Abstract
The CATS protein was recently identified as a novel CALM interacting protein. CATS increases the nuclear and specifically the nucleolar localization of the leukemogenic CALM/AF10 fusion protein. We cloned and characterized the murine Cats gene. Detailed analysis of murine Cats expression during mouse embryogenesis showed an association with rapidly proliferating tissues. Interestingly, the Cats transcript is highly expressed in murine hematopoietic cells transformed by CALM/AF10. The CATS protein is highly expressed in leukemia, lymphoma and tumor cell lines but not in non‐proliferating T‐cells or human peripheral blood lymphocytes. CATS protein levels are cell cycle dependent and it is induced by mitogens, suggesting a role of CATS in the control of cell proliferation and possibly CALM/AF10‐mediated leukemogenesis.
Keywords: CALM/AF10, Leukemia, Nucleolus, Proliferation, CATS
1. Introduction
The rare but recurring t(10;11)(p13;q14) translocation leads to the fusion of CALM (PICALM) with AF10 (Dreyling et al., 1996)
In patients, the occurrence of CALM/AF10 rearrangement has been observed in acute myeloid leukemia (AML), T‐cell acute lymphoblastic leukemia (ALL) and in malignant lymphoma and is associated with a poor prognosis (Dreyling et al., 1998; Kumon et al., 1999; Narita et al., 1999). In T‐ALL with T‐cell receptor (TCR) gamma/delta rearrangement, the CALM/AF10 fusion is found in up to 30% of cases and constitutes the most frequent rearrangement in this leukemia subtype (Asnafi et al., 2003, 2004, 2005). Moreover CALM/AF10 positive T‐ALL have been characterized by overexpression of the HOXA and BMI1 genes. The overexpression of the HOX genes is reminiscent of the HOX gene expression patterns observed in leukemias carrying MLL rearrangements (Dik et al., 2005; Krause et al., 2004; Soulier et al., 2005).
The CALM/AF10 fusion protein is the critical transcript in the leukemogenic process. It is a constant feature observed in all patients with a t(10;11)(p13;q14). Whereas the reciprocal AF10/CALM fusion mRNA can only be detected in about half of the patients with a CALM/AF10 fusion (Bohlander et al., 2000, 2000, 1996, 1998, 2004, 1999). Recently, the leukemogenic potential of the CALM/AF10 fusion gene to generate AML with lymphoid features was demonstrated in murine bone marrow transplantation (Deshpande et al., 2006) and transgenic mice models (Caudell et al., 2007).
The AF10 gene is one of the few MLL partner genes, which is independently rearranged with a third gene in leukemia (Ayton and Cleary, 2001; Dreyling et al., 1996). AF10 contains protein domains common to transcription factors and has been suggested to play a role in heterochromatin‐mediated transcriptional silencing (Debernardi et al., 2002, 2000, 2001, 2004, 2003, 1995). Structure function analyses of the MLL/AF10 fusion protein have shown that the critical region for transformation in AF10 is the octapeptide motif (OM) and the leucine zipper (LZ) of AF10 (DiMartino et al., 2002), which are also present in the CALM/AF10 fusion. The OM/LZ motif of AF10 interacts with GAS41, a component of a chromatin remodeling complex, with the histone methyltransferase hDOT1L and also with the lymphoid regulator Ikaros (Debernardi et al., 2002; Greif et al., 2008; Okada et al., 2005). Like in MLL fusions upregulation of Hoxa genes by hDOT1L is also involved in CALM/AF10‐mediated transformation (Okada et al., 2006).
CALM plays a pivotal role in clathrin‐mediated endocytosis (CME) (De Camilli et al., 2002; Evans and Owen, 2002; Ford et al., 2001; Harel et al., 2008; Kalthoff et al., 2002; Meyerholz et al., 2005; Tebar et al., 1999). CALM interacts with the clathrin heavy chain through its C‐terminal third and with phosphoinositides through its ANTH domain in the N‐terminal third, promoting assembly of clathrin triskelia into clathrin cages in vitro (Ford et al., 2001, 2002). Both overexpression or down regulation of CALM have been shown to inhibit receptor‐mediated endocytosis and impair endosome trafficking in the trans golgi network (Huang et al., 2004; Meyerholz et al., 2005; Tebar et al., 1999). Evidence that CALM and its involvement in the CME processes play an important role for hematopoiesis, endocytosis‐mediated iron transport and growth was provided by Klebig and co workers, who showed that point mutations in the mouse CALM homologue Picalm are responsible for abnormalities in hematopoiesis and iron metabolism (Klebig et al., 2003). An MLL/CALM fusion has been described in a case of childhood AML (Wechsler et al., 2003).
The involvement of CALM in two separate translocations with both MLL and AF10 as well as the fact that a number of other MLL and non‐MLL fusion partners found in leukemia are endocytic proteins suggests that disturbing CME may be important in leukemogenesis (Bohlander et al., 2000, 2001, 1998; Polo et al., 2004; Wechsler et al., 2003). Indeed, Chao and coworkers have reported perturbed endocytosis by CALM‐containing fusion proteins, associated with prolonged growth factor signaling and enhanced cellular proliferation (Chao et al., 2004, 2005).
We have recently reported the isolation of CATS in a yeast two hybrid screen as a CALM interacting protein expressed in thymus and spleen (Archangelo et al., 2006). In GenBank, CATS has been computer‐annotated as the Homo sapiens family with sequence similarity 64, member A (FAM64A; accession number NM_019013). The CATS gene is located on chromosome 17 band p13. The genomic locus spans approximately 7kb and contains six exons. At least three alternatively spliced versions of CATS could be identified which code for two protein isoforms of 238 or 248 amino acids. Expression of fluorescent‐tagged CATS proteins showed that CATS localize mainly to the nucleus where it shows preference for nucleoli and to a lesser extent in the cytoplasm and at the cell membrane. The CATS interaction region of CALM was mapped to amino acids 221–294 of CALM. This domain is contained in the CALM portion of both CALM/AF10 and the MLL/CALM fusion proteins. Expression of CATS markedly increased the nuclear localization of CALM and of the leukemogenic fusion protein CALM/AF10, and this effect of CATS was stronger for CALM/AF10 than for CALM. Moreover, we demonstrated that CATS acts as a transcriptional repressor when fused to a heterologous DNA‐binding domain in a GAL4‐based transactivation assay. In that report we proposed that CATS might be a critical modulator of normal CALM function in lymphoid cells and that CATS could be very important for understanding the leukemogenic potential of the CALM/AF10 fusion protein (Archangelo et al., 2006).
To further elucidate CATS function we performed an extensive analysis of CATS expression. We characterized the murine Cats gene and examined its expression in leukemic cells derived from a CALM/AF10 murine bone marrow transplant model in comparison to similar cell populations of a non‐leukemic mouse. A monoclonal antibody against CATS was generated, which confirmed the nucleolar localization of the endogenous CATS protein and its expression in human thymus. Moreover, we show that CATS is strongly expressed in cancer cell lines in a cell cycle dependent manner and is induced by mitogens. The strong upregulation of CATS protein upon mitogenic activation and its high expression in proliferating cells, but not in quiescent cells, suggest a role of CATS in the control of cell proliferation.
2. Results
2.1. The murine Cats gene
The murine Cats gene is located on mouse chromosome 11 (genomic contig NT_096135). The genomic locus spans approximately 6kb and similar to the human counterpart comprises six exons with a non‐coding first exon. The non‐canonical splice donor site, which is present in the human CATS gene is also found in the third intron of the murine Cats gene. The complete Cats cDNA clone IMAGp998H174619Q2 comprises 1613 nucleotides. The murine Cats gene encodes a 231 amino acid protein, which shares 76% homology to human isoform 1 and 71% to isoform 2.
2.2. Cats is mainly expressed in thymus and ovary of adult mice
Northern blot analysis of adult mouse tissues revealed a 1.5kb Cats transcript strongly expressed in thymus, ovary and to a lower extent in heart. Cats transcripts were absent in brain, kidney, liver, bone marrow and in spleen (Figure 1A).
Figure 1.
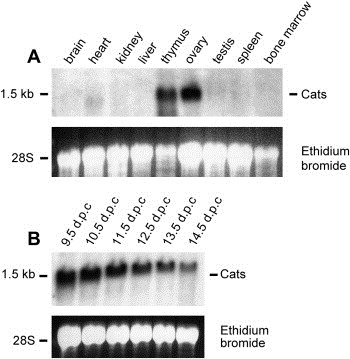
Expression analysis of the murine Cats gene. (A) Multiple tissue Northern blot of adult mouse tissues. (B) Developmental Northern blot from whole mouse embryos of day 9.5 to 14.5d.p.c. Blots were hybridized with a Cats cDNA probe. The ethidium bromide stained gel is shown as loading control.
2.3. Cats is strongly expressed throughout mouse embryogenesis
Northern blot analysis of Cats with total RNA from whole mouse embryos revealed a strong expression of Cats during embryogenesis. The Cats mRNA was detectable as early as 9.5d.p.c. and decreased gradually from stage 10.5 to 14.5d.p.c. (Figure 1B).
Whole mount in situ hybridization was performed to analyze the temporo‐spatial expression of Cats during mouse development. Hybridization with antisense probe revealed widespread expression of Cats at stage 9.5d.p.c. with prominent expression in the neural tube (Figures 2B,I; nt), somites (Figure 2B; sm), the posterior region of the midbrain (Figure 2D; pmb), olfactory placode (Figures 2A,B,D; olp) and the branchial arches (Figures 2A,H; bra) but no expression in the primitive heart (Figure 2A; he). At 10.5d.p.c. there is a reduction of the widespread expression and stronger Cats expression is seen in the neural tube (Figures 2F,G; nt), branchial arches (Figure 2E; mx and md), developing limbs (Figures 2E; fl and G; hl), telencephalon, nasal process (Figure 2E; te), lense vesicle (Figures 2P,Q; le), anterior and posterior regions of the mid and hindbrain (Figure 2E; amb, pmb and hb). From 11.5d.p.c. on, strong staining is also seen in the genital tubercle (Figure 2R; ge) and hair and vibrissae follicles (Figures 2S–V; hf and vf). A reduction of Cats expression is seen from stage 12.5d.p.c. onwards with a complete lack of Cats expression in the cephalic region and the neural tube in 14.5d.p.c. embryos.
Figure 2.
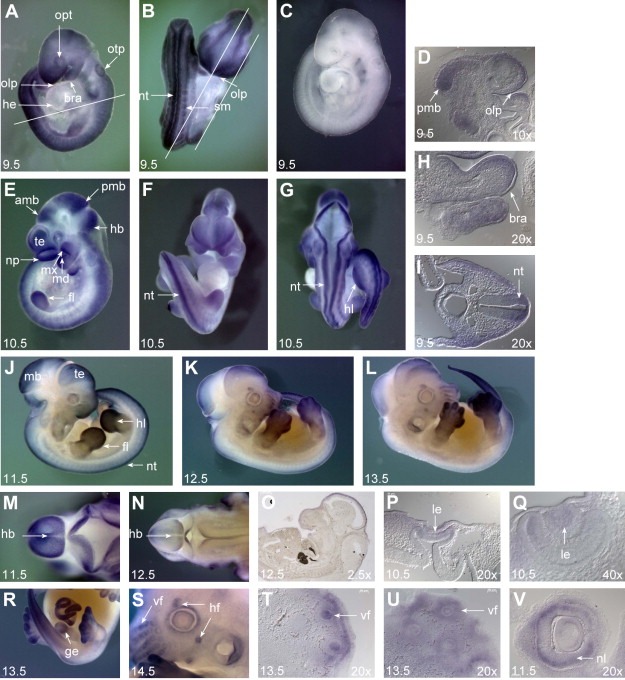
Whole mount in situ hybridization analysis during mouse embryonic development. Embryos were hybridized with the Cats antisense probe. (A, B, D, H, I) Expression of Cats at embryonic stage of 9.5d.p.c. Widespread transcript distribution with prominent expression in the neural tube (nt), somites (sm), posterior region of the midbrain (pmb), olfactory placode (olp) and in branchial arches (bra). Note the complete absence of Cats expression in the primitive heart (he). Otic pit: otp; optic vesicle: opt. Bars indicate the plane of section. (C) 9.5d.p.c. embryo hybridized with the Cats sense probe as a control for staining specificity. (E–G, P, Q) At 10.5d.p.c., there is a reduction of widespread Cats expression, transcripts are additionally detected in fore‐ and hindlimbs (fl and hl, respectively), telencephalon (te), nasal process (np), lens vesicle (le), anterior region of the midbrain (amb), hindbrain (hb) and mandibular and maxillary component of first branchial arch (md and mx, respectively). (J–L) Right lateral view of embryos from 11.5, 12.5 and 13.5d.p.c. Note reduction of Cats overall expression and prominent expression in the limbs. (M, N) Dorsal view of the cephalic region of 11.5 and 12.5d.p.c. embryos. Note the abrupt reduction of Cats expression in the hindbrain and neural tube from stage 11.5 to 12.5d.p.c. (O) Remaining Cats expression in the midbrain of a 12.5 embryo. (P–V) Transcripts detected in the lense vesicle (le), genital tubercle (ge), hair follicles (hf), vibrissae follicles (vf) and neural layer of optic cup (nl). (D, H, I, O–Q, T–V) Sagittal (D, H, O, T–V) and transverse (I, P, Q) sections from whole mount stained embryos. Stage of development and imaging magnification are indicated on the left and right bottom of the photographs, respectively.
Cats is strongly expressed during limb development. In general, stronger expression is detected in hindlimbs than in forelimbs and the expression is slightly more marked in the posterior region of the limb buds. At 11.5 and 12.5d.p.c., expression is observed at the distal domain and the underlying mesenchyme but it is not present in the apical ectodermal ridge (AER). Distally, expression of Cats becomes confined to the digits at stages 13.5 and 14.5d.p.c. (Figure 3).
Figure 3.
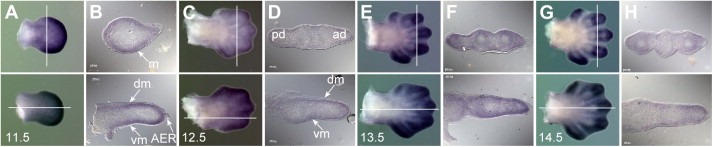
Cats expression during limb development. In all panels the dorsal side of the limb is facing, the anterior side is to the top. The upper half of each figure shows a forelimb, the lower a hindlimb. Sections were generated from whole mount stained limb buds with 10× magnification (B, D, F, H). Bars indicate the plane of section. Transverse sections (forelimbs) are orientated with posterior side to the left, sagittal sections (hindlimbs) with the proximal side to the left. (A) Limb buds of a 11.5d.p.c. embryo. The expression of Cats is more prominent in the distal part of the paddle‐shaped fore‐ and hindlimb buds. (B) The transverse section shows staining in the underlying mesenchyme (m) and the sagittal section in the dorsal and ventral mesenchyme (dm and vm, respectively) but not in the apical ectodermal ridge (AER). (C) At 12.5d.p.c. Cats expression is still detected in the distal part of the polygonal‐shaped fore‐ and hindlimb buds. (D) Note stronger staining in the posterior domain (pd) rather than in the anterior domain (ad). (E, G) Distally, expression of Cats becomes confined to the digits in stages 13.5 and 14.5d.p.c.
2.4. Cats is widely expressed in different hematopoietic cell subpopulations
RT‐PCR was performed in order to determine Cats expression in the murine hematopoietic compartment. Transcripts were observed in all T‐cell subpopulations isolated from the thymus (CD4+, CD8+ and CD4+/CD8+) with lower levels of transcripts in CD8+ cells (Figure 4A). Similarly, extremely low levels of Cats were observed in the CD8+ cells isolated from bone marrow. Except for the pro‐B cell subpopulation (B220+) where Cats expression was nearly absent, abundant levels of transcript were observed in macrophages (Mac1+), pro‐erythrocytes (Ter119+), granulocytes (Gr‐1+), mast cells (c‐kit+) and progenitor cells (Sca‐1+/c‐kit+) (Figure 4B).
Figure 4.
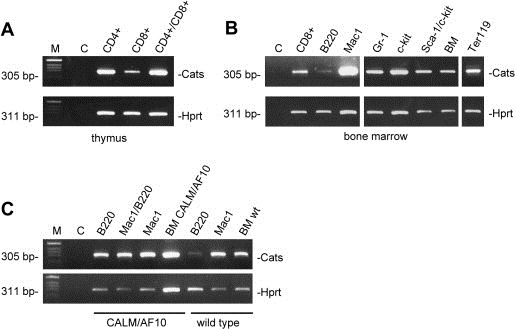
Expression analysis of Cats on hematopoietic compartment of a normal and a CALM/AF10 leukemic mouse. RT‐PCR on cell populations purified from the murine thymus (A) and bone marrow (B). Samples are: T‐cells (CD4+, CD8+ and CD4+/CD8+), pro‐B cells (B220+), macrophages (Mac1+), pro‐erythrocytes (Ter119+), granulocytes (Gr‐1+), mast cells (c‐kit+), progenitor cells (Sca‐1+/c‐kit+) and whole bone marrow. (C) Analysis of Cats expression in leukemic cells derived from a CALM/AF10 murine bone marrow transplant model. Cats is expressed in the B220+/Mac1−, B220+/Mac1+, B220−/Mac1+ populations and whole bone marrow from a CALM/AF10 leukemic mouse. The controls used were B220+, Mac‐1 and whole bone marrow from a non‐leukemic mouse. Note the different levels of Cats expression in B220+ cells derived from a non‐leukemic and a leukemic mouse. Amplification of murine Hprt was used as control for RNA and cDNA integrity. BM: bone marrow; M: DNA size marker; C: negative control: no template.
2.5. Cats is upregulated in B220+ leukemia cells from a CALM/AF10 mouse
Expression analysis was also performed on leukemic cells derived from a CALM/AF10 murine bone marrow transplant model (Deshpande et al., 2006). Cats expression was determined in B220+/Mac1−, B220+/Mac1+, B220−/Mac1+ populations and whole bone marrow from a CALM/AF10 leukemic mouse. Cats was strongly expressed in all four samples derived from the leukemic mouse. Moreover, a much higher level of Cats transcripts was observed in the B220+/Mac1− cell population in comparison to a B220+ population from a non‐leukemic mouse (Figure 4C).
2.6. CATS is a nuclear protein enriched at the nucleoli
Immunofluorescence was performed in order to determine the subcellular localization of endogenous human CATS. The α‐CATS 2C4 antibody detected the protein in the nucleus of U2OS cells. In these cells, different CATS expression levels were observed from cell to cell. Strong accumulation of CATS was also observed in the nucleolus of some cells as evidenced by the colocalization of CATS with the nucleolar marker Fibrillarin (Figure 5B). However, the nucleolar expression of CATS was not a constant feature suggesting a transient accumulation or sequestration of CATS in the nucleolus (Figure 5).
Figure 5.
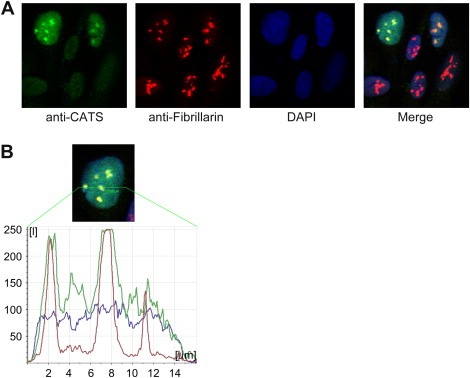
Subcellular localization of endogenous CATS. Endogenous CATS was visualized by confocal microscopy in non‐synchronized U2OS cells. (A) CATS is a nuclear protein localized to the nucleolus of some cells. Note the different levels of CATS expression from cell to cell. (B) Confocal line scan through the nucleus of the upper left cell from panel A. The picture shows a cell with all three channels (Alexa 488 (CATS), Cy3 (Fibrillarin) and DAPI) merged. Note that the peak intensity of the CATS signal correlates with Fibrillarin staining of the nucleolar structures.
2.7. CATS expression in human thymus
Western blot with cellular extract from human thymus was probed with the α‐CATS 2C4 antibody. The antibody clearly recognized the 28kDa CATS protein, confirming CATS expression in the human thymus (Figure 6A).
Figure 6.
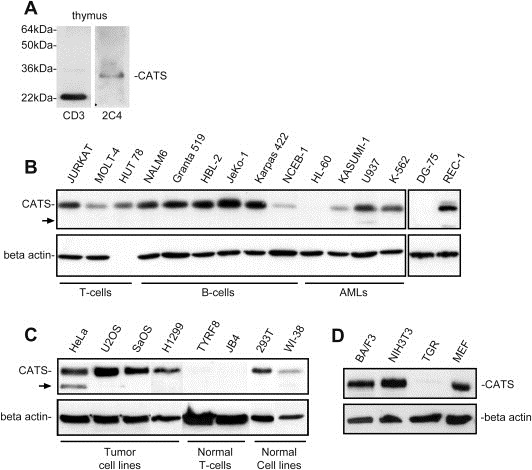
Western blot analysis of endogenous CATS using the α‐CATS 2C4 mAb. (A) CATS protein is expressed in human thymus. The α‐CD3 12 mAb, which detects CD3 epsilon in the cytoplasm, was used as a positive control to demonstrate the integrity of proteins in the thymus lysate. (B) CATS is highly expressed in leukemia and lymphoma cell lines. (C) CATS expression in normal and tumor cell lines. (D) Expression of Cats protein in murine cell lines (BA/F3, NIH3T3 and MEF) and a rat cell line (TGR). The α‐CATS 2C4 antibody recognizes the murine but not the rat protein. Arrows indicate the additional band found in cellular extracts of the CALM/AF10 fusion positive cell line U937 and in HeLa cell line. Blots were stripped and reprobed with an α‐beta actin antibody, which served as a control for the loading and integrity of the protein extracts.
2.8. CATS is strongly expressed in leukemia and lymphoma cell lines
Western blot analysis revealed that human CATS is highly expressed in nearly all leukemia and lymphoma cell lines tested (B‐ and T‐ALL, B‐ and T‐lymphoma and AML FAB M2, M4 and M5). The levels of CATS expression did not seem to correlate with the differentiation stage of the cells as expression levels were similar for immature and mature T and B cell lines. No CATS protein could be detected in the AML cell line HL‐60 and the Burkitt‐lymphoma cell line DG‐75. In the CALM/AF10 fusion positive cell line U937, an additional band of lower molecular weight was detected with the α‐CATS 2C4 antibody (Figure 6B).
2.9. CATS is strongly expressed in proliferating cells
Western blot analysis revealed high expression of the CATS protein in tumor cell lines (HeLa, U2OS, SaOS and H1299) and in the transformed cell lines HEK 293 and WI‐38 (embryonal kidney‐ and human fibroblast cell line, respectively). No CATS expression was detected in the non‐proliferating T‐cell lines TyrF8 and JB4 (Figure 6C). The fact that CATS was not expressed in quiescent cells but strongly expressed in highly proliferating cells, suggested that CATS might play a role in cell proliferation. Interestingly, an additional CATS band was also detected in protein extracts of HeLa cells. Additional Western blot analyses performed with lysates from highly proliferative murine and rat cell lines revealed a strong expression of Cats in murine pro B‐cells (BA/F3), fibroblasts (NIH3T3) and embryonic fibroblasts (MEF). Cats was not detected in the rat fibroblast cell line (TGR). The fact that the α‐CATS 2C4 antibody does recognize the rat protein was expected since the antibody was generated in this species (Figure 6D).
2.10. CATS is a marker for proliferation
We examined CATS expression levels after resting human peripheral blood lymphocytes (PBLs) were induced to proliferate with phytohemagglutinin (PHA). Western blot analysis revealed a clear upregulation of CATS in activated PBLs, whereas no CATS protein was observed in the quiescent cells (Figure 7A). Similarly, expression analysis of CATS in lysates of serum starved and serum stimulated T98G (glioblastoma cell line) cells revealed a strong correlation between CATS expression and proliferation. Low levels of CATS were observed in quiescent cells. Gradual and progressive upregulation of CATS expression is seen after serum stimulation, especially from hour 12 onwards (Figure 7B). These results indicate that CATS is linked to proliferation and is regulated in a cell cycle dependent manner (see also below).
Figure 7.
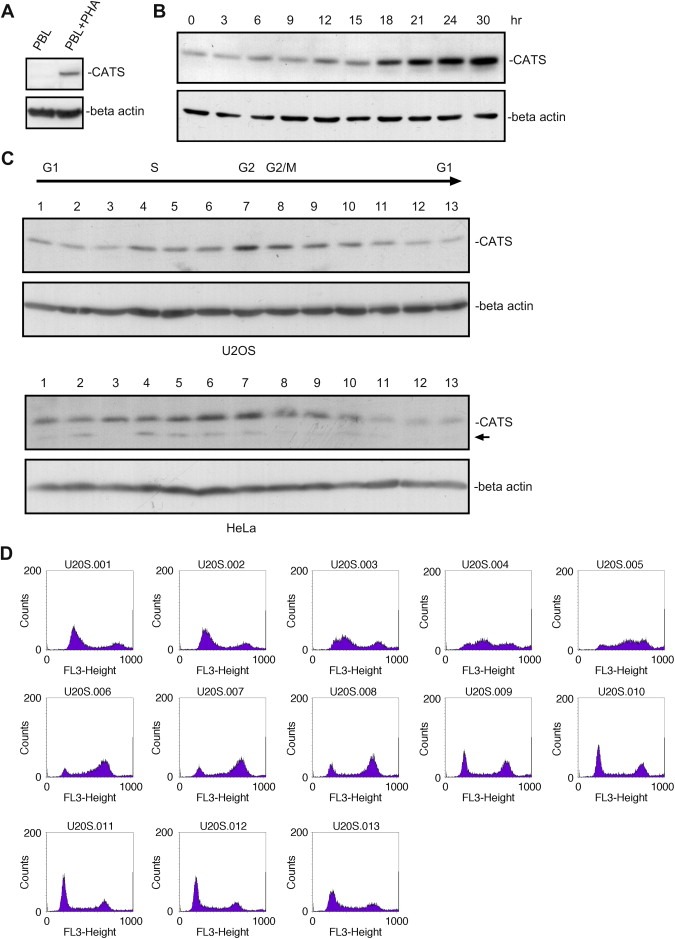
CATS expression is upregulated in proliferating cells and is cell cycle dependent. (A) Primary human PBLs were stimulated for clonal expansion with PHA for 3days. Cell lysates were analysed by Western blotting using α‐CATS 2C4 antibody. CATS is not expressed in quiescent PBLs whereas the protein clearly accumulates in the proliferating cells. (B) T98G cells were serum stimulated after 48h of serum starvation. Cell extracts, prepared every 3h for up to 30h after stimulation were blotted and probed with α‐CATS 2C4 antibody. CATS is strongly upregulated in proliferating cells. Numbers indicate hours after serum stimulation. (0) Starved cells. (C) U2OS and HeLa cells were synchronized by a double thymidine block. CATS expression varies in the different stages of the cell cycle. Upregulation of CATS protein was observed in cell extracts corresponding the S, G2 and G2/M phases of the cell cycle (sample 4–11 from U2OS and samples 5–10 from HeLa). (D) FACS analysis of the U2OS samples (1–13). Sample 1 from HeLa lysates is from a non‐synchronized cell population, the arrow indicates the smaller CATS isoform expressed also in U937 cells. Blots were stripped and reprobed with α‐beta actin antibody.
2.11. CATS is regulated in a cell cycle dependent manner
Western blot analysis of U2OS and HeLa cells, which had undergone cell cycle synchronization by a double thymidine block revealed that levels of CATS expression varies throughout the cell cycle and is regulated in a cell cycle dependent manner. CATS is upregulated in samples corresponding to S, G2 and G2/M phases. Samples in which most of the cells were found in G1 phase showed lower expression of CATS (Figure 7C).
Immunofluorescence experiments on synchronized U2OS cells, revealed that CATS transiently accumulates at the nucleoli in a cell cycle dependent manner. A homogenous nucleolar localization and similar expression levels were observed in cells blocked at the G1/S transition (Figure 8). However, both the nucleolar localization and the expression levels became heterogeneous after the release of the block as the cells progressed through the cell cycle and regained their asynchronous state (Suppl. Figure 2).
Figure 8.
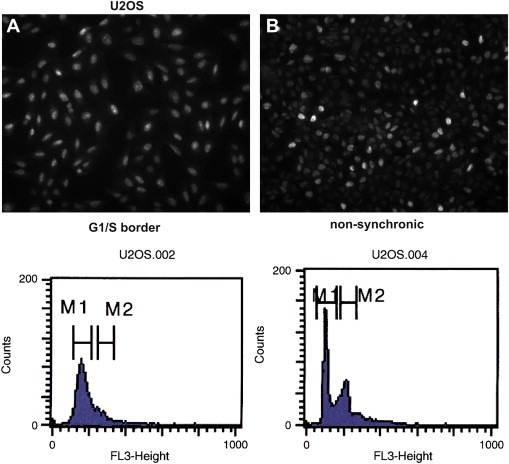
The nucleolar localization of CATS is cell cycle dependent. Immunofluorescence microscopy of synchronized U2OS cells. (A) CATS is homogeneously expressed and localized at the nucleolar structures of every cell from a population blocked the G1/S boundary. (B) Heterogeneous expression and localization of CATS in non‐synchronized cells. The lower panels show the flow cytometry analysis of the same populations.
3. Discussion
Recently, an increasing number of CALM and AF10 interacting proteins have been identified (Archangelo et al., 2006; Debernardi et al., 2002; Forissier et al., 2007; Greif et al., 2008; Okada et al., 2006; Pasalic et al., 2006), however their role in CALM/AF10‐dependent transformation remains mostly elusive. Here we present a detailed characterization of the human and murine CALM interactor CATS. We describe a monoclonal antibody (mAb) generated against the C‐terminus of CATS (α‐CATS 2C4), which recognizes both human CATS isoforms and the murine protein. Using this mAb we confirmed the nucleolar localization of the endogenous CATS and showed strong CATS expression in nearly all leukemia, lymphoma and tumor cell lines examined but not in non‐proliferating T‐cells. Primary quiescent PBLs, which have not been selected for growth in culture, do not express CATS protein but only when they are activated and start to proliferate. Similarly, in glioblastoma cells induced to proliferate, CATS showed an increasing and progressive upregulation with accumulation of CATS when cells entered S phase and maximum protein levels in G2 phase. Moreover expression analysis on synchronized cells revealed an evident cell cycle dependent accumulation of CATS protein in the S and G2 phase of the cell cycle and a transient localization of CATS at the nucleoli of cells at the G1/S border. Our data clearly correlate high levels of CATS protein with the proliferative state of the cell. Thus CATS can be viewed as a marker for proliferation and the amount of CATS protein is correlated to the cell cycle phases, with low levels in early G1, and high in the S–G2 phases. Moreover, it is tempting to speculate that the cell cycle dependent sequestration of CATS to the nucleolar structures might be associated with cell proliferation and/or inhibition of CATS function in the nucleoplasm. It should be kept in mind, however, that at the precise cellular function of the protein is unknown at present. Surprisingly, we failed to detect CATS protein in the leukemic cell line HL‐60 and in the Burkitt‐lymphoma cell line DG‐75, suggesting that CATS is not essential for proliferation in these cell lines. Intriguingly, we were able to amplify CATS transcripts from the HL‐60 cell line (data not shown).
Expression analysis of the murine Cats gene demonstrated upregulation of the Cats transcript in B220+ cells from a CALM/AF10 leukemic mouse in comparison to B220+ cells from a non‐leukemic mouse. In this murine leukemia model, the B220+/MAC1− cells are highly enriched for leukemia stem cells (Deshpande et al., 2006). These findings not only confirm CATS as a marker for proliferation but also suggest that CATS is an early player in CALM/AF10‐mediated transformation. Consistent with its correlation with cellular proliferation, Cats is highly expressed throughout murine embryogenesis but not in the differentiated cells of most adult tissues. Cats expression decreased gradually and proportionally as embryos develop to latter stages, possibly reflecting the increase in cells exiting the cell cycle.
Recently, Okada and colleagues have shown that DOT1L is capable of interacting and recruiting the CALM/AF10 fusion protein to the nucleus and consequently to activate the transcription of the CALM/AF10 target Hoxa5 (Okada et al., 2006). The fact that CATS is capable of recruiting the CALM/AF10 fusion protein to the nucleolus suggests that CALM/AF10 can possibly activate transcription in the vicinity of nucleolar structures and/or affect the function of nucleolar proteins. Nucleolar proteins have been shown to regulate cell proliferation and growth by controlling ribosome biogenesis and TP53 function and deregulation of these finely balanced mechanisms is a key event in the initiation and progression of malignant transformation (Maggi and Weber, 2005; Olson, 2004; Rubbi and Milner, 2003; Ruggero and Pandolfi, 2003). Conversely, since CATS function is connected to cellular proliferation, it is tempting to speculate that the disruption of normal CATS function through the interaction with the CALM/AF10 fusion protein contributes to the mechanism underlying the malignant transformation by CALM/AF10. Supporting this idea is the mounting evidence suggesting that as yet not defined secondary events increasing proliferation and/or apoptosis collaborate with the impaired differentiation caused by CALM/AF10 expression resulting in a completely transformed leukemia cells (Caudell and Aplan, 2007; Caudell et al., 2007).
The complete absence of CATS expression in resting T‐cells (JB4 and TYRF8) is very striking. This is in complete contrast to the high expression observed in highly proliferative T‐cell lines derived from leukemia patients. Similarly, quiescent PBLs do not express CATS protein but strongly express CATS when induced by mitogens (e.g. PHA). It is tempting to speculate that CALM/AF10 localization and function is in part modulated by the presence of CATS in these proliferating cells.
In summary, the fact that CATS changes the subcellular localization of CALM/AF10 as well as the specific CATS expression in lymphoid tissues and T‐lymphocytes induced to proliferate indicates that CATS function might be important to understand the malignant transformation mediated by CALM/AF10. This might be especially relevant since the CALM/AF10 fusion is the most frequent rearrangement found in T‐ALL with T‐cell receptor gamma/delta rearrangement (Asnafi et al., 2003).
4. Experimental procedures
4.1. Expression analysis of the murine Cats transcripts
Whole mount in situ hybridization: Balb/c embryos were dissected at E9.5–E14.5d.p.c. Sense and antisense probes were generated using the DIG RNA labeling mix (Roche, Mannheim, Germany) according to the manufacture's instructions. Whole mount in situ hybridization was performed as previously described (Sporle and Schughart, 1998). An antisense riboprobe of a 305bp Cats fragment corresponding to exons 2 and 3 was generated from mouse thymus cDNA using the primers mCATSF364 (5′‐TCCAGAGCCAGAGCAGCAAG‐3′) and mCATSR630 (5′‐CAGACTCCCTCCTTAGCCGC‐3′). Stained embryos were cryoprotected overnight at 4°C in 30% sucrose/PBS, embedded in Jung Tissue Freezing Medium™ (Leica Instruments, Nussloch, Germany), and sectioned at −25°C (35μm) with a Cryotome CM 1850 (Leica). Processed sections were mounted on slides with Kaisers Glyceringelatine (Merck, Darmstadt, Germany). For documentation, whole embryos were mounted on a thin layer of agarose under a MZ APO stereomicroscope (Leica) in dark field illumination with additional diffuse light from above and recorded on a JVC KY‐F70 digital camera using a Leica QWin software (Leica Microsystems Imaging Solutions, Cambridge, UK). Sections were observed under an automated Axioplan 2 Imaging microscope (Carl Zeiss, Jena, Germany) equipped with an AxioCam HR digital camera and images were recorded using the AxioVision 3.1 software (Carl Zeiss, Jena, Germany). Pictures were adjusted for brightness and contrast in Adobe Photoshop® 7.0 (Adobe Systems, Mountain View, USA).
4.2. Northern blot and RT‐PCR
Total RNA were isolated using the RNeasy® Mini Kit (Qiagen, Hilden, Germany) and QIAshredder™ column, according to the manufacturer's instructions. Northern blot analysis was performed with 10μg total RNA following standard procedures. A 850‐bp 32P‐labeled probe corresponding to the Cats ORF was generated with the Megaprime™ DNA labeling system (Amersham Biosciences, Freiburg, Germany).
For RT‐PCR analysis, a total of 1μg of DNAse I treated RNA was reverse transcribed with oligo dT primers and SuperScript II enzyme (Invitrogen™ Life Technologies, Karlsruhe, Germany).
cDNA from leukemic cells derived from a CALM/AF10 murine bone marrow transplant model were prepared from highly purified B220+/Mac1−, B220+/Mac1+ and B220−/Mac1+ cells obtained after propagation of a single sorted B220+/myeloid marker− cell from a bulk leukemic cell population grown in IL3 supplemented medium as previously described (Deshpande et al., 2006). For amplification of Cats transcripts, PCR reactions were carried out in 20μl with 1μl of cDNA, 0.16μl of PAN Script DNA Polymerase (PAN™ Biotech, Aidenbach, Germany) and 0.5μM of Cats specific primers mCATSF364 and mCATSR630 under stringent conditions (1.2mM MgCl). The annealing temperature used for the PCR reaction was 60°C in a 30 cycle program. The murine Hprt primer pair (5′‐GGGGGCTATAAGTTCTTTGC‐3′ and 5′‐TCCAACACTTCGAGAGGTCC‐3′) was used as control to amplify the same cDNA.
4.3. Production of CATS mAb
Approximately 50μg of purified GST‐CATSv2CT fusion protein (Archangelo et al., 2006) was injected both i.p. and subcutaneously into Lou/C rats using CPG2006 (TIB MOLBIOL, Berlin, Germany) as adjuvant. The mAb was generated as previously described (Holzel et al., 2005). The hybridoma designated CATS 2C4 was stably subcloned and used for further analysis. The immunoglobulin isotype was determined using mAbs against the rat IgG heavy and light chains. The α‐CATS 2C4 mAb has the IgG subclass IgG2a and recognizes both the human and mouse CATS proteins.
4.4. Immunobloting and immunofluorescence
Cellular extracts were lysed in RIPA buffer as previously described (Archangelo et al., 2006). Total cell lysates were separated on SDS‐PAGE gels and transferred onto nitrocellulose membrane (Hybond™ ECL™, Amersham Pharmacia biotech). Immunodetection was performed with the α‐CATS 2C4 mAb.
For immunofluorescence, U2OS cells were grown on coverslips, fixed in 2% paraformaldehyde (PFA) for 10min and permeabilized with PBS/0.1% Triton X for 15min at RT. Unspecific binding was blocked with PBS/10% FCS for 1h. Coverslips were incubated with CATS2C4 mAb (diluted 1:10) alone or with rabbit anti‐Fribillarin (Abcam) diluted 1:100 at 4°C overnight in a humidified chamber. Secondary Alexa 488 (Invitrogen) and Cy™3‐conjugated (Jackson) antibodies diluted 1:500 were used for detection of the primary antibodies for 1h at RT. Nuclei were counterstained with DAPI (Sigma‐Aldrich) and cells were mounted on slides using Cytomat medium (DAKO). Immunostained specimens were analyzed in a confocal fluorescence laser scanning system (TCS‐SP2 scanning system and DM IRB inverted microscope, Leica, Solms, Germany). Alternatively, immunofluorescence of synchronized cells was analyzed with an automated Axiovert 200M microscope (Zeiss, Jena, Germany) equipped with single‐band pass filter sets for visualization of Alexa 488 fluorescence. Images were recorded using the Openlab 3.08 software (Improvision, Coventry, UK).
4.5. FACS‐sorting of murine hematopoietic cell subpopulation
Primary cells from bone marrow and thymus were obtained from 4weeks old donor mice. For cellular suspension derived from bone marrow, ammonium chloride treatment was used to lyse the red blood cells. Cellular suspensions were incubated with phycoerythrin‐labeled Gr‐1, Sca‐1, Ter119, CD4, and allophycocyanin‐labeled Mac1, c‐kit, B220 or CD8 (BD Pharmingen, Heidelberg, Germany) for 30min on ice. Cells were sorted using FACSVantage SE System (BD Biosciences, Palo Alto, CA) and cell subpopulations were collected in DMEM medium supplemented with 50% FBS. Sorted cells were used for RNA extraction.
4.6. Cells
Human PBLs were prepared from peripheral blood of healthy donors by standard Ficoll/Hypaque gradient centrifugation and subsequently depleted of CD14+ monocytes. Cells were cultured in RPMI160 medium supplemented with 10% FCS, 1% penicillin/streptomycin and 1% glutamine. PBLs were stimulated for 3days with phytohemagglutinin (PHA, 1μg/ml).
T‐cell clones: The HLA‐A*0201‐restricted tyrosinase peptide tyr368–376 (YMNGTMSQV)‐specific cytotoxic T‐cell clone TyrF8 (Visseren et al., 1995) and JB4 (Milani et al., 2005) a T‐cell clone recognizing HLA‐A2‐expressing cell lines were kindly provided by Dr. Elfriede Nössner (Institute of Molecular Immunology, GSF, Munich, Germany). The T‐cell clones were cultured and stimulated as previously described (Milani et al., 2005).
The cell lines used for expression analysis of the native CATS protein were: HEK 293, WI‐38, HeLa, U2OS, SaOS, T98G and H1299; Burkitt lymphoma: DG75; B‐cell leukemia and B‐cell lymphoma cell lines: NALM6, Granta 519, HBL‐2, JeKo‐1, Karpas 422, NCEB‐1 and REC‐1; T‐cell leukemia and T‐cell lymphoma cell lines: Jurkat, MOLT‐4 and HUT 78; myelocytic cell lines: HL‐60 and KASUMI‐1; the monocytic cell line U937 and the erythrocytic cell line K‐562.
Murine and rat cell lines: NIH3T3, BA/F3, MEF and TGR.
Tumor and transformed cell lines were obtained from the American Type Culture Collection (ATCC, Manassas, USA) or the German Collection of Microorganisms and Cell Cultures (DSMZ, Braunschweig, Germany) and grown according to the suppliers' recommendations.
4.7. Serum stimulation and cell synchronization
T98G human glioblastoma cells were serum starved (DMEM/0.1% FBS) for 72h. Cells were then stimulated with DMEM/10% FBS and cell lysates were prepared at the indicated time points. Cell cycle progression was monitored by FACS analysis of the DNA content (see below). Cell cycle synchronization of HeLa and U2OS cells was performed by a double thymidine block as previously described (Whitfield et al., 2000). Briefly, cells were treated with 2mM thymidine for 18h, then incubated in regular medium for 9h and followed by 18h in the presence of 2mM thymidine. Finally, cells were released from cell cycle block by several washes with PBS and subsequent incubation in regular culture medium. Cells were lysed and prepared for immunostaining at the indicated time points. Cell cycle synchronization and release was controlled by FACS analysis of DNA content. Cells were washed in PBS and then fixed in ice‐cold 70% ethanol and incubated for 30min on ice. Afterwards, cells were washed two times with PBS and resuspended in PBS containing RNAse A. Prior to FACS analysis, propidium iodide was added at a final concentration of 20μg/ml.
Supporting information
Supplementary data
Supplementary data
Acknowledgements
We thank Dr. Elfriede Nössner for kindly providing the T‐cell clones JB4, TyrF8, T‐cell lines HUT 78, MOLT‐4 and the human PBLs (CD14−). This research was supported by the Deutsche José Carreras Leukämie‐Stiftung e.V. (DJCLS) grants F02/02 and F05/06 (to LFA and PAG, respectively), the Frauenbeauftragte‐LMU, Munich, Germany (grant to LFA), the Fundação de Amparo à Pesquisa de São Paulo (FAPESP), Brazil grants 07/08019‐1 and 07/54870‐5 (to LFA and STOS, respectively), as well as a National Genome Research Network (NGFN) grant from the German Ministry of Education and Research (BMBF) to SKB (N1KR‐S31T15) and a grant from the Deutsche Forschungsgemeinschaft (DFG) to SKB (SFB 684, A6).
Supplementary materials 1.
1.1.
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.molonc.2008.08.001.
Archangelo Leticia Fröhlich, Greif Philipp A., Hölzel Michael, Harasim Thomas, Kremmer Elisabeth, Przemeck Gerhard K.H., Eick Dirk, Deshpande Aniruddha Jayant, Buske Christian, Hrabé de Angelis Martin, Olalla Saad Sara Teresinha, Bohlander Stefan K., (2008), The CALM and CALM/AF10 interactor CATS is a marker for proliferation, Molecular Oncology, 2, doi: 10.1016/j.molonc.2008.07.006.
References
- Archangelo, L.F. , Glasner, J. , Krause, A. , Bohlander, S.K. , 2006. The novel CALM interactor CATS influences the subcellular localization of the leukemogenic fusion protein CALM/AF10. Oncogene 25, (29) 4099–4109. [DOI] [PubMed] [Google Scholar]
- Asnafi, V. , Radford-Weiss, I. , Dastugue, N. , Bayle, C. , Leboeuf, D. , Charrin, C. , Garand, R. , Lafage-Pochitaloff, M. , Delabesse, E. , Buzyn, A. , 2003. CALM-AF10 is a common fusion transcript in T-ALL and is specific to the TCRgammadelta lineage. Blood 102, (3) 1000–1006. [DOI] [PubMed] [Google Scholar]
- Asnafi, V. , Beldjord, K. , Libura, M. , Villarese, P. , Millien, C. , Ballerini, P. , Kuhlein, E. , Lafage-Pochitaloff, M. , Delabesse, E. , Bernard, O. , 2004. Age-related phenotypic and oncogenic differences in T-cell acute lymphoblastic leukemias may reflect thymic atrophy. Blood 104, (13) 4173–4180. [DOI] [PubMed] [Google Scholar]
- Asnafi, V. , Buzyn, A. , Thomas, X. , Huguet, F. , Vey, N. , Boiron, J.M. , Reman, O. , Cayuela, J.M. , Lheritier, V. , Vernant, J.P. , 2005. Impact of TCR status and genotype on outcome in adult T-cell acute lymphoblastic leukemia: a LALA-94 study. Blood 105, (8) 3072–3078. [DOI] [PubMed] [Google Scholar]
- Ayton, P.M. , Cleary, M.L. , 2001. Molecular mechanisms of leukemogenesis mediated by MLL fusion proteins. Oncogene 20, (40) 5695–5707. [DOI] [PubMed] [Google Scholar]
- Bohlander, S.K. , Muschinsky, V. , Schrader, K. , Siebert, R. , Schlegelberger, B. , Harder, L. , Schemmel, V. , Fonatsch, C. , Ludwig, W.D. , Hiddemann, W. , 2000. Molecular analysis of the CALM/AF10 fusion: identical rearrangements in acute myeloid leukemia, acute lymphoblastic leukemia and malignant lymphoma patients. Leukemia 14, (1) 93–99. [DOI] [PubMed] [Google Scholar]
- Carlson, K.M. , Vignon, C. , Bohlander, S. , Martinez-Climent, J.A. , Le Beau, M.M. , Rowley, J.D. , 2000. Identification and molecular characterization of CALM/AF10fusion products in T cell acute lymphoblastic leukemia and acute myeloid leukemia. Leukemia 14, (1) 100–104. [DOI] [PubMed] [Google Scholar]
- Caudell, D. , Aplan, P.D. , 2007. The role of CALM-AF10 gene fusion in acute leukemia Leukemia PMID: 18094714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caudell, D. , Zhang, Z. , Chung, Y.J. , Aplan, P.D. , 2007. Expression of a CALM-AF10 fusion gene leads to Hoxa cluster overexpression and acute leukemia in transgenic mice. Cancer Res. 67, (17) 8022–8031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao, M.M. , Walker, A.C. , Pendergast, M.B. , Bohlander, S.K. , Wechsler, D.S. , 2004. Perturbed endocytosis by CALM-containing fusion proteins: a leukemogenic mechanism in AML. Blood 104, (11) 922a–923a. [Google Scholar]
- Chao, M.M. , Erichsen, D.A. , Krajewski, M.L. , Bohlander, S.K. , Wechsler, D.S. , 2005. Perturbed endocytosis by leukemogenic CALM-containing fusion proteins is associated with prolonged growth factor signaling and enhanced cellular proliferation. Blood 106, (11) 696a [Google Scholar]
- De Camilli, P. , Chen, H. , Hyman, J. , Panepucci, E. , Bateman, A. , Brunger, A.T. , 2002. The ENTH domain. FEBS Lett 513, (1) 11–18. [DOI] [PubMed] [Google Scholar]
- Debernardi, S. , Bassini, A. , Jones, L.K. , Chaplin, T. , Linder, B. , de Bruijn, D.R. , Meese, E. , Young, B.D. , 2002. The MLL fusion partner AF10 binds GAS41, a protein that interacts with the human SWI/SNF complex. Blood 99, (1) 275–281. [DOI] [PubMed] [Google Scholar]
- Deshpande, A.J. , Cusan, M. , Rawat, V.P. , Reuter, H. , Krause, A. , Pott, C. , Quintanilla-Martinez, L. , Kakadia, P. , Kuchenbauer, F. , Ahmed, F. , 2006. Acute myeloid leukemia is propagated by a leukemic stem cell with lymphoid characteristics in a mouse model of CALM/AF10-positive leukemia. Cancer Cell 10, (5) 363–374. [DOI] [PubMed] [Google Scholar]
- Di Fiore, P.P. , De Camilli, P. , 2001. Endocytosis and signaling. an inseparable partnership. Cell 106, (1) 1–4. [DOI] [PubMed] [Google Scholar]
- Dik, W.A. , Brahim, W. , Braun, C. , Asnafi, V. , Dastugue, N. , Bernard, O.A. , van Dongen, J.J. , Langerak, A.W. , Macintyre, E.A. , Delabesse, E. , 2005. CALM-AF10+T-ALL expression profiles are characterized by overexpression of HOXA and BMI1 oncogenes. Leukemia 19, (11) 1948–1957. [DOI] [PubMed] [Google Scholar]
- DiMartino, J.F. , Ayton, P.M. , Chen, E.H. , Naftzger, C.C. , Young, B.D. , Cleary, M.L. , 2002. The AF10 leucine zipper is required for leukemic transformation of myeloid progenitors by MLL-AF10. Blood 99, (10) 3780–3785. [DOI] [PubMed] [Google Scholar]
- Dreyling, M.H. , Martinez-Climent, J.A. , Zheng, M. , Mao, J. , Rowley, J.D. , Bohlander, S.K. , 1996. The t(10;11)(p13;q14) in the U937 cell line results in the fusion of the AF10 gene and CALM, encoding a new member of the AP-3 clathrin assembly protein family. Proc. Natl. Acad. Sci. U.S.A. 93, (10) 4804–4809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyling, M.H. , Schrader, K. , Fonatsch, C. , Schlegelberger, B. , Haase, D. , Schoch, C. , Ludwig, W. , Loffler, H. , Buchner, T. , Wormann, B. , 1998. MLL and CALM are fused to AF10 in morphologically distinct subsets of acute leukemia with translocation t(10;11): both rearrangements are associated with a poor prognosis. Blood 91, (12) 4662–4667. [PubMed] [Google Scholar]
- Evans, P.R. , Owen, D.J. , 2002. Endocytosis and vesicle trafficking. Curr. Opin. Struct. Biol. 12, (6) 814–821. [DOI] [PubMed] [Google Scholar]
- Floyd, S. , De Camilli, P. , 1998. Endocytosis proteins and cancer: a potential link?. Trends Cell Biol. 8, (8) 299–301. [DOI] [PubMed] [Google Scholar]
- Ford, M.G. , Pearse, B.M. , Higgins, M.K. , Vallis, Y. , Owen, D.J. , Gibson, A. , Hopkins, C.R. , Evans, P.R. , McMahon, H.T. , 2001. Simultaneous binding of PtdIns(4,5)P2 and clathrin by AP180 in the nucleation of clathrin lattices on membranes. Science 291, (5506) 1051–1055. [DOI] [PubMed] [Google Scholar]
- Ford, M.G. , Mills, I.G. , Peter, B.J. , Vallis, Y. , Praefcke, G.J. , Evans, P.R. , McMahon, H.T. , 2002. Curvature of clathrin-coated pits driven by epsin. Nature 419, (6905) 361–366. [DOI] [PubMed] [Google Scholar]
- Forissier, S. , Razanajaona, D. , Ay, A.S. , Martel, S. , Bartholin, L. , Rimokh, R. , 2007. AF10 dependent transcription is enhanced by its interaction with FLRG. Biol. Cell 99, (10) 563–571. [DOI] [PubMed] [Google Scholar]
- Greif, P.A. , Tizazu, B. , Krause, A. , Kremmer, E. , Bohlander, S.K. , 2008. The leukemogenic CALM/AF10 fusion protein alters the subcellular localization of the lymphoid regulator Ikaros. Oncogene 27, (20) 2886–2896. [DOI] [PubMed] [Google Scholar]
- Harel, A. , Wu, F. , Mattson, M.P. , Morris, C.M. , Yao, P.J. , 2008. Evidence for CALM in directing VAMP2 trafficking. Traffic 9, (3) 417–429. [DOI] [PubMed] [Google Scholar]
- Holzel, M. , Rohrmoser, M. , Schlee, M. , Grimm, T. , Harasim, T. , Malamoussi, A. , Gruber-Eber, A. , Kremmer, E. , Hiddemann, W. , Bornkamm, G.W. , 2005. Mammalian WDR12 is a novel member of the Pes1-Bop1 complex and is required for ribosome biogenesis and cell proliferation. J. Cell Biol. 170, (3) 367–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, F. , Khvorova, A. , Marshall, W. , Sorkin, A. , 2004. Analysis of clathrin-mediated endocytosis of epidermal growth factor receptor by RNA interference. J. Biol. Chem. 279, (16) 16657–16661. [DOI] [PubMed] [Google Scholar]
- Kalthoff, C. , Alves, J. , Urbanke, C. , Knorr, R. , Ungewickell, E.J. , 2002. Unusual structural organization of the endocytic proteins AP180 and epsin 1. J. Biol. Chem. 277, (10) 8209–8216. [DOI] [PubMed] [Google Scholar]
- Klebig, M.L. , Wall, M.D. , Potter, M.D. , Rowe, E.L. , Carpenter, D.A. , Rinchik, E.M. , 2003. Mutations in the clathrin-assembly gene Picalm are responsible for the hematopoietic and iron metabolism abnormalities in fit1 mice. Proc. Natl. Acad. Sci. U.S.A. 100, (14) 8360–8365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause, A. , Kohlmann, A. , Haferlach, T. , Schoch, C. , Schnittger, S. , Mecucci, C. , Ludwig, W.D. , Bohlander, S.K. , 2004. The CALM/AF10 fusion: molecular analysis of the fusion transcripts in 13 cases of AML and ALL; gene expression profiling reveals HOX gene deregulation. Blood 104, (11) 791a [Google Scholar]
- Kumon, K. , Kobayashi, H. , Maseki, N. , Sakashita, A. , Sakurai, M. , Tanizawa, A. , Imashuku, S. , Kaneko, Y. , 1999. Mixed-lineage leukemia with t(10;11)(p13;q21): an analysis of AF10-CALM and CALM-AF10 fusion mRNAs and clinical features. Genes Chromosomes Cancer 25, (1) 33–39. [DOI] [PubMed] [Google Scholar]
- Linder, B. , Newman, R. , Jones, L.K. , Debernardi, S. , Young, B.D. , Freemont, P. , Verrijzer, C.P. , Saha, V. , 2000. Biochemical analyses of the AF10 protein: the extended LAP/PHD-finger mediates oligomerisation. J. Mol. Biol. 299, (2) 369–378. [DOI] [PubMed] [Google Scholar]
- Linder, B. , Gerlach, N. , Jackle, H. , 2001. The Drosophila homolog of the human AF10 is an HP1-interacting suppressor of position effect variegation. EMBO Rep 2, (3) 211–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maggi, L.B. , Weber, J.D. , 2005. Nucleolar adaptation in human cancer. Cancer Invest 23, (7) 599–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerholz, A. , Hinrichsen, L. , Groos, S. , Esk, P.C. , Brandes, G. , Ungewickell, E.J. , 2005. Effect of clathrin assembly lymphoid myeloid leukemia protein depletion on clathrin coat formation. Traffic 6, (12) 1225–1234. [DOI] [PubMed] [Google Scholar]
- Milani, V. , Frankenberger, B. , Heinz, O. , Brandl, A. , Ruhland, S. , Issels, R.D. , Noessner, E. , 2005. Melanoma-associated antigen tyrosinase but not Melan-A/MART-1 expression and presentation dissociate during the heat shock response. Int. Immunol 17, (3) 257–268. [DOI] [PubMed] [Google Scholar]
- Narita, M. , Shimizu, K. , Hayashi, Y. , Taki, T. , Taniwaki, M. , Hosoda, F. , Kobayashi, H. , Nakamura, H. , Sadamori, N. , Ohnishi, H. , 1999. Consistent detection of CALM-AF10 chimaeric transcripts in haematological malignancies with t(10;11)(p13;q14) and identification of novel transcripts. Br. J. Haematol 105, (4) 928–937. [DOI] [PubMed] [Google Scholar]
- Okada, Y. , Feng, Q. , Lin, Y. , Jiang, Q. , Li, Y. , Coffield, V.M. , Su, L. , Xu, G. , Zhang, Y. , 2005. hDOT1L links histone methylation to leukemogenesis. Cell 121, (2) 167–178. [DOI] [PubMed] [Google Scholar]
- Okada, Y. , Jiang, Q. , Lemieux, M. , Jeannotte, L. , Su, L. , Zhang, Y. , 2006. Leukaemic transformation by CALM-AF10 involves upregulation of Hoxa5 by hDOT1L. Nat. Cell Biol. 8, (9) 1017–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson, M.O. , 2004. Sensing cellular stress: another new function for the nucleolus?. Sci. STKE 224, pe10 2004 [DOI] [PubMed] [Google Scholar]
- Pasalic, Z. , Tizazu, B. , Archangelo, L. , Krause, A. , Greif, P.A. , Bohlander, S.K. , 2006. The four and a half LIM domain protein 2 (FHL2) interacts with CALM and is highly expressed in AML with complex aberrant karyotpes. Blood 108, (11) 4337 [Google Scholar]
- Perrin, L. , Dura, J.M. , 2004. Molecular genetics of the Alhambra (Drosophila AF10) complex locus of Drosophila. Mol. Genet. Genomics 272, (2) 156–161. [DOI] [PubMed] [Google Scholar]
- Perrin, L. , Bloyer, S. , Ferraz, C. , Agrawal, N. , Sinha, P. , Dura, J.M. , 2003. The leucine zipper motif of the Drosophila AF10 homologue can inhibit pre-mediated repression: implications for leukemogenic activity of human MLL-AF10 fusions. Mol. Cell. Biol. 23, (1) 119–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo, S. , Pece, S. , Di Fiore, P.P. , 2004. Endocytosis and cancer. Curr. Opin. Cell Biol. 16, (2) 156–161. [DOI] [PubMed] [Google Scholar]
- Rubbi, C.P. , Milner, J. , 2003. Disruption of the nucleolus mediates stabilization of p53 in response to DNA damage and other stresses. Embo J 22, (22) 6068–6077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggero, D. , Pandolfi, P.P. , 2003. Does the ribosome translate cancer?. Nat. Rev. Cancer 3, (3) 179–192. [DOI] [PubMed] [Google Scholar]
- Saha, V. , Chaplin, T. , Gregorini, A. , Ayton, P. , Young, B.D. , 1995. The leukemia-associated-protein (LAP) domain, a cysteine-rich motif, is present in a wide range of proteins, including MLL, AF10, and MLLT6 proteins. Proc. Natl. Acad. Sci. U.S.A. 92, (21) 9737–9741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soulier, J. , Clappier, E. , Cayuela, J.M. , Regnault, A. , Garcia-Peydro, M. , Dombret, H. , Baruchel, A. , Toribio, M.L. , Sigaux, F. , 2005. HOXA genes are included in genetic and biologic networks defining human acute T-cell leukemia (T-ALL). Blood 106, (1) 274–286. [DOI] [PubMed] [Google Scholar]
- Sporle, R. , Schughart, K. , 1998. Paradox segmentation along inter- and intrasomitic borderlines is followed by dysmorphology of the axial skeleton in the open brain (opb) mouse mutant. Dev. Genet. 22, (4) 359–373. [DOI] [PubMed] [Google Scholar]
- Tebar, F. , Bohlander, S.K. , Sorkin, A. , 1999. Clathrin assembly lymphoid myeloid leukemia (CALM) protein: localization in endocytic-coated pits, interactions with clathrin, and the impact of overexpression on clathrin-mediated traffic. Mol. Biol. Cell 10, (8) 2687–2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visseren, M.J. , van Elsas, A. , van der Voort, E.I. , Ressing, M.E. , Kast, W.M. , Schrier, P.I. , Melief, C.J. , 1995. CTL specific for the tyrosinase autoantigen can be induced from healthy donor blood to lyse melanoma cells. J. Immunol 154, (8) 3991–3998. [PubMed] [Google Scholar]
- Wechsler, D.S. , Engstrom, L.D. , Alexander, B.M. , Motto, D.G. , Roulston, D. , 2003. A novel chromosomal inversion at 11q23 in infant acute myeloid leukemia fuses MLL to CALM, a gene that encodes a clathrin assembly protein. Genes Chromosomes Cancer 36, (1) 26–36. [DOI] [PubMed] [Google Scholar]
- Whitfield, M.L. , Zheng, L.X. , Baldwin, A. , Ohta, T. , Hurt, M.M. , Marzluff, W.F. , 2000. Stem-loop binding protein, the protein that binds the 3′ end of histone mRNA, is cell cycle regulated by both translational and posttranslational mechanisms. Mol. Cell. Biol. 20, (12) 4188–4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data
Supplementary data


