Abstract
Our limited understanding of the biological impact of the whole spectrum of early breast lesions together with a lack of accurate molecular‐based risk criteria for the diagnosis and assignment of prognostic significance to biopsy findings presents an important problem in the clinical management of patients harboring precancerous breast lesions. As a result, there is a need to identify biomarkers that can better determine the outcome of early breast lesions by identifying subpopulations of cells in breast premalignant disease that are at high‐risk of progression to invasive disease. A first step towards achieving this goal will be to define the molecular phenotypes of the various cell types and precursors – generated by the stem cell hierarchy – that are present in normal and benign conditions of the breast. To date there have been very few systematic proteomic studies aimed at characterizing the phenotypes of the different cell subpopulations present in normal human mammary tissue, partly due to the formidable heterogeneity of mammary tissue, but also due to limitations of the current proteomic technologies. Work in our laboratories has attempted to address in a systematic fashion some of these limitations and here we present our efforts to search for biomarkers using normal fresh tissue from non‐neoplastic breast samples. From the data generated by the 2D gel‐based proteomic profiling we were able to compile a protein database of normal human breast epithelial tissue that was used to support the biomarker discovery program. We review and present new data on the putative cell‐progenitor marker cytokeratin 15 (CK15), and describe a novel marker, dihydropyriminidase‐related protein 3 (DRP3) that in combination with CK15 and other well known proteins were used to define molecular phenotypes of normal human breast epithelial cells and their progenitors in resting acini, lactating alveoli, and large collecting ducts of the nipple. Preliminary results are also presented concerning DRP3 positive usual ductal hyperplasias (UDHs) and on single cell layer columnar cells (CCCs). At least two bona fide biomarkers of undifferentiated ERα/PgR negative luminal cells emerged from these studies, CK15 and c‐KIT, which in combination with transformation markers may lead to the establishment of a protein signature able to identify breast precancerous at risk of progressing to invasive disease.
Keywords: Normal breast epithelium, Proteomics, DRP3, CK15, Progenitors-like cells, UDH, CCL
Abbreviations
- IHC
immunohistochemistry
- 2D PAGE
two-dimensional polyacrylamide gel electrophoresis
1. Introduction
Breast cancer is by far the most common cancer among women, both in developed and developing regions, with an estimated 1.38 million new cases diagnosed in 2008, and the global incidence rates are increasing (Ferlay et al., 2010). In Denmark, the number of new cases diagnosed per year in the period 2004–2008 was approximately 4200, corresponding to an age‐standardized rate (W) of 88.6 new cases per 100,000 persons per year. Also, here the incidence trend is upward‐going with an estimated annual increase in the past 10 years of 1.0% per annum (Engholm et al., 2010).
The prominent burden of breast cancer in the female population, prompted by a steady rise in many countries in the female lifetime risk of developing breast cancer, has increased alarmingly over the last two decades (Héry et al., 2008) and breast cancer is now the leading cause of cancer death in women worldwide and the fifth cause of death from cancer overall (458,000 deaths). Survival analysis by stage at diagnosis for 1999–2006 from 17 SEER geographic areas, showed that for patients presenting with localized (confined to primary site) disease the 5‐year relative survival was 98%, with regional (spread to regional lymph nodes) disease was 83.6%, whereas for patients where cancer had metastasized survival was 23.4% (Altekruse et al., 2008). Early diagnosis of breast cancer is therefore essential for reducing both mortality and morbidity of the disease (Berry et al., 2005, 2005, 1998, 2007, 2005, 2005).
Refinements in mammographic breast screening and the use of even more sensitive imaging technologies have resulted in detection and diagnosis of breast cancer in increasingly early stages, which combined with the introduction of newer and improved therapies has led to a decrease risk of dying from breast cancer (Bray et al., 2004). But mammographic breast screening has also brought about a dramatic increase in the number of patients that present with proliferative changes in the breast, which has lead to the emergence of benign breast disease as an important therapeutic and prognostic challenge (Ernst et al., 2004; Tavassoli, 2009). The advent of even more sensitive imaging technologies such as tomosynthesis, color Doppler ultrasonography, contrast enhanced MRI, and positron emission mammography will only heighten this problem (Fass, 2008).
The earliest recognizable precursor of invasive carcinoma of the breast is atypical hyperplasia, ductal (ADH) or lobular (ALH), the diagnosis of which is based solely on pathological criteria as there are no molecular markers able to single out these lesions (Boecker et al., 2006, 2007, 2006, 2006, 2003, 2009). In addition, because the molecular determinants of progression are largely unknown, the clinical management of women with pre‐invasive lesions is largely based on the histological classification of such lesions. Patients presenting with ADH have an increased risk for invasive breast cancer, which is about four to five times that of the broad population, reaching nearly a tenfold risk if the patient has a first‐degree family member with breast cancer. Cellular hyperplasia is also observed in columnar cell lesions (CCLs), also known as flat epithelial atypia (Ellis, 2010; Haupt et al., 2010; Jara‐Lazaro et al., 2009; Schnitt and Vincent‐Salomon, 2003; Wellings et al., 1975), and CCLs with atypia have been proposed to be one of the earliest recognizable lesions in the progression to invasive carcinoma of the breast (Abdel‐Fatah et al., 2007; Feeley and Quinn, 2008; Simpson, 2009; Simpson et al., 2005). Both loss of heterozygosity and comparative genomic hybridization have shown molecular similarities between these lesions and invasive breast carcinomas (Dabbs et al., 2006; Schnitt and Vincent‐Salomon, 2003; Simpson et al., 2005).
Our limited understanding of the biological significance of the whole spectrum of early precancerous lesions as well as the lack of accurate molecular‐based risk criteria for the diagnosis and assignment of prognostic significance to biopsy findings are underlying causes of our continued dependency on morphology‐based classification of tumors (Pinder, 2010; Ellis, 2010; Nofech‐Mozes et al., 2005; Cornfield et al., 2004). Precise non‐surgical diagnosis based on histopathological features supplemented with molecular data, in combination with targeted therapies, are expected to lead to a predictive, and individualized approach to cancer care (Kim, 2010; Mueller et al., 2010; Nelson et al., 2010; Pierotti et al., 2010; Podo et al., 2010; Toft and Cryns, 2010). Consequently, there is an urgent need to develop complementary diagnostic methods to detect and classify breast cancer in a very early stage, as well as molecular markers that can better determine the outcome of early breast lesions and the field of clinical proteomics is well poised to address these issues (Bohndiek and Brindle, 2010, 2009 Dec 1, 2010, 2010, 2009). A first step towards achieving this goal will be the definition in greater detail of the molecular phenotypes of the various cell types and precursors that exist in normal and benign conditions of the breast (Lopez‐Garcia et al., 2010; Muggerud et al., 2010).
To date there have been very few systematic proteomic studies aimed at characterizing the phenotype of the different cell subpopulations present in normal human mammary tissue in vivo, partly due to the formidable heterogeneity of mammary tissue, but also due to limitations of the current proteomic technologies (Bini et al., 1997; Brennanet al., 2010; Findeisen and Neumaier, 2009; Fink‐Retter et al., 2009; Franzen et al., 1996; Gast et al., 2009; Jain, 2008; Wulfkuhleet al., 2002).
Work in our laboratories has attempted to address in a systematic fashion some of these limitations (Celis et al., 2009, 2007, 2006, 2007, 2010) and here we present our strategies to search for biomarkers using 2D PAGE‐based proteomic profiling of normal fresh tissue from non‐neoplastic breast, review and present new data on the putative progenitor marker CK15, and describe a novel marker, dihydropyriminidase‐related protein 3 (DRP3) that in combination with CK15 and other well known proteins may contribute to define the molecular phenotypes of normal human breast epithelial cells and their progenitors.
2. Experimental procedures
2.1. Sample collection and handling
Tissue samples from clinical high‐risk patients (high‐risk definition according to the Danish Breast Cooperative Group; www.dbcg.dklast accessed 22.10.09) that underwent mastectomy between 2003 and 2008 were provided by the Department of Pathology at the Copenhagen University Hospital. Normal tissue biopsies sampled from discarded anonymous excess tissue from reduction mammoplasties were also procured (Erichsens Privathospital, Denmark). Tissue samples for gel analysis were flash‐frozen in liquid nitrogen and were rapidly transported to the Institute of Cancer Biology where they were stored at −80 °C; on average no more than 15 min elapsed from tissue excision to freezing (Celis et al., 2003). Samples for interstitial fluid recovery were kept in PBS at 4 °C and were routinely prepared within a maximum of 30–45 min from the time of surgical excision as previously described (Celis et al., 2004a). The project was approved by the Scientific and Ethical Committee of the Copenhagen and Frederiksberg Municipalities (KF 01‐069/03).
2.2. Sample preparation for two‐dimensional polyacrylamide gel electrophoresis (2D PAGE)
Twenty to thirty, six‐μm cryostat sections of frozen tissues were resuspended in 0.1 ml lysis solution (O'Farrell, 1975) or CLB1 buffer (Gromov et al., 2008). The resulting lysates were frozen and kept at −20 °C until used, usually within 24–48 h (Celis et al., 2004a). Twenty to forty μl were applied to the gels and each sample was run at least in duplicate. The first and last sections of each sample were used for immunofluorescence analysis using cytokeratin 19 (CK19) antibodies as this epithelial marker is ubiquitously expressed by mammary epithelial cells (Bartek et al., 1985). The availability of these pictures greatly facilitated the interpretation of the gel data as it gave a rough estimate of the ratio of glands/tumour cells to stromal tissue. After running the second dimension, gels were placed in 7.5% acetic acid, 50% ethanol, and 0.05% formalin for 1 h, washed 3 × 30 min in 7.5% acetic acid, 10% ethanol and stained with silver nitrate according to a procedure compatible with mass spectrometry (Gromova and Celis, 2004).
2.3. 2D PAGE and Western immunoblotting
2D PAGE (isoelectric focusing, IEF) and 2D gel Western blotting were performed as previously described (Celis et al., 2004b).
2.4. Protein spot handling and mass spectrometry analysis
Protein spots were excised from silver stained dry gels and the gel pieces were rehydrated in water. Gel pieces were detached from the cellophane film and cut into 1 mm2 pieces followed by “in‐gel” digestion as previously described (Shevchenko et al., 1996) followed by a procedure that has been reported previously (Gromov et al., 2010). Briefly, MALDI‐TOF‐TOF data were acquired using an Ultraflex ™ III 200 time‐of‐flight mass spectrometer (Bruker Daltonik, Germany) equipped with a Smart beam™ laser and a LIFT‐TOF/TOF unit. Data acquisition and data processing were performed by the Flex Control 3.0 and Flex Analysis 3.0 software (Bruker Daltonik, Germany). All of the spectra were obtained using reflector positive mode with an acceleration voltage of 25 kV, reflector voltage of 26.38 kV and detection suppressed up to 450 Da. A total of 2000 shots in steps of 200 shots were added to one spectrum in the mass range of m/z 600–4000.
2.5. Spectral analysis and protein identification
Post‐acquisition two step calibration was automatically performed in Flex Analysis using standard peptide calibration mixture (Bruker Daltonics, Germany) for external calibration followed by an additional post‐acquisition internal calibration step to obtain better mass accuracies. Ubiquitous presented auto‐digested tryptic mass values visible in all the spectra were used for internal calibration. The background masses (matrix, metal adducts, tryptic peptides from keratins) were automatically subtracted from finally generated pick list and were excluded from the further analysis. For protein identification, peptide masses were transferred to the BioTools 3.2 interface (Bruker Daltonik, Germany) to search in the National Center for Biotechnology non‐redundant NCBInr (20090905) database using in house MASCOT search engine (version 2.2, released 28.08.2009, Matrix Science Ltd.). No restriction on the protein molecular mass and taxonomy was applied as a first step. A number of fixed (acrylamide modified cystein, i.e. propionamide/carbamidomethylation) and variable modifications (methionine oxidation and protein N‐terminus acetylation) were included in the search parameters. The peptide tolerance did not exceed 30 ppm and a maximum of one trypsin missed cleavage was allowed. Protein identifications were considered to be confident when the protein score of the hit exceeded the threshold significance score of 65 (p < 0.05) and no less than 4 peptides were recognized. If necessary, sequence analysis was performed to confirm the protein identity.
2.6. Antibodies
Commercially available antibodies against the following antigens were used in this study (stated working dilutions are for IHC): CK5 (rabbit monoclonal clone EP 1601 Y; used at a dilution of 1:100) from EPITOMICS; CK8 (mice culture supernatant M20; used at a dilution of 1:200) from Monosan; CK14 (clone LL002; used at a dilution of 1:200), CK19 (mouse monoclonal; used at a dilution of 1:1000) from Labvision‐ThermoScientific; CK15 (rabbit polyclonal anti‐peptide; used at a dilution of 1:12000) from AVIVA Systems Biology; c‐Kit (rabbit polyclonal anti‐peptide A4502; used at a dilution of 1:500), ERα (mouse monoclonal clone 1D5; used at a dilution of 1:500), PgR (mouse monoclonal clone PgR 636; used at a dilution of 1:200), p63 (mouse monoclonal clone AA4; used at a dilution of 1:200), vimentin (mouse monoclonal V9; used at a dilution of 1:500) and CD44 (mouse monoclonal clone DF 1485; used at a dilution of 1: 200) from Dako (Glostrup, Denmark); TUC‐4 protein (rabbit polyclonal anti‐peptide AB5454; used at a dilution of 1:2000), from Millipore, phospho STAT5a –Y694 (rabbit polyclonal antibody; used at a dilution of 1:300) from ABGENT; beta‐casein (used at a dilution of 1200) from AbD Serotec; thymidine phosphorylase (mouse monoclonal clone P‐GF.44C; used at a dilution of 1:300) from IMGENEX, DRP3 (anti‐DYSPL3, HPA010948; used at a dilution of 1:500) from Atlas Antibodies AB, and CRMP1 (rabbit polyclonal anti‐peptide used at a dilution of 1:200) from Abcam. Rabbit polyclonal anti‐peptide antibodies EP071757 [C‐term peptide inmunogen: RIVAPPGGRSNITSLS (555‐570)] and EP071758 [N‐term peptide immunogen: SYQGKKNIPRITSDR (2‐15)] raised against DRP3 were generated by Eurogentec (Belgium).
2.7. Immunohistochemistry
Following surgery, fresh tissue blocks were immediately fixed in neutral buffered formalin and paraffin embedded for archival use. Five‐μm sections were cut from the tissue blocks and mounted on Super Frost Plus slides (Menzel‐Gläser, Braunschweig, Germany), baked at 60 °C for 60 min, deparaffinized, and rehydrated through graded alcohol rinses (Celis et al., 2007a). Heat induced antigen retrieval was performed by immersing slides immersing the slides in Tris/EDTA pH 9.0 buffer (10 mM Tris, 1 mM EDTA) and microwaving in a 750 W microwave oven for 10 min. The slides were then cooled at room temperature for 20 min and rinsed abundantly in tap water. Non‐specific staining of slides was blocked (10% normal goat serum in PBS buffer) for 15 min, and endogenous peroxidase activity quenched using 0.3% H2O2 in methanol for 30 min. Antigen was detected with a relevant primary antibody, followed by a suitable secondary antibody conjugated to a peroxidase complex (HRP conjugated goat anti‐rabbit or anti‐mouse antibody) (DakoCytomation, Glostrup, Denmark). Finally, color development was done with 3, 3′‐diaminobenzidine (Pierce, IL, USA) as a chromogen to detect bound antibody complex. Slides were counterstained with hematoxylin. Standardization of the dilution, incubation, and development times appropriate for each antibody allowed an accurate comparison of expression levels in all cases.
2.8. Tissue microarrays (TMAs)
Breast cancer tissue microarray slides were obtained from Pantomics (BRC1501, BRC1502 and BRC1503 – Pantomics Inc., CA, USA). The TMAs contained a total of 210 non‐overlapping breast tumours. We also used a TMA prepared at the Department of Pathology, University of Valencia and Instituto Valenciano de Oncologia that contained 133 breast carcinomas. The slides were stained as above using an appropriate primary antibody. For detection of immune complexes we used a horseradish peroxidase‐labeled polymer (Envision+ detection kit, DAKO, Denmark) as a secondary antibody. All slides were independently reviewed by three of the authors (JEC, TC, and JMAM) and in the few discrepant cases a consensus was reached after joint review.
2.9. Indirect immunofluorescence analysis
Five‐μm formalin‐fixed paraffin‐embedded (PFFE) sections were prepared as above. Following antigen retrieval, sections were treated with Image‐iT FX™ signal enhancer (Molecular Probes, OR, USA) to block non‐specific staining and subsequently incubated sequentially with the two first species‐diverse primary antibodies (e.g. one mouse and one rabbit antibodies) at the appropriate dilution. Detection of immune complexes was done with anti‐ideotypic secondary antibodies conjugated to Alexa Fluor® 488 and Alexa Fluor® 594 (Molecular Probes, USA). As a final step, a third antigen was detected using an antibody directly conjugated to Alexa Fluor® 633. Nuclear material was counterstained with DAPI. The sections were washed three times with cold phosphate‐buffered saline (PBS) between incubations. Normal rabbit and/or mouse sera instead of primary antibodies were used as a negative control. Sections were imaged using a Zeiss LSM510Meta confocal laser scanning microscope (Carl Zeiss MicroImaging Gmbh, Germany).
3. Results and discussion
3.1. Discovery of specific markers of human breast luminal and progenitor‐like cells
For the past years our laboratory has carried out a systematic and comprehensive proteomic profiling of benign and malignant breast tissue in a search for differentially expressed markers for early detection and stratification of patients, and novel targets for therapeutic intervention in breast cancer. Our research activities are part of long‐term ongoing strategies and have resulted in the identification of specific markers that can classify some subtypes of breast cancer, markers that can categorize tumor cellular phenotypes, as well as candidate serological biomarkers for early detection (Celis et al., 2004, 2006, 2005, 2007, 2006, 2009, 2007, 2010).
Recognizing the potential role of biomarkers in predicting progression of premalignant lesions to cancer and the clinical implications for management of breast disease that such biomarkers of malignant potential could ultimately have, we have focused our latest efforts on a discovery‐driven analysis of normal breast tissue and benign breast conditions. Accordingly, we have used two different strategic approaches to search for markers of progenitor‐like cells. One approach was based on the comparison of the protein expression profiles of normal as opposed to benign conditions, using matched pairs of samples collected from the same patient. We applied this strategy to the analysis of benign hyperproliferative conditions such as sclerosing adenosis with apocrine differentiation (Celis et al., 2007b), which allowed us to identify a novel marker of normal luminal cells and their putative progenitors, cytokeratin 15 (CK15) (Celis et al., 2007a).
The second approach entailed a slightly different experimental methodology, whereby we compared the protein expression profiles of various normal human breast tissue biopsies exhibiting contrasting ratios of usual ductal hyperplasia (UDH) lesions, low versus high. We surmised that some of the proteins upregulated in the latter samples may represent a potential source of new biomarkers of very early events. For these studies we have used 2D PAGE‐based proteomic profiling of numerous tissue samples collected distal to tumors and containing histologically normal‐looking breast epithelia. We identified one protein(s), dihydropyrimidinase‐related protein 3 (DRP3), reported here, that showed increased expression in some UDH lesions.
3.1.1. Stem cell‐like marker CK15 in breast epithelial cells
As mentioned above, our laboratories have previously applied a biomarker discovery strategy based on the proteomic analysis of matched control/diseased pairs of samples collected from the same patient to the analysis of benign hyperproliferative conditions such as sclerosing adenosis with apocrine differentiation (Celis et al., 2007a). We identified CK15 as a novel marker that defined subpopulations of epithelial cells some of which may correspond to progenitor/stem‐like cells. We detected both ERα−/PgR−CK15+/CK19+ luminal cells and ERα−/PgR−/CK15+/CK19− epithelial cells that were either positive or negative for the myoepithelial marker p63 (Barbareschi et al., 2001), implying that the expression of CK15 may be permissive in progenitor cells giving raise to both luminal and myoepithelial lineages. These data were supported by findings from an independent study carried out by Villadsen and colleagues and reported concurrently with ours. These researchers showed, using a different experimental approach from us, that a candidate stem cell zone resides in ducts that are enriched in cells identified as being SSEA‐4hi/CK5+/CK6a+/CK15+/Bcl‐2+ cells (Villadsen et al., 2007) strengthening the proposition that CK15 may be a stem cell marker in breast epithelial cells.
Ours studies also uncovered four CK15/CK19 luminal phenotypes (CK15+/CK19+, CK15+/CK19−, CK15−/CK19+, CK15−/CK19−) that were often observed within the same tissue sample (illustrated in Supplementary Figure 1), revealed expression of CK15 by luminal cells present in some UDH lesions, documented the loss of this cytokeratin as a result of tumour progression in a p53 positive tumour in which UDH, ADH, carcinoma in situ (CIS) and invasive disease coexisted in the same lesion, and highlighted the expression of CK15 by a few breast carcinomas (4.2%; 5 out of 120) in a set of prospectively collected tumours from high‐risk patients (Celis et al., 2007a).
Given the fact that the patients represented in the above set of tumours were selected based on specific criteria and were collected prospectively (Celis et al., 2007a), we reevaluated and report here the expression of CK15 by breast carcinomas in a larger number of tumours using two independent sets of non‐selected samples: one set composed of three, commercially available, non‐overlapping breast cancer tissue microarrays (TMA's) (BRC1501, BRC1502 and BRC1503; Pantomics, USA) and another set comprising one single TMA containing samples collected at the Instituto Valenciano de Oncologia. In total, the four TMAs included 343 independent breast tissue samples. The set of three Pantomics TMAs contained 210 carcinomas of which 49 were triple negative breast cancers (defined as ERα−, PgR−, and Her2‐neu negative), 31 were Her2‐neu, 118 were Luminal A, and 12 were Luminal B. 26 of the lesions were CK15 positive (12.38%) and of these 11 were triple negatives, 4 were Her2‐neu and 11 were Luminal A (Supplementary Tables 1A–C). The Valencia TMA, on the other hand, contained 133 carcinomas of which 26 were triple negatives, 13 were Her2‐neu, 66 were Luminal A, and 28 were Luminal B. 20 of the lesions were CK15 positive (15%) and of these 9 were triple negatives, 2 were Her2‐neu, 6 were Luminal A, and 3 were Luminal B (Supplementary Table 2). Taken together these results showed that the number of CK15 positive carcinomas was underestimated in our original study, most likely due to patient bias, and that a more accurate proportion of CK15 positive carcinomas would be 12–15%. Moreover, we found that the CK15+ phenotype cut across subtypes defined either by classical histopathological or molecular parameters.
Considering that the expression of CK15 has been shown to be dependent on TP53 at both the transcriptional (Kostic and Shaw, 2000) and protein level (Cha et al., 2010) and taking into consideration our previous results that indicated that p53 positive ADHs in a well‐defined tumour did not expressed CK15, we correlated the CK15 expression data with the p53 status in the Pantomics BRC1501, BRC1502 and BRC1503 TMAs as the p53 information was available from the vendor. The results showed that out of the 26 breast carcinomas that were positive for CK15 in these TMAs, only 6 were positive for p53 and that these corresponded to 4 triple negatives and 2 Her‐2 carcinomas, supporting the notion that CK15 expression may be under the control of p53, but in some tumours this control seems to be lost (Supplementary Tables 1A–C).
We concluded from the above results that CK15 positive carcinomas arise across various breast tumour subtypes and that some CK15+ precursor lesions may under certain conditions progress to invasive disease (Celis et al., 2007a). Understanding the molecular mechanisms underlying the expression of CK15 as well as characterizing the malignant potential of CK15+ luminal cells and precursor cells may help predict patients at risk of developing breast cancer and guide their clinical management. It may also identify specific cell subpopulations that can be isolated and used to derive novel therapeutic targets.
3.1.2. Identification of dihydropyrimidinase‐related protein 3 (DRP3) as a molecular marker defining a subpopulation of normal breast epithelial cells
To complement the above studies, we have started to search for protein biomarkers of early events by comparing the protein expression profiles of breast samples exhibiting contrasting ratios of UDH lesions, low versus high. We surmised that some of the proteins upregulated in the latter samples may represent a potential source of new biomarkers. For these studies we have analyzed 32 samples containing histologically normal‐looking breast epithelia obtained from areas distal to malignant tumours (see Experimental Procedures). These analyses allowed us to compile a 2D gel protein database of normal human breast epithelial tissue that comprises so far a total of 338 identified protein spots (Supplementary Figure 2 and Supplementary Table 3). Out of several potential biomarkers identified so far in this study, we describe below the characterization of one, potentially useful biomarker and exemplify the challenges one faces when dealing with members of protein families in the context of heterogeneous tissues.
3.1.2.1. Mass spectrometry identification of a “DRP3” isoform
Figure 1 depicts a selected area of 2D gels showing protein expression profiles of normal breast tissue biopsies exhibiting low (Figure 1A) or higher levels of UDHs (Figure 1B), pinpointing an upregulated protein in the UDH‐enriched preparations (indicated with a red arrow) that was unknown in our database of normal breast proteins. Using MALDI‐TOF‐MS mass spectrometry (Supplementary Figure 3) the protein spot was identified, with 32% sequence coverage scattered throughout, as dihydropyrimidinase‐related protein 3 (DRP3, SwissProt number Q14195), a protein also known as collapsin response mediator protein 4 (CRMP4) or Unc‐33‐like phosphoprotein 1 (Ulip‐1). DRP3 is a member of the DRP/CRMP family of proteins and is thought to play a role in neuronal differentiation, axonal outgrowth, and possibly in neuronal regeneration. The protein family consists of 5 closely related phosphoproteins (DRP‐1 through 5) of similar molecular size (60–66 kDa) and relatively high (50–90%) amino acid sequence homology (Figure 2). DRP isoforms are capable of mutual heteromerization and have been shown to undergo multiple phosphorylations as well as exhibiting differential tissue distribution (Hamajima et al., 1996; Charrier et al., 2003; Quinn et al., 1999). In addition, mammalian DRP‐1, ‐2, and ‐4 isoforms appear to undergo alternative splicing (Leung et al., 2002; Quinn et al., 2003).
Figure 1.
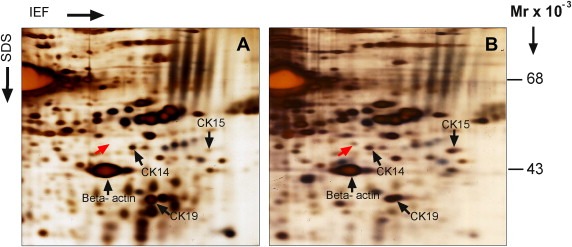
Illustration of differential protein expression in samples with opposite relative amounts of UDH lesions. Magnified sections of representative 2‐D PAGE gels run with lysates from non‐malignant tissue samples containing (A) low or (B) higher relative levels of UDHs are shown. Gels were visualized by silver nitrate staining. Arrows indicate the positions of a differentially expressed protein (red arrow) and reference proteins (CK14, CK19, CK15 and β‐actin; black arrows).
Figure 2.
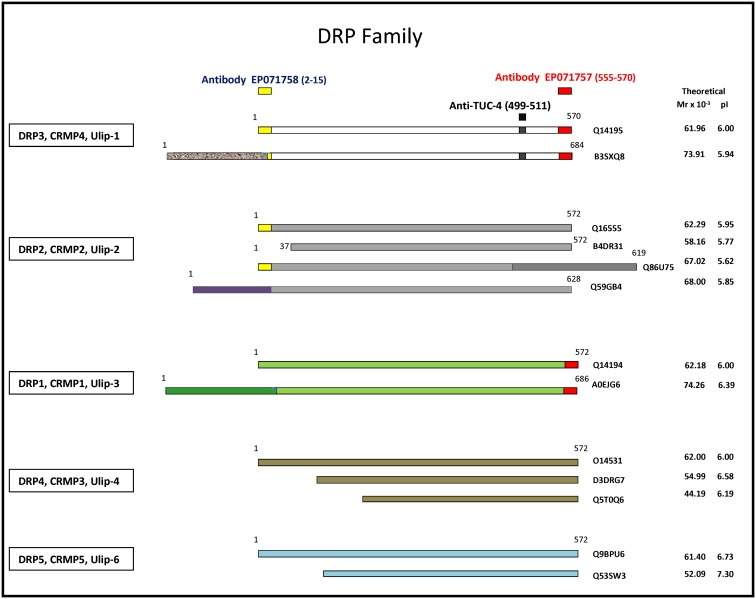
Schematic representation of DRP protein family members. DRP proteins −1 to −5 are presented in different colors with known variants (truncated and long variants) shown. In each case differences in domain structure in the long variants are indicated with colored boxes. The location of the immunogens for antibodies EP071758 (yellow box), anti‐TUC‐4 (black box), and EP07175 (red box) are presented and the aa positions indicated. In each case the calculated Mr and pI is indicated as well as the SwissProt accession number.
Despite the high sequence homology between DRP3 and the three most closely related members of the DRP/CRMP protein family (86% with DRP1, 90% with DRP2, and 86% with DRP4), and their very close molecular masses and pI's (Figure 2), the MASCOT search results revealed that only the DRP3 isoform was present in the analyzed spot (Supplementary Figure 3). The apparent Mr and pI of the identified polypeptide (51 kDa, pI of 5.4), however, did not correspond to that of any of the variants of DRP3 deduced from the known sequence (Figure 2), suggesting that it may correspond to a, yet unknown, processed variant of DRP3. Given the occurrence of stimuli‐dependent variants such as the retinoic acid‐induced 58 kDa hUlip protein isoform identified in neuroblastoma cells (Gaetano et al., 1997), and the calpain‐dependent cleavage of DRP3 (60 kDa) observed in primary rat cortical neurons exposed to NMDA and oxidative stress (H2O2) (Kowara et al., 2005, 2006) it is likely that a smaller, processed isoform of DRP3 could occur under certain biological circumstances.
3.1.2.2. Generation and 2D Western blot characterization of anti‐DRP3 antibodies
Peptide‐specific rabbit polyclonal antibodies were raised against two selected peptides located at the N‐terminal (SYQ GKK NIP RIT SDR, indicated with a yellow box in Figure 2; antibody EP071758) and C‐terminal (RIV APP GGR SNI TSL S, indicated with a red box in Figure 2; antibody EP071757) regions of DRP3 (Figure 2; Q14195), respectively. These two antibodies were expected, by sequence analysis, to allow the identification of both full length and truncated DRP3 forms, although some cross‐reactivity to DRP2 and DRP1 was predicted for EP071758 and for EP071757, respectively. To determine the specificity of the two antibodies, we performed 2D gel Western blotting using tissue extracts of normal breast samples that expressed the MS identified protein spot.
3.1.2.2.1. EPO71758 antibody detects the MS identified “DRP3” truncated form and the 62 kDa full‐length DRP3 isoform in extracts of normal breast tissue
As illustrated in panel 1 of Figure 3 with extracts from normal tissue specimen 83 (N83), the N‐terminal antibody (EP071758) reacted with the original protein identified from the 2D gels (“DRP3”, red arrow in Figure 3 Panel 1 sub‐figure A), and in addition with two higher molecular weight variants termed “α” and “β” (yellow and blue arrows, respectively). The variant “α” has an apparent Mr of 58 kDa and pI of 5.8 (Figure 3 Panel 1 sub‐figure A, variant “α”, yellow arrow). Variant “β” on the other hand has an apparent Mr of 62 kDa and a pI of 6.0 (Figure 3 Panel 1 sub‐figure A, variant “β”; blue arrow), and may correspond to the full‐length DRP3 form Q14195 (Figure 2). The C‐terminal antibody (EP071757), on the other hand, did not react with the original “DRP3” protein spot – indicating that the “DRP3” variant may not have the C‐terminal sequence recognized by this antibody – but identified a single polypeptide (termed “γ”) with an apparent molecular weight of 74 kDa and a pI of about 5.2 (Figure 3 Panel 1 sub‐figure B, variant “γ”, black arrow). The “γ” polypeptide recognized by the EP071757 antibody was observed in immunoblots of several normal tissue samples, and was also detected in immunoblots of normal breast interstitial fluid (NIF) (data not shown) indicating that is externalized to the microenvironment (Celis et al., 2004a). This polypeptide, however, was not recognized by the N‐terminal EP071758 antibody (Figure 3 Panel 1 sub‐figure A). MALDI‐TOF mass spectrometry analysis of several proteins recovered from areas of silver stained 2D gels of normal breast biopsies where the spot corresponding to the “γ” polypeptide was expected to migrate failed to identify the protein, most likely due to its low abundance.
Figure 3.
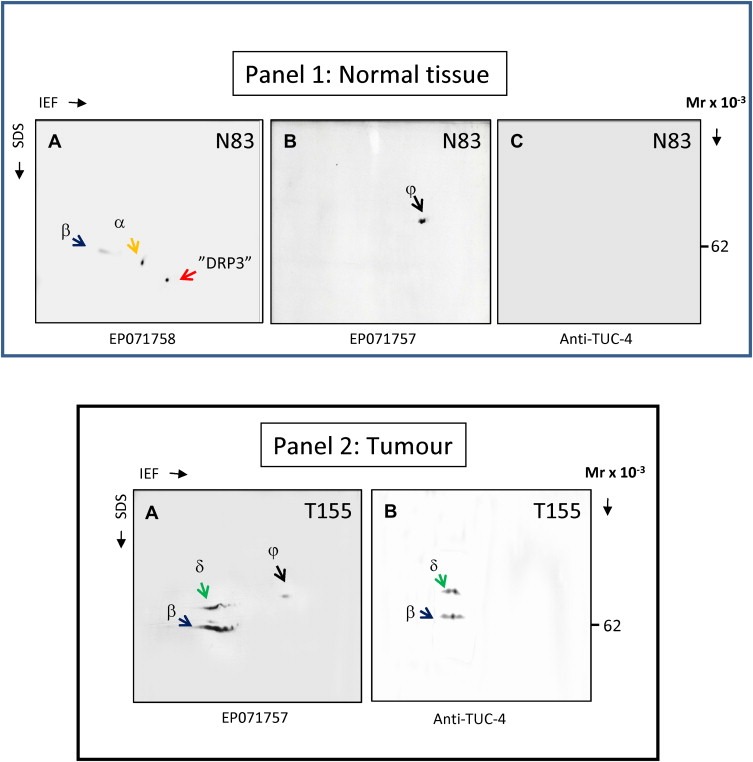
Determination of antibody specificity by 2D immunoblot analysis. Lysates of tissue specimens were resolved by 2D PAGE (IEF). The resolved proteins were blotted onto a nitrocellulose membrane, and DRP3 protein(s) detected with EP071758 (Panel 1 sub‐figure A), EP071757 (Panel 1 sub‐figure B, and Panel 2 sub‐figure A) or anti‐TUC‐4 antibody (Panel 1 sub‐figure C, and Panel 2 sub‐figure B), respectively. The immunoblot protein patterns varied in normal (Panel 1) or tumor tissues (Panel 2). Different variants are indicated with color‐coded arrows and identified with captions “α, β, γ, and δ”.
Given the multiple and non‐overlapping specificity patterns generated by the two antibodies, we tried to gain some additional insight into the identity of the protein(s) recognized by these antibodies. To this end, we acquired a commercially available antibody previously reported to be specific for DRP3 (anti‐TUC4, Millipore) (Quinn et al., 2003, 1999). The position of the peptide used to generate this antibody is represented schematically in Figure 2 (C‐terminal peptide aa 499‐511; black box). As illustrated in this figure, the TUC‐4 antibody should only recognize the two DRP3 variants Q14195 and B3SXQ8, while the EP071757 antibody may recognize in addition the DRP1 isoforms Q14194 and A0EJG6. 2D gel blot analysis of extracts from N83 with the TUC‐4 antibody yielded a negative result (Figure 3 Panel 1 sub‐figure C), a fact that may be due to low affinity of the antibody (see below).
3.1.2.2.2. EPO71757 antibody detects the 62 kDa full‐length DRP3 and a higher molecular weight variant in tumour extracts
Western 2D gel blot analysis of tumour extracts of breast carcinoma specimen T155 probed with the EP071757 and TUC‐4 antibodies gave almost identical reactivity and highlighted the same two proteins, polypeptide “β” also detected in normal tissue by antibody EP071758 and that may correspond to full‐length DRP3 form Q14195 and polypeptide “δ” (Figure 3 Panel 2 sub‐figures A and B, green arrows), which may correspond to the long variant of DRP3 B3SXQ8 (Figure 2). Western 2D gel blot analysis of 10 other breast carcinoma specimens with the EP071757 antibody yielded essentially the same results. Even though we were unable to unequivocally identify these polypeptides in tissue extracts, MALDI‐TOF‐MS mass spectrometry analysis of 2D gels of cellular extracts from U251MG glioma cells (known to express DRP3; Hiratsuka et al., 2003) allowed us to positively identify the two polypeptides “β” and “δ” as DRP3 variants (Q14195; data not shown).
Taken together, the results of the tumour immunoblots confirmed that the TUC‐4 and EP071757 antibodies specifically react with variants of the DRP3 family. It is possible, however, that polypeptide “γ” may correspond to the long DRP1 variant, although IHC analysis of tumours with a CRMP1 (DRP1, Figure 2) specific rabbit polyclonal anti‐peptide antibody revealed nuclear staining of epithelial cells in some tumours (results not shown), a staining pattern that was never observed with EP071757. None of the antibodies we tested (including two additional commercially available antibodies‐ DPYSL3, Sigma–Aldrich and 23951, Abcam) were able to exclusively recognize the original “DRP3” protein spot identified from the 2D gels.
3.1.2.3. IHC characterization of the anti‐DRP3 antibodies
Given that the reagents at our disposal were specific for DRP3, although recognizing different variants of this protein than the isoform we originally pinpointed as an upregulated protein in the UDH‐enriched preparations, we proceeded to determine if the DRP3 variants recognized by our EP071757 and EP071758 antibodies were differentially expressed in UDH lesions.
Exploratory IHC analyses of mammary tissues with the EP071757 and EP071758 antibodies showed that only the EP071757 probe generated a strong signal‐to‐noise ratio and well‐defined tissue staining and therefore only this probe was used for further IHC‐based studies. As illustrated in Figure 4, panel A, the EP071757 antibody regularly stained myoepithelial cells (Figure 4, Panel A, sub‐figure A) luminal epithelial cells in some UDHs (Figure 4, Panel A, sub‐figure B), lactating cells (Figure 4, Panel A, sub‐figure C), and some sporadic luminal epithelial cells in collecting ducts (Figure 4, Panel A, sub‐figure D), as well as in a few resting ductular and acinar structures (Figure 4, Panel A, sub‐figure E, red arrow). Out of a grand total of 2156 acini analyzed in 32 tissue samples, 95 were found positive for DRP3. The EP071757 antibody also showed moderate staining of fibroblasts in the connective tissue (Figure 4, Panel A, sub‐figure E, blue arrow) and endothelial cells in the vasculature (Figure 4 Panel A, sub‐figure E, green arrow).
Figure 4.
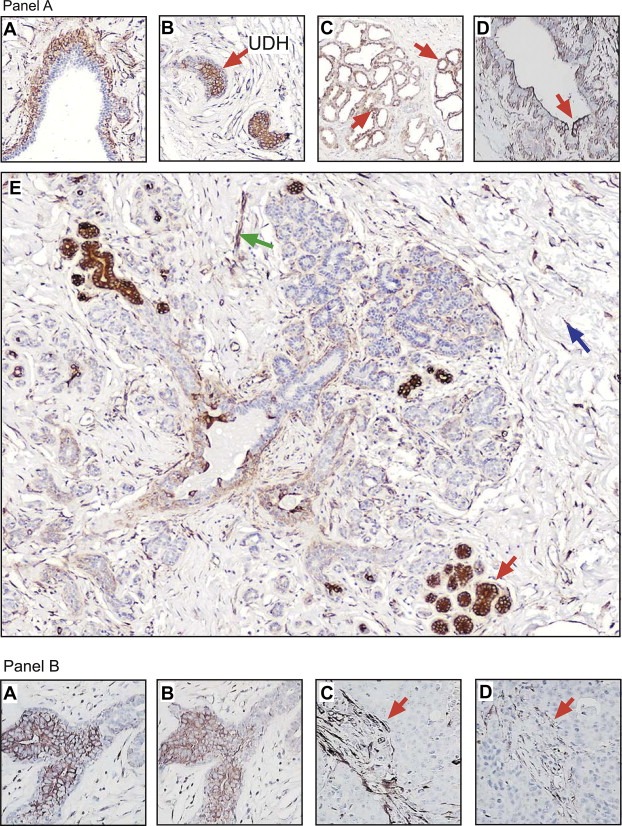
Expression analysis of DRP3 in formalin‐fixed paraffin embedded breast tissue samples. (Panel A) Immunohistochemical staining of DRP3 protein in normal breast tissue samples demonstrated the presence of the DRP antigen in (sub‐figure A) myoepithelial cells lining normal ducts, (sub‐figure B) luminal cells within some UDHs, (sub‐figure C) lactating cells, (sub‐figure D) collecting ducts, and in some (sub‐figure E) resting acinar structures. Cells indicated with red arrows show illustrative DRP3 immunoreactivity. (Panel B) IHC analysis of tandem tissue sections showed that immunoreactivity of EP071757 (sub‐figure A) and anti‐TUC‐4 antibodies (sub‐figure B) was comparable. Immunoreactivity of DRP3 was enhanced in tumor samples but predominantly located in stromal cells both with EP071757 (sub‐figure C) and anti‐TUC‐4 antibodies (sub‐figure D).
IHC staining of tandem sections from normal biopsies with the EP071757 and the TUC‐4 antibodies yielded very similar results (compare Figure 4, panel B, sub‐figures A and B, respectively), although the signal contrast between positive and negative staining in luminal cells was much stronger in the samples reacted with the EP071757 antibody. Interestingly, in tumour sections, we observed a generalized lack of staining of epithelial cells with both antibodies (illustrated in Figure 4, Panel B, sub‐figures C and D), but instead detected a strong staining of fibroblasts in the connective tissue in contrast to what was observed for normal tissues (red arrows in Figure 4, Panel B, sub‐figures C and D). Taken together, the results suggest that cellular expression of DRP3 may change both at the level of variant composition, but also in a cell‐type specific manner, with changes in the stromal compartment occurring during breast cancer progression. Analysis of three, commercially available, non‐overlapping breast cancer tissue microarrays (TMA's) (BRC1501, BRC1502 and BRC1503; Pantomics, USA) with the EP071757 antibody indicated that of the 210 tumours contained in these TMAs 191 (91%) showed enhanced staining in the connective tissue. Only 17 carcinomas (8%) were positive for DRP3 in the tumour epithelial cells and of these 10 were triple negative, 2 were Luminal A, 1 was Luminal B, and 4 were Her2‐neu.
Even though the antibodies we had at our disposal showed similar cross‐specificity towards the DRP3 isoforms it is currently not possible to identify with certainty which DRP3 variant(s) is/are expressed in a particular cell type/state. Clearly, the immunogens used to raise the various antibodies have different properties, the affinity of the antibodies for their antigen varies, and there are important differences in the sample processing methodologies used in immunoblotting and IHC. These considerations may explain why in some cases, in particular normal samples, we observed IHC staining but lack of reactivity in immunoblots. Notwithstanding, the remarkable similarity of IHC staining patterns and the differential patterns of expression observed suggest that these reagents may be of practical use for cellular immunophenotyping studies, and since our EP071757 antibody performed best in IHC, we selected this probe for the studies described below.
3.2. Defining the populations of cells present in normal mammary epithelia
The human breast is a complex branching tree‐like structure in which the entire mammary branching duct tree is lined by an inner layer of luminal epithelial cells, which in turn is encompassed by an outer layer of myoepithelial cells that is in direct contact with the basement membrane (Supplementary Figure 4) (Boecker et al., 2006; Moffat and Going, 1996). Any increase in cell number within the ductal space is defined as epithelial hyperplasia. Lobular (glandular) structures form at the ends of the terminal ducts lobular units (TDLU's) where most types of breast cancer are derived from (Wellings et al., 1975; Moffat and Going, 1996). TDLU's include the terminal duct and its corresponding lobule, which contains the alveoli/acini (Russo and Russo, 2004). The latter, being the milk producing units of the breast, empty into small lactiferous ducts that converge in each lobule, with several lobules forming a lobe.
The epithelium of the mammary gland exists in a highly dynamic state, and undergoes dramatic morphological changes. The profound expansion of mammary epithelium that occurs during puberty and pregnancy provides strong support for the existence of a cell with proliferative and regenerative potential characteristic of stem cells and today there is growing evidence for the existence of a differentiation hierarchy in the adult mammary gland with stem‐like cells at the apex of the differentiation hierarchy (Stingl et al., 2001; Smith and Chepko, 2001; Visvader, 2009 and references therein). These cells are believed to give rise to mature epithelium of either the luminal or myoepithelial lineage via a series of lineage‐restricted intermediates or progenitors (Booth et al., 2008; Visvader, 2009; Visvader and Lindeman, 2010). Recent work by Molyneux et al. (2010) showed that removing Brca1 in mouse mammary epithelial luminal progenitors produces tumors that phenocopy the majority of sporadic basal‐like breast tumors, whereas directing Brca1 deficiency to basal cells generates tumors that express molecular markers of basal breast cancers but that do not histologically resemble sporadic basal‐like breast tumors. These findings parallel previous results by Goldstein and colleagues in prostate cancer (Goldstein et al., 2010, 2010) demonstrating that histological characterization of cancers does not necessarily correlate with the cellular origins of the disease. Thus, elucidating the functional and molecular properties of discrete subpopulations of cells and how they relate to the normal cellular hierarchy in breast tissue is a pre‐requisite to understanding the mechanisms germane to tumor initiation and propagation.
3.2.1. Immunophenotype of luminal cells and progenitor‐like cells in normal human breast tissues
Having identified two novel candidate biomarkers, CK15 and DRP3, which showed differential expression in normal breast epithelial cells, early lesions, and invasive disease in a non‐ubiquitous manner we proceeded to ascertain the immunophenotype of normal human luminal epithelial cells using these two markers as fulcrum for our analyses. Since expression of CK15 and DRP3 only occurred in a subset of cells we used both markers in combination with other previously reported markers in order to identify and characterize discrete cell populations that may have different biological potential. To date, mostly hormone receptors (Hammond et al., 2010; Thorpe, 1988), CK5/6 (Bánkfalvi et al., 2004; Böcker et al., 2002), CK19 (Bartek et al., 1985), CK14 (Böcker et al., 2002; Bánkfalvi et al., 2004), CK8 and CK18 (Moll et al., 2008), and the receptor tyrosine kinase c‐KIT (Lim et al., 2009; Natali et al., 1992) have been used to assess the phenotype of luminal cells in a non‐systematic fashion. To be as inclusive as possible in our analyses, we immunophenotyped normal human luminal epithelial cells, not only in resting acini, but also in lactating alveoli and large collecting ducts of the nipple. For this, we used a selected battery of antibodies that comprised probes against CK15, DRP3 (EP071757 antibody), CK19, ERα, PgR, c‐KIT, CK5, CK14, as well as vimentin, a protein that we have found to be expressed by some breast precursor cells (Celis et al., 2007a). In addition, in some specific cases we used antibodies against a few additional markers such as CD44 (Naor et al., 1997; Shipitsin et al., 2007) and p63, a marker of myoepithelial cells (Barbareschi et al., 2001).
The expression of the various markers was assessed by a combined approach using IHC and multiple indirect‐label immunofluorescence analysis of serial sections of histologically normal breast epithelia obtained from normal breast reductions, as well as from numerous tissue samples collected from regions distal to the tumour (see Experimental Procedures). Below we describe the extended immunophenotype of luminal cells present in resting and lactating breast luminal cells as well as in large collecting ducts of the nipple (Table 1) and describe our preliminary efforts to analyze more complex lesions such as UDHs and CCLs.
Table 1.
Immunophenotype of luminal cells in normal breast structures.
| Breast structure | Phenotype | Marker | ||||||
|---|---|---|---|---|---|---|---|---|
| c‐KIT | CK5 | CK14 | Vim | ERα | PgR | CD44 | ||
| Resting acini | CK15+/CK19+/DRP3− | Pos | Pos & Nega | Neg | Neg | Neg | Neg | Neg |
| CK15+/CK19+/DRP3+ | Pos attenuated | Pos & Negb | Negc | Neg | Neg | Neg | Neg | |
| CK15+/CK19−/DRP3− | Pos & Negb | Pos & Negb | Neg | Neg | Neg | Neg | Neg | |
| CK15+/CK19−/DRP3+ | Pos attenuated | Pos & Negb | Neg | Neg | Neg | Neg | Neg | |
| CK15−/CK19+/DRP3− | Neg | Neg | Neg | Neg | Pos | Posd | Neg | |
| CK15−/CK19+/DRP3+ | Pos attenuated | Pos & Negb | Neg | Neg | Neg | Neg | Neg | |
| Lactating alveoli | CK15−/CK19+/DRP3+ | Neg | Neg | Neg | Neg | Neg | Neg | Neg |
| Collecting ducts | CK15+/CK19+/DRP3− | Pose & Neg | Pos & Negb | Pos & Negb | Neg | Neg | Neg | Neg |
| CK15+/CK19+/DRP3+ | Neg | Pos | Pos | Neg | Neg | Neg | Neg | |
| CK15−/CK19+/DRP3+ | Neg | Pos | Pos | Neg | Neg | Neg | Neg | |
Either negative acini within positive lobules or negative acini in the whole lobule.
Positive and negative luminal cells are observed.
Occasionally we observed CK14 positive cells but we cannot exclude the possibility that they arise as a result of hyperplasia.
There are also negative cells.
The staining could be attenuated in same cases.
3.2.1.1. Immunophenotype of luminal cells in resting acini
As pointed out earlier a significant outcome of our prior studies was the identification of all four possible CK15/CK19 luminal cell phenotypes (CK15+/CK19+, CK15+/CK19−, CK15−/CK19+, and CK15−/CK19−) in both acini and ducts, and that were often present within the same tissue section or even in the same lobule (Supplementary Figure 1) (Celis et al., 2007a). An extended immunophenotype of the main CK15/CK19 types observed in normal resting acini and gathered from our present work is described below:
3.2.1.1.1. CK15+/CK19+ luminal cells
Given the striking variation in the levels of the ERα and PgR as well as the number of cells expressing these receptors in normal breast specimens, and considering that is becoming increasingly clear that the evolution of ERα‐positive and ‐negative breast lesions follows different molecular pathways (Lopez‐Garcia et al., 2010) it was necessary first to determine the receptor status of the various CK15/CK19 phenotypes using quadruple immunostaining of consecutive normal breast tissue sections reacted with antibodies against CK15, CK19, and ERα (Figure 5A) and CK15, CK19 and PgR (Figure 5B), respectively. In line with our previous studies we found the CK15+/CK19+ luminal cells in acinar structures to be ERα and PgR negative (yellow arrows in Figure 5A and B; Celis et al., 2007a), a phenotype shared by CK15+/CK19− luminal cells (green arrows in Figure 5A and B). As shown below, cells with these phenotypes may exhibit low expression of ERα. Differentiated CK15−/CK19+ on the other hand were receptor permissive and, as expected, could express ERα and PgR (white arrows in Figure 5A and B).
Figure 5.
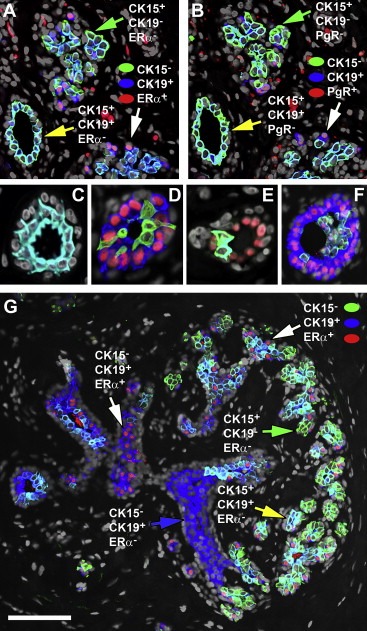
Nuclear receptor status in the various CK15/CK19 cell populations. Indirect four color immunofluorescence analysis of serial tissue sections from a normal breast specimen reacted with antibodies against cytokeratin 15 (CK15, Alexa Fluor® 488; green channel), cytokeratin 19 (CK19, Alexa Fluor® 594; red channel), and (A) ERα or (B) PgR (ER or PgR, Alexa Fluor® 633; blue channel), and counterstained with the nuclear stain DAPI (grey channel). (C–F) Illustrative images of acinar structures stained with CK15 (Alexa Fluor® 488; green channel), ERα (Alexa Fluor® 594; red channel), and cytokeratin 19 (Alexa Fluor® 633; blue channel) showing varying degrees of hyperplasia. (G) A low magnification view of a ductal tree illustrating the various cellular phenotypes observed and the negative correlation between ERα and CK15. Scale bar, 100 μm. In all cases only merged images are shown.
CK15+/CK19+ cells were generally found in a single luminal layer (Figure 5A–C) above the myoepithelial cells, although often we observed elongated CK15+ cells (CK19+ or CK19−) located in between and above CK15−/ERα+ cells generating a partial (Figure 5D and E) or complete second layer and sometimes a mild hyperplasia (Figure 5F).
Thorough analysis of the immunofluorescence images suggested that CK15 and the ERα and PgR receptors may in fact be negatively associated (Figure 5G, compare CK15+/ERα− phenotypes with CK15−/ERα+; green and yellow arrows with white arrows respectively), very much as it has been reported for ERα and the proliferation marker Ki67 (Shoker et al., 2000). This relationship seems to hinge on receptor expression thresholds rather than cell lineage permissiveness. Indeed, semiquantitative assessment of receptor expression levels using immunofluorescence analysis of several normal biopsies, taking advantage of the wider dynamic range of this technique in comparison to IHC, revealed that luminal cells exhibiting high staining levels of ERα or PgR were CK15 negative (Figure 6A and B, respectively; white arrows), and only cells that were negative or expressed low levels of the receptors were CK15 positive (Figure 6A and B, yellow arrows), a phenomenon that was also observed in the case of c‐KIT (results not shown). The latter result is noteworthy as Westbury and colleagues recently reported differential expression of c‐KIT and ERα in breast tissue in response to therapeutic radiation (Westbury et al., 2009).
Figure 6.
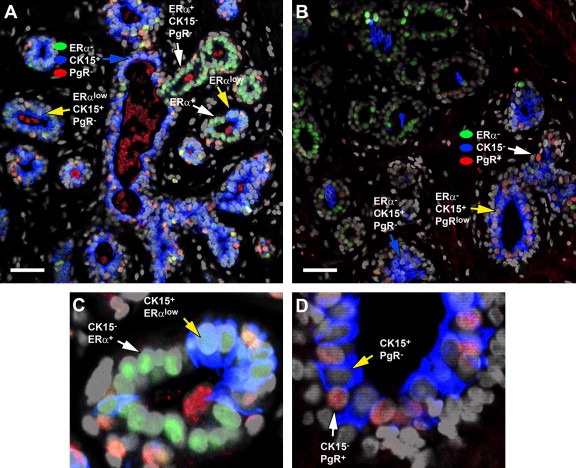
Negative correlation of nuclear receptor status and CK15 expression. Indirect four color immunofluorescence analysis of a normal breast specimen reacted with antibodies against ERα (ER, Alexa Fluor® 488; green channel), PgR (PgR, Alexa Fluor® 594; red channel), and cytokeratin 15 (CK15, Alexa Fluor® 633; blue channel), counterstained with the nuclear stain DAPI (grey channel). (C and D) Enlarged regions of the sections presented in A and B panels, respectively, illustrating the negative correlation between (C) ERα or (D) PgR, and CK15. Scale bar, 50 μm. In all cases only merged images are shown.
Figure 7, which presents consecutive sections of normal human breast tissue collected from a 39‐year‐old woman (N81) and stained with the battery of antibodies illustrates the inferred extended immunophenotype of CK15+/CK19+/ERα−/PgR−/DRP3− acinar luminal cells (Table 1). These cells are c‐KIT+ (Figure 7D), CK5+ or − (Figure 7E; negative acini in positive lobules or negative acini in the whole lobule; red arrows indicate negative acini), and negative for CK14 (Figure 7F), vimentin, and CD44 (not shown; Table 1). The cells stained only weakly with CK8 (not shown). A similar overall phenotype was observed in the ducts, but in this case it was easier to identify single layers of luminal cells in contact with myoepithelial cells.
Figure 7.
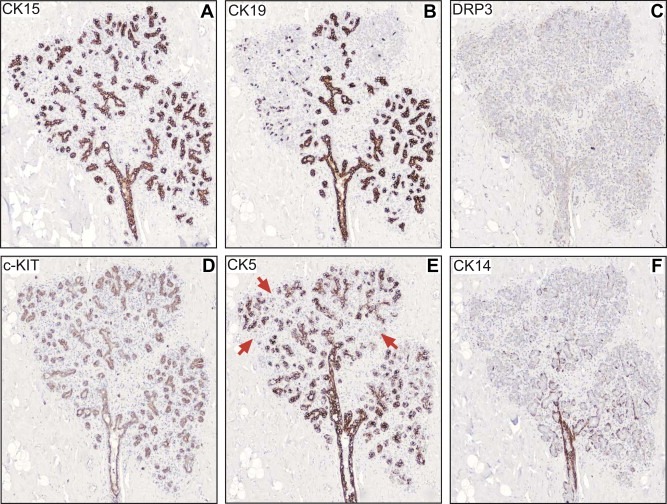
IHC analysis of serial sections of paraffin‐embedded normal breast tissue stained with antibodies against (A) CK15, (B) CK19, (C) DRP3, (D) c‐KIT, (E) CK5 and (F) CK14. The red arrows in (E) indicate CK5 negative acini.
Only sporadic CK15+/CK19+/DRP3+ luminal cells were encountered in acini with a single luminal layer (Supplementary Figure 5, panel A, sub‐figures A‐C) and these were c‐KIT+ attenuated (not shown), CK5+ and − (Supplementary Figure 5, panel A, sub‐figure D; mainly negative cells are observed in this picture), CK14− (Supplementary Figure 5, panel A, sub‐figure E), vimentin− (Supplementary Figure 5, panel A, sub‐figure F), and ERα, PgR, and CD44 negative (not shown) (Table 1).
3.2.1.1.2. CK15+/CK19− luminal cells
Cells exhibiting this phenotype were in general ERα and PgR –, c‐KIT+ and −, CK5+ and −, and CK14, vimentin, and CD44 negative (Table 1; see also CK19 negative area in Figure 7B). DRP3+/CK15+/CK19− cells (Supplementary Figure 5, panel B, sub‐figures A–C) were infrequent in acini containing a single luminal layer and these were c‐KIT+ attenuated, CK5 + and – and CK14, vimentin, and CD44 negative (Table 1).
3.2.1.1.3. CK15−/CK19+ luminal cells
Cells with this phenotype were ERα+ and PgR+ or −, c‐KIT‐, CK5−, CK14−, vimentin−, CD44− (Table 1) and stained strongly with CK8 (not shown). Only a few acinar structures exhibited cells that were DRP3+/CK15−/CK19+ (Supplementary Figure 5, panel C, sub‐figures A‐C) and these were c‐KIT+ attenuated, CK5+ or −, and CK14, vimentin and CD44 negative (Table 1).
3.2.1.2. Immunophenotype of luminal cells in lactating alveoli
Analysis of samples from lactating breast (6 weeks and longer) with the antibody battery showed that luminal cells in lactating breast were ERα and PgR negative (Holdaway et al., 1984), expressed DRP3 (Figure 4, panel A, sub‐figure C) and CK19 (not shown), but were negative for CK15 (Figure 8A and B, green signal), c‐KIT, CK14, CK5, vimentin, and CD44 (results not shown). As expected from published studies, the lactating cells expressed casein (Figure 8A and B, blue signal) as well as STAT5a, a transcription factor that is a central determinant of gland development and that promotes alveolar differentiation (Hynes et al., 1997; Miyoshi et al., 2001) (Figure 8A and B; red signal). A few undifferentiated CK15 positive cells were present adjacent to lactating alveoli or occasionally in lactating alveoli, but these had a phenotype similar to that of resting acini and were negative for DRP3, casein, and P‐STAT5a (transition to lactating alveoli indicated with white arrow in Figure 8A). In a few cases, however, we found CK15+/P‐STAT5a+/casein‐ undifferentiated cells at the interphase between casein negative and positive cells (yellow arrow in Figure 8B) suggesting that these cells give rise to the differentiated counterparts.
Figure 8.
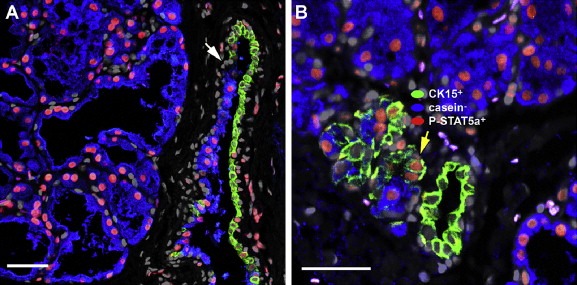
Indirect four color immunofluorescence analysis of lactating alveoli reacted with antibodies against cytokeratin 15 (CK15, Alexa Fluor® 488; green channel) P‐Stat5a (P‐STAT5a, Alexa Fluor® 594; red channel), casein (casein, Alexa Fluor® 633; blue channel), counterstained with the nuclear stain DAPI (grey channel). Lactating breast tissue samples presented (A) undifferentiated CK15+/P‐Stat5a−/casein− cells present occasionally within lactating alveoli or in transitional structures. In a few cases (B) one could observe CK15+/P‐Stat5a+/casein− cells.
3.2.1.3. Large central collecting ducts of the nipple
Considering that the lactiferous ducts participate in the formation of the developing breast (Fata et al., 2004, 2004) and that lobules and large ducts of normal breast tissue may be derived from the same stem cell (Tsai et al., 1996) it is likely that these structures represent a potential source of progenitor cells. It is well known that carcinomas of the breast arise as a result of mutations and/or epigenetic changes in cells originating in the TDLU's rather than in the large ducts (Ohuchi et al., 1984, 2004), a fact that implies that the collecting duct progenitor cells may be refractory to carcinogenesis. Indeed, only benign adenomas of the nipple have been reported, and even these are very rare (Sugai et al., 2002). Work by Russo et al. (2005) on the other hand, have presented evidence for the protective effect of pregnancy on cancer development. Their studies suggest that putative stem cells 1, which are the targets of neoplastic events, shift towards stem cells 2 during parity leading to differentiation of the mammary gland, a process that is believed makes the gland refractory to carcinogenesis. Even though the authors have found differences in the molecular signature of the stem 1 and 2 cells, it is unclear what the molecular mechanisms that underlie the phenomenon are (Russo et al., 2007).
Recently, we reported the presence of luminal cells in the collecting ducts that had the phenotype CK15+/CK19+/CK14+/vimentin− and that may correspond to putative progenitors. Our studies also revealed CK15+/CK19−/CK14+ cells, some of which located to the basal layer (Celis et al., 2007a) and that may correspond to a lineage‐restricted myoepithelial progenitor generated by loss of CK19 and possibly loss and gain of other markers (Adriance et al., 2005; Deugnier et al., 2002; Gudjonsson et al., 2005). Here we have re‐examined the phenotypes of the luminal cells in the large collecting ducts using the extended battery of antibodies. Besides confirming previous results the new data extended these observations by adding additional markers, in particular DRP3 and c‐KIT.
The major phenotypes of luminal cells in the collecting ducts included: (a) CK15+/CK19+/DRP3−/c‐KIT+ and −/CK5+ and −/CK14+ and −/vimentin−/ERα−/PgR−/CD44− (Figure 9 A–F, green arrows); (b) CK15+/CK19+/DRP3+/c‐KIT−/CK5+/CK14+/vimentin−/ERα−/PgR−/CD44− (Figure 9A–F, red arrows), and (c) CK15−/CK19+/DRP3+/c‐KIT−/CK5+/CK14+/vimentin−/ERα−/PgR−/CD44− (Figure 9A–F, blue arrows) (Table 1). It is noteworthy that the ratio of DRP3 positive cells was greatly elevated in collecting ducts as compared to resting acini. In a single section, 8 out of 17 large collecting ducts analyzed contained DRP3 positive cells. In contrast, out of a grand total of 2156 resting acini analyzed in 32 tissue samples, only 95 were found positive for DRP3.
Figure 9.
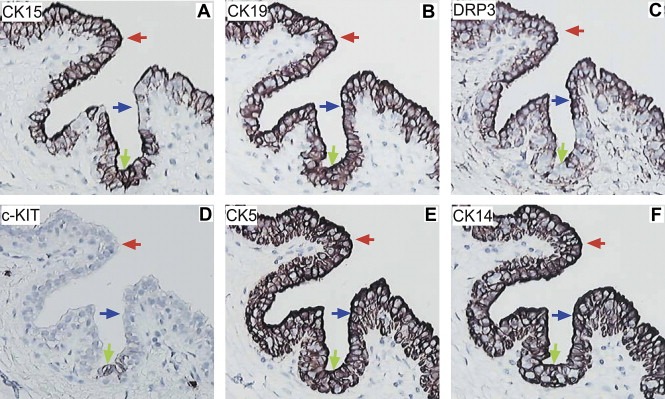
Immunohistochemical analysis of large collecting ducts of the nipple in normal breast tissue. Serial tissue sections of normal breast specimens were stained with antibodies against (A) CK15, (B) CK19, (C) DRP3, (D) c‐KIT, (E) CK5, and (F) CK14 allowing the immunophenotyping of cells present at the collecting ducts.
3.3. Towards defining the phenotype of complex benign mammary lesions
The immunophenotyping of complex benign breast lesions such as UDHs (Ellis, 2010; Pinder and Ellis, 2003), and CCLs (Pinder and Reis‐Filho, 2007) proved to be more difficult than we originally anticipated. This was partly due to the high cellular complexity and intricate structure of some of these lesions and partly because of the inherent limitations of our combined IHC and immunofluorescence approach to determine the expression of multiple markers in a given cell type in the background of complex lesions. Usually, with IHC one can use 3 to 4 serial sections to follow marker expression in simple structures; our immunofluorescence based approach allows a resolution improvement with a maximum of 3 markers (in addition to nuclear counterstaining) resolved at the single cell level. However, to address more complex lesions additional marker resolution at the single cell level is required to unequivocally define the cellular phenotypes of these lesions. In spite of these limitations, however, we have gathered some preliminary novel data on UDHs, in particular DRP3 positive UDHs, as well as on single cell layer CCLs. The data is briefly summarized below.
3.3.1. DRP3 positive UDHs
Epithelial hyperplasia is the most common form of proliferative breast disease and perhaps one of the most difficult to diagnose accurately (Ellis, 2010; Pinder and Ellis, 2003). Proliferating epithelial cells in UDHs can comprise three‐ to four‐cell layers above the basement membrane (mild) (Elston and Ellis, 1998) or five or more (moderate), often with accompanying bridging of the luminal space. In florid hyperplasia the changes are more marked and frequently these lesions contain a mixture of cells comprising luminal cells, myoepithelial cells, and metaplastic apocrine cells.
Presently, there is very little known about the phenotype of luminal cells in UDHs, although Böcker and colleagues have shown that cytokeratin 5/6 is expressed by cells in benign proliferative breast disease (Böcker et al., 2002) and our own studies have shown that CK15 and DRP3 are expressed by some hyperplasias (see above). Since DRP3 positive UDHs were easily identifiable using the EP071757 antibody, we focused only on these lesions as a first approximation.
DRP3 positive ductal hyperplasias (DHs) with mild and moderate atypia were identified in 21 out of 26 independent biopsies analyzed, which were derived from women with ages ranging from 38 to 71 years old. Three main types of DRP3 positive UDHs were identified with luminal cells having the following phenotypes: (i) DRP3+/CK15+/CK19+ (Figure 10, panel A, sub‐figures A–C), (ii) DRP3+/CK15+/CK19− (Figure 10, panel B, sub‐figures A–C), and (iii) DRP3+/CK15−/CK19+ (Figure 10, panel C, sub‐figures A–F); it should be noted that in some lesions we observed very low staining intensity with CK15, indicative of a possible attenuated phenotype. The latter DRP3+/CK15−/CK19+ UDHs were more common both within a single section of a given biopsy as well between biopsies from different individuals. The extended phenotype of the luminal cells in these lesions was found to be c‐KIT+ but attenuated, CK5+, CK14+, and ERα, PgR, p63, and CD44 negative. Obviously, not all the cells in the hyperplastic epithelium presented in panel C in Figure 10 have the same phenotype, particularly in the case of CK's 5 and 14, but the majority does. Work is currently underway to determine the phenotype of more complex florid lesions as well as sclerosing adenosis with apocrine differentiation.
Figure 10.
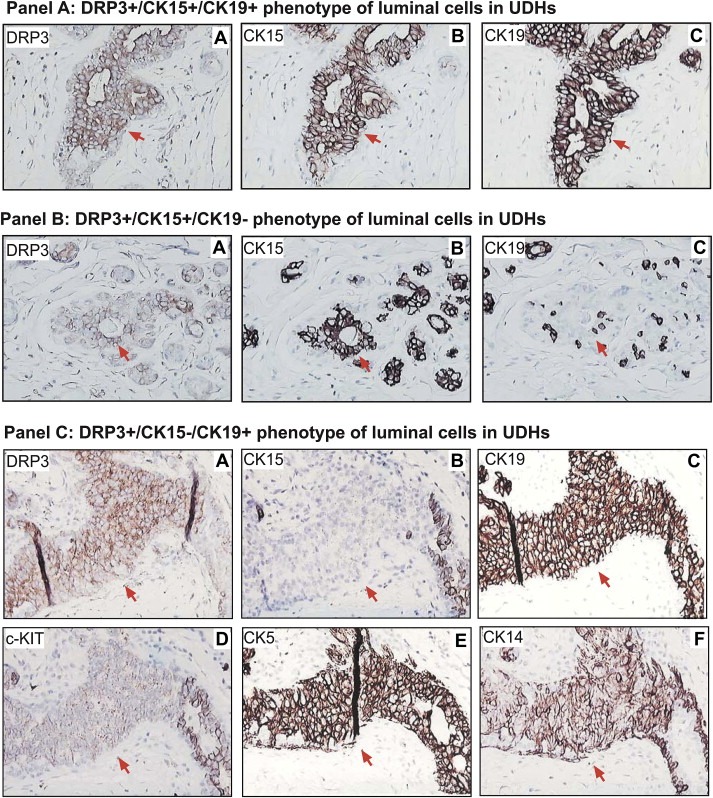
Immunophenotyping of luminal cells in UDHs. IHC analysis of serial tissue sections of normal breast tissue containing DRP3 positive UDHs. Serial tissue sections were stained with antibodies against DRP3, CK15 and CK19, allowing identification of three major DRP3+ cellular phenotypes: (panel A) DRP3+ (sub‐figure A)/CK15+ (sub‐figure B)/CK19+ (sub‐figure C), (panel B) DRP3+ (sub‐figure A)/CK15+ (sub‐figure B)/CK19− (sub‐figure C), and (panel C) DRP3+ (sub‐figure A)/CK15− (sub‐figure B)/CK19+ (sub‐figure C). Red arrows indicate cells with these phenotypes.
3.3.2. Columnar cell lesions (CCLs)
Columnar cell lesions of the breast include a morphological spectrum of alterations of the luminal cells lining the ducts and dilated acini of the TDLUs (Pinder and Reis‐Filho, 2007). CCL's are composed of cuboidal to tall columnar cells and are being encountered with increasing incidence in breast screening biopsies as they are associated with microcalcifications. CCLs can exhibit a single cell layer (columnar cell change, CCC) or multiple layers (columnar cell hyperplasia, CCH) and these are considered benign lesions. Columnar cell lesions are thought to be the earliest recognizable histological features that are a non‐obligate, intermediary step in the development of invasive carcinoma of the breast (Feeley and Quinn, 2008; Simpson et al., 2005). Published studies have shown that CCLs exhibit strong nuclear reactivity for ERα and PgR (Simpson et al., 2005; Tremblay et al., 2005) and are CK5/6 and CK14 negative (Simpson et al., 2005).
Given the complexity of these lesions we have as a first approximation focused on single cell layer columnar cells (CCCs). Figure 11A shows indirect immunofluorescence staining of breast tissue sections reacted with antibodies against CK15 (green signal), CK19 (blue signal) and ERα (red signal). Besides confirming previous studies concerning the positive receptor status of the CK19 positive columnar cells our observations revealed sporadic, elongated CK15+/CK19−/ERα−/PgR− luminal cells (seen as green cells in Figure 11A) that in some cases were seen entering the lumen of the ducts (indicated with white arrow, Figure 11A). The phenotype of such sporadically occurring cells could not be defined with certainty although IHC analysis of consecutive sections from several biopsies exhibiting CCCs suggested that some of the CK15+/CK19−/ERα−/PgR− luminal cells may be c‐KIT+ (Figure 11D) (Polat, 2007), CK5+ (Figure 11E), and CK14+ (Figure 11F). Independent immunofluorescence analysis indicated that there are ERα−/PgR− vimentin positive cells among the CCCs as exemplified in Figure 11B with a tissue preparation reacted with antibodies against vimentin, c‐KIT and CK19 (Figure 11B). Many of the vimentin positive cells were CK19 and c‐KIT negative (white arrow in Figure 11B), although a few were c‐KIT positive (yellow arrows in Figure 11B). We have also detected DRP3+/ERα−/PgR− luminal cells in CCCs (data not shown). Clearly, the immunophenotype of the various ERα negative cells in CCCs is quite complex and further studies using multiple‐label immunofluorescence on serial sections are needed in order to assess the extended phenotype of individual cells.
Figure 11.
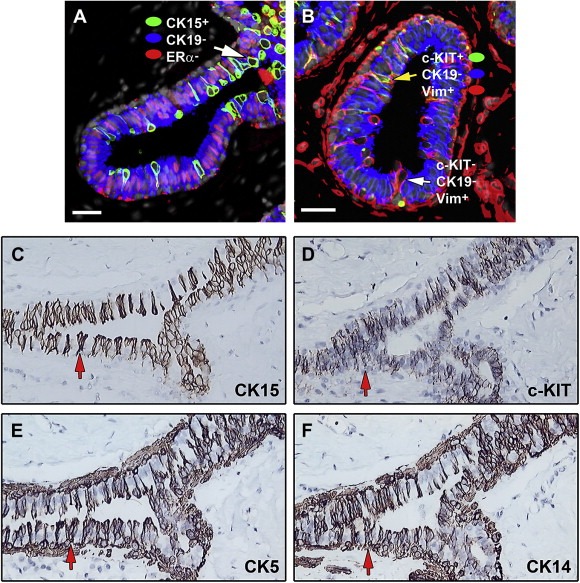
Immunophenotyping of columnar cell changes. (A and B) Indirect four color immunofluorescence analysis of tissue sections containing CCCs with antibodies against (A) cytokeratin 15 (CK15, Alexa Fluor® 488; green channel), ERα (ER, Alexa Fluor® 594; red channel), and cytokeratin 19 (CK19, Alexa Fluor® 633; blue channel), counterstained with the nuclear stain DAPI (grey channel) and (B) against c‐KIT (c‐KIT, Alexa Fluor® 488; green channel), vimentin (Vim, Alexa Fluor® 594; red channel), and cytokeratin 19 (CK19, Alexa Fluor® 633; blue channel), counterstained with the nuclear stain DAPI (grey channel). (C–F) Immunohistochemical analysis of CCCs. Serial tissue sections of normal breast specimens were stained with antibodies against (C) CK15, (D) c‐KIT, (E) CK5, and (F) CK14 to immunophenotype cells present at CCCs.
3.4. Concluding remarks
Even though our discovery‐driven biomarker research program is still at the very beginning, we have been able up till now to devise strategies that allowed the identification of novel luminal markers, such as CK15 (Celis et al., 2007a; Villadsen et al., 2007) and DRP3, which in combination with other known proteins have provided a more comprehensive picture of the immunophenotype of the various luminal cell types present in the normal human breast. Of all the markers analyzed, however, CK15 seems to be central as this protein is expressed by undifferentiated luminal cells in resting acini and ducts as well as by cells that may give rise to the differentiated lactating cells. Moreover, we have previously shown that CK15 positive cells are present at the intersection between undifferentiated luminal cells and differentiated apocrine cells (Celis et al., 2009) suggesting that their phenotype can be readily modified by the action of external cues. CK15 positive luminal cells in acini co‐express CK19, c‐KIT and sometimes CK5, while in structures like large collecting ducts of the nipple luminal cells can express in addition CK14. Complex phenotypes have also been observed in CK15 positive UDHs as well as in CCCs, with some cells in the latter structures being also vimentin and DRP3 positive. Further studies of CCC lesions may reveal more complex phenotypes of precursor cells and may shed some additional light as to their relation with early precancerous lesions.
Our studies have also shown that the expression of CK15 may be lost during progression. Indeed, we have presented evidence that p53 mutations may abrogate the expression of CK15 as ascertained in a well characterized tumour in which UDH, ADH, CIS and invasive disease coexisted in the same lesion (Celis et al., 2007a). Moreover, we have observed down regulation of CK15 in ERα positive ADHs as exemplified in Figure 12 which show serial sections of a breast tissue biopsy stained with antibodies against ERα (Figure 12A), CK15 (Figure 12B), c‐KIT (Figure 12C), and CK19 (Figure 12D). As depicted in Figure 12B, ERα positive luminal cells in the ADHs are negative for CK15, while morphologically normal appearing epithelial cells present in the surrounding acini express normal levels of this protein (blue arrows). All luminal cells in the preparation, however, express CK19, a cytokeratin that is found in most breast carcinomas (Bartek et al., 1985). Interestingly, loss of CK15 expression is accompanied by loss of c‐KIT expression (Figure 12C), a fact that is in line with our observation that the expression of both proteins is negatively affected by the expression levels of ERα and PgR. Given that most breast carcinomas arise from TDLUs, and considering the expression characteristics of both CK15 and c‐KIT, we consider these proteins as bona fide biomarkers of normal undifferentiated luminal cells in resting acini.
Figure 12.
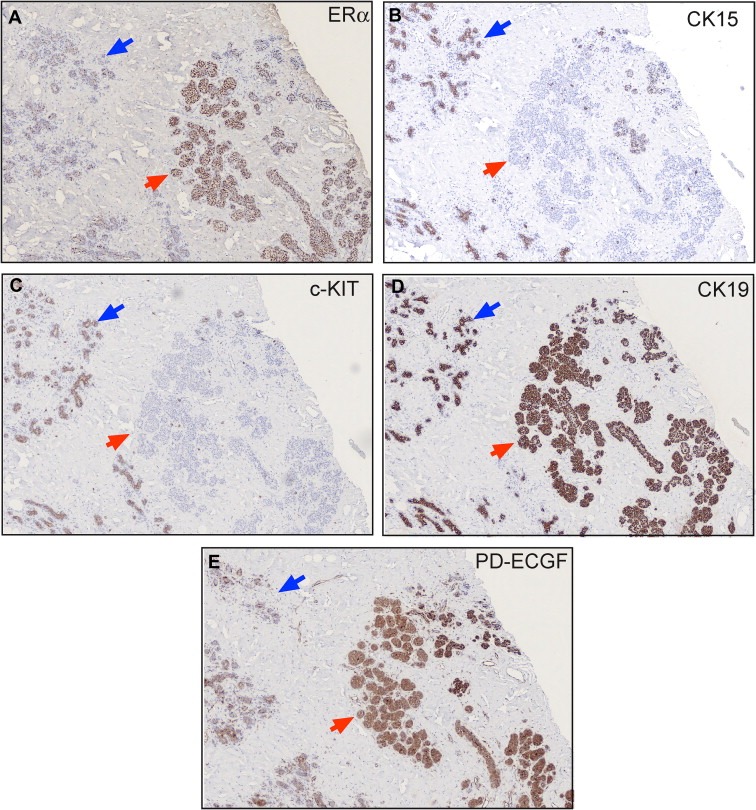
IHC analysis of serial sections of paraffin‐embedded normal breast tissue stained with antibodies against (A) ERα, (B) CK15, (C) c‐KIT, (D) CK19 and (E) PD‐ECGF, also known as endothelial cell growth factor (ECGF).
Ultimately, our aim is to derive a protein signature that can be used to identify breast precancerous lesions at risk of progressing to invasive disease. But application of such a protein signature for cellular phenotyping in a clinical setup would require prior detection of precancerous lesions and tissue acquisition. A better scenario would be to use biomarkers that can identify the presence of precancerous lesions in a non‐invasive manner, such as a blood test. Towards this end we have searched for adequate protein biomarkers that are present in tissue interstitial fluid (Celis et al., 2004a; Gromov et al., 2010) as these may represent potential candidates for developing a blood test for the early diagnosis of breast cancer. To be of potential value, these biomarkers must be expressed already in early lesions as exemplified in Figure 12E with the staining of a precancerous lesion with an antibody against one such marker (thymidine phosphorylase, also known as endothelial cell growth factor). As additional markers with similar expression characteristics are identified it should be possible to establish a signature that could be used to evaluate retrospective samples with a long‐term clinical follow‐up. The latter may not be an easy task as it will require collaboration between several groups and infrastructures such a biobanks (Riegman et al., 2008).
Another corollary of our studies is that the identification of molecular markers defining cellular phenotypes that are present in human breast tissue will allow the development of in vitro cultures (2D/3D organotypic co‐cultures, tissue slices) that resemble more closely the various phenotypes we have described and that could be used as model systems to study breast cancer initiation and progression as well as to validate novel therapeutic targets (Burdett et al., 2010; Hodgson et al., 2009; Jing et al., 1996; Kenny et al., 2007; Olsen et al., 2010; Parajuli and Doppler, 2009; Vaira et al., 2010; van der Kuip et al., 2006).
It should be stressed that we have found very few tumours expressing either CK15, c‐KIT, or DRP3. In the case of CK15 only 12–15% of breast carcinomas analysed expressed this protein and these correspond mainly to triple negatives, Her2‐neu, and the Luminal A subtype. Likewise, of the 343 tumours analyzed in the Pantomics and Valencia TMAs only 5 were c‐KIT positive and of these 3 were triple negatives, one was Her2‐neu, and 1 was Luminal A. Finally, of the 210 Pantomic breast carcinomas analysed only 17 were positive for DRP3 in the tumour cells and of these 10 were triple negative, 2 were Luminal A, 1 was Luminal B, and 4 were Her2‐neu. Work is currently underway to establish some prognostic correlation between the expression of these markers, the molecular profile of the carcinoma cases, and the clinical data in order to determine if they offer any prognostic value.
The expression pattern of DRP3 is particularly interesting as it uncovers a very dynamic behavior of this protein during cancer progression. DRP3 shows increased epithelial cell expression in a subset of UDHs but only a few invasive carcinomas show tumor‐cell expression of the protein; instead most tumors displayed increased stromal‐cell expression of DRP3. Recent work by Gao and colleagues showed that expression of CRMP4/DRP3 was inversely associated with the lymph node metastasis of pancreatic cancer and that overexpression of CRMP4 in pancreatic cancer cell lines reduced its in vitro invasive activity (Gao et al., 2010). Furthermore, these authors also demonstrated experimentally that forced expression of CRMP4/DRP3 in pancreatic cancer cell lines induces a decreased metastatic state in a mouse model of pancreatic cancer metastasis. These data together with the results presented in this report underline the need for further research into the role of DRP3 in breast cancer. Dissecting the various regulatory networks controlling DRP3 expression and the molecular mechanisms underlying disease pathogenesis might provide a clearer understanding of the function of this protein, a fact that will enhance its usefulness as an early biomarker and potential prognostic factor.
Supporting information
The following are the Supplementary data related to this article:
Supplementary Figure 1 Immunohistochemical analysis of tandem tissue sections showing normal breast epithelium stained with antibodies against (A) CK15 and (B) CK19. In one single field of vision one can identify all four possible cellular phenotypes.
Supplementary Figure 2 Silver stained IEF 2D gel pattern of Normal 95 specimen. Identified spots are indicated with arrows and a number corresponding to their ID number on Supplementary Table 2.
Supplementary Figure 3 DRP3 protein identification by MALDI‐TOF‐MS.
Supplementary Figure 4 Morphology of mammary gland. View of a ductal tree present in a normal tissue specimen with epithelial cells stained with CK19. A schematic depiction of a TDLU is shown.
Supplementary Figure 5 Immunophenotyping of luminal cells in resting acini. Serial tissue sections were stained with antibodies against DRP3, CK15 and CK19, allowing identification of three major DRP3+ cellular phenotypes: (panel A) DRP3+ (sub‐figure A)/CK15+ (sub‐figure B)/CK19+ (sub‐figure C) with CK5+ or − (sub‐figure D), CK14− (sub‐figure E) and vimentin− (sub‐figure F), (panel B) DRP3+ (sub‐figure A)/CK15+ (sub‐figure B)/CK19− (sub‐figure C), and (panel C) DRP3+ (sub‐figure A)/CK15− (sub‐figure B)/CK19− (sub‐figure C). Red arrows indicate cells with these phenotypes.
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Acknowledgements
We would like to thank Kitt Christensen, Dorte Holm, Lene Jørgensen, Sofia Svensson, and Signe Trentemøller for expert technical assistance. This work was supported by grants from the John and Birthe Meyer Foundation, the Danish Cancer Society, Novo Nordisk, the Kai Lange and Gundhild Kai Lange Fond, the Saint Albans Church, the Lisa and Gudmund Jørgensens Fond and the “Race against Breast Cancer”. The earmarked support of the Marketing Department at the Danish Cancer Society through their fundraising activities on behalf of DCTB is greatly appreciated.
Supplementary data 1.
1.1.
Supplementary data related to this article can be found online at doi:10.1016/j.molonc.2010.09.005.
Moreira José M.A., Cabezón Teresa, Gromova Irina, Gromov Pavel, Timmermans-Wielenga Vera, Machado Isidro, Llombart-Bosch Antonio, Kroman Niels, Rank Fritz, Celis Julio E., (2010), Tissue proteomics of the human mammary gland: Towards an abridged definition of the molecular phenotypes underlying epithelial normalcy, Molecular Oncology, 4, doi: 10.1016/j.molonc.2010.09.005.
Contributor Information
José M.A. Moreira, Email: jom@cancer.dk
Julio E. Celis, Email: jec@cancer.dk
References
- Abdel-Fatah, T.M.A. , Powe, D.G. , Hodi, Z. , Lee, A.H.S. , Reis-Filho, J.S. , Ellis, I.O. , 2007. High frequency of coexistence of columnar cell lesions, lobular neoplasia, and low grade ductal carcinoma in situ with invasive tumular carcinoma and invasive lobular carcinoma. Am. J. Surg. Pathol. 31, 417–426. [DOI] [PubMed] [Google Scholar]
- Adriance, M.C. , Inman, J.L. , Petersen, O.W. , Bissell, M.J. , 2005. Myoepithelial cells: good fences make good neighbors. Breast Cancer Res. 7, 190–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- In Altekruse S.F., Kosary C.L., Krapcho M., Neyman N., Aminou R., Waldron W., Ruhl J., Howlader N., Tatalovich Z., Cho H., Mariotto A., Eisner M.P., Lewis D.R., Cronin K., Chen H.S., Feuer E.J., Stinchcomb D.G., Edwards B.K.(Eds.), SEER Cancer Statistics Review, 1975–2007. National Cancer Institute; Bethesda, MD: [Google Scholar]
- Bánkfalvi, A. , Ludwig, A. , De-Hesselle, B. , Buerger, H. , Buchwalow, I.B. , Boecker, W. , 2004. Different proliferative activity of the glandular and myoepithelial lineages in benign proliferative and early malignant breast diseases. Mod. Pathol. 17, 1051–1061. [DOI] [PubMed] [Google Scholar]
- Barbareschi, M. , Pecciarini, L. , Cangi, M.G. , Macrì, E. , Rizzo, A. , Viale, G. , Doglioni, C. , 2001. p63, a p53 homologue, is a selective nuclear marker of myoepithelial cells of the human breast. Am. J. Surg. Pathol. 25, 1054–1060. [DOI] [PubMed] [Google Scholar]
- Bartek, J. , Taylor-Papadimitriou, J. , Miller, N. , Millis, R. , 1985. Patterns of expression of keratin 19 as detected with monoclonal antibodies in human breast tissues and tumours. Int. J. Cancer. 36, 299–306. [PubMed] [Google Scholar]
- Berry, D.A. , Cronin, K.A. , Plevritis, S.K. , Fryback, D.G. , Clarke, L. , Zelen, M. , Mandelblatt, J.S. , Yakovlev, A.Y. , Habbema, J.D. , Feuer, E.J. , 2005. Cancer Intervention and Surveillance Modeling Network (CISNET) Collaborators: effect of screening and adjuvant therapy on mortality from breast cancer. N. Engl. J. Med. 353, 1784–1792. [DOI] [PubMed] [Google Scholar]
- Bini, L. , Magi, B. , Marzocchi, B. , Arcuri, F. , Tripodi, S. , Cintorino, M. , Sanchez, J.C. , Frutiger, S. , Hughes, G. , Pallini, V. , Hochstrasser, D.F. , Tosi, P. , 1997. Protein expression profiles in human breast ductal carcinoma and histologically normal tissue. Electrophoresis. 18, 2832–2841. [DOI] [PubMed] [Google Scholar]
- Böcker, W. , Moll, R. , Poremba, C. , Holland, R. , Van Diest, P.J. , Dervan, P. , Bürger, H. , Wai, D. , Ina Diallo, R. , Brandt, B. , Herbst, H. , Schmidt, A. , Lerch, M.M. , Buchwallow, I.B. , 2002. Common adult stem cells in the human breast give rise to glandular and myoepithelial cell lineages: a new cell biological concept. Lab. Invest. 82, 737–746. [DOI] [PubMed] [Google Scholar]
- Boecker, W. , Buerger, H. , Herbst, H. , Decker, T. , 2006. Basic principles of precursor lesions of breast cancer. In Boecker W., Preneoplasia of the Breast: a New Conceptual Approach to Proliferative Breast Disease. Saunders; Munich, Germany: 337–368. [Google Scholar]
- Bohndiek, S.E. , Brindle, K.M. , 2010. Imaging and ‘omic’ methods for the molecular diagnosis of cancer. Expert Rev. Mol. Diagn. 10, 417–434. [DOI] [PubMed] [Google Scholar]
- Booth, B.W. , Boulanger, C.A. , Smith, G.H. , 2008. Stem cells and the mammary microenvironment. Breast Dis. 29, 57–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray, F. , McCarron, P. , Parkin, D.M. , 2004. The changing global patterns of female breast cancer incidence and mortality. Breast Cancer Res. 6, (6) 229–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan, D.J. , O'Connor, D.P. , Rexhepaj, E. , Ponten, F. , Gallagher, W.M. , 2010. Antibody-based proteomics: fast-tracking molecular diagnostics in oncology. Nat. Rev. Cancer. 10, 605–617. [DOI] [PubMed] [Google Scholar]
- Burdett, E. , Kasper, F.K. , Mikos, A.G. , Ludwig, J.A. , 2010. Engineering tumors: a tissue engineering perspective in cancer biology. Tissue Eng. Part B. Rev. 16, 351–359. [DOI] [PubMed] [Google Scholar]
- Celis, J.E. , Cabezón, T. , Moreira, J.M. , Gromov, P. , Gromova, I. , Timmermans-Wielenga, V. , Iwase, T. , Akiyama, F. , Honma, N. , Rank, F. , 2009. Molecular characterization of apocrine carcinoma of the breast: validation of an apocrine protein signature in a well-defined cohort. Mol. Oncol. 3, 220–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celis, J.E. , Gromov, P. , Cabezón, T. , Moreira, J.M. , Ambartsumian, N. , Sandelin, K. , Rank, F. , Gromova, I. , 2004. Proteomic characterization of the interstitial fluid perfusing the breast tumor microenvironment: a novel resource for biomarker and therapeutic target discovery. Mol. Cell. Proteomics. 3, 327–344. [DOI] [PubMed] [Google Scholar]
- Celis, J.E. , Gromov, P. , Gromova, I. , Moreira, J.M. , Cabezón, T. , Ambartsumian, N. , Grigorian, M. , Lukanidin, E. , Thor Straten, P. , Guldberg, P. , Bartkova, J. , Bartek, J. , Lukas, J. , Lukas, C. , Lykkesfeldt, A. , Jäättelä, M. , Roepstorff, P. , Bolund, L. , Ørntoft, T. , Brünner, N. , Overgaard, J. , Sandelin, K. , Blichert-Toft, M. , Mouridsen, H. , Rank, F.E. , 2003. Integrating proteomic and functional genomic technologies in discovery-driven translational breast cancer research. Mol. Cell. Proteomics. 2, 369–377. [DOI] [PubMed] [Google Scholar]
- Celis, J.E. , Gromova, I. , Cabezón, T. , Gromov, P. , Shen, T. , Timmermans-Wielenga, V. , Rank, F. , Moreira, J.M. , 2007. Identification of a subset of breast carcinomas characterized by expression of cytokeratin 15: relationship between CK15+ progenitor/amplified cells and pre-malignant lesions and invasive disease. Mol. Oncol. 1, 321–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celis, J.E. , Gromova, I. , Gromov, P. , Moreira, J.M. , Cabezón, T. , Friis, E. , Rank, F. , 2006. Molecular pathology of breast apocrine carcinomas: a protein expression signature specific for benign apocrine metaplasia. FEBS Lett. 580, 2935–2944. [DOI] [PubMed] [Google Scholar]
- Celis, J.E. , Moreira, J.M. , Cabezón, T. , Gromov, P. , Friis, E. , Rank, F. , Gromova, I. , 2005. Identification of extracellular and intracellular signaling components of the mammary adipose tissue and its interstitial fluid in high risk breast cancer patients: toward dissecting the molecular circuitry of epithelial-adipocyte stromal cell interactions. Mol. Cell. Proteomics. 4, 492–522. [DOI] [PubMed] [Google Scholar]
- Celis, J.E. , Moreira, J.M. , Gromova, I. , Cabezón, T. , Gromov, P. , Shen, T. , Timmermans, V. , Rank, F. , 2007. Characterization of breast precancerous lesions and myoepithelial hyperplasia in sclerosing adenosis with apocrine metaplasia. Mol. Oncol. 1, 97–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celis, J.E. , Trentemøller, S. , Gromov, P. , 2004. Gel-based proteomics: high-resolution two-dimensional gel electrophoresis of proteins. Isoelectric Focusing (IEF) and Nonequilibrium pH Gradient Electrophoresis (NEPHGE) In Celis J.E., Carter N., Hunter T., Shotton D., Simons K., Small J.V.(Eds.), Cell Biology. A Laboratory Handbook. vol. 4, Elsevier; San Diego: [Google Scholar]
- Cha, S. , Imielinski, M.B. , Rejtar, T. , Richardson, E.A. , Thakur, D. , Sgroi, D.C. , Karger, B.L. , 2010. In situ proteomic analysis of human breast cancer epithelial cells using laser capture microdissection (LCM)-LC/MS: annotation by protein set enrichment analysis (PSEA) and gene ontology (GO). Mol. Cell. Proteomics. 10.1074/mcp.M110.000398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrier, E. , Reibel, S. , Rogemond, V. , Aguera, M. , Thomasset, N. , Honnorat, J. , 2003. Collapsin response mediator proteins (CRMPs): involvement in nervous system development and adult neurodegenerative disorders. Mol. Neurobiol. 28, 51–64. [DOI] [PubMed] [Google Scholar]
- Cornfield, D.B. , Palazzo, J.P. , Schwartz, G.F. , Goonewardene, S.A. , Kovatich, A.J. , Chervoneva, I. , Hyslop, T. , Schwarting, R. , 2004. The prognostic significance of multiple morphologic features and biologic markers in ductal carcinoma in situ of the breast: a study of a large cohort of patients treated with surgery alone. Cancer. 100, 2317–2327. [DOI] [PubMed] [Google Scholar]
- Dabbs, D.J. , Carter, G. , Fudge, M. , Peng, Y. , Swalsky, P. , Finkelstein, S. , 2006. Molecular alterations in columnar cell lesions of the breast. Mod. Pathol. 19, 344–349. [DOI] [PubMed] [Google Scholar]
- Deugnier, M.A. , Teulière, J. , Faraldo, M.M. , Thiery, J.P. , Glukhova, M.A. , 2002. The importance of being a myoepithelial cell. Breast Cancer Res. 4, 224–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Early Breast Cancer Trialists' Collaborative Group (EBCTCG) 2005. Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomised trials. Lancet. 365, 1687–1717. [DOI] [PubMed] [Google Scholar]
- Early Breast Cancer Trialists' Collaborative Group 1998. Tamoxifen for early breast cancer: an overview of the randomised trials. Lancet. 351, 1451–1467. [PubMed] [Google Scholar]
- Ellis, I.O. , 2010. Intraductal proliferative lesions of the breast: morphology, associated risk and molecular biology. Mod. Pathol. (23 Suppl. 2) S1–S7. [DOI] [PubMed] [Google Scholar]
- Elston, C.W. , Ellis, I.O. , 1998. Assessment of histological grade In Elston C.W., Ellis I.O.(Eds.), The Breast. vol. 13, Churchill Livingstone; Edinburgh/New York: 356–384. [Google Scholar]
- Engholm, G. , Ferlay, J. , Christensen, N. , Bray, F. , Gjerstorff, M.L. , Klint, Å , Køtlum, J.E. , Ólafsdóttir, E. , Pukkala, E. , Storm, H.H. , 2010. NORDCAN: Cancer Incidence, Mortality, Prevalence and Prediction in the Nordic Countries, Version 3.6 Association of the Nordic Cancer Registries/Danish Cancer Society; http://www.ancr.nu [Google Scholar]
- Ernst, M.F. , Voogd, A.C. , Coebergh, J.W. , Roukema, J.A. , 2004. Breast carcinoma diagnosis, treatment, and prognosis before and after the introduction of mass mammographic screening. Cancer. 100, 1337–1344. [DOI] [PubMed] [Google Scholar]
- Fass, L. , 2008. Imaging and cancer: a review. Mol. Oncol. 2, 115–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fata, J.E. , Werb, Z. , Bissell, M.J. , 2004. Regulation of mammary gland branching morphogenesis by the extracellular matrix and its remodeling enzymes. Breast Cancer Res. 6, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feeley, L. , Quinn, C.M. , 2008. Columnar cell lesions of the breast. Histopathology. 52, 11–19. [DOI] [PubMed] [Google Scholar]
- Ferlay, J. , Shin, H.-R. , Bray, F. , Forman, D. , Mathers, C. , Parkin, D.M. , 2010. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int. J. Cancer. 10.1002/ijc.25516 [DOI] [PubMed] [Google Scholar]
- Findeisen, P. , Neumaier, M. , 2009. Mass spectrometry based proteomics profiling as diagnostic tool in oncology: current status and future perspective. Clin. Chem. Lab. Med. 47, 666–684. [DOI] [PubMed] [Google Scholar]
- Fink-Retter, A. , Czerwenka, K. , Gschwantler-Kaulich, D. , Hudelist, G. , Pischinger, K. , Manavi, M. , Kubista, E. , Singer, C.F. , 2009. Proteomics in mammary cancer research. Eur. J. Gynaecol. Oncol. 30, 635–639. [PubMed] [Google Scholar]
- Franzen, B. , Auer, G. , Alaiya, A.A. , Eriksson, E. , Uryu, K. , Hirano, T. , Okuzawa, K. , Kato, H. , Linder, S. , 1996. Assessment of homogeneity in polypeptide expression in breast carcinomas shows widely variable expression in highly malignant tumors. Int. J. Cancer. 69, 408–414. [DOI] [PubMed] [Google Scholar]
- Gaetano, C. , Matsuo, T. , Thiele, C.J. , 1997. Identification and characterization of a retinoic acid-regulated human homologue of the unc-33-like phosphoprotein (hUlip) from neuroblastoma cells. J. Biol. Chem. 272, 12195–12201. [DOI] [PubMed] [Google Scholar]
- Gao, X. , Pang, J. , Li, L.Y. , Liu, W.P. , Di, J.M. , Sun, Q.P. , Fang, Y.Q. , Liu, X.P. , Pu, X.Y. , He, D. , Li, M.T. , Su, Z.L. , Li, B.Y. , 2010. Expression profiling identifies new function of collapsin response mediator protein 4 as a metastasis-suppressor in prostate cancer. Oncogene. 29, 4555–4566. [DOI] [PubMed] [Google Scholar]
- Gast, M.C. , Schellens, J.H. , Beijnen, J.H. , 2009. Clinical proteomics in breast cancer: a review. Breast Cancer Res. Treat. 116, 17–29. [DOI] [PubMed] [Google Scholar]
- Goldstein, A.S. , Huang, J. , Guo, C. , Garraway, I.P. , Witte, O.N. , 2010. Identification of a cell of origin for human prostate cancer. Science. 329, 568–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein, A.S. , Stoyanova, T. , Witte, O.N. , 2010. Primitive origins of prostate cancer: in vivo evidence for prostate-regenerating cells and prostate cancer-initiating cells. Mol. Oncol. 4, 385–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein, N.S. , Kestin, L.J. , Vicini, F.A. , 2007. Monomorphic epithelial proliferations: characterization and evidence suggesting they are the pool of partially transformed lesions from which some invasive carcinomas arise. Am. J. Clin. Pathol. 128, 1023–1034. [DOI] [PubMed] [Google Scholar]
- Gromov, P. , Celis, J.E. , Gromova, I. , Rank, F. , Timmermans-Wielenga, V. , Moreira, J.M. , 2008. A single lysis solution for the analysis of tissue samples by different proteomic technologies. Mol. Oncol. 2, 368–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromov, P. , Gromova, I. , Bunkenborg, J. , Cabezon, T. , Moreira, J.M. , Timmermans-Wielenga, V. , Roepstorff, P. , Rank, F. , Celis, J.E. , 2010. Up-regulated proteins in the fluid bathing the tumour cell microenvironment as potential serological markers for early detection of cancer of the breast. Mol. Oncol. 4, 65–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromova, I. , Celis, J.E. , 2004. Protein detection in gels by silver staining: a procedure compatible with mass-spectrometry In Celis J.E., Carter N., Hunter T., Shotton D., Simons K., Small J.V.(Eds.), Cell Biology. A Laboratory Handbook. vol. 4, Elsevier; San Diego: [Google Scholar]
- Gudjonsson, T. , Adriance, M.C. , Sternlicht, M.D. , Petersen, O.W. , Bissell, M.J. , 2005. Myoepithelial cells: their origin and function in breast morphogenesis and neoplasia. J. Mammary Gland Biol. Neoplasia. 10, 261–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamajima, N. , Matsuda, K. , Sakata, S. , Tamaki, N. , Sasaki, M. , Nonaka, M. , 1996. A novel gene family defined by human dihydropyrimidinase and three related proteins with differential tissue distribution. Gene. 180, 157–163. [DOI] [PubMed] [Google Scholar]
- Hammond, M.E. , Hayes, D.F. , Dowsett, M. , Allred, D.C. , Hagerty, K.L. , Badve, S. , Fitzgibbons, P.L. , Francis, G. , Goldstein, N.S. , Hayes, M. , Hicks, D.G. , Lester, S. , Love, R. , Mangu, P.B. , McShane, L. , Miller, K. , Osborne, C.K. , Paik, S. , Perlmutter, J. , Rhodes, A. , Sasano, H. , Schwartz, J.N. , Sweep, F.C. , Taube, S. , Torlakovic, E.E. , Valenstein, P. , Viale, G. , Visscher, D. , Wheeler, T. , Williams, R.B. , Wittliff, J.L. , Wolff, A.C. , American Society of Clinical OncologyCollege of American Pathologists2010. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer (unabridged version). Arch. Pathol. Lab. Med. 134, e48–72. [DOI] [PubMed] [Google Scholar]
- Haupt, B. , Schwartz, M.R. , Xu, Q. , Ro, J.Y. , 2010. Columnar cell lesions: a consensus study among pathology trainees. Hum. Pathol. 41, 895–901. [DOI] [PubMed] [Google Scholar]
- Héry, C. , Ferlay, J. , Boniol, M. , Autier, P. , 2008. Changes in breast cancer incidence and mortality in middle-aged and elderly women in 28 countries with Caucasian majority populations. Ann. Oncol. 19, 1009–1018. [DOI] [PubMed] [Google Scholar]
- Hiratsuka, M. , Inoue, T. , Toda, T. , Kimura, N. , Shirayoshi, Y. , Kamitani, H. , Watanabe, T. , Ohama, E. , Tahimic, C.G. , Kurimasa, A. , Oshimura, M. , 2003. Proteomics-based identification of differentially expressed genes in human gliomas: down-regulation of SIRT2 gene. Biochem. Biophys. Res. Commun. 309, 558–566. [DOI] [PubMed] [Google Scholar]
- Hodgson, D.R. , Whittaker, R.D. , Herath, A. , Amakye, D. , Clack, G. , 2009. Biomarkers in oncology drug development. Mol. Oncol. 3, 24–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holdaway, I.M. , Mason, B.H. , Kay, R.G. , 1984. Steroid hormone receptors in breast tumours presenting during pregnancy or lactation. J. Surg. Oncol. 25, 38–41. [DOI] [PubMed] [Google Scholar]
- Hynes, N.E. , Cella, N. , Wartmann, M. , 1997. Prolactin mediated intracellular signaling in mammary epithelial cells. J. Mammary Gland Biol. Neoplasia. 2, 19–27. [DOI] [PubMed] [Google Scholar]
- Jain, K.K. , 2008. Innovations, challenges and future prospects of oncoproteomics. Mol. Oncol. 2, 153–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jara-Lazaro, A.R. , Tse, G.M.-K. , Tan, P.H. , 2009. Columnar cell lesions of the breast: an update and significance on core biopsy. Pathology. 41, 18–27. [DOI] [PubMed] [Google Scholar]
- Jing, Y. , Xu, X.C. , Lotan, R. , Waxman, S. , Mira-y-Lopez, R. , 1996. Human breast carcinoma slice cultures retain retinoic acid sensitivity. Braz. J. Med. Biol. Res. 29, 1105–1108. [PubMed] [Google Scholar]
- Jotwani, A.C. , Gralow, J.R. , 2009 Dec 1. 2010 Early detection of breast cancer: new biomarker tests on the horizon?. Mol. Diagn. Ther. 13, (6) 349–357. 10.2165/11318220-000000000-00000 [DOI] [PubMed] [Google Scholar]
- Karagiannis, G.S. , Pavlou, M.P. , Diamandis, E.P. , 2010. Cancer secretomics reveal pathophysiological pathways in cancer molecular oncology. Mol. Oncol. 4, (6) 496–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenny, P.A. , Lee, G.Y. , Myers, C.A. , Neve, R.M. , Semeiks, J.R. , Spellman, P.T. , Lorenz, K. , Lee, E.H. , Barcellos-Hoff, M.H. , Petersen, O.W. , Gray, J.W. , Bissell, M.J. , 2007. The morphologies of breast cancer cell lines in three-dimensional assays correlate with their profiles of gene expression. Mol. Oncol. 1, 84–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S.Y. , 2010. A new paradigm for cancer therapeutics development. BMB Rep. 43, 383–388. [DOI] [PubMed] [Google Scholar]
- Kostic, C. , Shaw, P.H. , 2000. Isolation and characterization of sixteen novel p53 response genes. Oncogene. 19, 3978–3987. [DOI] [PubMed] [Google Scholar]
- Kowara, R. , Chen, Q. , Milliken, M. , Chakravarthy, B. , 2005. Calpain-mediated truncation of dihydropyrimidinase-like 3 protein (DPYSL3) in response to NMDA and H2O2 toxicity. J. Neurochem. 95, 466–474. [DOI] [PubMed] [Google Scholar]
- Kowara, R. , Moraleja, K.L. , Chakravarthy, B. , 2006. Involvement of nitric oxide synthase and ROS-mediated activation of L-type voltage-gated Ca2+ channels in NMDA-induced DPYSL3 degradation. Brain Res. 1119, 40–49. [DOI] [PubMed] [Google Scholar]
- Larson, P.S. , de Las, M.A. , Cerda, S.R. , 2006. Quantitative analysis of allele imbalance supports atypical ductal hyperplasia lesions as direct breast cancer precursors. J. Pathol. 209, 307–316. [DOI] [PubMed] [Google Scholar]
- Leung, T. , Ng, Y. , Cheong, A. , Ng, C.H. , Tan, I. , Hall, C. , Lim, L. , 2002. p80 ROKα binding protein is a novel splice variant of CRMP-1 which associates with CRMP-2 and modulates RhoA-induced neuronal morphology. FEBS Lett. 532, 445–449. [DOI] [PubMed] [Google Scholar]
- Levi, F. , Lucchini, F. , Negri, E. , La Vecchia, C. , 2007. Continuing declines in cancer mortality in the European Union. Ann. Oncol. 18, 593–595. [DOI] [PubMed] [Google Scholar]
- Lim, E. , Vaillant, F. , Wu, D. , Forrest, N.C. , Pal, B. , Hart, A.H. , Asselin-Labat, M.L. , Gyorki, D.E. , Ward, T. , Partanen, A. , Feleppa, F. , Huschtscha, L.I. , Thorne, H.J. , Fox, S.B. , Yan, M. , French, J.D. , Brown, M.A. , Smyth, G.K. , Visvader, J.E. , Lindeman, G.J. , kConFab2009. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat. Med. 15, 907–913. [DOI] [PubMed] [Google Scholar]
- Lopez-Garcia, M.A. , Geyer, F.C. , Lacroix-Triki, M. , Marchió, C. , Reis-Filho, J.S. , 2010. Breast cancer precursors revisited: molecular features and progression pathways. Histopathology. 57, 171–192. [DOI] [PubMed] [Google Scholar]
- McCarthy, J.R. , Bhaumik, J. , Karver, M.R. , Sibel Erdem, S. , Weissleder, R. , 2010. Targeted nanoagents for the detection of cancers. Mol. Oncol. 4, (6) 511–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi, K. , Shillingford, J.M. , Smith, G.H. , Grimm, S.L. , Wagner, K.U. , Oka, T. , Rosen, J.M. , Robinson, G.W. , Hennighausen, L. , 2001. Signal transducer and activator of transcription (Stat) 5 controls the proliferation and differentiation of mammary alveolar epithelium. J. Cell Biol. 155, 531–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffat, D.F. , Going, J.J. , 1996. Three dimensional anatomy of complete duct systems in human breast: pathological and developmental implications. J. Clin. Pathol. 49, 48–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moll, R. , Divo, M. , Langbein, L. , 2008. The human keratins: biology and pathology. Histochem. Cell Biol. 129, 705–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molyneux, G. , Geyer, F.C. , Magnay, F.A. , McCarthy, A. , Kendrick, H. , Natrajan, R. , Mackay, A. , Grigoriadis, A. , Tutt, A. , Ashworth, A. , Reis-Filho, J.S. , Smalley, M.J. , 2010. BRCA1 basal-like breast cancers originate from luminal epithelial progenitors and not from basal stem cells. Cell Stem Cell. 7, 403–417. [DOI] [PubMed] [Google Scholar]
- Mueller, C. , Liotta, L.A. , Espina, V. , 2010. Reverse phase protein microarrays advance to use in clinical trials. Mol. Oncol. 4, (6) 461–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muggerud, A.A. , Hallett, M. , Johnsen, H. , Kleivi, K. , Zhou, W. , Tahmasebpoor, S. , Amini, R.M. , Botling, J. , Børresen-Dale, A.L. , Sørlie, T. , Wärnberg, F. , 2010. Molecular diversity in ductal carcinoma in situ (DCIS) and early invasive breast cancer. Mol. Oncol. 4, 357–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naor, D. , Sionov, R.V. , Ish-Shalom, D. , 1997. CD44: structure, function, and association with the malignant process. Adv. Cancer Res. 71, 241–319. [DOI] [PubMed] [Google Scholar]
- Natali, P.G. , Nicotra, M.R. , Sures, I. , Mottolese, M. , Botti, C. , Ullrich, A. , 1992. Breast cancer is associated with loss of the c-kit oncogene product. Int. J. Cancer. 52, 713–717. [DOI] [PubMed] [Google Scholar]
- Nelson, A.L. , Dhimolea, E. , Reichert, J.M. , 2010. Development trends for human monoclonal antibody therapeutics. Nat. Rev. Drug Discov. 10.1038/nrd3229 [DOI] [PubMed] [Google Scholar]
- Nofech-Mozes, S. , Spayne, J. , Rakovitch, E. , Hanna, W. , 2005. Prognostic and predictive molecular markers in DCIS: a review. Adv. Anat. Pathol. 12, 256–264. [DOI] [PubMed] [Google Scholar]
- O'Farrell, P.H. , 1975. High resolution two-dimensional electrophoresis of proteins. J. Biol. Chem. 250, 4007–4021. [PMC free article] [PubMed] [Google Scholar]
- Ohuchi, N. , Abe, R. , Takahashi, T. , Tezuka, F. , 1984. Origin and extension of intraductal papillomas of the breast: a three-dimensional reconstruction study. Breast Cancer Res. Treat. 4, 117–128. [DOI] [PubMed] [Google Scholar]
- Olivotto, I.O. , Bajdik, C.D. , Ravdin, P.M. , Speers, C.H. , Coldman, A.J. , Norris, B.D. , Davis, G.J. , Chia, S.K. , Gelmon, K.A. , 2005. Population-based validation of the prognostic model ADJUVANT! for early breast cancer. J. Clin. Oncol. 23, 2716–2725. [DOI] [PubMed] [Google Scholar]
- Olsen, C.J. , Moreira, J. , Lukanidin, E.M. , Ambartsumian, N.S. , 2010. Human mammary fibroblasts stimulate invasion of breast cancer cells in a three-dimensional culture and increase stroma development in mouse xenografts. BMC Cancer. 19, 444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parajuli, N. , Doppler, W. , 2009. Precision-cut slice cultures of tumors from MMTV-neu mice for the study of the ex vivo response to cytokines and cytotoxic drugs. In Vitro Cell Dev. Biol. Anim. 45, 442–450. [DOI] [PubMed] [Google Scholar]
- Pierotti, M.A. , Negri, T. , Tamborini, E. , Perrone, F. , Pricl, S. , Pilotti, S. , 2010. Targeted therapies: the rare cancer paradigm. Mol. Oncol. 4, 19–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinder, S.E. , 2010. Ductal carcinoma in situ (DCIS): pathological features, differential diagnosis, prognostic factors and specimen evaluation. Mod. Pathol. (23 Suppl. 2) S8–S13. [DOI] [PubMed] [Google Scholar]
- Pinder, S.E. , Ellis, I.O. , 2003. The diagnosis and management of pre-invasive breast disease: ductal carcinoma in situ (DCIS) and atypical ductal hyperplasia (ADH)–current definitions and classification. Breast Cancer Res. 5, 254–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinder, S.E. , Reis-Filho, J.S. , 2007. Non-operative breast pathology: columnar cell lesions. J. Clin. Pathol. 60, 1307–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podo, F. , Buydens, L.M. , Degani, H. , Hilhorst, R. , Klipp, E. , Gribbestad, I.S. , Van Huffel, S. , van Laarhoven, H.W. , Luts, J. , Monleon, D. , Postma, G.J. , Schneiderhan-Marra, N. , Santoro, F. , Wouters, H. , Russnes, H.G. , Sørlie, T. , Tagliabue, E. , Børresen-Dale, A.L. , FEMME Consortium2010. Triple-negative breast cancer: present challenges and new perspectives. Mol. Oncol. 4, 209–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polat, A. , 2007. c-KIT expression in columnar cell lesions of the breast accompanied by benign and malignant breast diseases. Pathol. Res. Pract. 203, 765–769. [DOI] [PubMed] [Google Scholar]
- Quinn, C.C. , Chen, E. , Kinjo, T.G. , Kelly, G. , Bell, A.W. , Elliott, R.C. , McPherson, P.S. , Hockfield, S. , 2003. TUC-4b, a novel TUC family variant, regulates neurite outgrowth and associates with vesicles in the growth cone. J. Neurosci. 23, 2815–2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn, C.C. , Gray, G.E. , Hockfield, S. , 1999. A family of proteins implicated in axon guidance and outgrowth. J. Neurobiol. 41, 158–164. [PubMed] [Google Scholar]
- Riegman, P.H. , Morente, M.M. , Betsou, F. , de Blasio, P. , Geary, P. , Marble Arch International Working Group on Biobanking for Biomedical Research2008. Biobanking for better healthcare. Mol. Oncol. 2, 213–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo, J. , Balogh, G. , Russo, I.H. , 2007. Breast cancer prevention. Climacteric. (10 Suppl. 2) 47–53. [DOI] [PubMed] [Google Scholar]
- Russo, J. , Moral, R. , Balogh, G.A. , Mailo, D. , Russo, I.H. , 2005. The protective role of pregnancy in breast cancer. Breast Cancer Res. 7, 131–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo, J. , Russo, I.H. , 2004. Development of the human breast. Maturitas. 49, 2–15. [DOI] [PubMed] [Google Scholar]
- Schiess, R. , Wollscheid, B. , Aebersold, R. , 2009. Targeted proteomic strategy for clinical biomarker discovery. Mol. Oncol. 3, 33–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnitt, S.J. , 2006. Flat epithelial atypia. In Boecker W., Preneoplasia of the Breast: a New Conceptual Approach to Proliferative Breast Disease. Saunders; Munich, Germany: 369–378. [Google Scholar]
- Schnitt, S.J. , 2003. The diagnosis and management of pre-invasive breast disease: flat epithelial atypia: classification, pathologic features and clinical significance. Breast Cancer Res. 5, 263–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnitt, S.J. , Vincent-Salomon, A. , 2003. Columnar cell lesions of the breast. Adv. Anat. Pathol. 10, 113–124. [DOI] [PubMed] [Google Scholar]
- Shen, Y. , Yang, Y. , Inoue, L.Y. , Munsell, M.F. , Miller, A.B. , Berry, D.A. , 2005. Role of detection method in predicting breast cancer survival: analysis of randomized screening trials. J. Natl. Cancer Inst. 97, 1195–1203. [DOI] [PubMed] [Google Scholar]
- Shevchenko, A. , Wilm, M. , Vorm, O. , Mann, M. , 1996. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68, 850–858. [DOI] [PubMed] [Google Scholar]
- Shipitsin, M. , Campbell, L.L. , Argani, P. , Weremowicz, S. , Bloushtain-Qimron, N. , Yao, J. , Nikolskaya, T. , Serebryiskaya, T. , Beroukhim, R. , Hu, M. , 2007. Molecular definition of breast tumor heterogeneity. Cancer Cell. 11, 259–273. [DOI] [PubMed] [Google Scholar]
- Shoker, B.S. , Jarvis, C. , Clarke, R.B. , Anderson, E. , Munro, C. , Davies, M.P. , Sibson, D.R. , Sloane, J.P. , 2000. Abnormal regulation of the oestrogen receptor in benign breast lesions. J. Clin. Pathol. 53, 778–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson, J.F. , 2009. Update on atypical epithelial hyperplasia and ductal carcinoma in situ. Pathology. 41, 36–39. [DOI] [PubMed] [Google Scholar]
- Simpson, P.T. , Gale, T. , Reis-Filho, J.S. , Jones, C. , Parry, S. , Sloane, J.P. , Hanby, A. , Pinder, S.E. , Lee, A.H. , Humphreys, S. , Ellis, I.O. , Lakhani, S.R. , 2005. Columnar cell lesions of the breast: the missing link in breast cancer progression? A morphological and molecular analysis. Am. J. Surg. Pathol. 29, 734–746. [DOI] [PubMed] [Google Scholar]
- Smith, G.H. , Chepko, G. , 2001. Mammary epithelial stem cells. Microsc. Res. Tech. 52, 190–203. [DOI] [PubMed] [Google Scholar]
- Stingl, J. , Eaves, C.J. , Zandieh, I. , Emerman, J.T. , 2001. Characterization of bipotent mammary epithelial progenitor cells in normal adult human breast tissue. Breast Cancer Res. Treat. 67, 93–109. [DOI] [PubMed] [Google Scholar]
- Sugai, M. , Murata, K. , Kimura, N. , Munakata, H. , Hada, R. , Kamata, Y. , 2002. Adenoma of the nipple in an adolescent. Breast Cancer. 9, 254–256. [DOI] [PubMed] [Google Scholar]
- Tavassoli, F.A. , 2009. Challenges in breast pathology: new twists on old problems. Arch. Pathol. Lab. Med. 133, 852–854. [DOI] [PubMed] [Google Scholar]
- Thorpe, S.M. , 1988. Estrogen and progesterone receptor determinations in breast cancer. Technology, biology and clinical significance. Acta Oncol. 27, 1–19. [DOI] [PubMed] [Google Scholar]
- Toft, D.J. , Cryns, V.L. , 2010. Minireview: basal-like breast cancer: from molecular profiles to targeted therapies. Mol. Endocrinol. 10.1210/me.2010-0164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay, G. , Deschênes, J. , Alpert, L. , Quenneville, L.A. , 2005. Overexpression of estrogen receptors in columnar cell change and in unfolding breast lobules. Breast J. 11, 326–332. [DOI] [PubMed] [Google Scholar]
- Tsai, Y.C. , Lu, Y. , Nichols, P.W. , Zlotnikov, G. , Jones, P.A. , Smith, H.S. , 1996. Contiguous patches of normal human mammary epithelium derived from a single stem cell: implications for breast carcinogenesis. Cancer Res. 56, 402–404. [PubMed] [Google Scholar]
- Vaira, V. , Fedele, G. , Pyne, S. , Fasoli, E. , Zadra, G. , Bailey, D. , Snyder, E. , Faversani, A. , Coggi, G. , Flavin, R. , Bosari, S. , Loda, M. , 2010. Preclinical model of organotypic culture for pharmacodynamic profiling of human tumors. Proc. Natl. Acad. Sci. U.S.A. 107, 8352–8356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Kuip, H. , Mürdter, T.E. , Sonnenberg, M. , McClellan, M. , Gutzeit, S. , Gerteis, A. , Simon, W. , Fritz, P. , Aulitzky, W.E. , 2006. Short term culture of breast cancer tissues to study the activity of the anticancer drug taxol in an intact tumor environment. BMC Cancer. 7, 86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villadsen, R. , Fridriksdottir, A.J. , Rønnov-Jessen, L. , Gudjonsson, T. , Rank, F. , LaBarge, M.A. , Bissell, M.J. , Petersen, O.W. , 2007. Evidence for a stem cell hierarchy in the adult human breast. J. Cell Biol. 177, 87–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvader, J.E. , 2009. Keeping abreast of the mammary epithelial hierarchy and breast tumorigenesis. Genes Dev. 23, 2563–2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvader, J.E. , Lindeman, G. , 2010. Stem cells and cancer – the promise and puzzles. Mol. Oncol. 4, 369–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellings, S.R. , Jensen, H.M. , Marcum, R.G. , 1975. An atlas of subgross pathology of the human breast with special reference to possible precancerous lesions. J. Natl. Cancer Inst. 55, 231–233. [PubMed] [Google Scholar]
- Westbury, C.B. , Reis-Filho, J.S. , Dexter, T. , Mahler-Araujo, B. , Fenwick, K. , Iravani, M. , Grigoriadis, A. , Parry, S. , Robertson, D. , Mackay, A. , Ashworth, A. , Yarnold, J.R. , Isacke, C.M. , 2009. Genome-wide transcriptomic profiling of microdissected human breast tissue reveals differential expression of KIT (c-Kit, CD117) and oestrogen receptor-alpha (ERalpha) in response to therapeutic radiation. J. Pathol. 219, 131–140. [DOI] [PubMed] [Google Scholar]
- Wulfkuhle, J.D. , Sgroi, D.C. , Krutzsch, H. , McLean, K. , McGarvey, K. , Knowlton, M. , Chen, S. , Shu, H. , Sahin, A. , Kurek, R. , Wallwiener, D. , Merino, M.J. , Petricoin, E.F. , Zhao, Y. , Steeg, P.S. , 2002. Proteomics of human breast ductal carcinoma in situ. Cancer Res. 62, 6740–6749. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The following are the Supplementary data related to this article:
Supplementary Figure 1 Immunohistochemical analysis of tandem tissue sections showing normal breast epithelium stained with antibodies against (A) CK15 and (B) CK19. In one single field of vision one can identify all four possible cellular phenotypes.
Supplementary Figure 2 Silver stained IEF 2D gel pattern of Normal 95 specimen. Identified spots are indicated with arrows and a number corresponding to their ID number on Supplementary Table 2.
Supplementary Figure 3 DRP3 protein identification by MALDI‐TOF‐MS.
Supplementary Figure 4 Morphology of mammary gland. View of a ductal tree present in a normal tissue specimen with epithelial cells stained with CK19. A schematic depiction of a TDLU is shown.
Supplementary Figure 5 Immunophenotyping of luminal cells in resting acini. Serial tissue sections were stained with antibodies against DRP3, CK15 and CK19, allowing identification of three major DRP3+ cellular phenotypes: (panel A) DRP3+ (sub‐figure A)/CK15+ (sub‐figure B)/CK19+ (sub‐figure C) with CK5+ or − (sub‐figure D), CK14− (sub‐figure E) and vimentin− (sub‐figure F), (panel B) DRP3+ (sub‐figure A)/CK15+ (sub‐figure B)/CK19− (sub‐figure C), and (panel C) DRP3+ (sub‐figure A)/CK15− (sub‐figure B)/CK19− (sub‐figure C). Red arrows indicate cells with these phenotypes.
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data


