of mice injected with AAV-GFP or -Ezh1. G. Heat map showing H3K27me3, H3K27ac, EZH1 and H3K4me3 ChIP-seq signals at ± 5 kb of TSS of upregulated genes in AAV-Ezh1 compared to AAV-GFP.
Scale bar = 500 μm; *, P<0.05. There were 4–6 replicates per group. Numbers in parentheses indicate non-significant P-values.
To examine the cellular mechanism, we measured CM proliferation by EdU staining. We found that EdU+ CM frequency in the border zone of AAV-Ezh1 hearts was significantly greater than AAV-GFP, and AAV-Ezh2 did not have a significant effect (Online Figure VI A,B). To study the effect of Ezh1 over-expression on apoptosis, we measured the fraction of CMs positive for TUNEL staining, a marker for apoptosis. Compared to the AAV-Ezh2 group, TUNEL+ CMs were 10-fold less frequent in the AAV-Ezh1 group (Online Figure VI C,D), suggesting that EZH1 also enhances CM survival. Border zone CMs in AAV-Ezh1 hearts were significantly smaller than in AAV-GFP or AAV-Ezh2 (Online Figure VI E,F), which likely reflects amelioration of compensatory hypertrophy in AAV-Ezh1, as well as smaller size of proliferative CMs.
To dissect the molecular mechanism underlying enhanced cardiac myocyte proliferation and survival induced by Ezh1 over-expression, we profiled gene expression by RNA-seq and genomic occupancy of EZH1, H3K27me3, H3K27ac and H3K4me3 by ChIP-seq. Compared with AAV-GFP mice, 597 and 182 differentially expressed genes were significantly up- and down-regulated, respectively, in AAV-Ezh1 heart after MI (Online Figure VIIA). Upregulated genes were enriched in GO terms related to embryonic morphogenesis and cardiac muscle growth (Figure 4E), while down-regulated genes did not yield any significantly enriched terms. qRT-PCR validated 7 out 11 upregulated genes involved in fetal heart development (Online Figure VIIB).
EZH1 deposits H3K27me3, canonically a repressive histone mark9. A distinct EZH1-containing protein complex has been shown to activate genes in association with accumulation of H3K4me3 and H3K27me110, 11, histone marks which has been linked to gene activation16. To examine the mechanism of gene dysregulation, we measured H3K27me1/2/3 bulk levels by immunoblotting. Surprisingly, compared to AAV-GFP treatment, AAV-Ezh1 caused a reduction in overall H3K27me3 and a clear increase in H3K27me1 (Figure 4F). This result was further reinforced by genome-wide profiling of EZH1, H3K27me3, and H3K27ac chromatin occupancy by ChIP-seq. This showed that overexpressed EZH1 was found at the TSS of most upregulated genes (Figure 4G). Consistent with bulk levels, H3K27me3 occupancy of many upregulated genes was reduced by AAV-Ezh1 (Figure 4G). Furthermore, only 5535/31124 (17.8%) of EZH1 peaks overlapped with H3K27me3 peaks (Online Figure VIIC). These data suggest that EZH1 has alternate mechanism that is not dependent upon deposition of H3K27me3. In agreement with the reported activation role of EZH110, 11, we observed substantial overlap between EZH1 and H3K4me3 occupied chromatin regions (Online Figure VIID), including at promoters (±5 kb of TSS) of upregulated genes (Figure 4G). However, H3K27ac, a marker of active enhancers and promoters, did not obviously change at these promoters (Figure 4G). Together, these data suggest that EZH1 upregulates genes by increasing H3K27me1 levels and activating heart growth related genes, leading to augmented proliferation and survival during heart regeneration after injury.
DISCUSSION
Recent studies highlight strategies that redeploy developmental signaling pathways to achieve the goals of cardiac regeneration4 and point to epigenetic barriers to effective regeneration in mammalian hearts17 Previous work by us and others suggested that Ezh1 is functionally redundant with Ezh2 in differentiated cardiac myocytes13, 18, 19. Here, we provide direct evidence that Ezh1 and Ezh2 complement each other in heart development. Furthermore, we extended the analysis of Ezh1 and Ezh2 function to heart regeneration and made the surprising discovery that these genes have distinct activities in regeneration compared to development. Specifically, whereas Ezh2 was functionally dominant in development but dispensable for regeneration, Ezh1 contributed to heart development and was essential for heart regeneration. Indeed, Ezh1 was sufficient to enhance regeneration in 10 day old hearts, which normally lack regenerative capacity. It will be important to evaluate the ability of Ezh1 overexpression to improve myocardial outcome after adult heart injury.
The molecular mechanisms underlying EZH1/2 activity in development and regeneration were also distinct. As shown in ref. 13 and this study, during development Ezh2 and, to a lesser (but functionally sufficient) degree Ezh1 deposited H3K27me3 and repressed non-cardiac gene programs. In regeneration, EZH1 bound and upregulated genes related to cardiac growth and marked by H3K4me3, and globally increased H3K27me1. This suggests that EZH1 functions in regeneration in an EZH1-SUZ12 complex lacking EED that has been linked to transcriptional activation and associated with H3K4me3 and H3K27me110. Further experiments will be required to further define the EZH1-containing protein complex required for heart regeneration.
Collectively, our work illuminates a novel role for EZH1 in heart regeneration and illustrates the divergent role of epigenetic regulators in heart development and regeneration, as summarized in the working model (Online Figure VIII). Further interrogation of the factors that enhance or decrease competence for neonatal cardiac regeneration may provide a framework for resetting the epigenetic landscape to enhance myocardial outcome after heart injury.
Supplementary Material
Online table I. Antibodies used in this study
Online Table II. Primers used in this study
NOVELTY AND SIGNIFICANCE.
What Is Known?
Polycomb repressive complex 2 (PRC2) subunits EZH1 and EZH2 catalyze trimethylation of histone H3 lysine 27 (H3K27me3) to repress gene expression.
PRC2 regulates chromatin function during normal heart development.
Developmental pathways are often reactivated to drive organ regeneration.
What New Information Does This Article Contribute?
EZH1 and EZH2 are functionally redundant in differentiated cardiac myocytes for heart development.
EZH1 but not EZH2 is essential for neonatal heart repair.
EZH1 promotes heart repair by activating genes involved in cardiac growth.
The adult mammalian heart has limited regenerative capacity after injury. Epigenetic marks such as covalent histone modifications are important regulators of gene expression. However, little is known about the role of the epigenetic regulators in heart development vs regeneration. We investigated the function of EZH1 and EZH2, alternative subunits of PRC2, a major epigenetic repressor complex, in heart development and regeneration. Our data show that in developing heart, EZH1 and EZH2 function redundantly to silence ectopic gene transcription by depositing repressive histone mark H3K27me3. Surprisingly, EZH1, but not EZH2, was required for heart regeneration after neonatal myocardial infarction (MI). Moreover, over-expression of EZH1 but not EZH2 improved myocardial function after MI at postnatal stages with poor innate regenerative capacity. EZH1 activated cardiac growth genes to promote heart repair by enhancing the deposition of active histone marks. These findings suggest that heart regeneration requires epigenetic regulators and mechanisms that differ from those involved in heart development.
Acknowledgments
SOURCES OF FUNDING
AH was supported by a startup fund from Peking-Tsinghua Center for Life Sciences, Peking University, the 1000 Youth Talents Program of China, the National Natural Science Foundation of China (31571487). WTP was supported by funding from the National Heart Lung and Blood Institute (2UM1 HL098166 and HL116461) and by an Established Investigator Award from the American Heart Association.
Nonstandard Abbreviations and Acronyms
- qRT-PCR
quantitative RT-PCR
- ChIP-Qpcr
quantitative chromatin immunoprecipitation
- ChIP-seq
chromatin immunoprecipitation followed by next generation sequencing
- RNA-seq
RNA analysis by next generation sequencing
- E2KO
mutant Ezh2fl/fl::Tg(cTNT-Cre+) genotype
- E1KO
mutant Ezh1−/− genotype
- DKO
double mutants Ezh1−/−::Ezh2fl/fl::Tg(cTNT-Cre+) genotype
- PH3
histone H3 phosphorylated on serine 10, a marker of M phase of the cell cycle
Footnotes
DISCLOSURES
The authors have no conflicts of interest to disclose.
References
- 1.Bergmann O, Bhardwaj RD, Bernard S, Zdunek S, Barnabe-Heider F, Walsh S, Zupicich J, Alkass K, Buchholz BA, Druid H, Jovinge S, Frisen J. Evidence for cardiac myocyte renewal in humans. Science. 2009;324:98–102. doi: 10.1126/science.1164680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahuja P, Sdek P, MacLellan WR. Cardiac myocyte cell cycle control in development, disease, and regeneration. Physiol Rev. 2007;87:521–44. doi: 10.1152/physrev.00032.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Porrello ER, Mahmoud AI, Simpson E, Hill JA, Richardson JA, Olson EN, Sadek HA. Transient regenerative potential of the neonatal mouse heart. Science. 2011;331:1078–80. doi: 10.1126/science.1200708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Galdos FX, Guo Y, Paige SL, VanDusen NJ, Wu SM, Pu WT. Cardiac Regeneration: Lessons From Development. Circ Res. 2017;120:941–59. doi: 10.1161/CIRCRESAHA.116.309040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin Z, von Gise A, Zhou P, Gu F, Ma Q, Jiang J, Yau AL, Buck JN, Gouin KA, van Gorp PR, Zhou B, Chen J, Seidman JG, Wang DZ, Pu WT. Cardiac-specific YAP activation improves cardiac function and survival in an experimental murine MI model. Circ Res. 2014;115:354–63. doi: 10.1161/CIRCRESAHA.115.303632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tao G, Kahr PC, Morikawa Y, Zhang M, Rahmani M, Heallen TR, Li L, Sun Z, Olson EN, Amendt BA, Martin JF. Pitx2 promotes heart repair by activating the antioxidant response after cardiac injury. Nature. 2016;534:119–23. doi: 10.1038/nature17959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xiang FL, Guo M, Yutzey KE. Overexpression of Tbx20 in Adult Cardiac myocytes Promotes Proliferation and Improves Cardiac Function After Myocardial Infarction. Circulation. 2016;133:1081–92. doi: 10.1161/CIRCULATIONAHA.115.019357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang CP, Bruneau BG. Epigenetics and cardiovascular development. Annu Rev Physiol. 2012;74:41–68. doi: 10.1146/annurev-physiol-020911-153242. [DOI] [PubMed] [Google Scholar]
- 9.Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–43. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 10.Xu J, Shao Z, Li D, Xie H, Kim W, Huang J, Taylor JE, Pinello L, Glass K, Jaffe JD, Yuan GC, Orkin SH. Developmental Control of Polycomb Subunit Composition by GATA Factors Mediates a Switch to Non-Canonical Functions. Mol Cell. 2015 doi: 10.1016/j.molcel.2014.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mousavi K, Zare H, Wang AH, Sartorelli V. Polycomb protein Ezh1 promotes RNA polymerase II elongation. Mol Cell. 2012;45:255–62. doi: 10.1016/j.molcel.2011.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ezhkova E, Lien WH, Stokes N, Pasolli HA, Silva JM, Fuchs E. EZH1 and EZH2 cogovern histone H3K27 trimethylation and are essential for hair follicle homeostasis and wound repair. Genes Dev. 2011;25:485–98. doi: 10.1101/gad.2019811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He A, Ma Q, Cao J, von Gise A, Zhou P, Xie H, Zhang B, Hsing M, Christodoulou DC, Cahan P, Daley GQ, Kong SW, Orkin SH, Seidman CE, Seidman JG, Pu WT. Polycomb Repressive Complex 2 Regulates Normal Development of the Mouse Heart. Circ Res. 2012;110:406–15. doi: 10.1161/CIRCRESAHA.111.252205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiao K, Kulessa H, Tompkins K, Zhou Y, Batts L, Baldwin HS, Hogan BL. An essential role of Bmp4 in the atrioventricular septation of the mouse heart. Genes Dev. 2003;17:2362–7. doi: 10.1101/gad.1124803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prendiville TW, Guo H, Lin Z, Zhou P, Stevens SM, He A, VanDusen N, Chen J, Zhong L, Wang DZ, Gao G, Pu WT. Novel Roles of GATA4/6 in the Postnatal Heart Identified through Temporally Controlled, Cardiac myocyte-Specific Gene Inactivation by Adeno-Associated Virus Delivery of Cre Recombinase. PLoS One. 2015;10:e0128105. doi: 10.1371/journal.pone.0128105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–37. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 17.Oyama K, El-Nachef D, Zhang Y, Sdek P, MacLellan WR. Epigenetic regulation of cardiac myocyte differentiation. Front Genet. 2014;5:375. doi: 10.3389/fgene.2014.00375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Delgado-Olguin P, Huang Y, Li X, Christodoulou D, Seidman CE, Seidman JG, Tarakhovsky A, Bruneau BG. Epigenetic repression of cardiac progenitor gene expression by Ezh2 is required for postnatal cardiac homeostasis. Nat Genet. 2012;44:343–7. doi: 10.1038/ng.1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen L, Ma Y, Kim EY, Yu W, Schwartz RJ, Qian L, Wang J. Conditional ablation of Ezh2 in murine hearts reveals its essential roles in endocardial cushion formation, cardiac myocyte proliferation and survival. PLoS One. 2012;7:e31005. doi: 10.1371/journal.pone.0031005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Online table I. Antibodies used in this study
Online Table II. Primers used in this study