Abstract
It is known that noncationic porphyrins such as Photofrin (PF) are effective in mediating antimicrobial photodynamic inactivation (aPDI) of Gram-positive bacteria or fungi. However, the aPDI activity of PF against Gram-negative bacteria is accepted to be extremely low. Here we report that the nontoxic inorganic salt potassium iodide (KI) at a concentration of 100 mM when added to microbial cells (108/mL) + PF (10 μM hematoporphyrin equivalent) + 415 nm light (10 J/cm2) can eradicate (>6 log killing) five different Gram-negative species (Escherichia coli, Pseudomonas aeruginosa, Klebsiella pneumoniae, Proteus mirabilis, and Acinetobacter baumannii), whereas no killing was obtained without KI. The mechanism of action appears to be the generation of microbicidal molecular iodine (I2/I3−) as shown by comparable bacterial killing when cells were added to the mixture after completion of illumination and light-dependent generation of iodine as detected by the formation of the starch complex. Gram-positive methicillin-resistant Staphylococcus aureus is much more sensitive to aPDI (200–500 nM PF), and in this case potentiation by KI may be mediated mainly by short-lived iodine reactive species. The fungal yeast Candida albicans displayed intermediate sensitivity to PF-aPDI, and killing was also potentiated by KI. The reaction mechanism occurs via singlet oxygen (1O2). KI quenched 1O2 luminescence (1270 nm) at a rate constant of 9.2 × 105 M−1 s−1. Oxygen consumption was increased when PF was illuminated in the presence of KI. Hydrogen peroxide but not superoxide was generated from illuminated PF in the presence of KI. Sodium azide completely inhibited the killing of E. coli with PF/blue light + KI.
Keywords: antimicrobial photodynamic inactivation, Photofrin, Gram-negative bacteria, potassium iodide, reactive iodine species, singlet oxygen
Graphical abstract
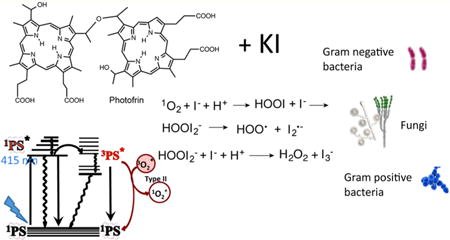
Antimicrobial photodynamic inactivation (aPDI) is a new approach to killing pathogenic microbes such as bacteria and fungi1 and is gaining popularity in the present age of antibiotic resistance.2 aPDI involves the use of dyes called photosensitizers (PS) in combination with visible light to produce reactive oxygen species (ROS) that kill the microbial cells.3 The photochemical mechanism is broadly divided according to the type of ROS generated: type 1 (superoxide, hydrogen peroxide, and hydroxyl radicals) and type II (singlet oxygen).4
It has long been known that anionic or neutrally charged PS have little to no activity in mediating aPDI of Gram-negative bacteria,5 unless methods are employed to permeabilize the outer membrane,6 for instance, by combination with polymyxin nonapeptide7 or ethylenediaminetetraacetic acid (EDTA).8 This is especially true for PS that mainly operate by the type II photochemical mechanism; that is, the excited triplet state PS undergo an energy transfer reaction with ground state triplet oxygen to produce excited state singlet oxygen.9 The explanation that is normally given is that the PS needs to bind strongly to the negatively charged Gram-negative outer membrane, because singlet oxygen has such a short diffusion distance.10 This strong binding between bacteria and PS can best be achieved with PS that have a pronounced cationic charge, but not with neutral/anionic PS such as Photofrin (PF).11 Moreover, there is evidence12 that cationic PS can also penetrate into the Gram-negative cells by the “self-promoted uptake pathway”.13 Due to the short diffusion length of singlet oxygen, it is hypothesized that extracellular generated singlet oxygen cannot easily cause fatal oxidative damage to Gram-negative cells from the outside in.14
Our laboratory has published papers showing that aPDI using several different PS can be potentiated by the addition of the nontoxic inorganic salt potassium iodide. The well-known cationic phenothiazium salt, methylene blue,15 cationic fullerenes,16 and UVA-mediated titanium dioxide photocatalysis17 all demonstrated significant potentiation of killing (up to 6 log extra). We generally used concentrations of KI up to 10 mM15 but recently used concentrations as high as 100 mM in studies with TiO2/UVA.17
We had previously discovered that aPDI using phenothiazium salts could be paradoxically potentiated using sodium azide.18,19 This potentiation was termed “paradoxical” because sodium azide is widely employed as a physical quencher of singlet oxygen and has previously been employed to inhibit aPDI.20 The hypothesis to explain this observation was that an electron transfer (via type 1 photochemistry) was important for potentiation by azide. Indeed, azide radicals were detected by electron spin resonance spin-trapping and could also be produced in the absence of oxygen.18 Addition of azide was proposed as a test to distinguish between type 2 (inhibition) and type 1 (potentiation) photochemical mechanisms in aPDI.21
Partly as a result of our studies with azide, we had formed the idea that potentiation of aPDI by the addition of iodide would also be an electron transfer process from I− to the excited state PS, giving iodine radicals (I•/I2•−) that carried out the antimicrobial killing. In the case of TiO2/UVA/KI the mechanism of potentiation was shown to include an additional two-electron oxidation of I− to give hypoiodite as well as a one-electron oxidation to give iodine.17
However, there is a study in the literature22 in which a chemical assay using KI (with the addition of ammonium molybdate as a catalyst) was proposed as an iodometric assay for PDT activity. Because our study with TiO2 had shown that a higher concentration of KI (100 mM) gave a better effect than 10 mM KI, we decided to test whether KI could potentiate aPDI, mediated by a traditional type 2 PS such as the clinically approved PS, Photofrin.
Results and Discussion
Potassium Iodide Potentiates aPDI Using Photofrin against Gram-Negative Bacteria
We initially tested the Gram-negative bacterial species Escherichia coli with 10 μM Photofrin and 10 J/cm2 of 415 nm blue light. We added different concentrations of KI (1, 10, 25, 50, 100 mM) into the solution during the illumination. There was no detectable killing without KI (data not shown) or indeed with 10 or 25 mM KI. However, to our surprise we found that the addition of 50 mM KI led to eradication (>6 log of killing) (Figure 1). To explore the generality of this finding, we repeated the experiment with four different Gram-negative species (Acinetobacter baumanii, Klebsiella pneumoniae, Proteus mirabilis, and Pseudomonas aeruginosa). As can be seen in Figure 1 all five species were eradicated by the addition of KI at concentrations up to 100 mM. There were minor differences in susceptibility, with the order being A. baumannii > E. coli > K. pneumoniae > P. mirabilis ≈ P. aeruginosa.
Figure 1.
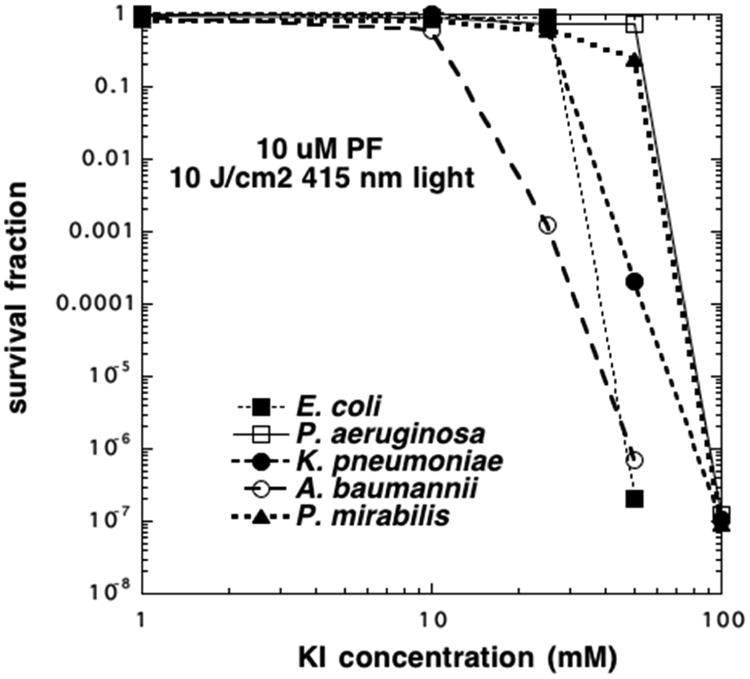
KI potentiates Photofrin-mediated aPDI of Gram-negative bacteria. Bacteria (108 cells/mL were incubated with 10 μM PF and exposed to 10 J/cm2 blue light in the presence of different concentrations of KI.
Concerning the possible toxicity of the KI, we had previously shown that KI concentrations up to 100 mM had no toxic effects on bacterial viability. We now tested concentrations much higher than 100 mM and found that KI concentrations up to 1.6 M (16 times higher) had no effect on bacterial viability. Only saturated KI (8.4 M) for 30 min killed all of the bacterial cells, presumably by osmotic stress (data not shown).
The bacterial suspension changed color during the light delivery period, and this observation led us to hypothesize that molecular iodine was generated during the aPDI reaction. If that was the case, the iodine generated could have been mainly/entirely responsible for the bacterial killing. This hypothesis was tested by adding the cells to the solution of PF and KI after the end of the illumination period and comparing the killing produced with that obtained when the cells were present during the illumination (Figure 2). As can be seen, eradication was achieved using both 50 and 100 mM KI in both cases. There was even some killing when cells were added after light using 25 mM KI.
Figure 2.
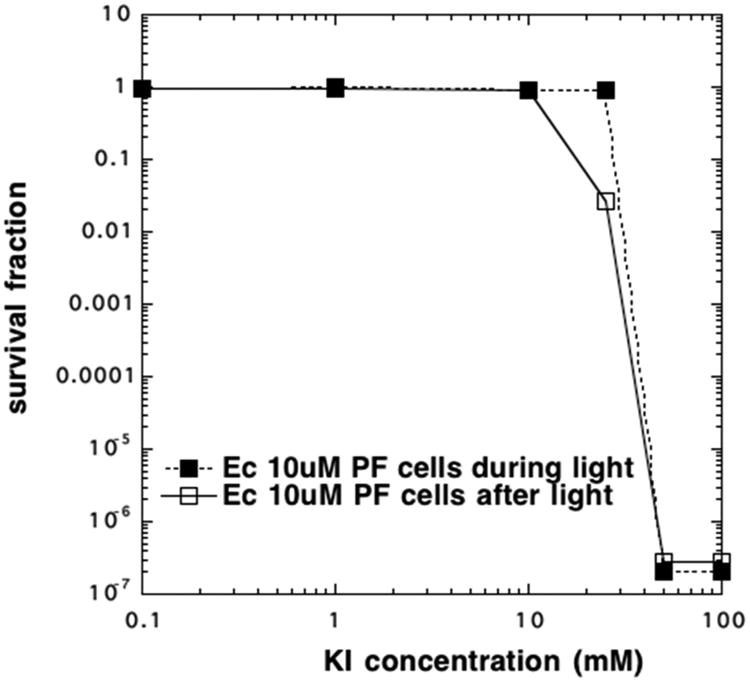
Comparison of aPDI killing with cells present and cells added after light. PF (10 μM) was exposed to 10 J/cm2 blue light in the presence of different concentrations of KI. E. coli cells (108/mL) were either present during light or added after light.
Effect of KI on aPDI against Gram-Positive Bacteria Using Photofrin
We next looked at Gram-positive bacteria, methicillin-resistant Staphylococcus aureus (MRSA). As Gram-positive species are much more sensitive to aPDI using all different types of PS, we selected a concentration of 200 nM PF (HP equivalent). In this case because PF binds to the bacterial cells, we also had a group with a “wash”, a centrifugation step between incubation of the cells with PF, the addition of KI solution, and subsequent illumination. Figure 3 shows the curves obtained with the addition of increasing concentrations of KI. As can be seen with a very low KI concentration (effectively zero), there was about 1.5 log of killing just with aPDI alone. There was about 1 log of potentiation with 10 mM KI and no wash and >2 log potentiation with 25 mM KI. There was significantly less potentiation with KI after a wash. When cells were added after light, there was no killing seen until the KI reached 100 mM, when eradication was obtained.
Figure 3.
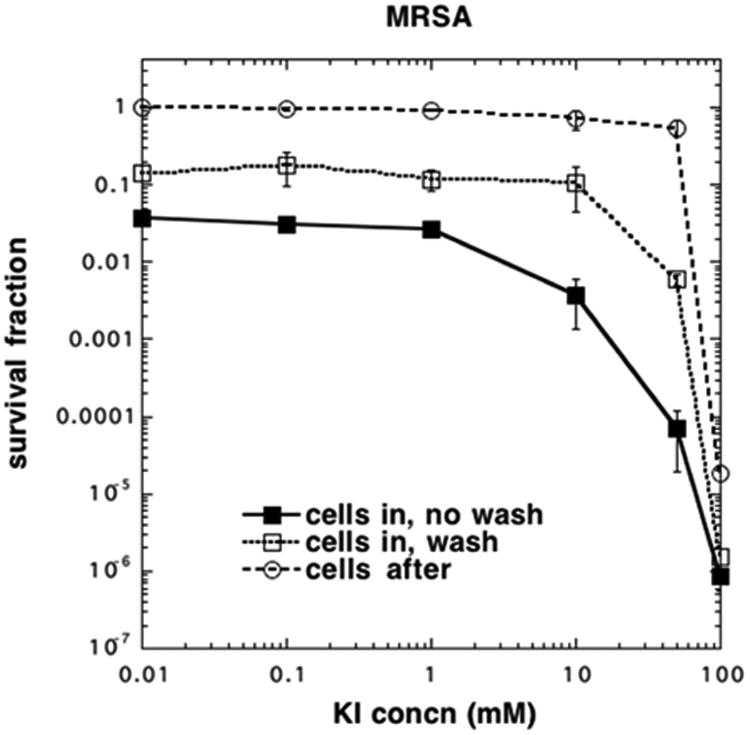
KI potentiates PF-mediated aPDI of Gram-positive MRSA. Cells (108/mL) were incubated with PF (200 nM) and then either centrifuged (wash) and resuspended or not (no wash), followed by addition of KI and illumination. In the “cells after” group, cells were added after completion of illumination.
Effect of KI on aPDI against Fungal Yeast Using Photofrin
Candida albicans was killed by Photofrin (10 μM) aPDI alone, with 2.5 log of killing without a wash, and there was still 2 log of killing remaining after a wash (see Figure 4). Potentiation of killing by addition of KI began to be evident at 1 mM with more additional killing with increasing KI concentrations and 1–2 log additional killing at 50 mM KI. When cells were added after light, eradication was obtained with 100 mM KI and a little killing (1 log) at 50 mM KI.
Figure 4.
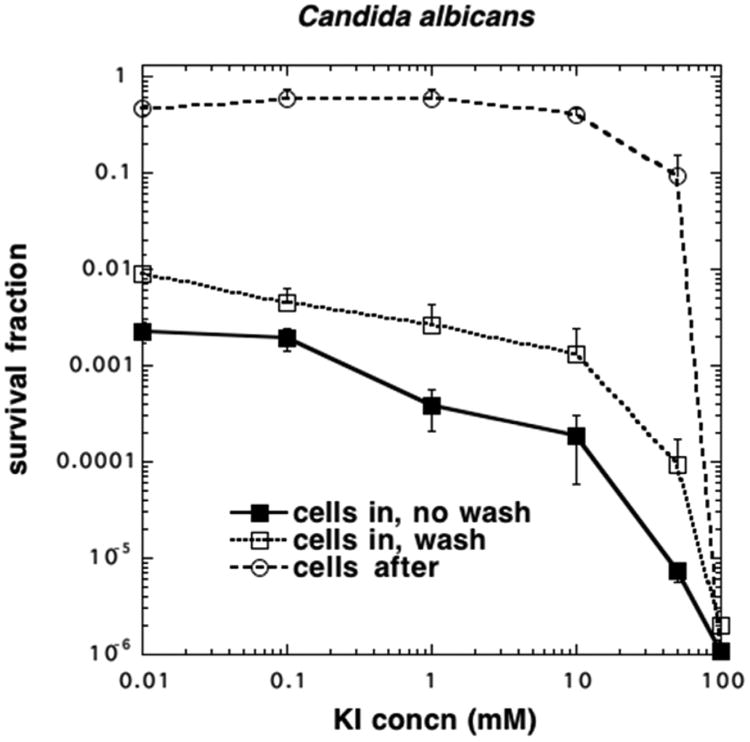
KI potentiates PF-mediated aPDI of fungal yeast C. albicans. Cells (107/mL) were incubated with PF (10 μM) and then either centrifuged (wash) and resuspended or not (no wash), followed by addition of KI and illumination. In the “cells after” group, cells were added after completion of illumination.
Mechanistic Studies
We formed the hypothesis that ROS produced during the photodynamic reaction were able to oxidize the iodide anion to molecular iodine. This was demonstrated by use of a starch–iodine assay as shown in Figure 5A. There was a linear increase in absorbance at 610 nm (starch iodine complex) with increasing fluences of blue light.
Figure 5.
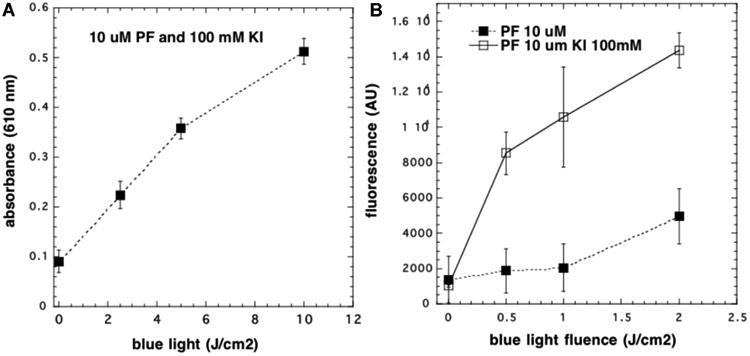
Formation of starch–iodine and hydrogen peroxide in solution (no cells): (A) PF (10 μM) and KI (100 mM) were illuminated with increasing fluence of 415 nm light and aliquots withdrawn and added to starch solution; (B) same as (A) but included PF without KI, and aliquots were withdrawn and added to Amplex Red reagent.
If the ROS that oxidized iodide was in fact singlet oxygen, then it would be expected that the oxygen would be chemically reduced to initially form superoxide and eventually would form hydrogen peroxide. Therefore, we expected more H2O2 to be formed in the presence of KI compared to its absence.
Figure 5B shows a linear increase in fluorescence from the Amplex Red assay with increasing fluences of blue light added to a mixture of PF (10 μM) and KI (100 mM), whereas in the absence of KI there was only a slight increase in hydrogen peroxide. However, when we attempted to measure superoxide using the nitroblue tetrazolium assay, no production of superoxide was observed either in the presence or in the absence of KI (data not shown).
To further corroborate the role of singlet oxygen in iodide oxidation, we asked whether KI could quench the luminescence at 1270 nm produced as singlet oxygen deactivates to its ground triplet state. The experiment was carried out in D2O to extend the lifetime of 1O2 and facilitate its measurement. Figure 6A illustrates this quenching with 10 mM KI and 100 mM KI. This experiment allowed us to calculate the rate constant for the reaction between singlet oxygen and iodide anion to be 9.2 × 105 M−1 s−1 as shown in Figure 6B. Because oxygen is being consumed during the reaction between singlet oxygen and iodide anion, it should be possible to measure this with electron paramagnetic resonance (EPR) oximetry using 4-hydro-3-carbamoyl-2,2,5,5-tetraperdeuteromethylpyrrolin-1-oxyl (mHCTPO) at a concentration of 0.1 mM as a dissolved oxygen-sensitive spin probe. Figure 6C shows that there was no detectable oxygen consumption with 10 mM KI, but at 50 mM KI, oxygen was indeed consumed, and the rate of consumption increased markedly at a KI concentration of 100 mM.
Figure 6.
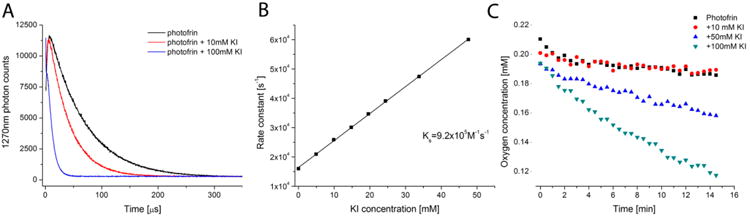
Quenching of singlet oxygen luminescence and oxygen consumption (no cells): (A) typical decay curves of 1270 luminescence signal; (B) plot of rate constant of singlet oxygen quenching against KI concentration; (C) oxygen consumption of PF illuminated in the presence of different concentrations of KI.
Finally, we wished to further confirm that singlet oxygen was responsible for the potentitation of aPDI killing of bacteria. We did this by adding 50 mM sodium azide to a mixture of 10 μM PF and 50 mM KI and compared the killing of E. coli, both when present during light and when added after the end of light delivery. Figure 7 shows that in both cases the killing of bacteria (eradication was obtained without azide) was completely inhibited.
Figure 7.
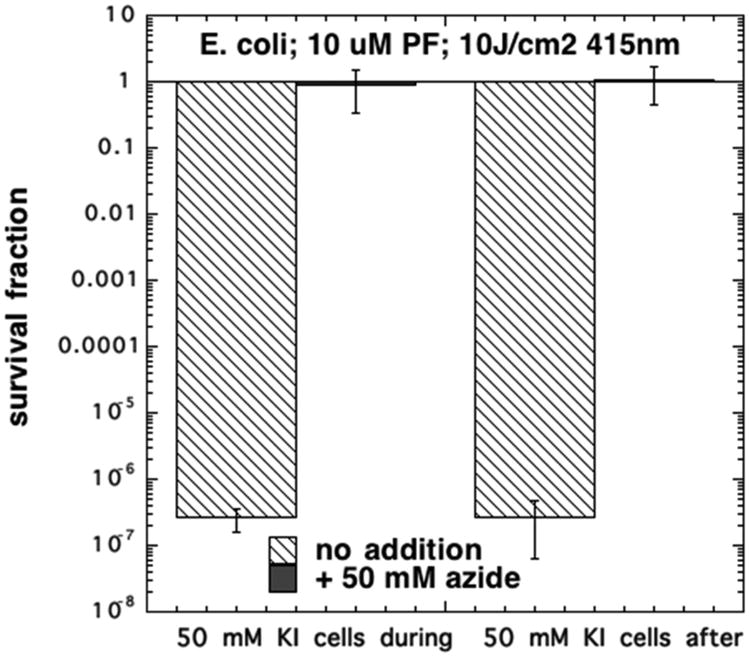
Inhibition of KI-potentiated aPDI of E. coli by addition of azide. PF (10 μM) and KI (50 mM) were illuminated with 415 J/cm2 either in presence or in the absence of NaN3 (50 mM) with E. coli (108 cells/mL) present or added after light.
As mentioned previously, it has long been known that PS such Photofrin are almost completely inactive in mediating aPDI killing of Gram-negative bacteria. Our data confirmed this assumption, with PF (10 μM) and 10 J/cm2 of blue light producing no killing at all of five different Gram-negative species. Maisch et al. were unable to detect any uptake of Photofrin (at concentrations up to 800 μg/mL equivalent to 1.3 μM HP equivalent) by E. coli.11 They also observed no bacterial killing and no singlet oxygen generated in washed cells.
Therefore, we were surprised to discover that after the addition of KI to PF excited by blue light, we were able to eradicate (>6 log of killing) five different Gram-negative bacterial species. Previously we had assumed that a type 1 photochemical process was required to oxidize iodide anions to iodine radicals. Although there are some reports of type 1 photochemical processes occurring with PF,23,24 by and large it is accepted to be a typical type 2 PS, which produces copious quantities of singlet oxygen upon illumination. It has been assumed that the reason why PF is inactive against Gram-negative bacteria is that neutral or slightly anionic PS such as PF do not bind to the Gram-negative outer membrane. Because there is no killing when the PF remains in solution, the logical conclusion is that Gram-negative bacteria are relatively protected against extracellular generated singlet oxygen.14 There have been some reports of Gram-negative bacteria being killed by extracellular singlet oxygen,25 but it is accepted that they are more resistant. However, because cationic PS (including cationic porphyrins26–28) can be exceptionally powerful PS (eradication at hundreds of nanomolar) against Gram-negative bacteria,29 it must be the case that singlet oxygen (if generated in close proximity to essential targets) is actually quite effective in killing Gram-negative bacterial cells. It is presently unclear just how much of the ineffectiveness of neutral/anionic PS in killing Gram-negative bacteria can be attributed to the inability of singlet oxygen to damage molecules in the outer structures (or indeed how lethal such damage may actually be) or else how much of the ineffectiveness is due to the inability of singlet oxygen to diffuse into the bacteria to reach interior sensitive sites. This stark difference between cationic PS and neutral/anionic PS in killing Gram-negative bacteria is not mirrored in the case of Gram-positive bacteria. Here both kinds of PS are equally effective in killing cells, and in fact there is some evidence that an excessively large number of cationic charges is actually counterproductive when it comes to killing Gram-positive cells.30 The porous nature of the Gram-positive cell wall has been credited with the ability of a wide variety of different PS structures to diffuse into the cells to reach sensitive sites. However, it is also indisputable that Gram-positive bacteria are also much more susceptible to extracellular generated singlet oxygen.14,25
As both neutral PS and singlet oxygen are uncharged molecules, it should not be surprising that both types of molecules can apparently diffuse readily into Gram-positive bacteria. Nevertheless, it is apparent that PF does bind to (not just passively diffuse into) Gram-positive bacteria, as shown by the retention of a considerable portion of the photosensitizing activity after a wash. When the higher aPDI activity without a wash is compared to that with a wash, it can be difficult to distinguish between easy diffusion of extracellular generated singlet oxygen into the cells and easy diffusion of the PF molecules into the cells that can be removed with a wash.
In the case of potentiation of PF-mediated aPDI by added iodide, several considerations are raised by our studies. First is the fact that the rate constant between singlet oxygen and iodide anion is rather low, being about 106 M−1 s−1. This explains why it is necessary to use as high a concentration of iodide as 100 mM to be able to generate a sufficient concentration of free iodine to kill Gram-negative cells.
There are two possible mechanisms by which singlet oxygen can oxidize iodide anion. The first is a one-electron transfer to produce iodide radicals (eq 1) (which will dimerize to give molecular iodine (eq 2)) and at the same time give superoxide anions that can undergo dismutation to produce hydrogen peroxide (eq 3).
| (1) |
| (2) |
| (3) |
Unfortunately, this reaction is not thermodynamically feasible: whereas the one-electron reduction potential of singlet oxygen (1O2/O•−2) is +0.65 V,31 the redox potential is as high as 1.2–1.4 V for iodide (I•/I−).32 Therefore, a one-electron oxidation of iodide by singlet oxygen is not realistic under most conditions.
The second possible mechanism is via an addition reaction of singlet oxygen to iodide.
| (4) |
| (5) |
| (6) |
| (7) |
| (8) |
As a result, tri-iodide anion and hydrogen peroxide are formed and, of course, oxygen consumption occurs. It should be noted that no iodide radicals are formed as erroneously suggested in reaction 1. Additional evidence in favor of the latter addition mechanism (eqs 4–8) is the fact that, although we were able to show increased formation of hydrogen peroxide (Figure 5B), we were unable to show any formation of superoxide (data not shown).
We therefore propose that the global reaction is as shown in eq 9.
| (9) |
Considering possible reactive intermediates in these reactions, an interesting paper by Dalmazio et al.33 from Brazil used mass spectrometry and ab initio free energy calculations to study the decomposition of hydrogen peroxide in the presence of iodide anions. They detected a species with m/z 287 that was proposed to be HOOI−2 and explained its formation as depicted in eq 10 and eq 5.
| (10) |
The free energy calculations revealed that the most thermodynamically favored decomposition pathway of HOOI−2 was via eq 11 to give two radicals I2•− and HOO•:
| (11) |
Competing decomposition pathways were energetically less favored by 25 kcal/mol for eq 12, by 28 kcal/mol for eq 13, and by 68 kcal/mol for eq 14.
| (12) |
| (13) |
| (14) |
The addition reaction between singlet oxygen and iodide has been reported by Mosinger and Mosinger in 199522 and has even been proposed as a method for indirect detection of singlet oxygen.34 A similar addition mechanism was proposed in our previous paper35 in which we suggested a similar mechanism for the oxidation of thiocyanate by singlet oxygen.
There is an equilibrium between I2 and I3− when iodine is dissolved in iodide solutions (eq 7). In fact, this equilibrium is why molecular iodine is soluble in aqueous potassium iodide to form Lugol's iodine solution. Lugol's solution consists of roughly equal parts of I2 and KI (15% solution consists of 5% (w/v) I2 and 10% (w/v) KI). In our case the KI is present in a large excess (necessitated by the unfavorable rate constant between singlet oxygen and iodide anion). It is estimated that roughly 1 mM (free iodine equivalent) in Lugol's solution is required to rapidly kill bacteria.36 However, because in our case the iodide is present in large excess, the equilibrium between I3− and I2 will be forced to the predominace of I3−, which is considered to be less bactericidal than I2.37 We consistently observed a better killing of Gram-negative bacteria when cells were added after light at 25 mM KI concentration (see Figure 2). The explanation for this is may be as follows. We know that surface iodination of Gram-negative cells (as might be produced by generation of iodine radicals in solution) does not lead to killing. However, this iodination will lead to less iodine radicals being available to form molecular iodine, so a slightly higher concentration of iodine is likely to be formed when the bacteria are absent. In fact, this can be observed as a darker color produced in the solution after illumination when the cells are absent.
It may well be claimed by critics “Why not just use Lugol's solution?” (which is actually better at killing bacteria because of the favorable equilibrium referred to above) instead of going through all this photodynamic “nonsense”. The answer to this criticism lies in the fact that the combination of aPDI and KI provides an “in situ” and “on demand” route to production of iodine together with hydrogen peroxide to kill Gram-negative bacteria. It is likely that the drawbacks of using topically applied iodine preparations such as povidone–iodine as a technique for treating localized infections,38–40 consist principally in the ability of molecular iodine to damage host mammalian cells at the same time as killing bacteria. Moreover, when iodine is added to infected tissue, there is a very short time available to allow it to penetrate to where the bacteria actually are located within the tissue. The iodine will be rapidly adsorbed by biomolecules (such as proteins) in the tissue. However, if the PS (such as PF) is allowed to penetrate into the tissue after topical application, and if the iodide (as a small anion) can also penetrate easily, then it may be possible to photogenerate molecular iodine and hydrogen peroxide exactly where it is needed. However, further work will be needed to examine whether the in situ produced iodine can cause any damage to host tissue as is seen with other iodine preparations.
In the case of Gram-positive bacteria, the situation is also quite interesting. Here it seems clear that potentiation of aPDI by the addition of KI must be due to photogeneration of reactive radicals such as or I2•− and HOO• or possibly by decomposition of HOOI−2.33 This conclusion is based upon the following evidence. There was potentiation of killing of MRSA with 10 mM KI in the case of no wash and with 25 mM KI in the case of a wash, whereas there was no potentiation of killing when bacteria were added after light until 100 mM KI is reached. PF does bind to some extent to the MRSA cells as shown by retention of aPDI effect after a wash. It is unknown what is the diffusion distance of iodine radicals, but it seems reasonable to suppose it is fairly short. Therefore, if the iodine radicals are iodinating the bacteria, then the radicals will have a better chance of reacting with bacteria when generated right next to the cells than they would have when generated in solution. Presumably, the iodine radicals generated in solution react with each other to form molecular iodine. It was somewhat surprising to us that as low a PF concentration as 200 nM was able to oxidize enough iodide to produce a bactericidal concentration of iodine. However, the need for a high concentration of iodide is chiefly governed by the short half-life of singlet oxygen, which is required to be trapped by iodide. Presumably, if the iodide concentration was not high enough (10 mM), then even a large concentration of PF would not produce a bactericidal concentration of iodine.
In the case of the fungal yeast C. albicans the situation is different yet again. Eukaryotic cells take up PS like PF inside themselves (or bound to the outside) as shown by the retention of most of the photosensitizing ability after a wash. Then the question is, does the iodide reach the same location as the PF, so it can be oxidized by the short-lived singlet oxygen? The answer would appear to be (at least partly) yes. There was potentiation of killing of C. albicans with 50 mM KI, whereas there was no real killing when cells were added after light until 100 mM KI was reached. Therefore, for Candida and for MRSA, the potentiation appears to be mediated by a combination of reactive radicals and molecular iodine.
Although there is a paper describing the use of an iodometric assay to compare the PDT abilities of different PS, we believe that the ability of singlet oxygen to oxidize iodide to iodine is not generally appreciated. Our data clearly show that in the case of PF photogenerated singlet oxygen is largely responsible for the oxidation of iodide to iodine. This conclusion may not apply to other PS. We originally described the potentiation of aPDI mediated by MB by addition of KI at concentrations up to 10 mM. Here there was about 1–2 log of extra killing, but our experiments showed that this extra killing was not due to production of iodine as there was no killing remaining when cells were added after light. This paper attributed the oxidation of iodide to a type 1 photochemical mechanism, although we now believe that it is possible that singlet oxygen played a role as well.
Next we studied the potentiation of aPDI by a cationic fullerene.16 Addition of KI (10 mM) potentiated the UVA or white light-mediated bacterial killing using a C60-fullerene bearing a decaquaternary chain and decatertiary amino groups (LC16). Killing of Gram-negative bacteria A. baumannii, Gram-positive MRSA, and fungal yeast C. albicans was potentiated by 1–2+ log. A mouse model infected with bioluminescent A. baumannii gave increased loss of bioluminescence when iodide (10 mM) was combined with LC16 and UVA/white light.
Titanium dioxide photocatalysis describes the combination of a heterogeneous system of TiO2 nanoparticles and UVA excitation. When used as an antimicrobial technique, it produces ROS including superoxide, hydrogen peroxide, and hydroxyl radicals. We reported that the addition of KI led to strong potentiation of killing.17 When the concentration of KI was kept at 10 mM, then hypoiodite was formed as a product, which could eradicate bacteria that were added immediately after light, but not when bacteria were added after a 2 h interval. However, when the KI concentration was increased to 100 mM, then the bactericidal activity in the solution remained after the light was switched off until 24 h later. We attributed the temporary antibacterial activity to the formation of hypoiodite and the permanent antibacterial activity to molecular iodine. It is known that hypoiodite has only a limited half-life, and after 2 h, it would have completely decomposed by disproportionation into iodide and iodate.41
We also found that TiO2 antimicrobial photocatalysis could be potentiated by the addition of sodium bromide (as well as iodide).42 Apparently, the photo-oxidation reaction produced by TiO2/UVA was strong enough to oxidize bromide to hypobromite. We have not observed that aPDI mediated by any traditional PS, such as a porphyrin, could be potentiated by the addition of bromide.
In conclusion, addition of KI at 100 mM can transform traditional anticancer PS such as Photofrin into powerful antimicrobial PS that can eradicate Gram-negative bacteria (as well as Gram-positives and fungi). The mechanism involves in situ photogeneration of molecular iodine. This may have clinical applications as PF has received regulatory approval all over the world. KI is known to be nontoxic and is used as a health-food supplement and as a treatment for fungal infections (especially sporotrichosis) when administered as oral saturated KI solution (>1 M concentration).43,44
Methods
Chemicals and Reagents
Photofrin was a kind gift from Lerdele Pharmaceuticals Inc., Carolina, Puerto Rico. Potassium iodide (KI) and sodium azide (NaN3) were purchased from Sigma-Aldrich (St. Louis, MO, USA). Starch indicator was purchased from RICCA Chemical Co. (Arlington, TX, USA). The Amplex Red hydrogen peroxide/peroxidase assay kit was purchased from Invitrogen (Carlsbad, CA, USA). PF stock solution was prepared in distilled H2O (dH2O) and stored at 4 °C in the dark for no more than 4 weeks prior to use. Concentrations of PF were expressed as molar equivalent of hematoporphyrin monomer (10 μM HP equiv = 600 μg/mL PF). KI solution was prepared in dH2O as required immediately before experimentation.
Cells and Culture Conditions
The following microbial strains were used: Gram-positive bacterium, methicillin-resistant Staphylococcus aureus (MRSA) US300; Gram-negative bacteria, Escherichia coli K-12 (ATCC 33780), Proteus mirabilis ATCC 51393 (Xen 44), Klebsiella pneumoniae, Acinetobacter baumannii ATCC BAA 747, and Pseudomonas aeruginosa ATCC 19660 (Xen 5P); fungal yeast, a luciferase-expressing Candida albicans strain (CEC 749). A colony of bacteria or fungal yeast was suspended in 20 mL of brain–heart infusion (BHI) broth for bacteria or yeast extract–peptone–dextrose (YPD) broth for C. albicans and grown overnight in a shaker incubator (New Brunswick Scientific, Edison, NJ, USA) at 120 rpm under aerobic conditions at 37 °C for bacteria and at 30 °C for C. albicans. For bacteria, an aliquot of 1 mL from an overnight bacterial suspension was refreshed in fresh BHI for 2–3 h at 37 °C to mid log phase. Cell concentration was estimated by measuring the optical density (OD) at 600 nm (OD of 0.6 = 108 CFU cells/mL). The C. albicans cell number was assessed with a hemocytometer.
aPDI Studies
Suspensions of bacteria (108 cells/mL) or C. albicans (107 cells/mL) were incubated in the dark at room temperature for 30 min with 10 μM PF (for Gram-negative bacteria and C. albicans) or 200 nM PF (for MRSA) and added a range of KI concentrations between 0 and 100 mM in pH 7.4 PBS. Centrifugation (5 min, 4000 rpm) of 1 mL aliquots was used to remove the excess of PF that was not taken up by the microbial cells when experiments required it. An aliquot of 100 μL was used as the dark control (DC) from each sample; another aliquot (200 μL) was transferred to a 96-well plate and illuminated from the top of the plates at room temperature with 10 J/cm2 of blue light. The light source we used was an Omnilux Clear-U light-emitting diode (LED) array (Photo Therapeutics, Inc., Carlsbad, CA, USA) that emitted blue light at a center wavelength of 415 nm to deliver 10 J/cm2 at an irradiance of 50 mW/cm2 as measured with a power meter (Coherent, Santa Clara, CA, USA). At the completion of illumination (or dark incubation), aliquots (100 μL) were taken from each well to determine CFU. Care was taken to ensure that the contents of the wells were mixed thoroughly before sampling, as bacteria can settle at the bottom. The aliquots were serially diluted 10-fold in PBS to give dilutions of 10−1–10−5 times in addition to the original concentration, and 10 μL aliquots of each of the dilutions were streaked horizontally on square BHI agar plates for bacteria or on YPD agar plates for Candida described by Jett et al.45 Plates were streaked in triplicate and incubated for 12–18 h at 37 °C (bacteria) or for 24–36 h at 30 °C (Candida) in the dark to allow colony formation. Each experiment was performed at least three times.
When experiments required it, 10 μM PF or 200 nM PF plus a range of KI concentrations between 0 and 100 mM in pH 7.4 PBS were exposed to 10J/cm2 blue light, then were added bacteria or C. albicans cells to the illuminated mixture of PF + KI. After 30 min of incubation, the aliquots were serially diluted as before. Each experiment was performed at least three times.
Suspensions of E. coli (108 cells/mL) were incubated in the dark at room temperature for 30 min with 10 μM PF plus 50 mM KI with or without added 50 mM NaN3 and were illuminated with 10 J/cm2 of 415 nm blue light. Alternatively, 10 μM PF plus 50 mM KI with or without 50 mM NaN3 was exposed to 10 J/cm2 of 415 nm blue light, and then 108 cells/mL E. coli was added and incubated 30 min. The aliquots were serially 10-fold diluted as before. Each experiment was performed at least three independent times.
A control group of cells treated with light alone (no PF added) showed the same number of CFU as absolute control (data not shown). Survival fractions were routinely expressed as ratios of CFU of microbial cells treated with light and PF (or PF in the absence of light) to CFUs of microbes treated with neither.
Amplex Red Assay for Hydrogen Peroxide
An Amplex Red hydrogen peroxide/peroxidase assay kit was used to detect the production of H2O2 from PF-mediated PDT. The colorless probe Amplex Red (10-acetyl-3,7-dihydroxyphenoxazine) reacts with H2O2 in the presence of peroxidase and forms resorufin (7-hydroxy-3H-phenoxazin-3-one).46 The detection process after PF mediated PDT was according to the manufacturer's instructions. The reaction systems containing 10 μM PF with or without added 100 mM KI were illuminated with increasing fluence of 415 nm light, and aliquots were withdrawn and added to 50 μM Amplex Red reagent and 0.1 U/mL horseradish peroxidase (HRP) in Krebs–Ringer phosphate (consists of 145 mM NaCl, 5.7 mM Na3PO4, 4.86 mM KCl, 0.54 mM CaCl2, 1.22 mM MgSO4, 5.5 mM glucose, pH 7.35). After 30 min of incubation, a fluorescence microplate reader (excitation, 530 nm; and emission, ∼590 nm) was used to measure incremental fluorescence after an incremental fluence of 415 nm light was delivered. Controls were (1) PF + KI in the dark, (2) KI + light, and (3) Amplex Red reagent alone. Each experiment was performed at least three times.
Iodine Starch Test
Both 10 μM PF and 100 mM KI were illuminated with increasing fluence of 415 nm light, and aliquots (50 μL) were withdrawn after different fluences and added to starch indicator (50 μL). A microplate reader (absorbance at 610 nm) was used to measure incremental absorbance after an incremental fluence of 415 nm light was delivered. Controls were (1) PF + KI in the dark, (2) KI + light, and (3) PF + light. Each experiment was performed at least three times.
Singlet Oxygen Detection
The photoreactivity of Photofrin was analyzed by determination of its ability to photogenerate singlet oxygen and induce photoconsumption of oxygen, employing the methods described elsewhere.47 In brief, time-resolved singlet oxygen detection was carried out as follows.
Phosphate-buffered (pH 7.2) D2O solutions of Photofrin (optical density ∼0.2 at 405 nm) in a 1 cm optical path quartz fluorescence cuvette (QA-1000; Hellma, Mullheim, Germany) were excited by pulses generated by an integrated nanosecond DSS Nd:YAG laser system equipped with a narrow bandwidth optical parametric oscillator (NT242-1k-SH/SFG; Ekspla, Vilnius, Lithuania), which delivered pulses at a repetition rate of 1 kHz, with energy up to several hundred microjoules in the visible region. The near-infrared luminescence (1270 nm) was measured perpendicularly to the 405 nm excitation beam in a photon-counting mode using a thermoelectric cooled NIR PMT module (H10330-45; Hamamatsu, Japan) equipped with a 1100 nm cutoff filter and an additional dichroic narrow-band filter NBP, selectable from the spectral range 1150–1355 nm (NDC Infrared Engineering Ltd., Bates Road, Maldon, Essex, UK). Data were collected using a computer-mounted PCI-board multichannel scaler (NanoHarp 250; PicoQuant GmbH, Berlin, Germany). Data analysis, including first-order luminescence decay fitted by the Levenberg–Marquardt algorithm, was performed by custom-written software. Typical acquisition time was 20 s. The effect of potassium iodide on singlet oxygen lifetime was examined in the concentration range of 0–50 mM.
Oxygen Photoconsumption Measurements
Time-dependent changes in oxygen concentration induced by light were determined by EPR oximetry using mHCTPO at concentration of 0.1 mM as the dissolved oxygen-sensitive spin probe. Samples containing 25 μM Photofrin in PBS, pH 7.2, were irradiated in EPR quartz flat cells in the resonant cavity with 402–508 nm (24 mW/cm2) light derived from a 300 W high-pressure compact arc xenon lamp (Cermax, PE300CE-13FM/Module300W; PerkinElmer Optoelectronics, GmbH, Wiesbaden, Germany) equipped with a water filter, heat-reflecting hot mirror, cutoff filter blocking light below 390 nm, and blue additive dichroic filter 505FD64-25 (Andover Corp., Salem, NC, USA). EPR samples were run using a microwave power of 1.06 mW, a modulation amplitude of 0.006 mT, a scan width of 0.3 mT, and a scan time of 21 s. Thirty subsequent scans every 30 s were acquired. EPR measurements were carried out using a Bruker EMX-AA EPR spectrometer (Bruker BioSpin, Rheinstetten, Germany).
Acknowledgments
This work was supported by U.S. NIH Grants R01AI050875 and R21AI121700. Research carried out at Jagiellonian University was sponsored in part by grants from the Poland National Science Center (2011/03/B/NZ1/00007 and 2013/08/W/NZ3/00700). Liyi Huang was supported by National Natural Science Foundation of China (81260239, 81472002), Guangxi Scientific and Technological Project (1355005-1-2), and Guangxi Natural Science Foundation (2016GXNSFAA380312).
Abbreviations
- aPDI
antimicrobial photodynamic inactivation
- BHI
brain–heart infusion
- CFU
colony-forming units
- DC
dark control
- EDTA
ethylenediaminetetraacetic acid
- EPR
electron paramagnetic resonance
- KI
potassium iodiode
- LED
light-emitting diode
- mHCTPO
4-hydro-3-carbamoyl-2,2,5,5-tetraperdeuteromethylpyrrolin-1-oxyl
- MRSA
methicillin-resistant Staphylococcus aureus
- OD
optical density
- PDT
photodynamic therapy
- PF
Photofrin
- PS
photosensitzer
- ROS
reactive oxygen species
- YPD
yeast peptone dextrose
Footnotes
Notes: The authors declare no competing financial interest.
References
- 1.Hamblin MR. Antimicrobial photodynamic inactivation: a bright new technique to kill resistant microbes. Curr Opin Microbiol. 2016;33:67–73. doi: 10.1016/j.mib.2016.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O'Neill J. The Review on Antimicrobial Resistance Chaired by Jim O'Neill. HM Government, Wellcome Trust; 2015. Tackling a Global Health Crisis: Initial Steps. [Google Scholar]
- 3.Hamblin MR, Hasan T. Photodynamic therapy: a new antimicrobial approach to infectious disease? Photochem Photobiol Sci. 2004;3(5):436–450. doi: 10.1039/b311900a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baptista MS, et al. Type I and II photosensitized oxidation reactions: guidelines and mechanistic pathways. Photochem Photobiol. 2017 doi: 10.1111/php.12716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nitzan Y, et al. Inactivation of Gram-negative bacteria by photosensitized porphyrins. Photochem Photobiol. 1992;55(1):89–96. doi: 10.1111/j.1751-1097.1992.tb04213.x. [DOI] [PubMed] [Google Scholar]
- 6.Malik Z, Ladan H, Nitzan Y. Photodynamic inactivation of Gram-negative bacteria: problems and possible solutions. J Photochem Photobiol, B. 1992;14(3):262–266. doi: 10.1016/1011-1344(92)85104-3. [DOI] [PubMed] [Google Scholar]
- 7.Nir U, et al. In vivo effects of porphyrins on bacterial DNA. J Photochem Photobiol, B. 1991;11(3–4):295–306. doi: 10.1016/1011-1344(91)80035-g. [DOI] [PubMed] [Google Scholar]
- 8.Bertoloni G, et al. Photosensitizing activity of water- and lipid-soluble phthalocyanines on Escherichia coli. FEMS Microbiol Lett. 1990;59(1–2):149–155. doi: 10.1111/j.1574-6968.1990.tb03814.x. [DOI] [PubMed] [Google Scholar]
- 9.Ogilby PR. Singlet oxygen: there is indeed something new under the sun. Chem Soc Rev. 2010;39(8):3181–3209. doi: 10.1039/b926014p. [DOI] [PubMed] [Google Scholar]
- 10.Klaper M, Fudickar W, Linker T. Role of distance in singlet oxygen applications: a model system. J Am Chem Soc. 2016;138(22):7024–7029. doi: 10.1021/jacs.6b01555. [DOI] [PubMed] [Google Scholar]
- 11.Maisch T, et al. The role of singlet oxygen and oxygen concentration in photodynamic inactivation of bacteria. Proc Natl Acad Sci USA. 2007;104(17):7223–7228. doi: 10.1073/pnas.0611328104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Minnock A, et al. Mechanism of uptake of a cationic water-soluble pyridinium zinc phthalocyanine across the outer membrane of Escherichia coli. Antimicrob Agents Chemother. 2000;44(3):522–527. doi: 10.1128/aac.44.3.522-527.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hancock RE, Bell A. Antibiotic uptake into Gram-negative bacteria. Eur J Clin Microbiol Infect Dis. 1988;7(6):713–720. doi: 10.1007/BF01975036. [DOI] [PubMed] [Google Scholar]
- 14.Valduga G, et al. Effect of extracellularly generated singlet oxygen on Gram-positive and Gram-negative bacteria. J Photochem Photobiol, B. 1993;21(1):81–86. doi: 10.1016/1011-1344(93)80168-9. [DOI] [PubMed] [Google Scholar]
- 15.Vecchio D, et al. Bacterial photodynamic inactivation mediated by methylene blue and red light is enhanced by synergistic effect of potassium iodide. Antimicrob Agents Chemother. 2015;59(9):5203–5212. doi: 10.1128/AAC.00019-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Y, et al. Potentiation of antimicrobial photodynamic inactivation mediated by a cationic fullerene by added iodide: in vitro and in vivo studies. Nanomedicine (London, U K) 2015;10(4):603–614. doi: 10.2217/nnm.14.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang YY, et al. Broad-spectrum antimicrobial effects of photocatalysis using titanium dioxide nanoparticles are strongly potentiated by addition of potassium iodide. Antimicrob Agents Chemother. 2016;60(9):5445–5453. doi: 10.1128/AAC.00980-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang L, et al. Paradoxical potentiation of methylene blue-mediated antimicrobial photodynamic inactivation by sodium azide: role of ambient oxygen and azide radicals. Free Radical Biol Med. 2012;53(11):2062–2071. doi: 10.1016/j.freeradbiomed.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kasimova KR, et al. Potentiation of photoinactivation of Gram-positive and Gram-negative bacteria mediated by six phenothiazinium dyes by addition of azide ion. Photochem Photobiol Sci. 2014;13(11):1541–1548. doi: 10.1039/c4pp00021h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spesia MB, Durantini EN. Photodynamic inactivation mechanism of Streptococcus mitis sensitized by zinc(II) 2,9,16,23-tetrakis[2-(N,N,N-trimethylamino)ethoxy]phthalocyanine. J Photochem Photobiol, B. 2013;125:179–187. doi: 10.1016/j.jphotobiol.2013.06.007. [DOI] [PubMed] [Google Scholar]
- 21.Garcia-Diaz M, Huang YY, Hamblin MR. Use of fluorescent probes for ROS to tease apart type I and type II photochemical pathways in photodynamic therapy. Methods. 2016;109:158–166. doi: 10.1016/j.ymeth.2016.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mosinger J, Mosinger B. Photodynamic sensitizers assay: rapid and sensitive iodometric measurement. Experientia. 1995;51(2):106–109. doi: 10.1007/BF01929349. [DOI] [PubMed] [Google Scholar]
- 23.Chekulayeva LV, et al. Hydrogen peroxide, superoxide, and hydroxyl radicals are involved in the phototoxic action of hematoporphyrin derivative against tumor cells. J Environ Pathol, Toxicol Oncol. 2006;25(1–2):51–77. doi: 10.1615/jenvironpatholtoxicoloncol.v25.i1-2.40. [DOI] [PubMed] [Google Scholar]
- 24.Athar M, Mukhtar H, Bickers DR. Differential role of reactive oxygen intermediates in Photofrin-I- and Photofrin-II-mediated photoenhancement of lipid peroxidation in epidermal microsomal membranes. J Invest Dermatol. 1988;90(5):652–657. doi: 10.1111/1523-1747.ep12560814. [DOI] [PubMed] [Google Scholar]
- 25.Dahl TA, Midden WR, Hartman PE. Comparison of killing of Gram-negative and Gram-positive bacteria by pure singlet oxygen. J Bacteriol. 1989;171(4):2188–2194. doi: 10.1128/jb.171.4.2188-2194.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Merchat M, et al. Meso-substituted cationic porphyrins as efficient photosensitizers of Gram-positive and Gram-negative bacteria. J Photochem Photobiol, B. 1996;32(3):153–157. doi: 10.1016/1011-1344(95)07147-4. [DOI] [PubMed] [Google Scholar]
- 27.Lambrechts SA, et al. Effect of monovalent and divalent cations on the photoinactivation of bacteria with meso-substituted cationic porphyrins. Photochem Photobiol. 2004;79(3):297–302. doi: 10.1562/sa-03-15.1. [DOI] [PubMed] [Google Scholar]
- 28.Pereira MA, et al. Influence of external bacterial structures on the efficiency of photodynamic inactivation by a cationic porphyrin. Photochem Photobiol Sci. 2014;13(4):680–690. doi: 10.1039/c3pp50408e. [DOI] [PubMed] [Google Scholar]
- 29.Huang L, et al. Stable synthetic mono-substituted cationic bacteriochlorins mediate selective broad-spectrum photo-inactivation of drug-resistant pathogens at nanomolar concentrations. J Photochem Photobiol, B. 2014;141C:119–127. doi: 10.1016/j.jphotobiol.2014.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang L, et al. Stable synthetic cationic bacteriochlorins as selective antimicrobial photosensitizers. Antimicrob Agents Chemother. 2010;54(9):3834–3841. doi: 10.1128/AAC.00125-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koppenol WH. Reactions involving singlet oxygen and the superoxide anion. Nature. 1976;262(5567):420–421. doi: 10.1038/262420a0. [DOI] [PubMed] [Google Scholar]
- 32.Wardman P. Reduction Potentials of One • Electron Couples Involving Free Radicals in Aqueous Solution. J Phys Chem Ref Data. 1989;18(4):1637–1755. [Google Scholar]
- 33.Dalmaźio I, et al. The iodide-catalyzed decomposition of hydrogen peroxide: mechanistic details of an old reaction as revealed by electrospray ionization mass spectrometry monitoring. J Braz Chem Soc. 2008;19(6):1105–1110. [Google Scholar]
- 34.Mosinger J, et al. Light-induced aggregation of cationic porphyrins. J Photochem Photobiol, A. 2006;181:283–289. [Google Scholar]
- 35.St Denis TG, et al. Thiocyanate potentiates antimicrobial photodynamic therapy: In situ generation of the sulfur trioxide radical anion by singlet oxygen. Free Radical Biol Med. 2013;65C:800–810. doi: 10.1016/j.freeradbiomed.2013.08.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Choi S, et al. Use of elemental iodine for shunt infection prophylaxis. Neurosurgery. 2003;52(4):908–912. doi: 10.1227/01.neu.0000053371.86661.94. discussion 912–913. [DOI] [PubMed] [Google Scholar]
- 37.Atemnkeng MA, Plaizier-Vercammen J, Schuermans A. Comparison of free and bound iodine and iodide species as a function of the dilution of three commercial povidone-iodine formulations and their microbicidal activity. Int J Pharm. 2006;317(2):161–166. doi: 10.1016/j.ijpharm.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 38.Burks RI. Povidone-iodine solution in wound treatment. Phys Ther. 1998;78(2):212–218. doi: 10.1093/ptj/78.2.212. [DOI] [PubMed] [Google Scholar]
- 39.Goldenheim PD. An appraisal of povidone-iodine and wound healing. Postgrad Med J. 1993;69(Suppl 3):S97–S105. [PubMed] [Google Scholar]
- 40.Khan MN, Naqvi AH. Antiseptics, iodine, povidone iodine and traumatic wound cleansing. J Tissue Viability. 2006;16(4):6–10. doi: 10.1016/s0965-206x(06)64002-3. [DOI] [PubMed] [Google Scholar]
- 41.Zhao X, et al. Kinetic and Mechanistic Aspects of the Reactions of Iodide and Hypoiodous Acid with Permanganate: Oxidation and Disproportionation. Environ Sci Technol. 2016;50(8):4358–4365. doi: 10.1021/acs.est.6b00320. [DOI] [PubMed] [Google Scholar]
- 42.Wu X, et al. Broad-spectrum antimicrobial photo-catalysis mediated by titanium dioxide and UVA is potentiated by addition of bromide ion via formation of hypobromite. Free Radical Biol Med. 2016;95:74–81. doi: 10.1016/j.freeradbiomed.2016.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Costa RO, et al. Use of potassium iodide in dermatology: updates on an old drug. An Bras Dermatol. 2013;88(3):396–402. doi: 10.1590/abd1806-4841.20132377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sandhu K, Gupta S. Potassium iodide remains the most effective therapy for cutaneous sporotrichosis. J Dermatol Treat. 2003;14(4):200–202. doi: 10.1080/09546630310020452. [DOI] [PubMed] [Google Scholar]
- 45.Jett BD, et al. Simplified agar plate method for quantifying viable bacteria. BioTechniques. 1997;23(4):648–650. doi: 10.2144/97234bm22. [DOI] [PubMed] [Google Scholar]
- 46.Driever SM, et al. Imaging of reactive oxygen species in vivo. Methods Mol Biol. 2009;479:109–116. doi: 10.1007/978-1-59745-289-2_7. [DOI] [PubMed] [Google Scholar]
- 47.Szewczyk G, et al. Aerobic photoreactivity of synthetic eumelanins and pheomelanins: generation of singlet oxygen and superoxide anion. Pigm Cell Melanoma Res. 2016;29(6):669–678. doi: 10.1111/pcmr.12514. [DOI] [PubMed] [Google Scholar]


