Abstract
An important goal of contemporary HIV type 1 (HIV-1) research is to identify cellular cofactors required for viral replication. The HIV-1 Rev protein facilitates the cytoplasmic accumulation of the intron-containing viral gag-pol and env mRNAs and is required for viral replication. We have previously shown that a cellular protein, human Rev-interacting protein (hRIP), is an essential Rev cofactor that promotes the release of incompletely spliced HIV-1 RNAs from the perinuclear region. Here, we use complementary genetic approaches to ablate hRIP activity and analyze HIV-1 replication and viral RNA localization. We find that ablation of hRIP activity by a dominant-negative mutant or RNA interference inhibits virus production by mislocalizing Rev-directed RNAs to the nuclear periphery. We further show that depletion of endogenous hRIP by RNA interference results in the loss of viral replication in human cell lines and primary macrophages; virus production was restored to wild-type levels after reintroduction of hRIP protein. Taken together, our results indicate that hRIP is an essential cellular cofactor for Rev function and HIV-1 replication. Because hRIP is not required for cell viability, it may be an attractive target for the development of new antiviral strategies.
Keywords: Rev-directed RNA export, RNA localization
Replication of lentiviruses, such as HIV type 1 (HIV-1), entails a tightly regulated complex life cycle that depends upon specific interactions between viral RNAs and viral and cellular proteins. In principle, every step in the viral life cycle provides an opportunity for therapeutic intervention (1–3). Anti-HIV drugs that are currently available or in clinical development inhibit critical steps in viral replication such as reverse transcription, viral protein maturation, virus entry, and integration of the proviral DNA (4–7). However, therapeutic strategies that inactivate other critical steps in the viral life cycle remain unrealized (8–11).
An essential and characteristic step in HIV-1 replication is the export of the intron-containing gag-pol and env mRNAs from the nucleus to the cytoplasm. The viral regulatory protein Rev mediates this event, in conjunction with the cellular nuclear export machinery and several protein cofactors (12–15). Rev contains two functional domains: an arginine-rich RNA-binding motif, which specifically interacts with a cis-acting element within intron-containing viral RNAs designated the Rev-responsive element (RRE) (16, 17), and a leucine-rich “effector” domain that constitutes a nuclear export signal (14, 15, 17–19).
We have recently shown that the human Rev-interacting protein (hRIP) is a cellular cofactor required for Rev function (20). In the absence of functional hRIP, Rev-directed RNAs mislocalize and aberrantly accumulate at the nuclear periphery, where hRIP is localized. Importantly, the requirement for hRIP is highly specific, because comparable ablation of hRIP activity did not affect the intracellular distribution of cellular poly(A)+ mRNA, nuclear proteins, or other nuclear export signal-containing proteins. Collectively, these observations raise the possibility that hRIP could be a cellular cofactor for HIV-1 replication. Here, we use complementary genetic approaches to ablate hRIP activity and analyze viral replication and HIV-1 RNA localization.
Materials and Methods
Cell Lines. HeLa, HL2/3, and 293T fibroblasts and T-lymphoid cells were grown as described in ref. 20. Mononuclear leukocyte-rich cell preparations were obtained by leukapheresis from normal donors seronegative for HIV-1 and hepatitis B. Monocytes were isolated by countercurrent centrifugal elutriation and cultured as adherent monolayers as described in ref. 21. Monocyte-derived macrophages were infected 8 days after plating as described in ref. 22.
Plasmids. The hRIP derivative expression plasmids have been described (20). pRab-R-V5 contains the human Rab-related protein (Rab-R) cDNA (GenBank accession no. 4102708) cloned into pcDNA4C/V5-His (EcoRI–NotI). pFlag-hRIP contains nucleotides 12–1683 of the hRIP cDNA cloned into pCMV2Flag (Hin-dIII–XbaI). pFlag-hRIPRS was generated from pFlag-hRIP by using PCR-based site-directed mutagenesis. Silent mutations were introduced into the hRIP cDNA at positions 474G:C, 1605T:C, 1608A:C, and 1611T:C to render it resistant to RNA interference (RNAi) yet retain amino acid composition. All recombinant or mutagenized plasmids were analyzed by DNA sequencing. Protein levels in cell lysates were analyzed as described in ref. 20 by using antihemagglutinin (Roche Applied Science, Indianapolis; 1:500), anti-hRIP, or anti-Eps15 (SCB C19, 1:750; and C20, 1:750, respectively), anti-V5 (Invitrogen, 1:2,000), or anti-Flag (Kodak M5, 1:500) antibodies and visualized by enhanced chemiluminescence (Amersham Pharmacia). Mutant hRIP, Flag-hRIP, Flag-hRIPRS, or Rab-R protein levels were comparable to those of cellular hRIP and Rab-R, as determined by Western blotting with anti-hRIP or anti-Rab-R (AN1121, 1:500) antibodies.
DNA Transfections. 293T cells were transfected with 5 μg of pCMV, 5 μg of phRIP, 5 μg of phRIPΔN235, 5 μg of phRIPΔN360, 5 μg of phRIPΔC237, or 5 μg of phRIPΔC317 in the presence of 5 μg of pHIVLAI, as described in ref. 23. Control cells were transfected with 5 μg of pCMV, 5 μg of pCMV-yRIP, or 5 μg of pCMV-Eps15 in the presence of 5 μg of pHIVLAI. Transfection reactions contained 0.5 μg of pCMV β-galactosidase (β-gal) as an internal control for transfection efficiency, mRNA export, protein normalization, and nonspecific effects of the cytomegalovirus (CMV) promoter. Total DNA was adjusted to 15 μg using pCMV2Flag and pUC118.
HL2/3 cells were transfected with 5 μg of pCMV, 5 μg of phRIP, or 5 μg of phRIPΔN360 in the presence of 0.2 μg of pCMV β-gal by using geneporter 2 (Gene Therapy Systems, San Diego). Cell lysates were prepared at 48/h posttransfection and analyzed as described in ref. 20. Pr55gag and lamin A/C protein levels were analyzed by Western blotting with anti-HIV-1 Gag or anti-lamin A/C antibodies (SCB vT-20, 1:500; and N-18, 1:500, respectively) and visualized by using enhanced chemiluminescence.
Viral Replication Assays. Viral stocks and infections. 293T cells were cotransfected with 5 μg of pHIVADA and 1 μg of pCMV β-gal, as described in ref. 23. At 40 h posttransfection, culture supernatants were analyzed for reverse transcriptase (RT) activity and HIV-1 Gag p24 antigen, as described in refs. 24 and 25. For viral replication assays, nonadherent primary human macrophages grown in 24-well microtiter plates were treated with H1, M1, or L2 small interfering RNAs (siRNAs) (20 pM) at consecutive 24-hr intervals and infected with wild-type HIVADA (1 μg of Gag p24/0.7 × 106 cells per well) and incubated for 5 days. hRIP and clathrin levels in cell lysates prepared from the bulk culture were analyzed as described in ref. 20.
Viral RT activity and Gag p24 antigen production. Culture supernatants were passed through a 0.45-μm Acrodisc filter (Fisher Scientific) and frozen at –80°C until use. All samples were normalized to β-gal expression, and RT activity assays were performed in triplicate as described in ref. 24. HIV-1 Gag p24 antigen quantification was performed in duplicate by using a standard ELISA (IMB SoftMax Plus, Coulter).
RNA in Situ Hybridization and Immunofluorescence Microscopy. 293T cells were transfected with 3 μg of pCMV, 3 μg of phRIP, or 3 μg of phRIPΔN360 in the presence of 1.5 μg of pHIVLAI. The intracellular distributions of hRIP or HIV RNA in siRNA-treated cells were analyzed by using immunofluorescence microscopy and in situ hybridization, respectively, as described in ref. 20.
RNAi. hRIP-specific (H1, H2) and mutant (M1) siRNAs have been described (20). Human lamin A/C siRNAs were (L1) CUGGACUUCCAGAAGAACATT and UGUUCUUCUGGAAGUCCAGTT, and rat lamin A/C siRNAs were (L2) AAGAGCGGGAGAUGGCUGATT and UCAGCCAUCUCCCGCUCUUTT.
siRNA Transfections. HeLa cells were treated with H1, H2, M1, or L1 siRNAs, as described in ref. 20. After 24 h, cells were cotransfected with 5 μg of pHIVLAI and 0.5 μg of pCMV β-gal by using geneporter 2. Cell viability was assayed by staining with trypan blue and monitoring β-gal activity (≥97.8). Cells and culture supernatants were collected 72 h posttransfection; this time point represents the maximum levels of inhibition afforded by interfering with or ablating hRIP activity at the peak of virus production.
Jurkat cells (2 × 106 cells per ml) were electroporated with 5 μg of pHIVNL4.3-GFP (26) and 5 μg of pCMV β-gal or with 5 μg of pHIVNL4.3-GFP; 5 μg of pCMV β-gal; and H1, M1, T441 (27), or L1 siRNAs, as described in ref. 28. Electroporation was performed in duplicate in a Cell-PORATOR (GIBCO/BRL) set at 244V/1180μF by using a fitted cuvette with a 0.4-cm gap width. Cell viability (≥93.6%) was monitored as described above. After 72 h, cells were seeded on glass coverslips, then treated with 1.5% paraformaldehyde/0.14% glutaraldehyde for 15 min and mounted on slides by using Vectashield (Vector Laboratories) with 10 μg/ml DAPI. GFP-positive cells (≥200 cells per sample) were visualized with a Zeiss Axioplan 2 fluorescence microscope and the number of infected cells determined by multiple independent rounds of counting. Percentages of GFP-positive cells in Fig. 3B have been adjusted for transfection efficiency and background signals.
Fig. 3.
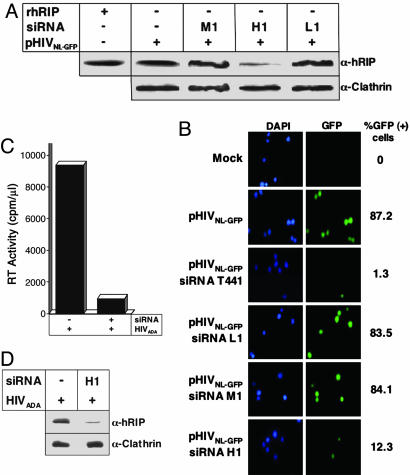
Ablation of hRIP activity inhibits viral replication in human T cell lines and primary macrophages. (A) Depletion of cellular hRIP by using RNAi. Jurkat cells were electroporated with an HIV-1 molecular clone, pHIVNL4.3/GFP, and the indicated siRNA. hRIP and clathrin levels in cell lysates were analyzed as in Fig. 2 A. –, absence; +, presence. (B) GFP-positive cells were visualized by using fluorescence microscopy and the number of infected cells determined by multiple independent rounds of counting. Percentages of GFP-positive cells indicated in the right column have been adjusted for transfection efficiency and background signal. The data depict a representative sampling of each bulk culture. Mock, no DNA or siRNA. (C and D) Human monocyte-derived macrophages were treated with the indicated siRNAs and infected with supernatants containing wild-type HIVADA virions (1 μg of Gag p24/0.7 × 106 cells). hRIP and clathrin levels in cell lysates were analyzed as in A; viral RT activity was analyzed as in Fig. 2C. The data shown are representative of four independent experiments performed in duplicate (SD, 4.03). –, absence; +, presence.
Reconstitution of hRIP Activity After RNAi. HeLa cells were treated with Oligofectamine (Invitrogen) alone or H1 or M1 siRNAs, as described in ref. 20. After 24 hr, cells were transfected by using geneporter 2 as follows: mixtures for H1 siRNA-treated cells contained 2.5 μg of pCMV, 2.5 μg of pRab-R-V5, 2.5 μg of pFlag-hRIP, or 2.5 μg of pFlag-hRIPRS in the presence of 2.5 μg of pHIVLAI; mixtures for M1 siRNA-treated cells contained 2.5 μg of pCMV and 2.5 μg of pHIVLAI; mixtures for Oligofectamine-treated cells contained 2.5 μg of pCMV. All DNA mixtures contained 0.5 μg of pCMV β-gal. Cell viability after siRNA treatment and DNA transfection (>95.6%) was monitored as described above. After 72 h, transfected cells and culture supernatants were collected and cell lysates prepared as in ref. 20. Cellular hRIP and clathrin and Flag-hRIP, Flag-hRIPRS, and Rab-R-V5 levels were analyzed sequentially by Western blotting by using anti-hRIP, anticlathrin, anti-Flag, and anti-V5 antibodies, respectively, and visualized by using enhanced chemiluminescence.
Results
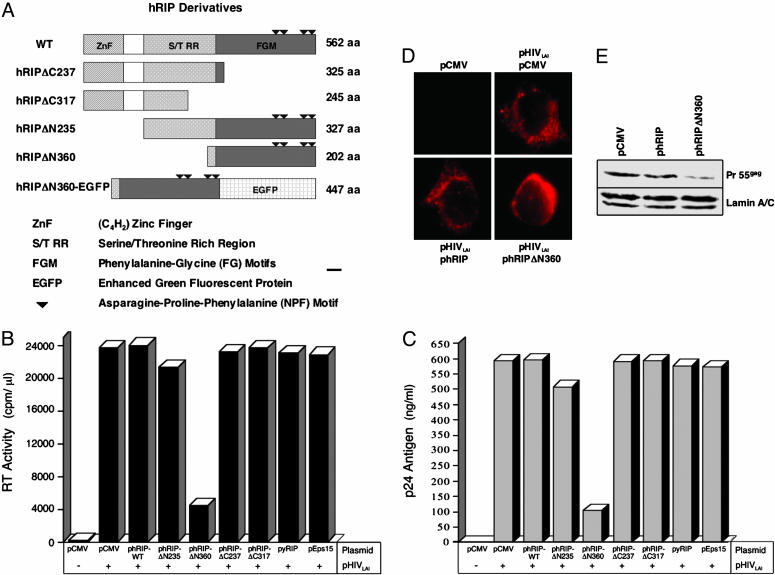
A Dominant-Negative hRIP Mutant Interferes with HIV-1 Production.Previous studies have shown that the hRIP mutant, hRIPΔN360, exerts a negative effect on Rev function in the presence of endogenous hRIP in transient transfection assays (20). The ability of hRIPΔN360 to act as a transdominant-negative inhibitor of Rev function suggested that it might also interfere with HIV-1 production. To explore this possibility, 293T cells were cotransfected with pHIVLAI, an HIV-1 molecular clone (29), and an empty vector (pCMV) or pHIVLAI and an hRIP derivative expression plasmid (Fig. 1A). To control for the specificity of the assay, cells were cotransfected with pHIVLAI and pCMV-yRIP, a plasmid that expresses Rip1p/Nup42, a yeast NPF-motif containing protein shown to interact with the Rev nuclear export signal in a genetic screen in yeast (30). Alternatively, cells were cotransfected with pHIVLAI and pCMV-Eps15 (31), a plasmid that expresses Eps15, an epsin-homology domain-containing protein reported to interact with hRIP and CRM1 in mammalian cells (32, 33). At 72 h posttransfection, culture supernatants were assayed for viral RT activity and Gag p24 antigen production, two standard parameters of viral replication (24, 25). Most of these hRIP derivatives had no discernible effect; we did, however, observe a marked reduction (5- to 6-fold) in RT activity and p24 levels in the presence of hRIPΔN360 (Fig. 1 B and C). Importantly, overexpression of Rip1p/Nup 42 or Eps15 had no discernible effect on virus production. These results indicate that the inhibitory activity of hRIPΔ360 on virus production is specific.
Fig. 1.
A dominant-negative hRIP mutant inhibits HIV-1 production by blocking release of Rev-directed RNAs from the nuclear periphery to the cytoplasm. (A) Schematic representation of N- and C-terminally truncated and fluorescent hRIP derivatives. ZnF, C4H2-type zinc finger domain; S/TRR, serine/threonine-rich region; FGM, phenylalanine-glycine motifs. ▾, asparagine-proline-phenylalanine (NPF) motifs. (B and C) Virus production in the presence of hRIP derivatives. 293T cells were cotransfected with an HIV-1 infectious molecular clone (pHIVLAI) and pCMV or pHIVLAI and the indicated hRIP expression plasmid. Control cells were transfected with pCMV or pHIVLAI and pCMV-yRIP or pHIVLAI and pCMV-Eps15. Virus production was assayed by measuring viral RT activity (cpm/μl) and Gag p24 antigen (ng/ml) in culture supernatants at 72 h posttransfection. Data shown in B and C are representative of six independent experiments, each performed in duplicate (SD, 3.80 and 2.88, respectively). –, absence; +, presence. (D) The hRIPΔN360 mutant causes HIV RNAs to mislocalize and aberrantly accumulate at the nuclear periphery. 293T cells were cotransfected with pHIVLAI and pCMV, or pHIVLAI and the indicated hRIP expression plasmid. Control cells were transfected with pCMV. The intracellular distribution of HIV RNA was analyzed by fluorescent RNA in situ hybridization by using a Cy3-labeled RRE-specific probe. (E) The hRIPΔN360 mutant inhibits Rev function in cells chronically infected with HIV. HL2/3 cells were transfected with the indicated plasmids. Pr55Gag and lamin A/C protein levels were analyzed by Western blotting by using anti-HIV-1 Gag or anti-lamin A/C antibodies, respectively, and visualized by enhanced chemiluminescence.
A prediction of these findings is that hRIPΔN360 blocks virus production by interfering with Rev function. To verify the nature of the inhibition exerted by hRIPΔN360, we analyzed the intracellular distribution of Rev-directed RNAs by fluorescent RNA in situ hybridization. 293T cells were cotransfected with pHIVLAI and pCMV, pHIVLAI and phRIP, or pHIVLAI and phRIPΔN360. The intracellular distribution of Rev-directed RNAs was visualized using a fluorochrome-labeled oligonucleotide probe complementary to stem-loop IIB of the HIV-1 RRE (20). Fig. 1D Upper Right shows that Rev-directed RNAs are efficiently exported from the nucleus and accumulate in the cytoplasm. Cells transfected with an empty DNA vector contained no fluorescent signals after hybridization with the probe, confirming that the detection was specific for HIV RNA (Fig. 1D Upper Left). Overexpression of hRIP had no discernible effect on the cytoplasmic accumulation of Rev-directed RNAs (Fig. 1D Lower Left). In contrast, Rev-directed RNAs were mislocalized and aberrantly accumulated at the nuclear periphery in the presence of hRIPΔN360 (Fig. 1D Lower Right). This RNA localization phenotype was strikingly similar to that observed with hRIPΔN360 in transient transfection assays of Rev function (20).
We also examined whether hRIPΔN360 could interfere with Rev function in cells stably expressing the hybrid infectious molecular clone, HXB2/3gpt (34). HL2/3 cells, which express high levels of HIV proteins, were transfected with pCMV, phRIP, or phRIPΔN360 in the presence of pCMV β-gal. Pr55gag expression levels were analyzed by Western blotting by using anti-HIV-1 Gag antibodies. We observed a marked reduction (5- to 6-fold) in the level of Pr55gag only in the presence of the hRIPΔN360, whereas the levels of the internal control protein, lamin A/C, or ectopically expressed β-gal were unchanged (Fig. 1E and data not shown). In situ hybridization analysis confirmed Rev-directed RNAs were mislocalized and accumulated at the nuclear periphery, where hRIPΔN360 is localized (data not shown). Additionally, hRIPΔN360 had no discernible effect on the ratio of unspliced: spliced HIV RNAs stably expressed in these cells (data not shown). Collectively, our results provide additional evidence that the inhibition exerted by hRIPΔN360 on Rev function is specific. Furthermore, we conclude that hRIPΔN360 inhibits HIV-1 replication by disrupting an intermediate step in Rev function, thereby causing Rev-directed RNAs to mislocalize and accumulate at the nuclear periphery.
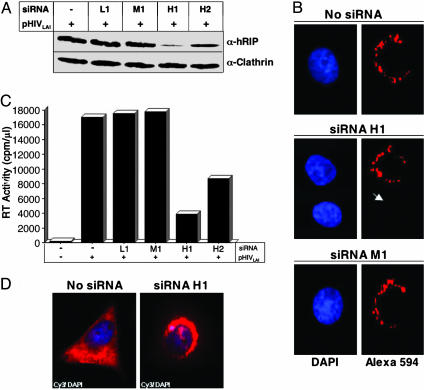
Ablation of hRIP Activity by RNAi Results in a Loss of Viral Replication. Previous studies have shown that ablation of hRIP activity by RNAi inhibits Rev function by mislocalizing RRE-containing RNAs at the nuclear periphery (20). A prediction of this finding is that loss of hRIP function could affect virus production. Using previously optimized conditions (20), we treated HeLa cells with siRNA duplexes homologous to hRIP coding sequences (hRIP-specific H1 or H2 siRNA) by using Oligofectamine. Alternatively, cells were treated with siRNAs harboring a single base-pair mismatch within the same hRIP sequences (mutant M1 siRNA) or homologous to human lamin A/C coding sequences (lamin A/C-specific L1 siRNA). The siRNA-treated cells were then cotransfected with pHIVLAI and pCMV β-gal by using cationic liposomes. Cellular hRIP levels were analyzed by Western blotting of cell extracts from the bulk culture and monitored by indirect immunofluorescence microscopy by using an anti-hRIP antibody. RT activity and p24 antigen in culture supernatants and cytosolic β-gal activity were quantified as in Fig. 1. The results shown in Fig. 2 A and B indicate that the level of cellular hRIP, which is localized predominantly at the nuclear periphery, was most effectively reduced by treatment with H1 siRNA. In contrast, treatment with M1 or L1 siRNA had no discernible effect on cellular hRIP levels (Fig. 2 A and B). Importantly, we observed a marked reduction (5- to 6-fold) in RT activity and p24 levels after depletion of cellular hRIP (Fig. 2C and data not shown). The RNAi appeared to be specific, because the level of cellular clathrin was unchanged by siRNA treatment (Fig. 2A). Most important, in situ hybridization analysis revealed that ablation of hRIP inhibits HIV production by blocking release of viral RNAs from the nuclear periphery to the cytoplasm (Fig. 2D). In the absence of functional hRIP, Rev-directed RNAs were mislocalized and accumulated aberrantly at the perinuclear region.
Fig. 2.
Ablation of hRIP activity inhibits HIV-1 production in mammalian cells. (A) Depletion of cellular hRIP using RNAi. HeLa cells were treated with hRIP-specific (H1 or H2), mutant (M1), or lamin A/C-specific (L1) siRNAs by using oligofectamine. Control cells were treated with Oligofectamine. After 24 h, siRNA-treated and control cells were transfected with pHIVLAI and pCMV β-gal by using cationic liposomes. hRIP and clathrin levels in cell lysates prepared from the bulk culture were analyzed at 48 h posttransfection by Western blotting using anti-hRIP or anti-clathrin antibodies, respectively, and visualized as in Fig. 1. –, absence; +, presence. (B) Efficient and specific depletion of cellular hRIP by using RNAi. Cells were treated with H1 or M1 siRNAs as in A. The intracellular distribution of hRIP was analyzed by indirect immunofluorescence microscopy by using anti-hRIP and Alexa 594 antibodies. The arrowhead indicates a representative H1 siRNA-treated cell depleted of cellular hRIP. (C) Ablation of hRIP activity inhibits virus production. Culture supernatants from siRNA-treated cells transfected in A were assayed for viral RT activity (cpm/μl) at 72 h posttransfection, as in Fig. 1. The data shown are representative of six independent experiments performed in duplicate (SD, 3.78). –, absence; +, presence. (D) Ablation of hRIP activity inhibits virus production by blocking release of Rev-directed RNAs from the perinuclear region. Cells were treated with Oligofectamine alone or H1 siRNAs and transfected as in A. The intracellular distribution of RRE-containing RNAs was analyzed as in Fig. 1D.
We next tested whether depletion of cellular hRIP could interfere with virus production in a more physiologically relevant context, a human T lymphocyte cell line. Jurkat cells were electroporated with H1, M1, or L1 siRNA, in the presence of pHIVNL4.3/GFP, an HIV-1 molecular clone with a GFP cDNA in place of Nef coding sequences (26). To control for the specificity of the assay, cells were mock electroporated or electroporated with pHIVNL4.3/GFP and an HIV-1 Vif-specific T441 siRNA (27) previously shown to specifically degrade genomic HIV RNA and abolish viral replication. The number of infected cells was determined by counting GFP-positive cells visualized by fluorescence microscopy (26). hRIP levels in cell lysates and RT activity in the culture supernatants were assayed as in Fig. 2. The results in Fig. 3 A and B indicate that cells depleted of hRIP by using the H1 siRNA failed to support virus production as monitored by the loss of GFP-positive cells. Consistent with previous reports (27), virus production was abolished in cells treated with the Vif-specific T441 siRNA (Fig. 3B), with no discernible effect on cellular hRIP levels (data not shown). We found that, in both instances, the absence of GFP-positive cells correlated with a marked or complete reduction in the levels of RT activity and p24 in culture supernatants (data not shown). In contrast, virus production was unchanged in cells treated with L1 or M1 siRNA (Fig. 3 A and B). Additionally, T441, L1, M1, or H1 siRNA had no discernible effect on cell viability or proliferation rates (data not shown). Furthermore, the RNAi appeared to be specific, because the level of clathrin was unchanged by siRNA treatment (Fig. 3A).
As a complementary approach, we tested whether ablation of hRIP activity could interfere with HIV replication in primary nondividing human macrophages. Adherent macrophages derived from peripheral blood monocytes were treated with H1, M1, or L2 siRNA at consecutive 24-hr intervals and infected with wild-type HIVADA virions (21, 22). After 5 days postinfection, RT activity in culture supernatants and hRIP levels in cell lysates were quantified as in Fig. 3. Consistent with our previous RNAi studies using cell lines, we found the level of virus replication was markedly reduced in macrophages depleted of cellular hRIP by using H1 siRNA (Fig. 3 C and D and data not shown).
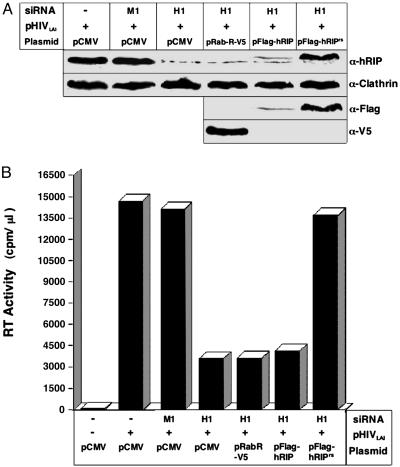
Our results indicate that ablation of hRIP activity using RNAi results in loss of HIV-1 replication. To confirm that the observed reduction in virus production was due specifically to down-regulation of hRIP expression, we reintroduced the hRIP protein into hRIP-depleted cells and monitored the level of virus production. HeLa cells were treated with H1 or M1 siRNA and transfected with pCMV, a plasmid that expresses an epitope-tagged hRIP (pFlag-hRIP) or a plasmid that expresses an epitope-tagged hRIP with silent mutations that render the gene resistant to RNAi (pFlag-hRIPRS), in the presence of pHIVLAI and pCMV β-gal. To control for the specificity of the assay, H1 siRNA-treated cells were transfected with pRab-R-V5, a plasmid that expresses an epitope-tagged human Rab-R in the presence of pHIVLAI and pCMV β-gal. Rab-R displays an overall relatedness of 71% and 46% identity with hRIP (32) and has been shown to enhance Rev activity in transient transfection assays (35). At 72 h posttransfection, RT activity and p24 levels in the culture supernatants and cytosolic β-gal activity were assayed as in previous experiments. Cellular hRIP, Flag-hRIP, Flag-hRIPRS, and Rab-R-V5 protein levels were analyzed by Western blotting of cell extracts from the bulk culture by using anti-hRIP, anti-Flag, and anti-V5 antibodies, respectively. Fig. 4A (first and third rows) shows that cellular hRIP and Flag-hRIP were efficiently depleted after treatment with H1 siRNA. In contrast, the Flag-hRIPRS protein levels were unaffected by H1 siRNA treatment and expressed at levels comparable to that of cellular hRIP (Fig. 4A, first and third rows). The level of cellular clathrin was unchanged by siRNA treatment, further demonstrating that the RNAi is specific (Fig. 4A, second row). Most important, virus production was restored to wild-type levels when the Flag-hRIPRS protein was reintroduced into hRIP-depleted cells (Fig. 4B), providing additional evidence for a functional role for hRIP in virus production. In contrast, expression of Flag-hRIP or Rab-R-V5 in hRIP-depleted cells had no discernible effect on virus production (Fig. 4), indicating that the ability of Flag-hRIPRS to rescue virus production is specific. Taken together, our results indicate that hRIP is an essential cellular cofactor for Rev function and HIV-1 replication.
Fig. 4.
Virus production is restored after reintroduction of hRIP protein into cells depleted of hRIP using RNAi. (A) HeLa cells treated with H1 siRNAs were cotransfected with pHIVLAI and pCMV or pHIVLAI and the indicated hRIP expression plasmid. Control cells were treated with M1 siRNAs and transfected with pCMV or treated with H1 siRNAs and transfected with pRab-R-V5. hRIP and clathrin, Flag-hRIP, Flag-hRIPRS, and Rab-R-V5 protein levels were analyzed by Western blotting of cell extracts from the bulk culture using anti-hRIP, anti-clathrin, anti-Flag, and anti-V5 antibodies, respectively, and visualized as in Fig. 3. –, absence; +, presence. (B) Culture supernatants from A were assayed for viral RT activity as in Fig. 2 A. Data are representative of three independent experiments (SD, 3.47). –, absence; +, presence.
Discussion
We have previously used transient transfection experiments to demonstrate that cellular hRIP is required for the ability of HIV-1 Rev to correctly localize viral RNAs in the cytoplasm (20). Here we confirm and extend this conclusion by demonstrating that hRIP is also essential for HIV-1 replication. Ablation of hRIP activity by a dominant-negative mutant or siRNAs mislocalizes Rev-directed RNAs to the nuclear periphery and, as a result, virus production is inhibited. Furthermore, we show that depletion of hRIP by RNAi results in the loss of viral replication in human cell lines and primary macrophages.
Human cells encode a protein, Rab-R, which is homologous to hRIP (32). It has been reported that overexpression of hRIP or Rab-R enhances Rev activity in mammalian cell-based transfection assays (35), raising the possibility that the function of these two proteins is redundant. However, we find that, after hRIP depletion, addition of hRIP but not Rab-R restores virus production, suggesting that these proteins have distinct nonoverlapping activities.
In conjunction with our previous study, the results presented here identify release of Rev-directed RNAs from the nuclear periphery as an essential step in HIV-1 replication. It is tempting to speculate that this perinuclear step links nuclear export of these viral RNAs with their cytoplasmic function, in particular virus assembly. It is well established that assembly and budding of HIV-1 virions is mediated by cellular vesicular protein sorting pathways (36–40). Several observations raise the possibility that hRIP is involved in directing genomic HIV-1 RNA from the perinuclear region to the appropriate cytoplasmic protein sorting pathway. First, hRIP activity is highly specific and is required for the proper cytoplasmic localization of RRE-containing RNAs but not typical cellular mRNAs (20). Second, hRIP is localized at the perinuclear region, where the machinery for vesicular trafficking is situated (35, 41, 42). Third, hRIP contains an N-terminal zinc finger motif that shares a high degree of sequence identity with the catalytic domain of ADP-ribosylation factor GTPase-activating proteins (Arf GAPs) (43). Arf GAPs comprise a large family of proteins that function as regulators of macromolecular assemblies involved in vesicular trafficking (44). Moreover, in the presence of hRIPΔN360, which lacks the zinc finger motif, Rev-directed RNAs mislocalize and accumulate at the nuclear periphery. Furthermore, it is significant that this RNA mislocalization phenotype requires both Rev and RRE-containing RNAs. Taken together, these findings suggest that the zinc finger motif of hRIP, and possibly a catalytic activity similar to that of Arf GAPs, may mediate release of Rev–RNA complexes from the perinuclear region. Fourth, hRIP is required for correct localization and function of acrosomal components during spermatogenesis, a process that depends upon vesicular trafficking (41, 42). Finally, recent studies have shown that murine leukemia virus RNA is transported to the plasma membrane through a vesicular trafficking pathway (45).
Rev is required for HIV-1 replication, and inhibition of Rev activity interferes with virus production. For these reasons, Rev has been considered an attractive target for therapeutic intervention. Several strategies have been attempted to reduce the level of Rev expression or to block its function (46). To date, however, these strategies have failed to produce an effective antiviral agent (11, 47). Cellular factors that are essential for Rev function offer alternate targets for inactivation and are potential candidates for drug development. The probability of selecting escape mutants in cellular genes, such as hRIP, is expected to be very low. Thus, antiviral agents directed against cellular proteins should avoid the development of drug-resistant HIV-1 variants that inevitably occurs when viral proteins are targeted (9, 40). hRIP is not required for cellular viability (41) and thus may be an attractive target for the development of new antiviral strategies.
Acknowledgments
We thank P. Di Fiore [Instituto FIRC di Oncologia Molecolare, Fondazione Italiana per la Ricerca sul Cancro (FIRC) Institute, Milan] and C. Fritz (Millenium Pharmaceuticals, Inc., Cambridge, MA) for plasmids. HL2/3 cells were obtained through the AIDS Research and Reference Reagent Program, National Institute of Allergy and Infectious Diseases, National Institutes of Health, from Drs. B. K. Felber and G. N. Pavlakis. We also thank N. Pouliot for expert technical assistance and J. M. Jacque and M. Stevenson for technical advice. We acknowledge the University of Massachusetts Center for AIDS Research Molecular Biology and Clinical Cores for oligonucleotide synthesis, DNA sequencing, and purified human mononuclear cells, respectively. We are grateful to K. Long and M. Green for valuable suggestions and critical review of the manuscript. This work was supported by the National Institute of Allergy and Infectious Diseases, the National Institutes of Health (Grant 5R01 AI43208), and the Center for AIDS Research Developmental Core (Grant 5P30 AI42845) to M.L.Z.
Author contributions: Z.Y. and M.L.Z. designed research; Z.Y., N.S.-V., I.E.C., E.L.K., and E.B.U. performed research; I.E.C. and E.L.K. contributed new reagents/analytic tools; E.L.K. and M.L.Z. analyzed data; and M.L.Z. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: HIV-1, HIV type 1; hRIP, human Rev interacting protein; RRE, Rev-responsive element; siRNAs, small interfering RNAs; RNAi, RNA interference; CMV, cytomegalovirus; β-gal, β-galactosidase; RT, reverse transcriptase; Rab-R, Rab-related protein.
References
- 1.Cohen, J. (1997) Science 277, 32–33. [DOI] [PubMed] [Google Scholar]
- 2.Richman, D. D. (2001) Nature 410, 995–1001. [DOI] [PubMed] [Google Scholar]
- 3.Tozser, J. (2003) Curr. Top. Med. Chem. 3, 1447–1457. [DOI] [PubMed] [Google Scholar]
- 4.Boyle, B. A. (2004) AIDS Read. 14, 412–416, 452. [PubMed] [Google Scholar]
- 5.De Clercq, E. (2004) Int. J. Biochem. Cell Biol. 36, 1800–1822. [DOI] [PubMed] [Google Scholar]
- 6.Johnson, A. A., Marchand, C. & Pommier, Y. (2004) Curr. Top. Med. Chem. 4, 1059–1077. [DOI] [PubMed] [Google Scholar]
- 7.Marks, K. & Gulick, R. M. (2004) Curr. Infect. Dis. Rep. 6, 333–339. [DOI] [PubMed] [Google Scholar]
- 8.Moore, J. P. & Stevenson, M. (2000) Nat. Rev. Mol. Cell. Biol. 1, 40–49. [DOI] [PubMed] [Google Scholar]
- 9.Ott, D. E. (2002) Rev. Med. Virol. 12, 359–374. [DOI] [PubMed] [Google Scholar]
- 10.Krogstad, P. (2003) Semin. Pediatr. Infect. Dis. 14, 258–268. [DOI] [PubMed] [Google Scholar]
- 11.Baba, M. (2004) Curr. Top. Med. Chem. 4, 871–882. [DOI] [PubMed] [Google Scholar]
- 12.Dayton A. I. (1996) J. Biomed. Sci. 3, 69–77. [DOI] [PubMed] [Google Scholar]
- 13.Kjems, J. & Askjaer, P. (2000) Adv. Pharmacol. 48, 251–298. [DOI] [PubMed] [Google Scholar]
- 14.Wodrich, H. & Krausslich, H. G. (2001) Results Probl. Cell. Differ. 34, 197–217. [DOI] [PubMed] [Google Scholar]
- 15.Cullen, B. R. (2003) Trends Biochem. Sci. 28, 419–424. [DOI] [PubMed] [Google Scholar]
- 16.Frankel, A. D. & Young, J. A. (1998) Annu. Rev. Biochem. 67, 1–25. [DOI] [PubMed] [Google Scholar]
- 17.Pollard, V. W. & Malim, M. H. (1998) Annu. Rev. Microbiol. 52, 491–532. [DOI] [PubMed] [Google Scholar]
- 18.Hope, T. J. (1999) Arch. Biochem. Biophys. 365, 186–191. [DOI] [PubMed] [Google Scholar]
- 19.Weis, K. (2002) Curr. Opin. Cell Biol. 14, 328–335. [DOI] [PubMed] [Google Scholar]
- 20.Sánchez-Velar, N., Udofia, E. B., Yu, Z. & Zapp, M. L. (2004) Genes Dev. 18, 23–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gendelman, H. E., Orenstein, J. M., Martin, M. A., Ferrua, C., Mitra, R., Phipps, T., Wahl, L. A., Lane, H. C., Fauci, A. S., Burke, D. S., et al. (1988) J. Exp. Med. 167, 1428–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Swingler, S., Mann, A., Jacque, J., Brichacek, B., Sasseville, V. G., Williams, K., Lackner, A. A., Janoff, E. N., Wang, R., Fisher, D. & Stevenson, M. (1999) Nat. Med. 5, 997–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bartz, S. R., Rogel, M. E. & Emerman, M. (1996) J. Virol. 70, 2324–2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goff, S., Traktman, P. & Baltimore, D. (1981) J. Virol. 38, 239–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burgard, M., Mayaux, M. J., Blanche, S., Ferroni, A., Guihard-Moscato, M. L., Allemon, M. C., Ciraru-Vigneron, N., Firtion, G., Floch, C., Guillot, F., et al. (1992) N. Engl. J. Med. 327, 1192–1197. [DOI] [PubMed] [Google Scholar]
- 26.Welker, R., Harris, M., Cardel, B. & Krausslich, H. G. (1998) J. Virol. 72, 8833–8840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jacque, J. M., Triques, K. & Stevenson, M. (2002) Nature 418, 435–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Melkonyan, H., Sorg, C. & Klempt, M. (1996) Nucleic Acids Res. 24, 4356–4357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peden, K., Emerman, M. & Montagnier, L. (1991) Virology 185, 661–672. [DOI] [PubMed] [Google Scholar]
- 30.Stutz, F., Neville, M. & Rosbash, M. (1995) Cell 82, 495–506. [DOI] [PubMed] [Google Scholar]
- 31.Benmerah, A., Lamaze, C., Begue, B., Schmid, S. L., Dautry-Varsat, A. & Cerf-Bensussan, N. (1998) J. Cell Biol. 140, 1055–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salcini, A. E., Confalonieri, S., Doria, M., Santolini, E., Tassi, E., Minenkova, O., Cesareni, G., Pelicci, P.G. & Di Fiore, P. P. (1997) Genes Dev. 11, 2239–2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Poupon, V., Polo, S., Vecchi, M., Martin, G., Dautry-Varsat, A., Cerf-Bensussan, N., DiFiore, P. P. & Benmerah, A. (2002) J. Biol. Chem. 277, 8941–8948. [DOI] [PubMed] [Google Scholar]
- 34.Ciminale, V., Felber, B. K., Campbell, M. & Pavlakis, G. N. (1990) AIDS Res. Hum. Retroviruses 6, 1281–1287. [DOI] [PubMed] [Google Scholar]
- 35.Doria, M., Salcini, A. E., Colombo, E., Parslow, T. G., Pelicci, P. G. & Di Fiore, P. P. (1999) J. Cell Biol. 147, 1379–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carter, C. A. (2002) Trends Microbiol. 10, 203–205. [DOI] [PubMed] [Google Scholar]
- 37.Amara, A. & Littman, D. R. (2003) J. Cell Biol. 162, 371–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.von Schwedler, U. K., Stuchell, M., Muller, B., Ward, D. M., Chung, H. Y., Morita, E., Wang, H. E., Davis, T., He, G. P., Cimbora, D. M., et al. (2003) Cell 114, 701–713. [DOI] [PubMed] [Google Scholar]
- 39.Sherer, N. M., Lehmann M. J., Jimenez-Soto, L. F., Ingmundson, A., Horner, S. M., Cicchetti, G., Allen, P. G., Pypaert, M., Cunningham, J. M. & Mothes, W. (2004) Traffic 4, 785–801. [DOI] [PubMed] [Google Scholar]
- 40.Freed, E. O. (2004) Trends Microbiol. 12, 170–177. [DOI] [PubMed] [Google Scholar]
- 41.Kang-Decker, N., Mantchev, G. T., Juneja, S. C., McNiven, M. A. & van Deursen, J. M. (2001) Science 294, 1531–1533. [DOI] [PubMed] [Google Scholar]
- 42.Kierszenbaum, A. L., Tres, L. L., Rivkin, E., Kang-Decker, N. & van Deursen, J. M. (2004) Biol. Reprod. 70, 1400–1410. [DOI] [PubMed] [Google Scholar]
- 43.Krishna, S. S., Majumdar, I. & Grishin, N. V. (2003) Nucleic Acids Res. 31, 532–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Randazzo, P. A. & Hirsch, D. S. (2004) Cell Signal. 16, 401–413. [DOI] [PubMed] [Google Scholar]
- 45.Basyuk, E., Galli, T., Mougel, M., Blanchard, J. M., Sitbon, M. & Bertrand, E. (2003) Dev. Cell 5, 161–174. [DOI] [PubMed] [Google Scholar]
- 46.Ptak, R. G. (2002) Exp. Opin. Invest. Drugs 11, 1099–1115. [DOI] [PubMed] [Google Scholar]
- 47.Dave, R. S. & Pomerantz, R. J. (2003) Rev. Med. Virol. 13, 373–385. [DOI] [PubMed] [Google Scholar]