Abstract
Purpose
Tumor relapse is the most frequent cause of death in stage 4 neuroblastomas. Since genomic information on the relapse precursor cells could guide targeted therapy, our aim was to find the most appropriate tissue for identifying relapse-seeding clones.
Experimental design
We analyzed 10 geographically and temporally separated samples of a single patient by SNP array and validated the data in 154 stage 4 patients.
Results
In the case study, aberrations unique to certain tissues and time points were evident besides concordant aberrations shared by all samples. Diagnostic bone marrow-derived DTCs (disseminated tumor cells) as well as the metastatic tumor and DTCs at relapse displayed a 1q deletion, not detected in any of the seven primary tumor samples. In the validation cohort, the frequency of 1q deletion was 17.8%, 10%, and 27.5% in the diagnostic DTCs, diagnostic tumors, and DTCs at relapse, respectively. This aberration was significantly associated with 19q and ATRX deletions. We observed a significant increased likelihood of an adverse event in the presence of 19q deletion in the diagnostic DTCs.
Conclusion
Different frequencies of 1q and 19q deletions in the primary tumors as compared to DTCs, their relatively high frequency at relapse, and their effect on event-free survival (19q deletion) indicate the relevance of analyzing diagnostic DTCs. Our data support the hypothesis of a branched clonal evolution and a parallel progression of primary and metastatic tumor cells. Therefore, searching for biomarkers to identify the relapse-seeding clone should involve diagnostic DTCs alongside the tumor tissue.
Keywords: Neuroblastoma, disseminated tumor cells, bone marrow, tumor evolution, tumor heterogeneity
Introduction
The clinical behavior of neuroblastomas is very heterogeneous ranging from spontaneous regression or maturation to highly malignant and metastatic disease (1–4). It is generally assumed that differences in the genetic background are, at least to a large degree, responsible for this clinical heterogeneity. Overall, low-risk neuroblastomas mostly have whole chromosomal gains without structural aberrations. High-risk neuroblastomas, i.e. mostly stage 4, on the contrary, present with different structural aberrations in the form of gains or losses of large chromosomal segments, referred to as segmental chromosomal aberrations (SCAs), MYCN amplification (MNA), and/or mutations as well as copy number alterations affecting certain genes or parts thereof (5–13). Whether genetic markers might help to identify the relapse-seeding clone and to categorize stage 4 patients into different prognostic subgroups is still a matter of debate.
Intra-tumor heterogeneity (ITH) in solid tumors can be responsible for treatment failure and drug resistance (14, 15). Despite the huge degree of inter-tumor genetic heterogeneity, studies on ITH in neuroblastoma have, thus far, mainly focused on MNA (16–21). As a single biopsy likely fails to capture the genetic complexity of a heterogeneous tumor, the relapse-seeding subclone may be overlooked (14).
Bone marrow (BM) is the most common site for disseminated tumor cells (DTCs) in neuroblastoma and its infiltration at diagnosis and relapse is a hallmark of the majority of stage 4 disease. Depending on the sensitivity of the detection technique, BM infiltration at diagnosis can be found in more than 90% of stage 4 patients (22). In previous studies, we showed that a full genomic and transcriptomic characterization of DTCs in neuroblastoma patients is feasible (23, 24). In the case of adult cancers, DTCs are generally considered to be metastasis precursor cells and may develop into a metastatic relapse after a period of dormancy (25–27). In this paper, we use the term DTCs for any BM-derived tumor cells regardless of the rate of BM infiltration.
In order to better understand tumor progression and how tumor ontogeny proceeds to metastasis and relapse, we analyzed different BM-derived DTC and tumor samples of a single neuroblastoma patient taken at diagnosis and at relapse, using high-density SNP array. For a more global conclusion on the clinical impact of genomic aberrations of DTCs, we compared the results of this patient with the genomic results obtained from samples at different time points and sites of 154 stage 4 neuroblastoma patients.
Materials and Methods
Clinical report of the case study
A four-year-old boy with a history of constipation, frequent infections, right knee pain, and a mass in the left middle abdomen was admitted to the St. Anna Children’s Hospital, Vienna, in 1993. CT scan detected a mass in the left retroperitoneum. Laboratory tests showed an increase in urinary catecholamines (VMA: 1.8 folds, HVA: 1.5 folds, and dopamine: 3 folds) and NSE in the blood. LDH was in the normal range. At this time, the patient had a BM involvement with 6% GD2-positive cells. He underwent surgery with diagnosis of stage 4 neuroblastoma. After resection of the para-aortic tumor, pathologic examination reported a 4.5×4.5×9.4 cm nodular neuroblastoma tumor. After four months of chemotherapy, abdominal CT and MIBG scans showed no evidence of a remaining tumor. The patient received high dose chemotherapy followed by an autologous peripheral stem cell transplantation and radiotherapy of the primary tumor site. The patient was in complete remission till 28 months after diagnosis, when he experienced a relapse with a tumor in the right frontoparietal region of the skull and BM involvement. Palliative chemo- and radiotherapy for the metastatic site resulted in an improvement in clinical and imaging signs. He experienced generalized pain 8.5 months after the relapse and an MRI showed different metastatic lesions in the paravertebral region of the thorax. Palliative radiotherapy was started for paravertebral site but had to be stopped due to the poor general condition of the patient. The patient remained on analgesic therapy and passed away 9.5 months after the relapse.
Patients and samples
For the case study, we analyzed 10 different samples of a stage 4 neuroblastoma patient including seven samples from different regions of the primary tumor and BM-derived DTCs at diagnosis as well as a metastatic tumor biopsy and BM-derived DTCs at the time of relapse (Supplementary Table S1). A tumor-free peripheral blood sample was used as reference.
For the validation cohort, we analyzed 122 BM-derived DTC samples at different time points (73 samples at diagnosis, 40 samples at relapse, and 9 samples during therapy or with unknown time point) and 81 primary tumors (50 tumors at diagnosis and 31 tumors after induction chemotherapy) of 154 stage 4 neuroblastoma patients. Only patients with positive BM were entered into the study. For 43 patients, more than one sample at different time points and/or different sites was analyzed (Supplementary Table S2). In cases where more than one primary tumor sample per patient was available, we combined the information of the different samples and counted shared aberrations as one per primary tumor.
Immunofluorescence detection
BM samples were analyzed by GD2 immunofluorescence staining for quantification of the tumor cell load using an automatic detection device, RCDetect/MetaCyte (MetaSystems GmbH, Germany) (28).
Sample preparation
Tumor and BM samples collected for the case study had been stored in the gas phase of liquid nitrogen. Bone marrow samples were enriched by applying a density gradient centrifugation (Lymphoprep™, AXIS-SHIELD PoC AS, Norway) followed by a magnetic bead-based enrichment technique using FITC-labeled anti-GD2 antibody (14.18 delta CH2 clone) and anti-FITC microbeads (Miltenyi Biotec, Germany) as already described in detail (23). The BM at diagnosis had a 6% DTC infiltration rate before and a 50% rate after enrichment. The tumor cell content of the BM sample at relapse was unknown; however, 45% DTC infiltration was obtained after enrichment. Cryosection slides were prepared from seven different regions of the primary tumor and from the tumor biopsy at relapse. After hematoxylin and eosin staining, the tumor cell rich regions were selected for DNA extraction. DNA was extracted using the high salt extraction method (29).
For the validation cohort, DNA was extracted from 122 BM-derived DTC samples. In case of BM samples with ≤50% tumor cell infiltration, tumor cells were enriched using the magnetic bead-based technique. In cases where no fresh or DMSO frozen BM was available, DNA was extracted from immunostained or unstained cytospin slides for samples with ≥30% tumor cell infiltration rate. Furthermore, tumor DNA was extracted from fresh or snap frozen tumor tissues and also from the tumor cell-free mononuclear cell fractions as reference.
Genomic analysis
Genomic profiles of tumor cells were generated using CytoScan™ HD Arrays (Affymetrix Inc., UK Ltd) and the data were analyzed using the ChAS software (Affymetrix Inc., UK Ltd) (23, 30). CytoScan HD has a genome-wide coverage with more than 2.6 million copy number and SNP markers. This array has more than 99% sensitivity, which can reliably detect copy number changes even as small as 25-50 kb size with a high specificity. Since it includes 750,000 SNP probes, it is possible to detect regions with copy neutral loss of heterozygosity (cnLOH) and low level of mosaicisms. Subclonal frequencies were calculated by evaluating the smooth signal level and the allele difference track pattern and by considering the tumor cell content of the samples as described before (23, 30, 31).
I-FISH
Interphase fluorescence in situ hybridization (I-FISH) experiments were performed on tumor touch imprints (TTIs) and BM cytospin slides for the detection of 1q deletion in the diagnostic DTCs and seven primary tumor samples of the case study. The cells were fixed in 4% formaldehyde at 4°C for 15 min and, after rinsing in PBS, were digested in 0.05% pepsin in 1 N HCL for 1-2 min. The slides were dehydrated in ethanol series: 70%, 95%, and 100%, 3 min each. Two probes (digoxin-labeled RP11-951L14 and biotin-labeled RP11-312G23, kindly provided by Dr. Mariano Rocchi, Bari, Italy) were added. Denaturation was performed on a heating plate at 78°C for 8 min and hybridization was carried out at 37°C overnight in a humid chamber. The slides were washed with formamide solution for 15 min at 42°C and with 2× standard saline citrate (SSC) for 7 min twice and incubated with mouse anti-biotin and sheep anti-digoxin antibodies at 37°C for 30 min in a humid chamber. After washing in 4× SSC/0.1 Tween 20 at 42°C for 7 min twice, the slides were incubated with goat anti-mouse Cy3 and rabbit anti-sheep FITC in 37°C for 30 min. Slides were covered with mounting medium plus DAPI after washing in Tween 20 and PBS.
Statistical analysis
To investigate the association between different aberrations and sample types, a generalized linear model was fitted using the logistic link function. The modelling of correlated data, i.e. samples from the same patient at different time points, was performed using the generalized estimating equation approach developed by Liang and Zeger (32). Fisher’s exact test was used to determine the association between different chromosomal aberrations. Kaplan-Meier curves were generated for EFS and OS. Curves were compared using a log-rank test. For measuring EFS, “time to event” was defined as the time from diagnosis to the time of the first relapse, progression, or death.
Ethics statement
Ethical approval was obtained from the St. Anna Children’s Hospital ethics committee and the patients’ parents gave their informed consent.
Data access
Affymetrix CytoScan HD array data of this study have been submitted to NCBI Gene Expression Omnibus under accession number GSE84291.
Results
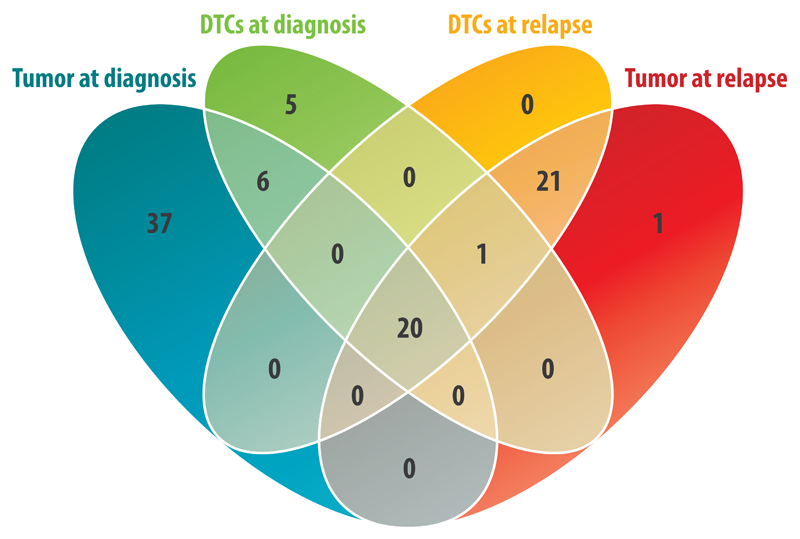
To study the clonal expansion during tumor development, we reconstructed the phylogenetic tree for the samples of a stage 4 patient by using chromosomal aberrations as markers of clonal origin (Fig. 1). For this purpose, we assumed that identical breakpoints occurred at the same time point during clonal evolution and that the number of aberrations increased with time. A schematic visualization of the way how the temporal sequence of different events was deduced from the appearance of different chromosomal aberrations is provided in the Supplementary Figure S1. In total, we found 91 breakpoints and three whole chromosome copy number changes in the 10 different samples of the patient (Supplementary Fig. S2, Supplementary Table S3). Eighteen breakpoints were homogeneously present in all cells of all samples, representing the genomic profile of the common ancestor clone. We detected 37, 5, and 21 breakpoints exclusively in the primary tumor, diagnostic DTCs, and relapse samples, respectively (Fig. 2). The primary tumor harbored genetically distinct subpopulations indicating extensive ITH. Except for a terminal deletion at 5p, which was found in 50% of cells of the relapse tumor, all other aberrations detected in the relapse samples were shared between tumor and DTCs.
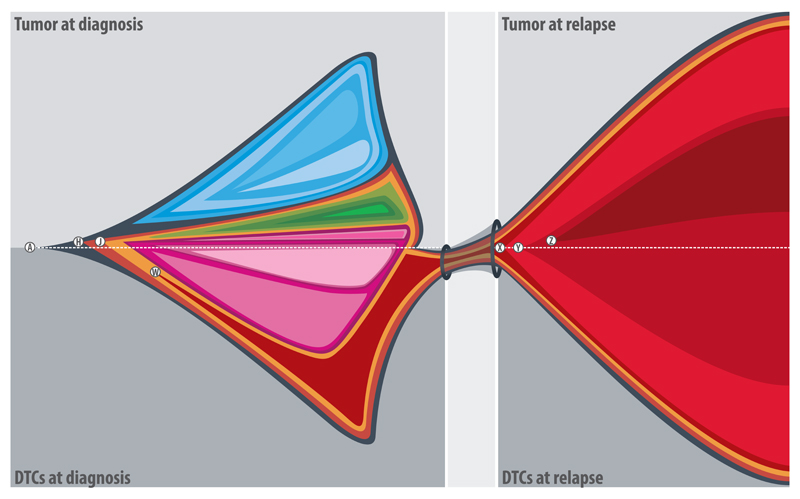
Figure 1.
Graphical representation of clonal expansion in a stage 4 neuroblastoma patient. Each quadrant represents the clonal architecture of a tissue/time point. Each color represents a group of chromosomal aberrations and the size of each colored area represents the proportion of cells with these aberrations within the analyzed samples (the detail of subclonal frequencies of different aberrations is provided in the supplementary table S3). The primary tumor displayed a branched clonal evolution leading to extensive intra-tumor heterogeneity. A 1q terminal deletion (W) which was present in the diagnostic DTCs and also in the both DTCs and metastatic tumor at relapse was not detected in any of the primary tumor samples. Certain chromosomal aberrations were acquired independently in geographically different tissues at diagnosis and at relapse, indicating parallel tumor progression. A: 18 breakpoints corresponding to 13 aberrations including ATRX deletion (exons: 2-8) present homogenously in all samples/time points; H: Chromosome Y loss and J: 1p33(49,772-49,927)×3 (affecting AGBL4 gene) are present homogenously in the DTCs at diagnosis and relapse samples and heterogeneously in the tumor at diagnosis; W: 1q42.2qter(231,262-249,250)×1 is present heterogeneously in the DTCs at diagnosis and homogenously in the relapse samples; X: 21 breakpoints corresponding to 13 aberrations (including 19q and PTPRD deletions) are homogenously present in the relapse samples; Y: duplication of chromosome 17 is heterogeneously present in the relapse samples; and Z: 5pterp15.2(1-12,709)×1 is heterogeneously present only in the metastatic tumor at relapse. Breakpoints are given in kb. The areas with blue, green and pink spectrums represent different groups of aberrations heterogeneously present in the tumor and DTCs at diagnosis (Supplementary Table S3).
Figure 2.
Venn diagram representing the number of breakpoints detected either homogenously or heterogeneously in different samples of one patient. Besides aberrations unique to certain tissues and time points, 20 breakpoints were shared between all samples consist of 18 breakpoints homogeneously present in all samples, representing the genomic profile of the common ancestor clone, and two breakpoints present homogenously in the diagnostic DTCs and relapse samples and heterogeneously in the primary tumor.
A terminal deletion affecting the long arm of chromosome 1 (231,262kb-qter) was homogenously present in the metastatic tumor and DTCs at relapse as well as in approximately 30% of the diagnostic DTCs. However, this aberration was undetectable in any of the seven analyzed primary tumor samples (Supplementary Fig. S3). Furthermore, chromosome Y loss (LOY) and a gain in the AGBL4 gene were homogenously present in the relapse samples and in the DTCs at diagnosis but were found in only about 35% of the primary tumor cells. The analyzed relapse samples showed a number of focal copy number changes affecting single genes or parts thereof, e.g. PTPRD and ATRX intragenic deletions, or affecting two to three adjacent genes (Supplementary Table S4). Deletions affecting ZMYM5, ARID2, ATRX, and RBMS3 were present in all samples. Deletions in PTPRD, RBMS3 (with different breakpoints from the deletion shared between all samples), and GALNT13 and gains in MIR1246, LOC375295, NRP2, PARD3B, DNAJC12, SIRT1, HERC4, and CADPS2 were present only in the relapse samples. GALNT13 was deleted in the DTCs at diagnosis with different breakpoints. AGBL4 was deleted in the relapse samples, DTCs at diagnosis and a subclone of the primary tumor.
The applied SNP array technique is able to detect copy number changes in samples with even less than 10% tumor cell content. In addition, in order to search for tumor cell clusters with 1q deletion, we applied I-FISH with probes flanking the breakpoint. The I-FISH pattern indicating a deletion of the terminal region of 1q (one single spot and one pair of signals) was found in none of the seven tumor touch imprints. However, this hybridization pattern indicating 1q deletion was found in the diagnostic DTCs (Supplementary Fig. S4).
To determine the general frequency of 1q terminal deletion, we analyzed the SNP array data of 203 BM-derived DTC and primary tumor samples of 154 stage 4 neuroblastoma patients (Supplementary Table S2).
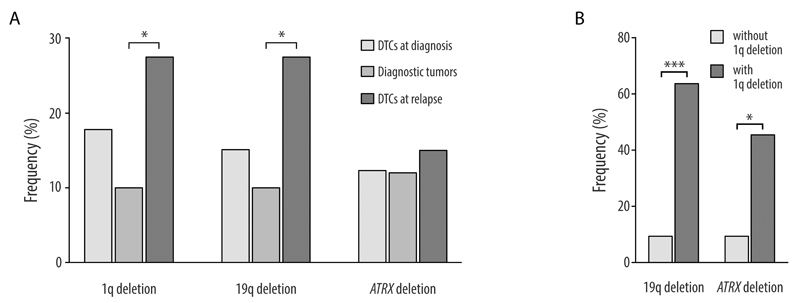
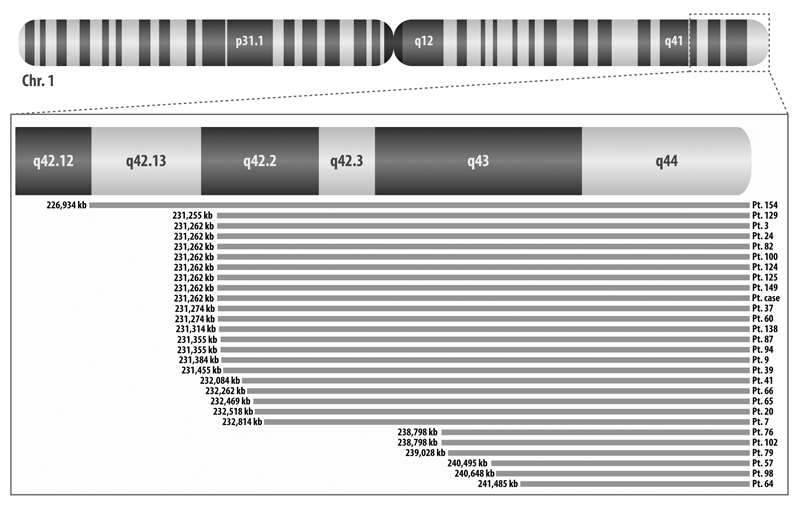
In the validation cohort, 17.5% of patients had at least one sample with 1q terminal deletion (Supplementary Table S5.A). In addition to the 1q terminal deletions resulting in partial monosomies in 22 samples, we detected a deletion in the trisomic q arm that resulted in a 1q imbalance with loss of the distal region in 11 samples. The frequency of this aberration was 17.8%, 10%, and 27.5% in the DTCs at diagnosis, diagnostic tumors, and DTCs at relapse, respectively (Fig. 3A). The breakpoints of all 33 samples clustered within a 14,551 kb region (226,934-241,485 kb). The smallest region of overlap (SRO) in samples with 1q deletion spanned the terminal 7,766 kb of the q arm (Fig. 4). This region consists of 93 genes including 80 protein-coding genes (Supplementary Table S6). By analyzing the R2 database (R2: Genomics Analysis and Visualization Platform, http://r2.amc.nl), we observed that among these genes, low expression of RGS7, AKT3, C1orf101, COX20, and CNST significantly decreased OS and/or EFS in neuroblastoma patients in many of datasets (9, 33–36) (Supplementary Table S7). The same result was observed for the FMN2 gene which was deleted in all samples but two.
Figure 3.
A. Frequencies of 1q and 19q terminal deletions and ATRX deletion in DTCs at diagnosis, diagnostic tumors, and DTCs at relapse. B. Frequencies of 19q terminal and ATRX deletions in the DTCs with and without 1q terminal deletions at diagnosis. Samples were restricted to patients ≥18 months without MNA.
Figure 4.
Deleted regions of 1q in different patients. For patients with more than one sample with this deletion, only one sample has been represented in the figure since deleted segments are the same for different samples. The deleted region in patient 64 represents the SRO.
For 10 patients (including the case study) with 1q deletions found in at least one of their samples, more than one analyzed sample from different time points and/or different sites was available. We observed spatial and/or temporal heterogeneity of 1q deletion between the different samples in six of these patients. In addition to the discrepancy observed in the studied case, we found 1q deletions in the diagnostic DTCs of two more patients, whereas this aberration was undetectable in the primary tumors. In two other patients, we found 1q deletions only in the DTCs at relapse but not in the corresponding primary tumors. In one case, a 1q deletion was present in the second relapse but neither in the primary tumor nor in the first relapse sample (Supplementary Table S5.A).
We also investigated the association between 1q deletion and other frequent aberrations in the DTCs at diagnosis and at relapse (Supplementary Fig. S5). Since we found 1q deletions only in patients ≥18 months and mainly in samples without MNA, we performed the statistical evaluation in patients ≥18 months without MNA. The 1q deletion was significantly associated with 19q and ATRX deletions at diagnosis (Fig. 3B). However, a significant correlation did not exist for DTCs at relapse. Both aberrations were also detected in all samples of the case study.
Terminal deletions of 19q were found in 18.2% of patients. The frequency of this aberration was 15.1% in the DTCs at diagnosis, 10% in the diagnostic tumors, and 27.5% in the DTC samples at relapse (Fig. 3A). We observed spatial and/or temporal heterogeneity of the 19q deletion in five out of nine patients with more than one analyzed sample (Supplementary Table S5.B). The SRO for samples with chromosome 19 deletions spanned from 57,633 kb to the terminal of the q arm (Supplementary Fig. S6). This region consists of 70 genes (43 of them are ZNF family genes) including 62 protein-coding genes (Supplementary Table 6). According to the R2, down-regulation of ZNF264 and AURKC decreases OS and/or EFS of neuroblastoma patients (Supplementary Table S7).
Intragenic deletions within the ATRX gene were observed in 13.6% of patients. We found this aberration exclusively in the samples without MNA of patients ≥18 months. The frequency of this deletion was about 12% in the diagnostic DTCs and primary tumors and 15% in the DTCs at relapse (Fig. 3A). In only one out of four patients with more than one analyzed samples, we observed spatial heterogeneity of the ATRX deletion. In this case the deletion was detected in the primary tumor but not in the DTCs at diagnosis (Supplementary Table S5.C).
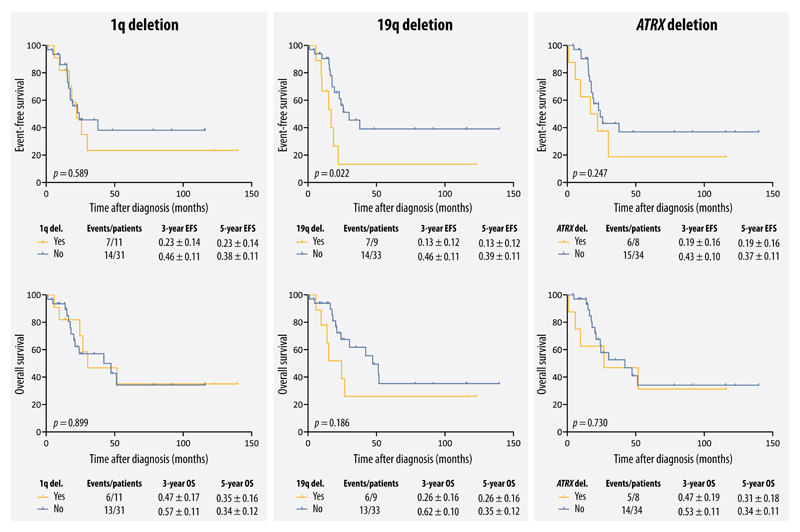
The three aberrations of interest were found exclusively (for ATRX deletions) or mainly (for 1q and 19 deletions) in the samples without MNA of patients ≥18 months. Therefore, in order to eliminate these confounding variables, the survival analysis was restricted to patients ≥18 months without MNA. Within this patient cohort, only 19q deletion found in diagnostic DTCs was considered to be a significant marker for EFS. 5-year EFS was 13±12% and 39±11% for 9 patients with and 33 patients without 19q deletions, respectively (p=0.022). Similarly, a non-significant effect of 19q deletions on OS was observed (5-year OS: 26±16% vs 35±12%, p=0.186). Although 1q and ATRX deletions in the diagnostic DTCs did not reach statistical significance, we observed an effect by these aberrations on EFS. 5-year EFS was 23±14% vs. 38±11% for 11 patients with 1q deletions compared to 31 patients without this aberration (p=0.589), and 19±16% vs. 37±11% for 8 patients with ATRX deletions compared to 34 patients without this aberration (p=0.247) (Fig. 5). Due to a low number of diagnostic tumors and tumors after induction therapy, we combined these two groups for survival analyses. Interestingly, 1q and ATRX deletions in the primary tumors showed no effect on OS and EFS rates. Although 19q deletions in the primary tumors decreased OS and EFS of patients, this effect was stronger when found in the diagnostic DTCs (Supplementary Fig. S7). To have a general overview of the effect of these aberrations on the survival of stage 4 patients regardless of age and the status of MYCN, we also performed the survival analysis for the whole population, without age and MNA adjustment. 1q, 19q and ATRX deletions in the diagnostic DTCs had comparable effects on EFS and OS compared with the presence of these aberrations in the diagnostic DTCs after eliminating age and MNA (Supplementary Fig. S8).
Figure 5.
Event-free and overall survival curves for patients with and without 1q, 19q, or ATRX deletions in the DTCs at diagnosis. Samples were restricted to patients ≥18 months without MNA.
Discussion
By comparing the genomic profile of DTCs and tumor samples at different time points, we were able to identify the evolutionary relationship of different clones, as well as the presumed order of chromosomal aberration events during tumor evolution in a neuroblastoma patient. Genomic data showed that the primary and metastatic tumor samples and DTCs at diagnosis and at relapse had a common ancestor. Despite this common origin, a parallel progression model of primary tumor and metastatic samples explains our data best. Presumably, tumor cells left the primary tumor site early and disseminated into the BM where they accumulated additional genomic aberrations independent of the primary tumor. At the primary tumor site, a branched clonal evolution of the cancer genome with stepwise accumulation of somatic aberrations resulted in extensive ITH.
Temporal heterogeneity concerning copy number changes and gene mutations was reported in neuroblastoma (37–39). In line with the studies performed on matched primary and relapse neuroblastomas, despite a decrease in subclonal copy number changes in the relapse samples compared to the primary tumors (39), a number of de novo aberrations were found in the relapse samples (37, 39). The question arises whether the accumulation of these new aberrations in the relapse clone was a result of selective pressure exerted by drug exposure or not. However, data on the genomic evolution of neuroblastomas do not necessarily support the view of drug mediated genomic evolution as sole cause for acquiring genomic aberrations. Schleiermacher and colleagues showed that accumulation of chromosomal aberrations occurs not only in patients receiving chemotherapy but also in patients who had not received chemotherapy (40). In addition, similar to previous studies (37, 39), we identified some unique aberrations in the primary tumor that were no longer present in the relapse samples. These aberrations may tell us about their minor impact concerning driving the tumor cell progression and chemoresistance. These data support the importance of performing biopsies for genomic analyses of relapsed neuroblastomas.
Most strikingly, our data indicate that the metastatic tumor and DTCs at relapse developed from a subclone that was already present in the BM at diagnosis but was either not detected or did not exist in the primary tumor. The aberration unique to this clone was a terminal deletion of 1q. This aberration, originally described by Fieuw et al. (41), was present in 30% of diagnostic DTCs and in all cells of the relapse samples but was not detected in any of the primary tumor subclones of the case study. Two additional aberrations (LOY and a gain within the AGBL4 gene) present in the relapse samples were detected in the diagnostic DTCs but these aberrations were only subclonal in the primary tumor and the detectability differed geographically within the tumor. Nevertheless, we cannot refute the possibility that the assumed relapse-seeding clone existed either in a region of the primary tumor that was not sampled or in a very low number of tumor cells and was therefore not detected by the applied techniques in the sampled regions.
Our data indicate that for two reasons the identification of the genomic profile of the relapse clone may be missed when analyzing only the primary tumor: first, sampling error due to ITH and second, independent evolution of the relapse clone(s) after dissemination. However, we showed that the relapse-seeding clone may be present in the metastatic tissue (BM) at diagnosis. Therefore, we hypothesize that by sampling and analyzing the BM at diagnosis in addition to the primary tumor, the chance for detecting relapse-specific aberrations at an early time point increases.
To verify which tissue (tumor vs. DTCs) harbors the 1q terminal deletion at a higher frequency, we searched in a large group of stage 4 patients. Fieuw and colleagues found 1q deletions in 7.8% of stage 4 primary tumors (41). In our cohort, the frequency of 1q deletions was 10% in tumors at diagnosis. In addition to the significantly higher frequency of this aberration in the relapse samples, the frequency of 1q deletion in the diagnostic DTCs was nearly double that of the primary tumors. In cases with spatial and/or temporal heterogeneity of 1q deletion, this aberration was present in the DTCs at diagnosis and/or relapse but was not found in the primary tumors. The presence of this aberration in the DTCs at diagnosis did not significantly increase the likelihood of an adverse event; therefore, further studies are needed to investigate the possible role of 1q deletion as a prognostic marker and to ravel the role of the genes in this region in tumor progression. There are some interesting genes in the SRO of this aberration. RGS7 (regulator of G-protein signaling 7) is a protein coding gene and the most proximal gene within the SRO. The regulators of G-protein signaling pathway play an important role in signaling transduction, cellular activities, and carcinogenesis (42, 43). This candidate tumor suppressor gene found to be methylated in renal cell carcinoma and associated with prognosis (44). Another interesting gene in this region is AKT3. The protein coded by this gene is a member of the serine/threonine protein kinase family. AKT kinases play a role as regulators of metabolism, translation, proliferation, survival, and angiogenesis (45). The AKT signaling pathway is important in tumor development, disease aggressiveness and drug resistance (46). AKT3 plays an important role in brain development and various mutations in this gene are linked to neurological disorders (45). Although AKT3 upregulation or increased copy number of its chromosomal region were reported in different cancers (46), recent data indicates that AKT3 inhibits cancer cell migration. It has been shown that knockdown of AKT3 increases the metastatic potential of tumor cells (47). Another gene is FMN2, a member of the formin homology protein family. Increased FMN2 protein level inhibits degradation of p21 and promotes cell cycle arrest (48, 49). Based on the R2 database, lower expression of these three genes decreases the survival of neuroblastoma patients.
19q terminal deletion, which was significantly associated with 1q deletion, could represent another potential marker for relapse-seeding clones since this aberration had a higher frequency in the relapse samples and significantly decreased the EFS of patients when found in the DTCs at diagnosis. Similar to our findings, this aberration was reported by Cobrinik et al. in non-MYCN-amplified tumors (50).
Deletion within the ATRX gene was also significantly associated with 1q deletion. Somatic alterations of the ATRX gene were reported to occur in 10-22% of stage 4 neuroblastoma tumors. Seventy percent of these alterations, which are exclusively present in patients ≥18 months with non-MNA tumors, were found in the form of deletions within this gene (6, 9–12). This aberration is proposed to define a subgroup of high-risk neuroblastomas resulting in elongation of telomeres by alternative lengthening of telomeres, ALT (10, 12). In line with these reports, we found intragenic ATRX deletions in 13.6% of stage 4 patients, exclusively older than 18 months without MNA. There was neither a considerable difference in the frequency between tumor and DTC samples nor between the different time points of sample acquisition, which suggests a driver role for this aberration.
Significant decreased EFS was observed only for patients with 19q deletion in the diagnostic DTCs. Although statistical significance was not achieved for decreased EFS of patients with 1q or ATRX deletions in the diagnostic DTCs and also not for decreased OS for patients with 19q deletion, we have to bear in mind that the survival analyses were performed for a prognostically highly unfavorable patient group (stage 4, ≥18 month). Larger studies are warranted to investigate and validate the independent prognostic importance of these aberrations.
In summary, we provide evidence for a branched clonal evolution with early dissemination and parallel evolution of the primary tumor and DTCs in a neuroblastoma patient. The relapse-associated aberration found in a single, well-characterized, neuroblastoma patient was verified in a large number of tumor and DTC samples. Our results indicate that the presumed relapse-seeding clone can be found with a different frequency in DTCs as compared to tumor biopsies. This study demonstrates the unmet need to study genetic changes in diagnostic DTCs alongside tumor tissues to better characterize and predict possible relapses.
Supplementary Material
Translational Relevance.
Since relapse is the major cause of death in neuroblastoma patients, early detection and characterization of the relapse-seeding clone would help to develop or choose an appropriate treatment. However, due to intra-tumor heterogeneity, single or even multiple tumor biopsies may fail to identify the relapse-seeding clone. In this study, we therefore focused on the genomic evolution of neuroblastoma tumors and bone marrow-derived disseminated tumor cells (DTCs) by analyzing geographically and temporally separated samples of 155 stage 4 neuroblastoma patients. Our data support a branched clonal evolution and a parallel progression of primary and metastatic tumor cells. We provide evidence that the analysis of DTCs at diagnosis besides the tumor biopsies may improve the probability for detecting the relapse-seeding clone. This highlights the unmet need to study genetic changes in diagnostic DTCs alongside tumor tissue to better characterize and predict possible relapses.
Acknowledgments
We would like to thank Dr. Christian Frech for bioinformatics consultations, Dr. Ingrid Pribill for providing clinical data, and Dr. Dominik Bogen and Marion Zavadil MSc for proof reading the manuscript. The research leading to these results has received funding from St. Anna Kinderkrebsforschung; Austrian National Bank (ÖNB), Grant Nos. 15114, 16611; Austrian Science Fund (FWF), Grant No. I 2799-B28; and the European Union’s Seventh Framework Program (FP7/2007-2013) under the project ENCCA, grant agreement HEALTH-F2-2011-261474.
Abbreviations
- ALT
alternative lengthening of telomeres
- BM
bone marrow
- cnLOH
copy neutral loss of heterozygosity
- CT
computational tomography
- DMSO
dimethylsulfoxide
- DTC
disseminated tumor cell
- EFS
event-free survival
- HVA
homovanillic acid
- ITH
intra-tumor heterogeneity
- LDH
lactic dehydrogenase
- LOH
loss of heterozygosity
- LOY
loss of chromosome Y
- Mb
megabase
- MIBG
metaiodobenzylguanidine
- MNA
MYCN amplification
- NSE
neuron-specific enolase
- OS
overall survival
- SCA
segmental chromosomal aberration
- SNP
single nucleotide polymorphism
- SRO
smallest region of overlap
- VMA
vanillylmandelic acid
Footnotes
The authors declare no potential conflicts of interest.
References
- 1.Brodeur GM, Bagatell R. Mechanisms of neuroblastoma regression. Nature reviews Clinical oncology. 2014;11:704–13. doi: 10.1038/nrclinonc.2014.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ambros IM, Zellner A, Roald B, Amann G, Ladenstein R, Printz D, et al. Role of ploidy, chromosome 1p, and Schwann cells in the maturation of neuroblastoma. N Engl J Med. 1996;334:1505–11. doi: 10.1056/NEJM199606063342304. [DOI] [PubMed] [Google Scholar]
- 3.D'Angio GJ, Evans AE, Koop CE. Special pattern of widespread neuroblastoma with a favourable prognosis. Lancet. 1971;1:1046–9. doi: 10.1016/s0140-6736(71)91606-0. [DOI] [PubMed] [Google Scholar]
- 4.Evans AE, Gerson J, Schnaufer L. Spontaneous regression of neuroblastoma. National Cancer Institute monograph. 1976;44:49–54. [PubMed] [Google Scholar]
- 5.Caren H, Kryh H, Nethander M, Sjoberg RM, Trager C, Nilsson S, et al. High-risk neuroblastoma tumors with 11q-deletion display a poor prognostic, chromosome instability phenotype with later onset. Proc Natl Acad Sci U S A. 2010;107:4323–8. doi: 10.1073/pnas.0910684107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheung NK, Zhang J, Lu C, Parker M, Bahrami A, Tickoo SK, et al. Association of age at diagnosis and genetic mutations in patients with neuroblastoma. JAMA. 2012;307:1062–71. doi: 10.1001/jama.2012.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohn SL, Pearson AD, London WB, Monclair T, Ambros PF, Brodeur GM, et al. The International Neuroblastoma Risk Group (INRG) classification system: an INRG Task Force report. J Clin Oncol. 2009;27:289–97. doi: 10.1200/JCO.2008.16.6785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martinsson T, Eriksson T, Abrahamsson J, Caren H, Hansson M, Kogner P, et al. Appearance of the novel activating F1174S ALK mutation in neuroblastoma correlates with aggressive tumor progression and unresponsiveness to therapy. Cancer Res. 2011;71:98–105. doi: 10.1158/0008-5472.CAN-10-2366. [DOI] [PubMed] [Google Scholar]
- 9.Molenaar JJ, Koster J, Zwijnenburg DA, van Sluis P, Valentijn LJ, van der Ploeg I, et al. Sequencing of neuroblastoma identifies chromothripsis and defects in neuritogenesis genes. Nature. 2012;483:589–93. doi: 10.1038/nature10910. [DOI] [PubMed] [Google Scholar]
- 10.Peifer M, Hertwig F, Roels F, Dreidax D, Gartlgruber M, Menon R, et al. Telomerase activation by genomic rearrangements in high-risk neuroblastoma. Nature. 2015 doi: 10.1038/nature14980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pugh TJ, Morozova O, Attiyeh EF, Asgharzadeh S, Wei JS, Auclair D, et al. The genetic landscape of high-risk neuroblastoma. Nat Genet. 2013;45:279–84. doi: 10.1038/ng.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Valentijn LJ, Koster J, Zwijnenburg DA, Hasselt NE, van Sluis P, Volckmann R, et al. TERT rearrangements are frequent in neuroblastoma and identify aggressive tumors. Nat Genet. 2015 doi: 10.1038/ng.3438. [DOI] [PubMed] [Google Scholar]
- 13.Matthay KK, Maris JM, Schleiermacher G, Nakagawara A, Mackall CL, Diller L, et al. Neuroblastoma. Nature reviews Disease primers. 2016;2:16078. doi: 10.1038/nrdp.2016.78. [DOI] [PubMed] [Google Scholar]
- 14.Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–92. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mroz EA, Tward AD, Pickering CR, Myers JN, Ferris RL, Rocco JW. High intratumor genetic heterogeneity is related to worse outcome in patients with head and neck squamous cell carcinoma. Cancer. 2013;119:3034–42. doi: 10.1002/cncr.28150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ambros PF, Ambros IM, Kerbl R, Luegmayr A, Rumpler S, Ladenstein R, et al. Intratumoural heterogeneity of 1p deletions and MYCN amplification in neuroblastomas. Med Pediatr Oncol. 2001;36:1–4. doi: 10.1002/1096-911X(20010101)36:1<1::AID-MPO1002>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 17.Berbegall AP, Villamon E, Piqueras M, Tadeo I, Djos A, Ambros PF, et al. Comparative genetic study of intratumoral heterogenous MYCN amplified neuroblastoma versus aggressive genetic profile neuroblastic tumors. Oncogene. 2015 doi: 10.1038/onc.2015.200. [DOI] [PubMed] [Google Scholar]
- 18.Bogen D, Brunner C, Walder D, Ziegler A, Abbasi R, Ladenstein RL, et al. The genetic tumor background is an important determinant for heterogeneous MYCN-amplified neuroblastoma. Int J Cancer. 2016 doi: 10.1002/ijc.30050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lundberg G, Jin Y, Sehic D, Ora I, Versteeg R, Gisselsson D. Intratumour diversity of chromosome copy numbers in neuroblastoma mediated by on-going chromosome loss from a polyploid state. PLoS One. 2013;8:e59268. doi: 10.1371/journal.pone.0059268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Villamon E, Berbegall AP, Piqueras M, Tadeo I, Castel V, Djos A, et al. Genetic instability and intratumoral heterogeneity in neuroblastoma with MYCN amplification plus 11q deletion. PLoS One. 2013;8:e53740. doi: 10.1371/journal.pone.0053740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marrano P, Irwin MS, Thorner PS. Heterogeneity of MYCN amplification in neuroblastoma at diagnosis, treatment, relapse, and metastasis. Genes Chromosomes Cancer. 2017;56:28–41. doi: 10.1002/gcc.22398. [DOI] [PubMed] [Google Scholar]
- 22.Mehes G, Luegmayr A, Ambros IM, Ladenstein R, Ambros PF. Combined automatic immunological and molecular cytogenetic analysis allows exact identification and quantification of tumor cells in the bone marrow. Clin Cancer Res. 2001;7:1969–75. [PubMed] [Google Scholar]
- 23.Abbasi MR, Rifatbegovic F, Brunner C, Ladenstein R, Ambros IM, Ambros PF. Bone marrows from neuroblastoma patients: An excellent source for tumor genome analyses. Molecular oncology. 2015;9:545–54. doi: 10.1016/j.molonc.2014.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rifatbegovic F, Abbasi MR, Taschner-Mandl S, Kauer M, Weinhausel A, Handgretinger R, et al. Enriched Bone Marrow Derived Disseminated Neuroblastoma Cells Can Be a Reliable Source for Gene Expression Studies-A Validation Study. PLoS One. 2015;10:e0137995. doi: 10.1371/journal.pone.0137995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kang Y, Pantel K. Tumor cell dissemination: emerging biological insights from animal models and cancer patients. Cancer cell. 2013;23:573–81. doi: 10.1016/j.ccr.2013.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klein CA. Parallel progression of primary tumours and metastases. Nat Rev Cancer. 2009;9:302–12. doi: 10.1038/nrc2627. [DOI] [PubMed] [Google Scholar]
- 27.Klein CA. Framework models of tumor dormancy from patient-derived observations. Current opinion in genetics & development. 2011;21:42–9. doi: 10.1016/j.gde.2010.10.011. [DOI] [PubMed] [Google Scholar]
- 28.Ambros PF, Mehes G, Hattinger C, Ambros IM, Luegmayr A, Ladenstein R, et al. Unequivocal identification of disseminated tumor cells in the bone marrow by combining immunological and genetic approaches--functional and prognostic information. Leukemia. 2001;15:275–7. doi: 10.1038/sj.leu.2402023. [DOI] [PubMed] [Google Scholar]
- 29.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ambros IM, Brunner C, Abbasi R, Frech C, Ambros PF. Ultra-High Density SNParray in Neuroblastoma Molecular Diagnostics. Front Oncol. 2014;4:202. doi: 10.3389/fonc.2014.00202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Conlin LK, Thiel BD, Bonnemann CG, Medne L, Ernst LM, Zackai EH, et al. Mechanisms of mosaicism, chimerism and uniparental disomy identified by single nucleotide polymorphism array analysis. Human molecular genetics. 2010;19:1263–75. doi: 10.1093/hmg/ddq003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liang K-Y, Zegner SL. Longitudinal data analysis using generalized linear models. Biometrika. 1986;73:13–22. [Google Scholar]
- 33.Asgharzadeh S, Pique-Regi R, Sposto R, Wang H, Yang Y, Shimada H, et al. Prognostic significance of gene expression profiles of metastatic neuroblastomas lacking MYCN gene amplification. Journal of the National Cancer Institute. 2006;98:1193–203. doi: 10.1093/jnci/djj330. [DOI] [PubMed] [Google Scholar]
- 34.Oberthuer A, Berthold F, Warnat P, Hero B, Kahlert Y, Spitz R, et al. Customized oligonucleotide microarray gene expression-based classification of neuroblastoma patients outperforms current clinical risk stratification. J Clin Oncol. 2006;24:5070–8. doi: 10.1200/JCO.2006.06.1879. [DOI] [PubMed] [Google Scholar]
- 35.Kocak H, Ackermann S, Hero B, Kahlert Y, Oberthuer A, Juraeva D, et al. Hox-C9 activates the intrinsic pathway of apoptosis and is associated with spontaneous regression in neuroblastoma. Cell death & disease. 2013;4:e586. doi: 10.1038/cddis.2013.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Su Z, Fang H, Hong H, Shi L, Zhang W, Zhang W, et al. An investigation of biomarkers derived from legacy microarray data for their utility in the RNA-seq era. Genome biology. 2014;15:523. doi: 10.1186/s13059-014-0523-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eleveld TF, Oldridge DA, Bernard V, Koster J, Daage LC, Diskin SJ, et al. Relapsed neuroblastomas show frequent RAS-MAPK pathway mutations. Nat Genet. 2015 doi: 10.1038/ng.3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schleiermacher G, Javanmardi N, Bernard V, Leroy Q, Cappo J, Rio Frio T, et al. Emergence of new ALK mutations at relapse of neuroblastoma. J Clin Oncol. 2014;32:2727–34. doi: 10.1200/JCO.2013.54.0674. [DOI] [PubMed] [Google Scholar]
- 39.Schramm A, Koster J, Assenov Y, Althoff K, Peifer M, Mahlow E, et al. Mutational dynamics between primary and relapse neuroblastomas. Nat Genet. 2015 doi: 10.1038/ng.3349. [DOI] [PubMed] [Google Scholar]
- 40.Schleiermacher G, Janoueix-Lerosey I, Ribeiro A, Klijanienko J, Couturier J, Pierron G, et al. Accumulation of segmental alterations determines progression in neuroblastoma. J Clin Oncol. 2010;28:3122–30. doi: 10.1200/JCO.2009.26.7955. [DOI] [PubMed] [Google Scholar]
- 41.Fieuw A, Kumps C, Schramm A, Pattyn F, Menten B, Antonacci F, et al. Identification of a novel recurrent 1q42.2-1qter deletion in high risk MYCN single copy 11q deleted neuroblastomas. Int J Cancer. 2012;130:2599–606. doi: 10.1002/ijc.26317. [DOI] [PubMed] [Google Scholar]
- 42.De Vries L, Zheng B, Fischer T, Elenko E, Farquhar MG. The regulator of G protein signaling family. Annual review of pharmacology and toxicology. 2000;40:235–71. doi: 10.1146/annurev.pharmtox.40.1.235. [DOI] [PubMed] [Google Scholar]
- 43.Hollinger S, Hepler JR. Cellular regulation of RGS proteins: modulators and integrators of G protein signaling. Pharmacological reviews. 2002;54:527–59. doi: 10.1124/pr.54.3.527. [DOI] [PubMed] [Google Scholar]
- 44.Ricketts CJ, Morris MR, Gentle D, Shuib S, Brown M, Clarke N, et al. Methylation profiling and evaluation of demethylating therapy in renal cell carcinoma. Clinical epigenetics. 2013;5:16. doi: 10.1186/1868-7083-5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cohen MM., Jr The AKT genes and their roles in various disorders. American journal of medical genetics Part A. 2013;161A:2931–7. doi: 10.1002/ajmg.a.36101. [DOI] [PubMed] [Google Scholar]
- 46.Cheung M, Testa JR. Diverse mechanisms of AKT pathway activation in human malignancy. Current cancer drug targets. 2013;13:234–44. doi: 10.2174/1568009611313030002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grottke A, Ewald F, Lange T, Norz D, Herzberger C, Bach J, et al. Downregulation of AKT3 Increases Migration and Metastasis in Triple Negative Breast Cancer Cells by Upregulating S100A4. PLoS One. 2016;11:e0146370. doi: 10.1371/journal.pone.0146370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yamada K, Ono M, Bensaddek D, Lamond AI, Rocha S. FMN2 is a novel regulator of the cyclin-dependent kinase inhibitor p21. Cell cycle. 2013;12:2348–54. doi: 10.4161/cc.25511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yamada K, Ono M, Perkins ND, Rocha S, Lamond AI. Identification and functional characterization of FMN2, a regulator of the cyclin-dependent kinase inhibitor p21. Molecular cell. 2013;49:922–33. doi: 10.1016/j.molcel.2012.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cobrinik D, Ostrovnaya I, Hassimi M, Tickoo SK, Cheung IY, Cheung NK. Recurrent pre-existing and acquired DNA copy number alterations, including focal TERT gains, in neuroblastoma central nervous system metastases. Genes Chromosomes Cancer. 2013;52:1150–66. doi: 10.1002/gcc.22110. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.