Abstract
Smallpox vaccine is highly effective, inducing protective immunity to smallpox and diseases caused by related orthopoxviruses. Smallpox vaccine efficacy was historically defined by the appearance of a lesion or “take” at the vaccine site, which leaves behind a characteristic scar. Both the take and scar are readily recognizable and were used during the eradication effort to indicate successful vaccination and to categorize individuals as “protected.” However, the development of a typical vaccine take may not equate to the successful development of a robust, protective immune response. In this report, we examined two large (>1,000) cohorts of recipients of either Dryvax® or ACAM2000 using a testing and replication study design and identified subgroups of individuals who had documented vaccine takes, but who failed to develop robust neutralizing antibody titers. Examination of these individuals revealed that they had suboptimal cellular immune responses as well. Further testing indicated these low responders had a diminished innate antiviral gene expression pattern (IFNA1, CXCL10, CXCL11, OASL) upon in vitro stimulation with vaccinia virus, perhaps indicative of a dysregulated innate response. Our results suggest that poor activation of innate antiviral pathways may result in suboptimal immune responses to the smallpox vaccine. These genes and pathways may serve as suitable targets for adjuvants in new attenuated smallpox vaccines and/or effective antiviral therapy targets against poxvirus infections.
Keywords: Smallpox, Smallpox Vaccine, Vaccinia virus, Antibody Formation, Immunity, Cellular, Immunity, Humoral, Antibodies
Introduction
Prior to the intensive eradication effort in the 1960s and 1970s, variola major virus, the causative agent of smallpox, killed hundreds of millions of individuals and left survivors with serious sequela, including extensive scarring and blindness [1]. A less severe strain, variola minor, began circulating in America/Africa in the early 1900s.[1] Early efforts to curb the disease through variolation, and later vaccination, led to the eradication of smallpox in 1980 [1, 2]. The potential for variola virus to be used as a biological weapon [3], as well as outbreaks of zoonotic poxviruses in the Americas, Africa, and Asia [4], has led to a resurgence of research aimed at early detection, the development of next-generation vaccines, and effective therapeutic agents [5].
Smallpox vaccine elicits robust adaptive immune responses in the majority of recipients [6]. CD4+ and CD8+ T cell responses peak weeks after immunization, while antibody (Ab) responses can be seen as early as four days after vaccination [7], but take several weeks to reach peak titers [1, 8]. Cellular immune responses to poxviruses slowly decline over a period of decades and are believed to play a greater role in resolution of the initial infection than protection upon subsequent exposure [9, 10]. Humoral immunity to poxviruses is remarkably long-lived, with evidence that antibody levels can be maintained for 60–90 years [11, 12]. Antibodies to both forms of viral particles, intracellular mature virions (IMV) and extracellular enveloped virions (EEV), are required for optimal protection [13, 14]. Upon revaccination, anamnestic antibody responses are seen within four days of immunization.[15]
Traditionally, the presence of a “take” (i.e., the formation of the classic Jennerian pustule at the vaccination site) was used as a marker for vaccine efficacy and taken as evidence of protection against smallpox [1]. However, the evidence that a local reaction to smallpox vaccine is absolute proof of immunity relies on historical anecdote and population-level epidemiology, rather than individual scientific substantiation.
Historically, neutralizing antibody titer has also been used as a marker of protection for many vaccines. Studies have documented very low serum titers of neutralizing Abs in fatal cases of smallpox [1, 16], with a definite relationship between Ab titer and clinical severity of illness [16]. Neutralizing antibody titers have also been used as a prognostic indicator of disease progression [15]. Prospective studies have attempted to define a neutralizing antibody titer that provides protection against smallpox [17, 18]. These studies involved small numbers of subjects, but a titer of >1:32 is commonly used as an estimate of protective immunity. It is possible that higher levels of humoral immunity would be necessary to protect someone from a high-dose pathogen exposure due to an act of bioterrorism.
The two historical definitions of protection (“take” and neutralizing Ab titer > 1:32) are not directly related, and the localized reaction at the vaccination site is not necessarily indicative of a systemic humoral immune response [19]. In a cohort of > 1,000 smallpox vaccine recipients, we identified a small subset of individuals with a documented vaccine take, but extremely low neutralizing Ab titers. A similar population was identified in a second cohort of 1,058 smallpox vaccine recipients. These findings indicate that vaccine take may not necessarily be an accurate representation of vaccine-induced immunity. The purpose of this study was to better characterize the immune responses (both humoral and cellular) of these individuals. A better understanding of how and why some individuals do not develop robust immune responses following smallpox vaccination may provide insights into the drivers of poxvirus immunity, provide possible biomarkers of response, and provide insight into the design of novel vaccine candidates.
Materials and Methods
Study Subjects
Details on the Dryvax cohort (n=1,076) and the ACAM2000 cohort (n=994) have been previously reported [20, 21]. We secured Institutional Review Board approval from both the Mayo Clinic and the Naval Health Research Center (NHRC) for all procedures. Each participant gave written informed consent prior to enrollment. All subjects were between 18 and 40 years of age and had received a single documented dose of Dryvax® or ACAM2000® at least 30 days, and not more than four years, prior to recruitment.
Virus Stocks and Inactivation
The NYCBOH strain of vaccinia virus (purchased from ATCC; Manassas, VA), the vSC56 strain of vaccinia virus (obtained from Dr. Bernard Moss, NIAID), and the IHD-J strain of vaccinia virus (obtained from Dr. Don Gammon, University of Massachusetts) were grown, purified, and titered according to established protocols [22]. The NYCBOH strain was then inactivated using psoralen and UV light, resulting in a six log reduction in viral infectivity [23].
Neutralization assay
Humoral immunity was measured using a vSC56 vaccinia-based assay as previously described [20]. Sera were tested three times (Dryvax cohort) or twice (ACAM2000 cohort). The EEV plaque reduction neutralization test was performed as described [13] using an anti-L1R Ab instead of the 2D5 MAb, and with a 36-hour incubation period.
Cytokine responses
ELISA assays were used to detect levels of cytokine secretion upon in vitro vaccinia stimulation, and were optimized and performed as described [24]. IFNγ secreting cells were detected using both total PBMC IFNγ ELISPOT and CD8+ IFNγ ELISPOT kits as previously described [25].
Microarray gene expression
Gene expression analysis was conducted as previously described [26]. Briefly, PBMCs were stimulated for 18 hours with or without inactivated vaccinia virus (MOI = 0.5). RNA Protect Reagent was added to cell cultures. RNA was isolated using the RNeasy Plus kits (Qiagen, Valencia, CA) and quantified using Nanodrop spectrophotometer and labeling and hybridization of the Affymetrix HG-U133Plus_2 chips took place at Mayo Clinic’s Medical Genome Facility. Arrays were scanned using a GeneChip Scanner 2000, data were extracted, quality controlled, and used for expression analysis.
Statistical methods
Categorical data were summarized using both frequencies and percentages, while continuous variables were described with medians and inter-quartile ranges (IQRs). Immune response outcomes were utilized as previously reported [26, 27].
We compared distributions of demographic and clinical characteristics across normal and low responders using Fisher’s exact tests. Associations of humoral protection with cytokine secretion and ELISPOT measures were evaluated using linear regression models. Formal evaluations used linear mixed effects models to simultaneously model all six observations per subject with an unstructured within-person variance-covariance matrix. These repeated measures models compare differences between the two stimulation states within each subject among groups of individuals categorized by immune status (“normal” and “low” response). Analyses were adjusted for gender, which was the only demographic and clinical variable found to be significantly associated with immune response. Inverse cumulative normal (probit) data transformations were utilized to correct for the skewness present in the cytokine and ELISPOT data. ELISPOT and ELISA responses are reported as the median number and interquartile ranges of spot forming units per well and pg/ml, respectively. Negative ELISA values indicate that, on average, unstimulated secretion values were higher than stimulated values.
Gene expression data quality-control and normalization methods were previously reported [26]. Per-gene linear mixed effects models [28] were used to assess significance response to stimulation for the normal responders relative to the low responders, using a random effect to account for the within subject correlation. Genes were ranked by p-values and false-discovery rate [29]. The statistical packages R and SAS® were used for analyses.
Pathway analysis was performed with MetaCorre using a reference gene list after filtering out unexpressed and probe-sets with “Absent calls” using Affymetrix microarray suite version 5 software, as implemented in dChip. Probesets not present in at least half of the samples were removed from analysis. Of the 31,728 probe sets (reference list) that passed filtering, 589 had p ≤ 0.25 and were used as focus genes.
Results
Demographic differences between subjects
1,076 recipients of a single dose of Dryvax®, who had a documented vaccine “take,” were recruited and neutralizing antibody titers and a panel of cytokine ELISA and ELISPOT assays were performed to characterize vaccinia-specific immune responses. Each subject had detectable Ab titers above the background level of response found in unvaccinated subjects, confirming successful vaccination. A small percentage (1.8%; 19/1,076) of the study population had neutralizing Ab titers below 1:32. For this study, these 19 subjects are labeled as “low responders,” according to the definition of Mack et al. [18], and have a median ID50 (serum dilution inhibiting 50% of viral activity) of 27.3 (IQR:24.2–29.5). The remaining 1,052 subjects were classified as “normal responders” and had a median ID50 of 134.5 (IQR: 80.5–207.3). Demographic variables for the two groups were broadly similar (Table 1) in terms of ethnicity, race, and age. Consistent with a previous report indicating that females exhibit higher antibody responses, [30] only one of the low responders was female. Gender was used as a covariate in all subsequent models.
Table 1.
Demographics of Normal and Low Responder Sub-cohorts.
| Cohort | Dryvax cohort | ACAM cohort | |||||
|---|---|---|---|---|---|---|---|
| Demographic Category | Subcategory | Low Responders1 n = 19 | Normal Responders1 n = 1,052 | p-value | Low Responders1 n = 38 | Normal Responders1 n = 956 | p-value |
| Gender | % Female | 5.3% | 26.6% | 0.04 | 10.5% | 10.3% | 1.0 |
| Race | Asian/American Indian | 0.0% | 7.0% | 0.5 | 0% | 4.6% | 0.3 |
| African American | 10.5% | 17.5% | 23.7% | 11.8% | |||
| White | 57.9% | 53.1% | 73.7% | 79.1% | |||
| More than one race | 15.8% | 8.0% | 0.0% | 1.5% | |||
| Other/Do not know | 15.8% | 14.4% | 2.6% | 3.0% | |||
| Ethnicity | Not Hispanic | 63.2% | 74.6% | 0.2 | 78.9% | 81.7% | 0.7 |
| Hispanic | 26.3% | 21.3% | 21.1% | 17.2% | |||
| Do not know | 10.5% | 4.1% | 0% | 1.2% | |||
| Age (years) | 19–21 | 15.8% | 22.7% | 0.7 | 7.9% | 15.2% | 0.2 |
| 22–23 | 36.8% | 24.0% | 34.2% | 27.8% | |||
| 24–27 | 26.3% | 28.3% | 23.7% | 33.5% | |||
| 28–40 | 21.1% | 25.0% | 34.2% | 23.5% | |||
| Time Since Vaccination (months) | 1–8 | 36.8% | 24.0% | 0.3 | 0% | 6.5% | 0.1 |
| 9–14 | 10.5% | 25.6% | 73.7% | 59.5% | |||
| 15–33 | 31.6% | 25.9% | 21.1% | 18.8% | |||
| 34–48 | 21.1% | 24.5% | 5.3% | 15.2% | |||
The low responder group includes all subjects with neutralizing antibody titers less than 1:32. The remaining subjects are classified as normal responders
Fisher’s exact test. Statistically significant differences are shown in bold font.
The neutralizing antibody assay measures neutralizing antibody titers to IMV rather than EEV. Our initial hypothesis was that a more complete measure of humoral immunity would correlate better with vaccine take. Therefore, we performed plaque reduction neutralization assays using EEV stocks of vaccinia IHD-J pre-treated with anti-L1R Ab as described in the Methods section. Sixteen individuals from the low responder group (those with sufficient serum to test) had a mean EEV ID50 of 17.2. Only 7 of those 16 (43.8%) subjects had detectable anti-EEV Ab. In contrast, in a random sampling of 62 normal responders, 50/62 (80.6%) of these subjects had detectable anti-EEV Ab. This group had an average EEV ID50 of 85.8, which is five times higher than the low responder group (p=0.005, Fisher’s Test).
Markers of cellular immunity in low responders
Although vaccine safety is dependent on cellular immunity,[31] T cell responses have been shown to be unnecessary for protection against orthopoxvirus challenge [32]. However, vaccine take has been correlated with the development of cellular immunity, [33] leading us to question whether or not our putative low responders had normal cellular immunity. Consequently, we examined cytokine secretion and IFNγ ELISPOT responses to enumerate cytokine-producing T cells (Figure 1 and Table 2). Of the cytokines showing significant differences between the two groups, IFNα is produced during early innate responses to viral infection, while IFNγ, IL-2, and IL-4 are closely associated with T cell function. Our IFNγ ELISPOT (both the total PBMC response and the CD8+ T cell response) results corroborated our cytokine secretion data, showing significantly fewer IFNγ producing cells (See Table 2) in the low responders.
Figure 1. Distribution of Cytokine Responses in Normal and Low Responders.
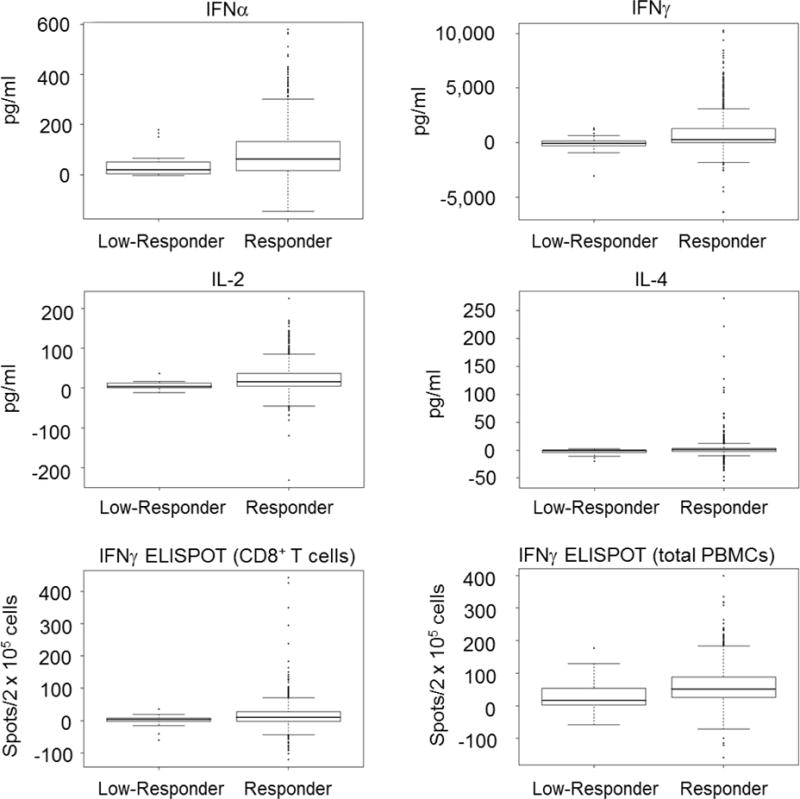
Box plots show secretion levels (in pg/ml) for the indicated cytokines. The thick horizontal bar inside the box represents the median level of cytokine secretion, the lower and upper limits of the box define the 25th and 75th percentile, respectively. The whiskers extending from the plots represent 1.5X the interquartile ranges with black dots representing data points outside of the 1.5X IQR limit. The bottom two panels show ELISPOT results (spot forming units per 2 × 105 cells) for both CD8+ T cells and total PBMCs.
Table 2.
Comparison of Vaccinia-Specific T Cell and Cytokine Responses in Sub-cohorts.
| Dryvax Cohort Low responder ELISPOT1 | Dryvax Cohort Normal responder ELISPOT1 | Dryvax Cohort p-value2 | ACAM Cohort Low responder ELISPOT 1 | ACAM Cohort Normal responder ELISPOT1 | ACAM Cohort p-value2 | |
|---|---|---|---|---|---|---|
| PBMCs | 16.0 (1, 57) | 52.0 (25, 89) | 0.007 | 7.0 (0, 30) | 18.0 (6, 37) | 0.615 |
| CD8+ T cells only | 4.0 (−2, 7) | 10.0 (−2, 27) | 0.027 | 1.0 (−1,3) | 4.0 (0,10) | 0.012 |
| Cytokine | Dryvax Cohort Low responder ELISA3 | Dryvax Cohort Normal responder ELISA3 | Dryvax Cohort p-value2 | ACAM Cohort Low responder ELISA3 | ACAM Cohort Normal responder ELISA3 | ACAM Cohort p-value 2 |
| IFNα | 20.8 (4, 58) | 63.5 (18, 133) | 0.007 | 206.9 (111, 279) | 179.7 (101, 282) | 0.4 |
| IFNβ | 1.8 (1, 5) | 1.6 (−3, 7) | 0.5 | NT | NT | – |
| IFNγ | −33.8 (−338, 269) | 294.4 (17.1, 1,273) | < 0.001 | 31.4 (−30, 612) | 292.2 (43, 986) | 0.05 |
| IL-1β | 45.2 (27, 172) | 50.8 (25, 122) | 0.9 | 36.3 (21, 159) | 65.1 (27, 159) | 0.3 |
| IL-2 | 4.2 (0, 13) | 16.4 (4, 37) | 0.003 | −0.9 (−9, 16) | 4.7 (−11, 35) | 0.06 |
| IL-4 | −1.3 (−5, 0) | 0.7 (−2, 4) | < 0.001 | NT | NT | – |
| IL-6 | 761.1 (461, 1488) | 1,076.1 (448, 1964) | 0.5 | 2,175 (731, 3,694) | 2,131 (543, 3,474) | 0.9 |
| IL-10 | −0.6 (−3, 2) | 2.8 (0, 11) | 0.2 | NT | NT | – |
| IL-12p40 | 38.2 (29, 88) | 62.9 (30, 122) | 0.3 | 79.2 (3, 176) | 93.1 (34, 210) | 0.7 |
| IL-12p70 | 2.2 (1, 8) | 3.0 (1, 6) | 0.6 | NT | NT | – |
| IL-18 | 0.1 (−3, 2) | 0.8 (−2, 3) | 0.1 | NT | NT | – |
| TNFα | 183.0 (93, 291) | 165.0 (91, 319) | 0.4 | 110.7 (43, 204) | 114.7 (50, 261) | 0.2 |
Bolded items are statistically significant at the 0.05 level.
ELISPOT response reported as the median number of spot forming units per well, with interquartile ranges displayed in parentheses.
p-value from repeated measures linear regression models adjusting for gender.
ELISA responses reported as pg/ml, with interquartile ranges displayed in parentheses. Negative values indicate that unstimulated secretion values were higher than stimulated values. NT = not tested.
Replication of study findings in an independent cohort
We examined an additional, separate cohort of 994 military personnel who had received the ACAM2000® smallpox vaccine (Table 1). Upon examination, this cohort also had a group of individuals (n=38) with neutralizing antibody titers below the 1:32 threshold. Upon comparison of this low responder group (with a median ID50 of 26.6) with the rest of their cohort (with a median ID50 of 120.9), we identified significantly fewer IFNγ producing CD8+ T cells (p=0.012) and lower IFNγ secretion levels (p=0.046). Although the ACAM2000 low responders exhibited the same trend for lower PBMC IFNγ ELISPOT counts, as well as reduced IL-1β, IL-2, IL-12p40 and TNFα secretion, the differences did reach statistical significance. All comparisons were from mixed models with an unstructured covariance matrix that adjusted for gender.
Evaluation of gene expression
Our next hypothesis was that the low responders had a deficiency in innate immune responses to vaccinia and that this deficiency interfered with the development of robust adaptive immunity following vaccination. A subset of the Dryvax cohort (those with the highest and lowest neutralizing Ab titers) had microarray data available that could be used to examine this question. [34] This subset included 11 of the low responders and 15 of the normal responders, providing an opportunity to search for differences in gene expression that might correlate with the lower immune responses. The data were first analyzed on an individual gene level with the most significant (p < 0.01) genes comprising chemokine genes (CXCL11, CXCL10, CCL2), antiviral genes (IFNA1, OASL), apoptosis genes (COP1, DDX17, NLRP1, TNFSF10), and genes involved in transcriptional and translational activity (EEF1A1, EIF3B, PNPT1, RPL3) all exhibiting greater upregulation (5 – 50%) in normal responders compared to low responders (Table 3).
Table 3.
Gene Expression Differences Between Normal and Low Responders.
| Gene Symbol1 | Gene Description | Delta (Low responder)2 | Delta (Normal responder) 3 | Interaction4 | P value5 |
|---|---|---|---|---|---|
| CXCL11 | chemokine (C-X-C motif) ligand 11 | 1.06 | 1.49 | 1.41 | 1.90E-06 |
| IFNA1 | interferon, alpha 1 | 1.18 | 1.43 | 1.20 | 0.001 |
| MALAT1 | metastasis associated lung adenocarcinoma transcript 1 (non-protein coding) | 1.06 | 0.80 | 0.75 | 0.002 |
| CXCL10 | chemokine (C-X-C motif) ligand 10 | 1.07 | 1.64 | 1.54 | 0.003 |
| OASL | 2′-5′-oligoadenylate synthetase-like | 1.15 | 1.41 | 1.23 | 0.003 |
| LDHB | lactate dehydrogenase B | 1.11 | 0.98 | 0.88 | 0.005 |
| COTL1 | coactosin-like 1 (Dictyostelium) | 1.15 | 0.93 | 0.81 | 0.006 |
| FOLR1 | folate receptor 1 (adult) | 1.35 | 0.95 | 0.70 | 0.006 |
| EEF1A1 | eukaryotic translation elongation factor 1 alpha 1 | 1.11 | 0.99 | 0.90 | 0.006 |
| LOC654007 | similar to Elongation factor 1-gamma (EF-1-gamma) (eEF-1B gamma) | 1.11 | 1.00 | 0.90 | 0.006 |
| CD38 | CD38 molecule | 1.03 | 1.23 | 1.19 | 0.006 |
| EEF1A1 | eukaryotic translation elongation factor 1 alpha 1 | 1.10 | 1.00 | 0.90 | 0.007 |
| RPL3 | ribosomal protein L3 | 1.13 | 1.00 | 0.88 | 0.009 |
| HSPB1 | heat shock 27kDa protein 1 | 0.89 | 1.02 | 1.14 | 0.009 |
HUGO gene name.
The mean fold change in gene expression between unstimulated and stimulated samples in the low responder group (subjects with neutralizing antibody titers ≤ 1:32).
The mean fold change in gene expression between unstimulated and stimulated samples in the normal responder group (subjects with neutralizing antibody titers > 1:32).
The interaction value reflects the change in gene expression in the normal responders relative to the change in gene expression in the low responders.
p-value from linear mixed effect models.
We performed a MetaCore pathway analysis and identified nine pathways with differential expression (Table 4), including Type I IFN signaling; proinflammatory (IL-1 and IL-6) pathways; CXCR4 signaling; IL-17 and apoptotic pathways. We also identified 10 GeneGo Disease categories with p < 0.05 (Table 4) that are primarily involved in the stimulation of innate immune pathways following viral infection.
Table 4.
Differential Pathway Expression in Normal and Low Responders.
| Pathway | p-value |
|---|---|
| Immune response – IL-17 signaling pathways | 0.0005 |
| Immune response – IFNα/β signaling pathway | 0.0006 |
| G-protein signaling – Proinsulin C peptide signaling | 0.001 |
| Immune response – IL-6 signaling pathway | 0.002 |
| Cytokine Production by Th17 cells in CF | 0.003 |
| Immune response – CXCR4 signaling via second messenger | 0.004 |
| Immune response – Signaling pathway mediated by IL6 and IL-1 | 0.003 |
| Glycolysis and gluconeogenesis | 0.003 |
| Apoptosis and survival – APRIL and BAFF signaling | 0.003 |
MetaCore pathway analysis performed as described in the Methods section.
Discussion
In our Dryvax® cohort (n=1,076), we identified 19 low responders with neutralizing Ab titers < 1:32, despite a documented vaccine “take.” We identified a similar group of low responders in recipients of ACAM2000®. Classically, this cutoff was used as a correlate of protection, and was developed from Mack et al. [18] The two different correlates of immune protection, vaccine take, and neutralizing antibody titers do not measure the same immune outcomes. In fact, relying solely on antibody titer may be misleading, as it ignores cellular immune processes that may contribute to protection from secondary exposure [35]. Our low responders also exhibited significantly lower levels of key cytokines involved in innate (IFNα) and cellular (INFγ, IL-2, IL-4) responses. It may be that these individuals have deficient innate responses to poxviruses that hamper optimal development of humoral and cellular immunity. The availability of a second cohort of smallpox vaccine recipients (n=994) allowed us to perform a replication study, wherein we identified an even larger group (n=38) of low responders subjects. As with the initial cohort, these low responders also had diminished CD8+ T cell ELISPOT responses and IFNγ secretion. Most of the other immune outcomes also exhibited a reduced response, but the reduction did not meet statistical significance (e.g., IL-2 secretion reduced by 50%: p=0.055). As noted earlier, we and others have described sex-based differences in humoral immunity to smallpox vaccination [30,36,37]. Sex differences have also been noted for numerous other vaccines as well [38, 39] and is an important factor that deserves to be studied in greater detail and taken into account when developing vaccination schedules and dosage recommendations, evaluating adverse event rates, and when planning clinical trials of new vaccines.
Our results (Table 4) indicate these low responders had a diminished innate antiviral gene expression pattern (IFNA1, CXCL10, CXCL11, OASL) upon in vitro viral stimulation, perhaps indicative of a dysregulated innate response. IP10 (CXCL10) levels have been shown to increase in primary smallpox vaccine responders 7–12 days after vaccination and were associated with fatigue and lymphadenopathy.[40] Vaccinia virus expressing the murine homolog of IP-10 (Crg-2) is attenuated in mouse models of infection,[41] which indicates that this chemokine mediates anti-poxvirus activity. In normal responders, CXCL11 exhibited a 49% increase in expression upon viral stimulation compared to a 6% increase in low responders (p = 1.9 × 10−6). Enhanced CXCL11 expression is indicative of more robust chemokine activity in response to viral infection; perhaps this is a result of the higher levels of IFNγ and IFNα seen in the normal responders. CD38, an activation marker integral to calcium signaling found on T cells, B cells, NK cells, and neutrophils [42], exhibited higher expression in normal responders (23% increase versus a 3% in low responders: p-value 0.006) and is also suggestive of greater immune activation after vaccinia virus stimulation, and may reflect the development of an early population of vaccinia-specific CCR5+CD38+ Th1 effector cells [43].
We also performed pathway analyses in order to gain insight into the functional effects of the differential gene expression patterns [44]. Pathways involved in IL-6, IL-17 signaling, IFNα/β signaling, CXCR4 and APRIL, and BAFF signaling also exhibited differential expression. Median IL-6 secretion in our low responders was only 70% of that seen in our normal responders; however, due to small subject numbers, this difference was not statistically significant. IL-17 induces production of proinflammatory mediators such as IL-6, TNFα, and MCP-1, and is involved in both resistance to poxvirus infection and viral clearance through NK and CD8+ cells [45, 46].
Perhaps most interesting was the differential expression of the IFNα/β pathway. This type I interferon pathway is crucial to immunity to poxviruses [47]. Type I IFNs promote robust humoral immunity through the induction of B cell activation, differentiation, isotype switching and the enhancement of immunologic memory and long-term antibody production [48], and contributes to CMI through the upregulation of costimulatory ligands and antigen presentation molecules on antigen presenting cells. The lower antibody titers and reduced markers of cellular immunity in our low responders may be a result of suboptimal activation of these critical immune pathways. Adjuvants or cytokine replacement therapies in conjunction with vaccination may serve to correct this low response phenotype. This is an especially attractive option with the safer, yet less immunogenic attenuate smallpox vaccines (MVA, LC16m8, NYVAC). Our data provide a tantalizing hint that several innate signaling and lymphocyte survival pathways are indeed responsible for the noted differences in immune outcomes among the normal and low responders to smallpox vaccine. Further studies focused on specific cell types isolated from these individuals may provide additional mechanistic details regarding the differential transcriptomic response contributing to the divergent vaccine outcome in these subjects.
From a population standpoint, only 1.8% (19/1,076) of the vaccinated individuals in the Dryvax cohort, and 4.3% (49/1,128) of the ACAM2000 cohort, had these extremely low humoral and cellular responses. Given the ease of visual identification of a vaccine “take” as an indicator of smallpox vaccine efficacy and the fact that the licensed smallpox vaccines were successful in eradicating smallpox, it is unlikely that this small number of low responders will alter public health policy regarding smallpox vaccination. Interestingly, the ACAM2000® subgroup was larger than and Dryvax low-responder group. Although the two vaccines contain the same strain of vaccinia virus and elicit a similar spectrum of immune response, they are not identical vaccines. In fact, ACAM2000 elicited lower neutralizing antibody titers and take rates than Dryvax during clinical trials of diluted vaccines [49, 50]. However, the results suggest that one must use caution in equating a vaccine take with “protection,” an area that warrants further investigation. When used outside of the healthy population that we studied, it is likely that larger “low-responder” rates would result. Furthermore, these results, obtained with vaccines based on the New York City Board of Health (NYCBOH) strain, may not necessarily hold true for other less immunogenic vaccine strains. In addition, some newer attenuated smallpox vaccines are injected and, therefore, do not elicit a vaccine take. Vaccine take can also be impaired in the presence of pre-existing poxvirus-specific Ab [51]. In these cases, relying on direct measures of humoral and/or cellular immunity may serve as better markers of vaccine immunogenicity and protection than simply relying on “take” rates.
The strengths of this study are the relatively large cohorts of vaccine recipients, the fairly comprehensive assessment of immune responses to smallpox vaccine (IFNg ELISPOT, secretion of Th1/Th2/pro-inflammatory cytokines, and humoral Ab measurements) and the available transcriptomic data that provided an opportunity to examine gene expression differences between low and normal responders. The availability of a second cohort in which we were able to identify a similar population allowed us to replicate our initial findings. The primary limitation is the small number of low responders. Even with cohorts of more than 1,000 subjects, we only identified 19 and 38 subjects in our respective cohorts. This small sample size affected our statistical power to detect biologically relevant effect sizes. Despite this, our data suggest that innate antiviral responses are critical for the development of robust adaptive immunity. Further investigation into the possible mechanisms involved in this extremely low immune responsiveness to smallpox vaccine may help elucidate important mechanisms of immune response and protection to poxviruses and may provide novel biomarkers for developing and testing new smallpox vaccines. In addition, it may provide quick, simple tests that predict non-response such that alternative vaccination schedules (increased dose, novel adjuvant use, prime-boost immunizations) could be employed to fully protect these individuals.
In conclusion, we identified a small population of individuals who develop inadequate humoral and cellular immune responses, despite the formation of the classical vaccine “take” following smallpox vaccination. These data highlight the incomplete understanding that we currently have regarding protective immunity to poxviruses. In order to better understand this low-responder phenomenon, we examined gene expression data to test the hypothesis that deficiencies in innate immune pathways might be responsible for the low responder phenotype. Our data indicate that the low-responder group has significantly diminished innate antiviral activity upon viral stimulation. This may correlate with the reduced adaptive immune responses to the vaccine. These genes or pathways may serve as useful early biomarkers capable of predicting response to the vaccine. Furthermore, they may provide targets for adjuvants that can be utilized in next-generation vaccines to appropriately enhance immune responses in vaccinated subjects. For example, vaccinia-stimulated IFNA expression is impaired in our low responders, a defect that may be rectified through the use of adjuvants targeting the STING pathway. STING has been shown to play an important role in antiviral responses to poxvirus infection [52]. Further investigation is likely to identify additional critical determinants of robust, durable immunity to the smallpox vaccine. Given the broad-spectrum, nonspecific nature of innate immunity, these finding may be applicable to other viral vaccines as well.
Acknowledgments
The authors thank the study participants and the research staff at the NHRC and Mayo Clinic that made this study possible – particularly, Drs. Meg Ryan and Kevin Russell. The authors also wish to recognize both Dave Watson and Megan O’Byrne for their statistical programming and analytical support. The microarray dataset is available through NIAID’s Immunology Database and Analysis Portal (ImmPort: https://immport.niaid.nih.gov).
Funding Statement
This work was supported by the NIH through the NIAID Population Genetics Analysis Program Contract No.HHSN266200400065C and Contract No. HHSN272201000025C, and by the National Center for Research Resources grant 1 UL1 RR024150-01. The funding agencies had no role in study design, data collection and analysis, decision to publish, or preparation of this manuscript.
Footnotes
Conflict of Interest/Disclosures
Dr. Poland is the chair of a Safety Evaluation Committee for novel investigational vaccine trials being conducted by Merck Research Laboratories. Dr. Poland offers consultative advice on vaccine development to Merck & Co. Inc., CSL Biotherapies, Avianax, Dynavax, Novartis Vaccines and Therapeutics, Emergent Biosolutions, Adjuvance, Microdermis, Seqirus, NewLink, Protein Sciences, GSK Vaccines, and Sanofi Pasteur. Dr. Kennedy has received grant funding from Merck Research Laboratories to study mumps vaccine responses. Drs. Poland and Ovsyannikova hold two patents related to vaccinia and measles peptide research. Dr. Jacobson is a member of a Safety Review Committee and a Data Monitoring Committee for several vaccine studies conducted by Merck Research Laboratories. The other authors do not have any conflict of interest. Dr. Poland’s consultant activities and this research have been reviewed by the Mayo Clinic Conflict of Interest Review Board and are conducted in compliance with Mayo Clinic Conflict of Interest policies.
References
- 1.Fenner F. Smallpox and its eradication. Geneva: World Health Organization; 1988. [Google Scholar]
- 2.Jenner E. An inquiry into the causes and effects of the variolae vaccinae, a disease discovered in some of the western counties of England, particularly Gloucestershire, and known by the name of the cow pox. London: Law; p. 1798. [Google Scholar]
- 3.Henderson DA, Inglesby TV, Bartlett JG, Ascher MS, Eitzen E, Jahrling PB, et al. Smallpox as a biological weapon: medical and public health management. Working Group on Civilian Biodefense. Jama. 1999 Jun 9;281(22):2127–37. doi: 10.1001/jama.281.22.2127. [DOI] [PubMed] [Google Scholar]
- 4.Frey SE, Belshe RB. Poxvirus zoonoses–putting pocks into context. The New England journal of medicine. 2004 Jan 22;350(4):324–7. doi: 10.1056/NEJMp038208. [DOI] [PubMed] [Google Scholar]
- 5.Kennedy RB, Ovsyannikova I, Poland GA. Smallpox vaccines for biodefense. Vaccine. 2009 Nov 5;27(Suppl 4):D73–9. doi: 10.1016/j.vaccine.2009.07.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kennedy RB, Ovsyannikova IG, Jacobson RM, Poland GA. The immunology of smallpox vaccines. Curr Opin Immunol. 2009 Jun;21(3):314–20. doi: 10.1016/j.coi.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moyron-Quiroz JE, McCausland MM, Kageyama R, Sette A, Crotty S. The smallpox vaccine induces an early neutralizing IgM response. Vaccine. 2009 Oct;:9. doi: 10.1016/j.vaccine.2009.09.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moller-Larsen A, Haahr S. Humoral and cell-mediated immune responses in humans before and after revaccination with vaccinia virus. Infection and immunity. 1978 Jan;19(1):34–9. doi: 10.1128/iai.19.1.34-39.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Amanna IJ, Slifka MK, Crotty S. Immunity and immunological memory following smallpox vaccination. Immunol Rev. 2006 Jun;211:320–37. doi: 10.1111/j.0105-2896.2006.00392.x. [DOI] [PubMed] [Google Scholar]
- 10.Combadiere B, Boissonnas A, Carcelain G, Lefranc E, Samri A, Bricaire F, et al. Distinct time effects of vaccination on long-term proliferative and IFN-gamma-producing T cell memory to smallpox in humans. J Exp Med. 2004 Jun 7;199(11):1585–93. doi: 10.1084/jem.20032083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Belshe RB, Newman FK, Frey SE, Couch RB, Treanor JJ, Tacket CO, et al. Dose-dependent neutralizing-antibody responses to vaccinia. J Infect Dis. 2004 Feb 1;189(3):493–7. doi: 10.1086/380906. [DOI] [PubMed] [Google Scholar]
- 12.Crotty S, Felgner P, Davies H, Glidewell J, Villarreal L, Ahmed R. Cutting edge: long-term B cell memory in humans after smallpox vaccination. J Immunol. 2003 Nov 15;171(10):4969–73. doi: 10.4049/jimmunol.171.10.4969. [DOI] [PubMed] [Google Scholar]
- 13.Law M, Smith GL. Antibody neutralization of the extracellular enveloped form of vaccinia virus. Virology. 2001 Feb 1;280(1):132–42. doi: 10.1006/viro.2000.0750. [DOI] [PubMed] [Google Scholar]
- 14.Kaufman DR, Goudsmit J, Holterman L, Ewald BA, Denholtz M, Devoy C, et al. Differential antigen requirements for protection against systemic and intranasal vaccinia virus challenges in mice. J Virol. 2008 Jul;82(14):6829–37. doi: 10.1128/JVI.00353-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Panchanathan V, Chaudhri G, Karupiah G. Correlates of protective immunity in poxvirus infection: where does antibody stand? Immunology and cell biology. 2008 Jan;86(1):80–6. doi: 10.1038/sj.icb.7100118. [DOI] [PubMed] [Google Scholar]
- 16.Sarkar JK, Mitra AC, Chakravarty MS. Relationship of clinical severity, antibody level, and previous vaccination state in smallpox. Transactions of the Royal Society of Tropical Medicine and Hygiene. 1972;66(5):789–92. doi: 10.1016/0035-9203(72)90095-8. [DOI] [PubMed] [Google Scholar]
- 17.Sarkar JK, Mitra AC, Mukherjee MK. The minimum protective level of antibodies in smallpox. Bull World Health Organ. 1975;52(3):307–11. [PMC free article] [PubMed] [Google Scholar]
- 18.Mack TM, Noble J, Jr, Thomas DB. A prospective study of serum antibody and protection against smallpox. Am J Trop Med Hyg. 1972 Mar;21(2):214–8. doi: 10.4269/ajtmh.1972.21.214. [DOI] [PubMed] [Google Scholar]
- 19.Lublin-Tennenbaum T, Katzenelson E, el-Ad B, Katz E. Correlation between cutaneous reaction in vaccinees immunized against smallpox and antibody titer determined by plaque neutralization test and ELISA. Viral immunology. 1990 Spring;3(1):19–25. doi: 10.1089/vim.1990.3.19. [DOI] [PubMed] [Google Scholar]
- 20.Kennedy R, Pankratz VS, Swanson E, Watson D, Golding H, Poland GA. Statistical approach to estimate vaccinia-specific neutralizing antibody titers using a high-throughput assay. Clinical and vaccine immunology : CVI. 2009 Aug;16(8):1105–12. doi: 10.1128/CVI.00109-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ovsyannikova IG, Jacobson RM, Ryan JE, Vierkant RA, Pankratz VS, Jacobsen SJ, et al. HLA class II alleles and measles virus-specific cytokine immune response following two doses of measles vaccine. Immunogenetics. 2005 Feb;56(11):798–807. doi: 10.1007/s00251-004-0756-0. [DOI] [PubMed] [Google Scholar]
- 22.Earl PL, Moss B, Wyatt LS, Carroll MW. Generation of recombinant vaccinia viruses. In: Coligan John E, et al., editors. Current protocols in protein science / editorial board. 2001. May, Chapter 5:Unit5 13. [DOI] [PubMed] [Google Scholar]
- 23.Tsung K, Yim JH, Marti W, Buller RM, Norton JA. Gene expression and cytopathic effect of vaccinia virus inactivated by psoralen and long-wave UV light. J Virol. 1996 Jan;70(1):165–71. doi: 10.1128/jvi.70.1.165-171.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ryan JE, Dhiman N, Ovsyannikova IG, Vierkant RA, Pankratz VS, Poland GA. Response surface methodology to determine optimal cytokine responses in human peripheral blood mononuclear cells after smallpox vaccination. J Immunol Methods. 2009 Feb 28;341(1–2):97–105. doi: 10.1016/j.jim.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haralambieva IH, Ovsyannikova IG, Kennedy RB, Vierkant RA, Pankratz VS, Jacobson RM, et al. Associations between single nucleotide polymorphisms and haplotypes in cytokine and cytokine receptor genes and immunity to measles vaccination. Vaccine. 2011 Oct 19;29(45):7883–95. doi: 10.1016/j.vaccine.2011.08.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oberg AL, Dhiman N, Grill DE, Ryan JE, Kennedy RB, Poland GA. Optimizing high dimensional gene expression studies for immune response following smallpox vaccination using Taqman(R) low density immune arrays. Journal of immunological methods. 2011 Mar 7;366(1–2):69–78. doi: 10.1016/j.jim.2011.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kennedy RB, Ovsyannikova IG, Pankratz VS, Haralambieva IH, Vierkant RA, Poland GA. Genome-wide analysis of polymorphisms associated with cytokine responses in smallpox vaccine recipients. Hum Genet. 2012 Sep;131(9):1403–21. doi: 10.1007/s00439-012-1174-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oberg AL, Mahoney DW. Linear Mixed Effects Models. In: Ambrosius WT, editor. Methods in Molecular Biology. Totowa, NJ: Humana Press; 2009. pp. 213–34. [DOI] [PubMed] [Google Scholar]
- 29.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society B. 1995;57:289–300. [Google Scholar]
- 30.Kennedy RB, Ovsyannikova IG, Pankratz VS, Vierkant RA, Jacobson RM, Ryan MA, et al. Gender effects on humoral immune responses to smallpox vaccine. Vaccine. 2009 Feb;:4. doi: 10.1016/j.vaccine.2009.01.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gordon SN, Cecchinato V, Andresen V, Heraud JM, Hryniewicz A, Parks RW, et al. Smallpox vaccine safety is dependent on T cells and not B cells. The Journal of infectious diseases. 2011 Apr 15;203(8):1043–53. doi: 10.1093/infdis/jiq162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Edghill-Smith Y, Golding H, Manischewitz J, King LR, Scott D, Bray M, et al. Smallpox vaccine-induced antibodies are necessary and sufficient for protection against monkeypox virus. Nat Med. 2005 Jul;11(7):740–7. doi: 10.1038/nm1261. [DOI] [PubMed] [Google Scholar]
- 33.Frey SE, Newman FK, Cruz J, Shelton WB, Tennant JM, Polach T, et al. Dose-related effects of smallpox vaccine. N Engl J Med. 2002 Apr 25;346(17):1275–80. doi: 10.1056/NEJMoa013431. [DOI] [PubMed] [Google Scholar]
- 34.Haralambieva IH, Oberg AL, Dhiman N, Ovsyannikova IG, Kennedy RB, Grill DE, et al. High-dimensional gene expression profiling studies in high and low responders to primary smallpox vaccination. The Journal of infectious diseases. 2012 Nov 15;206(10):1512–20. doi: 10.1093/infdis/jis546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kennedy JS, Frey SE, Yan L, Rothman AL, Cruz J, Newman FK, et al. Induction of human T cell-mediated immune responses after primary and secondary smallpox vaccination. J Infect Dis. 2004 Oct 1;190(7):1286–94. doi: 10.1086/423848. [DOI] [PubMed] [Google Scholar]
- 36.Haralambieva IH, Ovsyannikova IG, Kennedy RB, Larrabee BR, Shane Pankratz V, Poland GA. Race and sex-based differences in cytokine immune responses to smallpox vaccine in healthy individuals. Human immunology. 2013 Oct;74(10):1263–6. doi: 10.1016/j.humimm.2013.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Troy JD, Hill HR, Ewell MG, Frey SE. Sex difference in immune response to vaccination: A participant-level meta-analysis of randomized trials of IMVAMUNE smallpox vaccine. Vaccine. 2015 Oct 5;33(41):5425–31. doi: 10.1016/j.vaccine.2015.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Voysey M, Barker CI, Snape MD, Kelly DF, Truck J, Pollard AJ. Sex-dependent immune responses to infant vaccination: an individual participant data meta-analysis of antibody and memory B cells. Vaccine. 2016 Mar 29;34(14):1657–64. doi: 10.1016/j.vaccine.2016.02.036. [DOI] [PubMed] [Google Scholar]
- 39.Fink AL, Klein SL. Sex and Gender Impact Immune Responses to Vaccines Among the Elderly. Physiology (Bethesda) 2015 Nov;30(6):408–16. doi: 10.1152/physiol.00035.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cohen JI, Hohman P, Fulton R, Turk SP, Qin J, Thatcher K, et al. Kinetics of serum cytokines after primary or repeat vaccination with the smallpox vaccine. The Journal of infectious diseases. 2010 Apr 15;201(8):1183–91. doi: 10.1086/651453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mahalingam S, Farber JM, Karupiah G. The interferon-inducible chemokines MuMig and Crg-2 exhibit antiviral activity In vivo. Journal of virology. 1999 Feb;73(2):1479–91. doi: 10.1128/jvi.73.2.1479-1491.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Malavasi F, Deaglio S, Funaro A, Ferrero E, Horenstein AL, Ortolan E, et al. Evolution and function of the ADP ribosyl cyclase/CD38 gene family in physiology and pathology. Physiological reviews. 2008 Jul;88(3):841–86. doi: 10.1152/physrev.00035.2007. [DOI] [PubMed] [Google Scholar]
- 43.Zaunders JJ, Dyer WB, Munier ML, Ip S, Liu J, Amyes E, et al. CD127+CCR5+CD38+++ CD4+ Th1 effector cells are an early component of the primary immune response to vaccinia virus and precede development of interleukin-2+ memory CD4+ T cells. J Virol. 2006 Oct;80(20):10151–61. doi: 10.1128/JVI.02670-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nature genetics. 2003 Jul;34(3):267–73. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 45.Kohyama S, Ohno S, Isoda A, Moriya O, Belladonna ML, Hayashi H, et al. IL-23 enhances host defense against vaccinia virus infection via a mechanism partly involving IL-17. J Immunol. 2007 Sep 15;179(6):3917–25. doi: 10.4049/jimmunol.179.6.3917. [DOI] [PubMed] [Google Scholar]
- 46.Yeh N, Glosson NL, Wang N, Guindon L, McKinley C, Hamada H, et al. Tc17 cells are capable of mediating immunity to vaccinia virus by acquisition of a cytotoxic phenotype. J Immunol. 2010 Aug 15;185(4):2089–98. doi: 10.4049/jimmunol.1000818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Perdiguero B, Esteban M. The interferon system and vaccinia virus evasion mechanisms. J Interferon Cytokine Res. 2009 Sep;29(9):581–98. doi: 10.1089/jir.2009.0073. [DOI] [PubMed] [Google Scholar]
- 48.Le Bon A, Schiavoni G, D’Agostino G, Gresser I, Belardelli F, Tough DF. Type i interferons potently enhance humoral immunity and can promote isotype switching by stimulating dendritic cells in vivo. Immunity. 2001 Apr;14(4):461–70. doi: 10.1016/s1074-7613(01)00126-1. [DOI] [PubMed] [Google Scholar]
- 49.Handley L, Buller RM, Frey SE, Bellone C, Parker S. The new ACAM2000 vaccine and other therapies to control orthopoxvirus outbreaks and bioterror attacks. Expert review of vaccines. 2009 Jul;8(7):841–50. doi: 10.1586/erv.09.55. [DOI] [PubMed] [Google Scholar]
- 50.Artenstein AW, Johnson C, Marbury TC, Morrison D, Blum PS, Kemp T, et al. A novel, cell culture-derived smallpox vaccine in vaccinia-naive adults. Vaccine. 2005 May 9;23(25):3301–9. doi: 10.1016/j.vaccine.2005.01.079. [DOI] [PubMed] [Google Scholar]
- 51.Tan X, Chun S, Pablo J, Felgner P, Liang X, Davies DH. Failure of the smallpox vaccine to develop a skin lesion in vaccinia virus-naive individuals is related to differences in antibody profiles before vaccination, not after. Clinical and vaccine immunology : CVI. 2012 Mar;19(3):418–28. doi: 10.1128/CVI.05521-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dai P, Wang W, Cao H, Avogadri F, Dai L, Drexler I, et al. Modified vaccinia virus Ankara triggers type I IFN production in murine conventional dendritic cells via a cGAS/STING-mediated cytosolic DNA-sensing pathway. PLoS pathogens. 2014 Apr;10(4):e1003989. doi: 10.1371/journal.ppat.1003989. [DOI] [PMC free article] [PubMed] [Google Scholar]


