Abstract
The identification of the kinase or kinases targeted by protein kinase inhibitors is a critical challenge in validating their use as therapeutic agents or molecular probes. Here, to address this problem, we describe a chemical genomics strategy that uses a direct comparison between microarray transcriptional signatures elicited by an inhibitor of unknown specificity and those elicited by highly specific pharmacological inhibition of engineered candidate kinase targets. By using this approach, we have identified two cyclin-dependent kinases, Cdk1 and Pho85, as the targets of the inhibitor GW400426 in Saccharomyces cerevisiae. We demonstrate that simultaneous inhibition of Cdk1 and Pho85, and not inhibition of either kinase alone, by GW400426 controls the expression of specific transcripts involved in polarized cell growth, thus revealing a cellular process that is uniquely sensitive to the multiplex inhibition of these two kinases. Our results suggest that the cellular responses induced by multiplex protein kinase inhibitors may be an emergent property that cannot be understood fully by considering only the sum of individual inhibitor–kinase interactions.
Keywords: chemical genetics, genomics, target validation
Protein kinases are key regulators of most cellular signaling pathways in eukaryotic cells. Many protein kinase inhibitors have been developed to study specific functions of kinases in signaling pathways and as potential therapeutic agents (1). Because of the large size of the protein kinase superfamily (>500 human, >120 yeast) and the fact that most kinase inhibitors bind in the highly conserved ATP-binding pocket, it is widely accepted that kinase inhibitors inhibit more than one target (2). As a result, the inhibitors used as chemical tools to probe the often poorly understood roles of kinases in signaling pathways are paradoxically of incompletely characterized specificity. The lack of systematic methods to identify targets of kinase inhibitors accurately within cells has resulted in a situation in which it is equally difficult to rationalize why some kinase inhibitors become failed drugs, whereas others demonstrate surprising clinical efficacy (3).
To assess the full spectrum of cellular targets of kinase inhibitors, phenotypic information garnered from biological readouts such as genome-wide transcriptional profiles (4) and complex morphological screens (5) must be matched accurately to discrete interactions between the compound and the relevant protein targets. Current approaches to identify small molecule–target interactions provide either biochemical target information or rely on broad phenotypic outputs, such as cell death. For example, affinity purification using bead-immobilized kinase inhibitors results in the identification of both relevant and spurious targets (6). Cell-based high-throughput screening technologies, such as synthetic lethal (7) or haploinsufficiency (8, 9) screens, provide information about cellular pathways that control drug sensitivity but, because not all kinases are essential, do not necessarily identify all targets in a cell.
We have developed (10) a chemical genetic method that allows for the potent and monospecific pharmacological inhibition of individual kinases. A functionally silent mutation in the ATP active site sensitizes a protein kinase (an analog-sensitive allele) to specific inhibition by the small molecules 1-NA-PP1 or 1-NM-PP1. We envisioned identifying targets of kinase inhibitors by using a drug-to-drug comparative approach in which cellular effects caused by kinase inhibitors of incompletely characterized specificity could be matched to “reference profiles” of cellular effects elicited by specific inhibition of candidate analog-sensitive kinases. The advantage of such an approach is that, through iterative comparison with such reference profiles, targets that together account for all cellular effects of drug treatment could be identified (11). Here, we have used this strategy to identify the cellular targets in Saccharomyces cerevisiae of GW400426, a cyclin-dependent kinase (CDK) inhibitor of previously unknown specificity in this organism.
Materials and Methods
Chemical Synthesis. GW400426, 1-NA-PP1, and 1-NM-PP1 were synthesized as described (10, 19).
Strains and Plasmids. YRP1 was a gift from Karl Kuchler (Medical University of Vienna, Vienna). Pho85-as1 and Cdk1-as1 strains have been described (10, 12). The dual Cdk1-as1/Pho85-as1 strain was generated by integrating Cdk1-as1 into the Pho85-as1 strain by using standard pop-in/pop-out genetic techniques (13). Pho4-GFP strains were generated by transforming a GFP-Pho4::URA3 plasmid (14) into Pho85-as1 or YRP1 yeast and selecting on plates lacking uracil (-URA). Ipl1-as6 strain was created by first cloning, by means of homologous recombination, the Ipl1 ORF with 250 bp of upstream and downstream sequence into a pRS316 plasmid, simultaneously introducing the M181G (Ipl1-as1) mutation. The M181G T244G (Ipl1-as6) strain was created by QuikChange site-directed mutagenesis (Stratagene). The resulting plasmid was transformed into a diploid yeast strain with a heterozygous deletion of the IPL1 gene, the strain was sporulated, and the resulting spores were analyzed by tetrad dissection to identify haploid strains with both the IPL1 knockout and Ipl1-as6 plasmid.
Kinase IC50 Assays. Cdk1-His-6 and MBP-Clb2 were purified as described (10). Varying concentrations of GW400426 were incubated for 10 min at 23°C in a 25-μl reaction mixture containing 1 ng of Cdk1-His-6, 10 ng of MBP-Clb2, 5 μg of histone H1, 100 μM ATP, and 0.5 μCi (1 Ci = 37 GBq) of [γ-32P]ATP in kinase buffer (25 mM Hepes·NaOH, pH 7.4/10 mM NaCl/10 mM MgCl2/1mM DTT). Pho85 and Pho80 were purified recombinantly as a complex from Escherichia coli and used to monitor phosphorylation of Pho4 as described (15). Reactions included 100 pM of the kinase complex, 3 μM Pho4, 1 mM ATP, and 86 nM [γ-32P]ATP. All reaction products were analyzed by 12% SDS/PAGE, followed by autoradiography. For Cak1 IC50 determination, 10 ng of GST-Cak1 was incubated with 84 ng of GST-CDK2/10 μM ATP/5 μCi of [γ-32P]ATP as described (16), except in 5% DMSO because of the addition of inhibitor. All quantitation was performed with a Storm 860 PhosphorImager (Molecular Dynamics).
Orc6 Phosphorylation. Exponentially growing Cdk1-as1 or YRP1 cells were treated with DMSO, 1-NA-PP1, or GW400426 for 15 min. Cellular proteins were extracted into urea lysis buffer (20 mM Tris, pH 7.4/7 M urea/2 M thiourea/4% 3-[(3-cholamidopropy-l)dimethylammonio]-1-propanesulfonate/1% DTT/50 mM NaF/80 mM β-glycerophosphate/1 mM Na3VO4/1 mM PMSF), run out on SDS/PAGE, and blotted to nitrocellulose. The blot was probed with an mAb against Orc6 (SB49; 1:1,000) and visualized by ECL after probing with an horseradish peroxidase-conjugated goat anti-mouse Ab (Pierce; 1:1,500). Densitometry quantitation was done by using imagej software (available at: http://rsb.info.nih.gov/ij).
Pho4-GFP. Pho85-as1 or YRP1 cells carrying the Pho4-GFP plasmid were grown under selection to an OD600 of 0.5 and treated with 1% DMSO, 5 μM 1-NA-PP1 (Pho85-as1), or 20 μM GW400426 (YRP1). Samples were analyzed with static microscopy at 15 min after treatment. At least 100 cells were counted for each treatment.
Microarray Analysis. Microarrays containing ≈93% of yeast ORF full-length PCR products were fabricated as described (4). Yeast cells of the appropriate strain were grown to an OD600 of 0.7 and treated with either inhibitor or the equivalent volume of DMSO for 10 min. The cells were collected by filtration and flash-frozen in liquid nitrogen. Yeast total RNA preparation was carried out by using the hot acid phenol method (available at: www.microarrays.org). Selection for polyadenylated messenger RNA was carried out on 1 mg of total RNA by using the OligoTex kit (Qiagen). First-strand cDNA synthesis was carried out by using StrataScript reverse transcriptase (Stratagene) in the presence of a dNTP/amino-allyl-dUTP (Sigma) mixture. The cDNA from paired samples was then labeled with either Cy3 or Cy5 dyes and hybridized to the microarray as described (4). Fluorescence ratios were obtained with an Axon 4000A scanner. For experiments shown in Fig. 2a (except for lane 9), each experiment was done in replicate with Cy3 and Cy5 labeling reversed between inhibitor and DMSO treatments in the replicate experiments. “Dye-flipped” expression ratios were inverted and then averaged in log-space with their nonflipped counterparts. In Fig. 2a (lane 9) and for the time-course experiment shown in Fig. 2c, cells were treated with either inhibitor or DMSO, and hybridization was conducted between each sample and cDNA generated from a common reference RNA sample. Ratios of inhibitor/DMSO treatments at each time point were calculated in silico as described above.
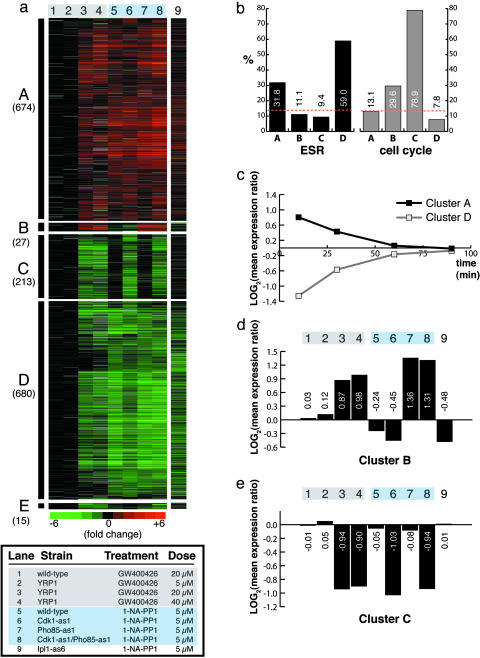
Fig. 2.
Hierarchical clustering of microarray data identifies gene expression clusters resulting from kinase inhibition. (a) Five robust clusters of genes (A–E) identified by hierarchical clustering of microarray expression data (lanes 1–8). Yeast cells were treated for 10 min. Log expression ratios for drug treatments compared with a DMSO control are shown. Each data point represents the geometric mean from two microarray experiments. Gray indicates incomplete data. Lane 9 shows data from genes in clusters A–E garnered from a 30-min treatment of Ipl1-as6 cells with 1-NA-PP1. (b) Percentage of genes within each cluster that are known to be cell-cycle-regulated or change in expression as part of the ESR. Orange dotted lines indicate the percentage of genes in the yeast genome that are involved in either process. Clusters A and D are enriched in induced and repressed ESR genes, respectively, demonstrating that these clusters correspond to the nonspecific yeast multidrug resistance response. Cluster C, corresponding to inhibition of Cdk1, is heavily enriched in cell-cycle-regulated genes. (c) Microarray data from a time course of WT cells treated with 5 μM of 1-NM-PP1. The log mean expression ratios for genes in cluster A or D are plotted at each time point. (d) Quantification of mean expression ratios from cluster B in a. (e) Quantification of mean expression ratios from cluster C in a.
To identify functional clusters of genes as shown in Fig. 2a, expression ratios were converted to log-space and the data set was filtered to include genes whose expression changed by ≥1.6-fold in at least two experiments. Genes were clustered with cluster 3.0 software by using average-linkage hierarchical clustering. For clarity, two columns of redundant data used in the initial clustering have been omitted from Figs. 2a and 3c. Cell-cycle-regulated genes were annotated based on the assignment of Spellman et al. (17). Environmental stress–response genes were annotated based on the assignment of Gasch et al. (18). Genes shown in Fig. 3c were identified by filtering in excel (Microsoft) by using a quantitative metric as follows: geometric mean of 20/40 μM GW400426 treatments >1.5-fold repressed, dual-inhibited strain >1.4-fold repressed, <1.67-fold repression in Pho85-as1 or WT treatments, (average of Cdk1-as1 inhibited)/(average GW400426 treatments) > 1.3. All raw and processed data are available as Data Sets 1–5, which are published as supporting information on the PNAS web site.
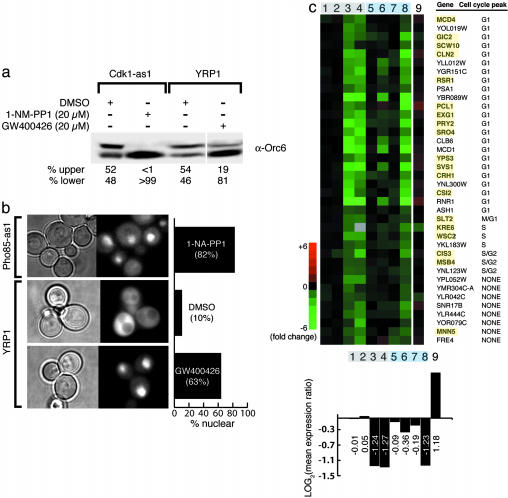
Fig. 3.
Functional validation of putative GW400426 targets. (a) Western blot analysis quantitating Orc6 phosphoisoforms present in Cdk1-as1 or YRP1 cells after treatment for 15 min with DMSO, 1-NA-PP1, or GW400426. (b) Representative fields of cells after treatment for 15 min of Pho85-as1 or YRP1 cells expressing a Pho4-GFP fusion construct with 5 μM 1-NA-PP1, 1% DMSO, or 20 μM GW400426. The percentage of fluorescent cells with nuclear-localized Pho4-GFP is quantified on the right. (c) Transcripts that are repressed most strongly by GW400426 or treatment of a Cdk1-as1/Pho85-as1 strain are highly enriched in G1 cell-cycle-regulated genes. Highlighted genes represent those with known roles in polarized cell growth. Quantification of mean expression ratios from each column are shown at the bottom (lanes are equivalent to those in Fig. 2a).
Supporting Information. For details on halo assays and doubling time measurements, see Supporting Text, which is published as supporting information on the PNAS web site.
Results
We chose to investigate the cellular effects of a class of highly potent and relatively specific inhibitors of mammalian CDK2 (19) (Fig. 1b). We sought to determine whether a representative compound of this class of inhibitors, oxindole GW400426 (Fig. 1a), would inhibit any of the 126 kinases that are present in the S. cerevisiae genome. To obtain a detailed picture of cellular effects elicited by GW400426, we conducted whole-genome microarray analysis on yeast cells treated with GW400426 at a concentration of 20 μM (see Fig. 5, which is published as supporting information on the PNAS web site). There were few significant (zero >1.5-fold) gene expression changes after a 10-min course of treatment and, notably, none in stress-response genes that normally respond to the presence of foreign molecules. GW400426 also fails to inhibit the growth of yeast cells at concentrations up to 50 μM (data not shown). These findings are consistent with the fact that many pharmacological agents are inert in yeast because of poor permeability across the yeast cell wall (20).
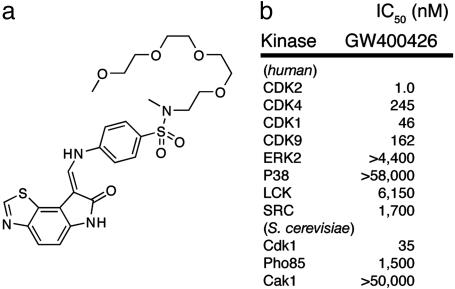
Fig. 1.
GW400426 is a potent in vitro CDK inhibitor. (a) Chemical structure of CDK inhibitor GW400426. (b)IC50 values for GW400426 against a spectrum of human and yeast kinases.
To circumvent the cell-permeability issue, we turned to a drug-sensitized yeast strain YRP1 (snq2Δpdr5Δerg6Δ). GW400426 inhibits the growth of YRP1 cells both on plates and in liquid culture (GI50 = 20 μM) (see Fig. 5). We conducted whole-genome microarray analysis (4) of YRP1 cells treated with GW400426 at three concentrations (5, 20, and 40 μM). At the two higher concentrations, we observed a large transcriptional response (Fig. 2a, lanes 3 and 4), with ≈1,600 induced or repressed genes.
Next, we sought to identify the genes, if any, whose expression had changed specifically in response to inhibition of protein kinases. The growth-inhibition data suggested that possibly one or more kinases essential for viability were being inhibited by GW400426. The yeast CDK2 homolog Cdk1 (essential) and the closely related CDK Pho85 (52% sequence identity to Cdk1, nonessential) were evaluated as likely candidate targets, because other mammalian CDK inhibitors have been shown to target these two kinases (21) in yeast. To construct reference profiles of transcriptional effects corresponding to specific inhibition of either kinase, we conducted microarray analysis on yeast strains carrying analog-sensitive alleles of Cdk1 and Pho85 (Cdk1-as1 and Pho85-as1, respectively) treated with 5 μM 1-NA-PP1. To determine whether inhibition of different kinases produces discernibly distinct transcriptional profiles, we also conducted microarray analysis on an analog-sensitive allele of the essential yeast mitotic kinase Ipl1 (Ipl1-as6) treated with 5 μM 1-NA-PP1.
We integrated the GW400426 and kinase-profile data sets and used hierarchical clustering combined with simple filtering criteria to ascertain common patterns in gene expression (22). In principle, our profiling strategy should result in the assembly of clusters of genes that could be correlated to specific inhibition of different kinases. In fact, we were able to identify five general clusters of genes (Fig. 2a) that emerged repeatedly despite the use of various clustering algorithms and filtering stringency.
Two of the clusters (A and D) included several hundred genes that were induced or repressed, respectively, across all drug treatments. We suspected that most, if not all, of these gene-expression changes were due to the yeast environmental stress–response (ESR) when challenged with foreign molecules or other stress conditions (18). Consistent with this hypothesis, these expression changes occurred even when we treated WT yeast with 5 μM 1-NA-PP1 (Fig. 2a, lane 5), a molecule that inhibits only sensitized kinases (10). These gene clusters are highly enriched in ESR genes (Fig. 2b) with >70% of the consensus yeast ESR genes represented in clusters A or D. Because of our choice to use 10-min inhibitor treatments to evaluate acute effects of kinase inhibition, our observations are subject to the peak of the ESR, a transient response that largely subsides after 1 h. To verify that this behavior applies to the genes in clusters A and D, we conducted a time course over 90 min of WT cells treated with 5 μM 1-NM-PP1. As shown in Fig. 2c, the average expression ratio of genes in clusters A and D approaches unity over this time span. Nevertheless, the use of chemical genetic-reference profiles and relatively simple clustering algorithms separates these general xenobiotic responses from responses due to kinase inhibition (clusters B, C, and E).
Examination of the 213 genes in cluster C reveals that this cluster of genes appears to represent those specifically regulated upon inhibition of Cdk1. Treating YRP1 cells with GW400426 or Cdk1-as1 cells with 1-NA-PP1 represses transcription of the genes in this cluster to a similar degree (Fig. 2e, lanes 3, 4, and 6). In contrast, the expression of these genes does not change in WT, Pho85-as1, or Ipl1-as6 cells treated with 1-NA-PP1 (Fig. 2e, lanes 5, 7, and 9). Consistent with the role of Cdk1 as the primary CDK responsible for driving cell-cycle progression, this cluster is heavily enriched (79%) (Fig. 2b) in genes whose transcription is known to be cell-cycle-regulated (17). Furthermore, highly represented in this cluster are entire suites of genes thought to be coordinately regulated by Cdk1 through control of specific transcription factor complexes [e.g., Swi five factor (SFF)-dependent CLB2 mitotic transcripts and MBF-dependent DNA repair/replication S-phase transcripts] (17, 23) (Fig. 4 Top).
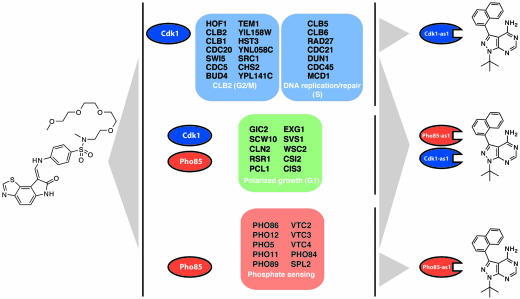
Fig. 4.
GW400426 elicits three distinct responses through inhibiton of yeast CDKs. Representative functional sets of genes (shown in Fig. 2a, clusters C and B, and Fig. 3c, respectively) corresponding to cellular responses elicited from inhibition of Cdk1 (Top), Pho85 (Bottom), or both simultaneously (Middle) by GW400426 are shown. Genes were validated by comparison with inhibition of the corresponding analog-sensitive strain with 1-NA-PP1 (as shown in Right).
GW400426 treatment also results in the induction (Fig. 2d, lanes 3 and 4) of a cluster of genes (Fig. 2a, cluster B) that appears to correspond to inhibition of Pho85. When we treated a Pho85-as1 strain with 5 μM 1-NA-PP1, we observed a robust increase in expression of these genes (Fig. 2d, lane 7), a response lacking in the WT, Cdk1-as1, or Ipl1-as6 strains (lanes 5, 6, and 9, respectively). Cluster B includes many genes that are involved in the yeast-sensory response to phosphate starvation (Fig. 4 Bottom) whose expression is controlled by Pho85 through its phosphorylation of the transcription factor Pho4 (14). The response to GW400426 is somewhat diminished in magnitude compared with the Pho85-as1 inhibited strain (discussed below).
We sought to independently confirm the microarray data identifying Cdk1 as a target of GW400426 by examining the phosphorylation state of a direct substrate of Cdk1 after inhibitor treatment. The origin of replication component Orc6 is phosphorylated by Cdk1 at the G1/S cell-cycle transition and dephosphorylated during mitosis, resulting in the presence of two phosphoisoforms in exponentially growing cells (24). Treating Cdk1-as1 cells with 20 μM 1-NM-PP1 results in the disappearance of the Cdk1-dependent phosphoisoform (Fig. 3a). Similarly, in YRP1 cells treated with 20 μM GW400426 the abundance of this phosphoisoform is greatly reduced (Fig. 3a). Thus, GW400426 blocks the phosphorylation of a direct substrate of Cdk1.
In principle, for a strictly linear pathway, both the microarray and Orc6 phosphorylation data could be the result of inhibition of either Cdk1 or a kinase upstream of Cdk1. In S. cerevisiae, Cdk1 is regulated by binding to its cyclin after phosphorylation by the CDK-activating kinase Cak1. Because Cak1 is known to be a branch point in activation of multiple kinases including the RNA polymerase II C-terminal domain kinase Kin28, we hypothesized that GW400426 was targeting Cdk1 and not the pleiotropic upstream kinase Cak1. Nonetheless, by using expressed and purified tagged versions of Cdk1 and Cak1 from baculovirus-infected insect cells, we evaluated the ability of GW400426 to inhibit these kinases in vitro. Strikingly, although GW400426 was a potent inhibitor of Cdk1 (IC50 = 35 nM; [ATP], 100 μM), it failed to inhibit Cak1 at concentrations up to 50 μM (Fig. 1b). Together, these data strongly support Cdk1 as being an in vivo target of GW400426.
Although the identification of Cdk1 as a target of GW400426 was not a surprise given its in vitro potency, GW400426 was only a modest inhibitor of Pho85 in vitro (IC50 = 1.5 μM; [ATP], 1 mM). To confirm Pho85 inhibition in cells, we took advantage of the fact that the direct Pho85 target, Pho4, is dephosphorylated upon Pho85 inhibition and shuttled from the cytoplasm to the nucleus (14). We transformed Pho85-as1 or YRP1 cells with an expression plasmid containing a Pho4-GFP fusion construct and monitored the cellular localization of Pho4 by using fluorescence microscopy. This test of Pho85 inhibition has been demonstrated to be more sensitive than a transcriptional readout (25). Treatment of YRP1 cells with GW400426 significantly increases the percentage of cells (>60%) with nuclear localization of Pho4-GFP compared with a DMSO control (10%) (Fig. 3b). In comparison, ≈80% of Pho85-as1 cells treated with 1-NA-PP1 had nuclear-localized Pho4-GFP. These results parallel the microarray data in suggesting that GW400426 inhibits Pho85 in cells to a lesser degree than that observed in the Pho85-as1 reference strain. Thus, for both Cdk1 and Pho85, the microarray signature proved to be a suitable readout for in vivo activities of these two kinases.
Having established that GW400426 inhibits both Cdk1 and Pho85 in cells, we sought to determine whether we had fully accounted for the transcriptional effects from GW400426 treatment. We queried the microarray data and identified a set of 76 genes repressed more strongly by GW400426 treatment than by inhibition of either Cdk1 or Pho85 alone. A subset of these genes are present in cluster E (Fig. 2a). We hypothesized that these genes might represent either a signature of a third target of GW400426 or genes uniquely regulated by multiplex inhibition of both Cdk1 and Pho85. The latter possibility was intriguing because some of the strongest signals in cluster E included genes encoding cyclins involved in the G1/S cell-cycle transition (PCL1 and CLN2) as well as those involved with cell-wall biogenesis (MCD4 and CSI2). Although Pho85 does not have a well defined role in the cell cycle, there is genetic evidence that it may contribute to the G1/S transition. In yeast strains in which the G1 cyclins Cln1 and Cln2 have been deleted, Pho85 and its associated cyclins Pcl1 and Pcl2 become essential for cell-cycle progression (26). Also, Pho85 has been implicated in monitoring cell-wall integrity (27).
To test whether the observed transcriptional effects were a result of a synthetic interaction between Cdk1 and Pho85, we constructed a yeast strain that allows for simultaneous inhibition of Cdk1 and Pho85. Treatment of this dual Cdk1-as1/Pho85-as1 strain with 5 μM 1-NA-PP1 (Fig. 2a, lane 8) results in a transcriptional profile that is a closer match to GW400426 treatment than inhibition of either kinase alone (see Fig. 6, which is published as supporting information on the PNAS web site). Furthermore, inhibition of the Cdk1-as1/Pho85-as1 strain results in the repression of the majority of the 70 genes identified above (Fig. 3c). Probing the functional relationships between these repressed genes, we found that 76% were known to be cell-cycle-regulated, and nearly 80% (23/29) of those were G1 transcripts. These genes are also highly enriched (18) in those that are known to be involved in yeast budding and morphogenesis. Thus, a portion of the biological response to GW400426 comes from an unexpected chemically induced epistasis between Cdk1 and Pho85 specifically at the G1 phase of the cell cycle. Although they do not reveal an exact mechanism, these results suggest an active, not purely redundant, role for Pho85 in the regulation of a specific, functional set of cell-cycle transcripts.
Discussion
We have identified the cellular targets of the small molecule kinase inhibitor GW400426 that together span the known biological activities of the inhibitor by matching biomarkers elicited by drug treatment to those elicited by specific pharmacological inhibition of candidate targets. This drug-to-drug reference profile strategy has revealed in detail how GW400426 acts through inhibition of the kinases Cdk1 and Pho85, both individually and in tandem, to regulate specific sets of cellular transcripts in yeast (Fig. 4).
We believe that this approach could be of general use in resolving the paradox between using kinase inhibitors of unknown specificity as biological probes to study specific cellular functions of those kinases. The crucial determination of whether a kinase-inhibitor interaction, identified either biochemically or from a genetic screen, is relevant under the conditions that prevail within a cell can be made by using the analog-sensitive version of the kinase, in the cell line or organism of interest, to link specific phenotypes from cell-based assays to inhibition of that kinase. In particular, the identification of Pho85 as an in vivo target of GW400426, despite its relatively poor in vitro potency, illustrates the necessity of using a cell-based readout of kinase inhibition to accurately evaluate cellular activity. This apparent difference in in vitro vs. in vivo susceptibility might be explained by the roles of Pho85 in nutrient sensing, functions that could require a pathway output that is highly sensitive to small modulations of kinase activity. In support of this hypothesis, the induction of Pho4-regulated transcripts by GW400426 is reversed within 10 min of washing out the drug (data not shown).
Analog-sensitive alleles offer an avenue to identify or to discount synthetic interactions arising from inhibition of multiple kinases within cells. The synthetic interaction between Cdk1 and Pho85 was revealed after our realization that the transcriptional effects elicited by GW400426 treatment were incompletely explained by inhibition of Cdk1 and Pho85 individually. Our results clearly illustrate the fundamental differences in this instance between the response of a cell to small molecule inhibition of two kinases in a pathway compared with inhibition of each kinase alone. This observation is relevant to the long-standing question of whether kinase inhibitors exert potent effects on cellular signaling pathways because of, or despite, their lack of specificity. For example, the drug Gleevec is a potent Bcr-Abl inhibitor used for the treatment of chronic myelogenous leukemia (CML) that has also shown potent in vitro inhibition of several other kinases including KIT (28). Because of the lack of Gleevec-resistant cases of CML observed clinically that arise from mutations in the KIT kinase (29), it has been assumed that Gleevec works exclusively through inhibition of Bcr-Abl. However, an analog-sensitive version of Bcr-Abl kinase has recently been used to demonstrate that if the kinase KIT is present, as in some populations of immature leukemic cells, simultaneous inhibition of both Bcr-Abl and KIT by Gleevec is required for effective suppression of cell growth (30). In contrast, inhibition of Bcr-Abl alone suppresses the growth of mature myeloid cells, which do not express KIT. Thus, analysis of the analog-sensitive allele of Bcr-Abl using growth as a biomarker suggests that Gleevec may exert some of its therapeutic effects through inhibition of both Bcr-Abl and Kit. Similarly, the prominence of kinases as key nodes in convergent signal transduction pathways in diseases such as EGF receptor (EGFR)/human EGFR (HER2)-mediated cancers suggests that multiplex inhibition of these kinases may be preferable, or even necessary, to block aberrant signaling (31).
We believe that analog-sensitive alleles will be useful in identifying biomarkers diagnostic of inhibition of individual kinases. These pharmacologically derived reference profiles, unlike reference profiles derived from genetic perturbations of candidate targets (11), use perturbations that share the same ATP-competitive mechanism of action as most of the kinase inhibitors that are used therapeutically or as molecular probes. Indeed, previous studies have demonstrated the incongruity of comparing transcriptional effects from chemical inhibition of the catalytic activity of Cdk1 (10) to heat-shift inactivation of a temperature-sensitive allele of Cdk1 (21), a kinase that functions through its catalytic activity as well as through protein–protein interactions (e.g., with FAR1, SIC1, CDC6, and CKS1) (32). With more wide-spread deployment of this technology using gene knock-ins to generate the necessary mice or cell lines carrying analog-sensitive alleles of the relevant kinases, these chemical tools should aid in both target validation and elucidation of the true mechanisms of action of protein kinase inhibitors.
Supplementary Material
Acknowledgments
We thank Karl Kuchler for providing the YRP1 yeast strain; Karen Kim and Shivkumar Venkatasubrahmanyam for help printing DNA microarrays; Mark Solomon (Yale University, New Haven, CT) for providing GST-Cak1 and GST-CDK2 proteins; Joe DeRisi, Dave Morgan, Erin O'Shea, and Audrey Gasch for critical review of the manuscript; Dustin Maly and Matt Simon for helpful discussions; Jeff Ubersax for advice regarding the Orc6 phosphorylation experiment; and Bruce Stillman (Cold Spring Harbor Laboratory, Cold Spring Harbor, NY) for providing the Orc6 Ab. This work was supported by GlaxoSmithKline and National Institutes of Health Grant AI-44009.
Author contributions: C.K., L.F.K., H.D.M., and K.M.S. designed research; C.K., D.M.K., S.H.D., and R.W.H. performed research; C.K. and D.M.K. analyzed data; and C.K. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: CDK, cyclin-dependent kinase; ESR, environmental stress–response.
References
- 1.Cohen, P. (2002) Nat. Rev. Drug Discov. 1, 309-315. [DOI] [PubMed] [Google Scholar]
- 2.Davies, S. P., Reddy, H., Caivano, M. & Cohen, P. (2000) Biochem. J. 351, 95-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang, G. J., Safran, M., Wei, W., Sorensen, E., Lassota, P., Zhelev, N., Neuberg, D. S., Shapiro, G. & Kaelin, W. G. (2004) Nat. Med. 10, 643-648. [DOI] [PubMed] [Google Scholar]
- 4.DeRisi, J. L., Iyer, V. R. & Brown, P. O. (1997) Science 278, 680-686. [DOI] [PubMed] [Google Scholar]
- 5.Sakowicz, R., Finer, J. T., Beraud, C., Crompton, A., Lewis, E., Fritsch, A., Lee, Y., Mak, J., Moody, R., Turincio, R., et al. (2004) Cancer Res. 64, 3276-3280. [DOI] [PubMed] [Google Scholar]
- 6.Knockaert, M., Gray, N., Damiens, E., Chang, Y. T., Grellier, P., Grant, K., Fergusson, D., Mottram, J., Soete, M., Dubremetz, J. F., et al. (2000) Chem. Biol. 7, 411-422. [DOI] [PubMed] [Google Scholar]
- 7.Parsons, A. B., Brost, R. L., Ding, H., Li, Z., Zhang, C., Sheikh, B., Brown, G. W., Kane, P. M., Hughes, T. R. & Boone, C. (2004) Nat. Biotechnol. 22, 62-69. [DOI] [PubMed] [Google Scholar]
- 8.Lum, P. Y., Armour, C. D., Stepaniants, S. B., Cavet, G., Wolf, M. K., Butler, J. S., Hinshaw, J. C., Garnier, P., Prestwich, G. D., Leonardson, A., et al. (2004) Cell 116, 121-137. [DOI] [PubMed] [Google Scholar]
- 9.Baetz, K., McHardy, L., Gable, K., Tarling, T., Reberioux, D., Bryan, J., Andersen, R. J., Dunn, T., Hieter, P. & Roberge, M. (2004) Proc. Natl. Acad. Sci. USA 101, 4525-4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bishop, A. C., Ubersax, J. A., Petsch, D. T., Matheos, D. P., Gray, N. S., Blethrow, J., Shimizu, E., Tsien, J. Z., Schultz, P. G., Rose, M. D., et al. (2000) Nature 407, 395-401. [DOI] [PubMed] [Google Scholar]
- 11.Marton, M. J., DeRisi, J. L., Bennett, H. A., Iyer, V. R., Meyer, M. R., Roberts, C. J., Stoughton, R., Burchard, J., Slade, D., Dai, H., et al. (1998) Nat. Med. 4, 1293-1301. [DOI] [PubMed] [Google Scholar]
- 12.Carroll, A. S., Bishop, A. C., DeRisi, J. L., Shokat, K. M. & O'Shea, E. K. (2001) Proc. Natl. Acad. Sci. USA 98, 12578-12583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guthrie, C. & Fink, G. R. (1991) Guide to Yeast Genetics and Molecular Biology (Academic, San Diego).
- 14.O'Neill, E. M., Kaffman, A., Jolly, E. R. & O'Shea, E. K. (1996) Science 271, 209-212. [DOI] [PubMed] [Google Scholar]
- 15.Jeffery, D. A., Springer, M., King, D. S. & O'Shea, E. K. (2001) J. Mol. Biol. 306, 997-1010. [DOI] [PubMed] [Google Scholar]
- 16.Tsakraklides, V. & Solomon, M. J. (2002) J. Biol. Chem. 277, 33482-33489. [DOI] [PubMed] [Google Scholar]
- 17.Spellman, P. T., Sherlock, G., Zhang, M. Q., Iyer, V. R., Anders, K., Eisen, M. B., Brown, P. O., Botstein, D. & Futcher, B. (1998) Mol. Biol. Cell 9, 3273-3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gasch, A. P., Spellman, P. T., Kao, C. M., Carmel-Harel, O., Eisen, M. B., Storz, G., Botstein, D. & Brown, P. O. (2000) Mol. Biol. Cell 11, 4241-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bramson, H. N., Corona, J., Davis, S. T., Dickerson, S. H., Edelstein, M., Frye, S. V., Gampe, R. T., Harris, P. A., Hassell, A., Holmes, W. D., et al. (2001) J. Med. Chem. 44, 4339-4358. [DOI] [PubMed] [Google Scholar]
- 20.Rogers, B., Decottignies, A., Kolaczkowski, M., Carvajal, E., Balzi, E. & Goffeau, A. (2001) J. Mol. Microbiol. Biotechnol. 3, 207-214. [PubMed] [Google Scholar]
- 21.Gray, N. S., Wodicka, L., Thunnissen, A. M., Norman, T. C., Kwon, S., Espinoza, F. H., Morgan, D. O., Barnes, G., LeClerc, S., Meijer, L., et al. (1998) Science 281, 533-538. [DOI] [PubMed] [Google Scholar]
- 22.Eisen, M. B., Spellman, P. T., Brown, P. O. & Botstein, D. (1998) Proc. Natl. Acad. Sci. USA 95, 14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Simon, I., Barnett, J., Hannett, N., Harbison, C. T., Rinaldi, N. J., Volkert, T. L., Wyrick, J. J., Zeitlinger, J., Gifford, D. K., Jaakkola, T. S. & Young, R. A. (2001) Cell 106, 697-708. [DOI] [PubMed] [Google Scholar]
- 24.Nguyen, V. Q., Co, C. & Li, J. J. (2001) Nature 411, 1068-1073. [DOI] [PubMed] [Google Scholar]
- 25.Springer, M., Wykoff, D. D., Miller, N. & O'Shea, E. K. (2003) PLoS Biol. 1, E28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Espinoza, F. H., Ogas, J., Herskowitz, I. & Morgan, D. O. (1994) Science 266, 1388-1391. [DOI] [PubMed] [Google Scholar]
- 27.Huang, D., Moffat, J. & Andrews, B. (2002) Mol. Cell. Biol. 22, 5076-5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wong, S. & Witte, O. N. (2004) Annu. Rev. Immunol. 22, 247-306. [DOI] [PubMed] [Google Scholar]
- 29.Druker, B. J. (2004) Adv. Cancer Res. 91, 1-30. [DOI] [PubMed] [Google Scholar]
- 30.Wong, S., McLaughlin, J., Cheng, D., Zhang, C., Shokat, K. M. & Witte, O. N. (2004) Proc. Natl. Acad. Sci. USA 101, 17456-17461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xia, W., Mullin, R. J., Keith, B. R., Liu, L. H., Ma, H., Rusnak, D. W., Owens, G., Alligood, K. J. & Spector, N. L. (2002) Oncogene 21, 6255-6263. [DOI] [PubMed] [Google Scholar]
- 32.Mendenhall, M. D. & Hodge, A. E. (1998) Microbiol. Mol. Biol. Rev. 62, 1191-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.