SUMMARY
The ESCRT-III complex induces outward membrane budding and fission through homotypic polymerization of its core component Shrub/CHMP4B. Shrub activity is regulated by its direct interaction with a protein called Lgd in flies, or CC2D1A or B in humans. Here, we report the structural basis for this interaction, and propose a mechanism for regulation of polymer assembly. The isolated third DM14 repeat of Lgd binds Shrub, and an Lgd fragment containing only this DM14 repeat and its C-terminal C2 domain is sufficient for in vivo function. The DM14 domain forms a helical hairpin with a conserved, positively charged tip, that, in the structure of a DM14 domain-Shrub complex, occupies a negatively charged surface of Shrub that is otherwise used for homopolymerization. Lgd mutations at this interface disrupt its function in flies, confirming functional importance. Together, these data argue that Lgd regulates ESCRT activity by controlling access to the Shrub self-assembly surface.
Graphical Abstract
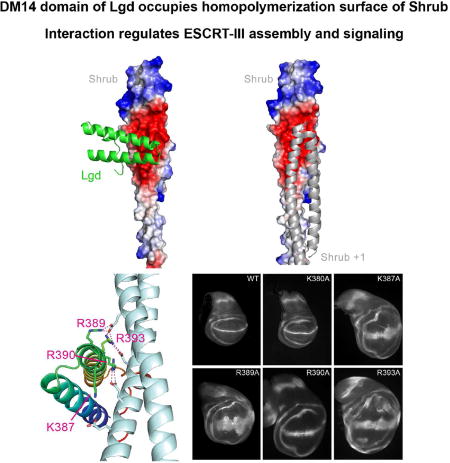
Shrub is a Drosophila ESCRT-III protein that self-associates to promote membrane budding and fission. Its activity is modulated by binding to Lgd, which suppresses self-association. McMillan et al. report the structural basis for masking of the Shrub self-association surface of Shrub by Lgd, which, together with functional studies in flies, suggests models for modulation of Shrub activity by Lgd.
INTRODUCTION
ESCRT (Endosomal Sorting Complex Required for Transport) complexes control the budding and abscission of membranes away from the cytosol (reviewed in (Hurley, 2015)). They are critical in many different cellular processes, such as formation of intraluminal vesicles (ILVs), membrane repair, and cytokinesis. The pathway is also co-opted by many membrane-enveloped viruses, such as HIV and Ebola virus, to exit the infected cell (Carlton and Martin-Serrano, 2007; Morita et al., 2007; Morita et al., 2011). The ESCRT-III complex functions via polymerization of Snf7 domain containing proteins at the cytosolic face of the membrane, and this process directly deforms and remodels the lipid bilayer (Carlson and Hurley, 2012; Saksena et al., 2009; Wollert et al., 2009).
Recently, the structural basis for Snf7-domain assembly was determined for homopolymers of the “core” Snf7 (yeast) and Shrub (fly) proteins (McMillan et al., 2016; Tang et al., 2016b), and for the related “accessory” CHMP1B/IST1 heteropolymer (McCullough et al., 2015). For Shrub and Snf7, staggered head-to-head polymerization is promoted by interaction of complementary surfaces at the “receiver” and “donor” faces of the helical hairpin structure. It is believed that ESCRT-III proteins cycle between a “closed” monomeric form in the cytosol and the active and “open”, polymeric form at the membrane surface, but the regulation and control of this process is poorly understood.
In animals, proper ESCRT-III function requires modulation of Shrub activity by the large multi-domain protein called Lethal (2) giant discs (Lgd) in flies, or Coiled-Coil and C2 Domain Containing proteins 1A and 1B (CC2D1A and B) in mammals (Drusenheimer et al., 2015; Troost et al., 2012). Loss of Lgd in flies results in aberrant sorting and signalling of membrane proteins such as Notch (Jaekel and Klein, 2006; Klein, 2003; Schneider et al., 2013), and produces defects in cell division (Matias et al., 2015; Morawa et al., 2015). In mice, loss of CC2D1A results in postnatal lethality from respiratory distress caused by central nervous system defects (Drusenheimer et al., 2015; Zhao et al., 2011). In humans, CC2D1A mutation is associated with a spectrum of neurological disorders, including intellectual disability, autism spectrum disorder, and seizures (Basel-Vanagaite et al., 2006; Manzini et al., 2014).
Lgd and CC2D1A/B contain four N-terminal repeats of a DM14 (Drosophila melanogaster 14) domain followed by a C2 domain at the C-terminus. There is substantial sequence divergence among the DM14 domains: the sequences of DM14-1 and DM14-3 are more similar to each other than to DM14-2 or DM14-4; likewise the sequences of DM14-2 and DM14-4 are more similar to each other than to DM14-1 and DM14-3. Our previous studies in flies established that truncated forms of Lgd that combine at least one “odd” DM14 domain with one “even” domain are sufficient to rescue lgd loss-of-function mutants (Troost et al., 2012). For the human homolog CC2D1A, the third DM14 repeat is sufficient for binding to CHMP4B monomers and this interaction readily inhibits CHMP4 polymerization (Martinelli et al., 2012).
How Lgd and CC2D1A regulate ESCRT-III function is not well understood. On the one hand, CC2D1A negatively regulates ESCRT-III polymerization in vitro (Martinelli et al., 2012) and prevents HIV budding in mammalian cell culture (Usami et al., 2012). Based on these observations, it was suggested that CC2D1A is an antagonist of ESCRT-IIII function. On the other hand, there is a strong genetic interaction between Lgd and Shrub in flies suggesting that Lgd is a positive regulator of Shrub (Troost et al., 2012). When one copy of shrub is removed, lgd mutants die in the early third instar larval stage, rather than in the pupal stage. The earlier time of death indicates an intimate relationship between Shrub and Lgd, and suggest that Lgd is a positive regulator of Shrub and ESCRT-III function. The apparent contradiction between the in vivo and cell culture results is reconciled when the observed dosage sensitivity of Lgd is also considered. When Lgd is over-expressed in Drosophila, ectopic activation of the Notch pathway also occurs, indicating attenuation of Shrub activity similar to that seen in lgd loss-of-function mutants (Gallagher and Knoblich, 2006; Jaekel and Klein, 2006).
The DM14 domains are critically important for the function of Lgd and unique to the Lgd protein family, but they have not been previously described structurally. Here, we show that introduction of a minimal Lgd construct containing only the third DM14 domain and the C-terminal C2 domain is sufficient for rescue of lgd phenotypes and interaction with Shrub, while even a variant with two even-numbered DM14 domains (2 and 4) fails to do so. We report the crystal structure of the third DM14 domain, in its apo form, as well as in complex with a truncated version of the Shrub monomer. These structures reveal the DM14 domain to be a short helical hairpin, which in the case of the third repeat, contains a conserved positively charged tip that directly interacts with a complementary electrostatic surface on Shrub. This surface on Shrub overlaps with the site used for homotypic interaction and filamentation. The Lgd-Shrub and Shrub-Shrub interactions are mutually exclusive and thus offer an explanation as to how Lgd directly regulates ESCRT function.
RESULTS
The third DM14 repeat of Lgd is sufficient for ESCRT-III binding and cellular function
Lgd and its homologs contain four tandem repeats of a DM14 (Drosophila melanogaster 14) domain and a C-terminal C2 domain (Figure 1A). The C2 domain is required for proper subcellular localization and stability of Lgd (Troost et al., 2012), but is not believed to directly interact with ESCRT family proteins. In contrast, the DM14 region is required for the interaction with Shrub/CHMP4B (Martinelli et al., 2012; McMillan et al., 2016). The DM14 modules within Lgd diverge substantially at the level of primary sequence (Figure S1A, B). The most similar repeats, DM14-1 and DM14-3, share 43% sequence identity (Figure S1A). The DM14-1 and DM14-3 modules, however, each share only 22% identity with the “even” numbered DM14-2 or DM14-4 modules.
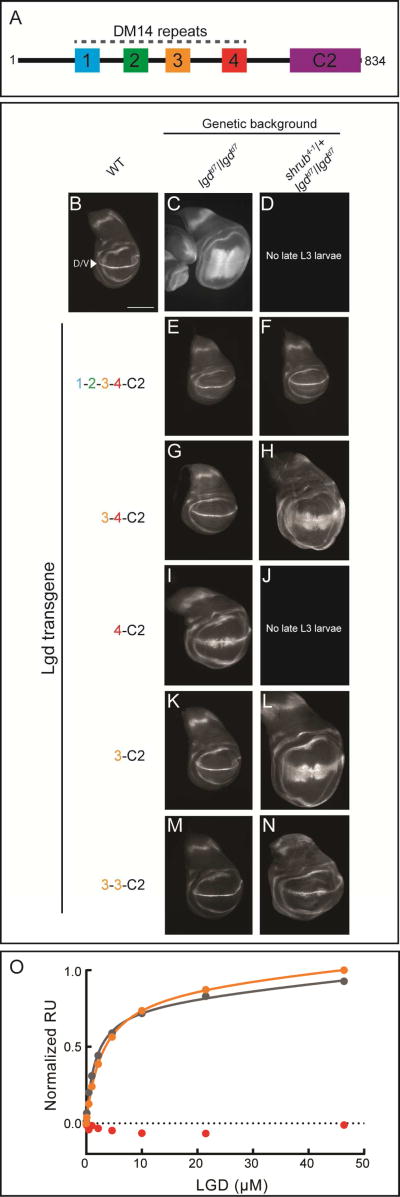
Figure 1.
A, Domain organization of Lgd, highlighting the four DM14 modules and C-terminal C2 domain. B-N, Wing imaginal discs of Drosophila late third instar larvae. B, a wildtype disc, stained for Wg. C-N, In each panel, the indicated Lgd construct was transgenically expressed at the same insertion site in lgd null (lgdd7/lgdd7), and lgd null/shrub heterozygous (lgdd7/lgdd7, shrub4-1/+) backgrounds. Each imaginal disc was stained for the Notch target gene, wingless (wg), as a surrogate for MVB dysfunction. Wingless expression is restricted to the dorsal/ventral (D/V) boundary line during normal development (arrowhead in B). Loss of ESCRT function results in ectopic broadening of the Wg D/V stripe, as seen in C. Lethality in early L3 stage of development was observed in parts D and J. Scale bar: 200 µm. O, Interaction of Shrub (6–106) with Lgd (DM14-3, orange; DM14-4, red; DM14-3+4, grey) detected using SPR. Response units are normalized to the molecular mass of each Lgd construct. See also Figures S1, S2, and Table S1.
To elucidate the role of the various DM14 repeats in Lgd function, we took advantage of the observation that lgd loss-of-function results in ectopic activation of the membrane receptor Notch in a ligand-independent manner in the developing fly wing disc (Jaekel and Klein, 2006; Klein, 2003). Indeed, a spectrum of lgd deficiencies can be distinguished by monitoring the relative degree of ectopic Notch signalling in this developmental context (Figure 1B–D): the lgd null (lgdd7/lgdd7) genotype results in ectopic activation of Notch away from the dorsal/ventral (D/V) boundary, as measured by the downstream target gene wingless (wg) (Figure 1C), and the lgd null background, when further sensitized to ESCRT perturbation (Figure 1D) by loss of a single functional copy of shrub (shrub4-1/+, lgdd7/lgdd7, hereafter, “sensitized null background”), results in lethality at the beginning of the L3 stage of development (Troost et al. 2012; Figure 1D).
To assess the functionality of various forms of Lgd, therefore, we probed for their ability to suppress the simple and sensitized lgd null genetic backgrounds in a rescue assay. Each Lgd protein was transgenically expressed under control of the endogenous lgd promoter, and all constructs were inserted in the 86Fb genomic landing site. Introduction of the wildtype Lgd protein rescues the Notch phenotype in both the lgd null and sensitized null backgrounds (Figure 1E–F). Introduction of a truncated Lgd protein containing only the 3rd and 4th DM14 repeats and the C-terminal C2 (3-4-C2) rescued the lgd null phenotype (Figure 1G). The sensitized null background was partially rescued, allowing development until the pupal stages and permitting the analysis of wing discs, but ectopic Notch activation was still observed, suggesting less activity than the full-length construct (Figure 1H). Further truncation of the 3rd DM14 repeat (4-C2) resulted in ectopic Notch activation and pupal lethality even in the lgd null background (Figure 1I–J), consistent with a critical role for DM14-3. A protein containing only the DM14-3 domain and the C-terminal C2 domain (3-C2) was sufficient for rescue of the lgd null phenotype, and development of the shrub4-1 sensitized background beyond the L3 stage (Figure 1K–L). Moderate ectopic activation of the Notch pathway, however, was still observed in the null sensitized background, comparable to that seen with the 3-4-C2 protein (compare Figures 1H and 1L). A second copy of the DM14-3 module (3-3-C2) further attenuated the shrub sensitized, lgd null phenotype (Figure 1M–N). These results indicate that the minimal DM14-3 module is capable of regulating ESCRT function within the cell, likely through its direct interaction with Shrub (Troost et al., 2012).
To begin investigating the biochemical basis for modulation of Shrub activity by Lgd, we analyzed the binding of a polymerization-competent fragment of Shrub (6–106) to the DM14-3 domain (3596–423) using surface plasmon resonance (SPR). For comparison, the binding of Lgd fragments either containing both the 3rd and 4th (3596–560) or only the 4th DM14 domain (DM14-4, 496–560) to Shrub were also measured. Whereas both proteins containing DM14-3 bound to Shrub and exhibited similar affinities (Figure 1O, Table S1), binding of the isolated DM14-4 module to Shrub was not detected (Figure 1O). The DM14-3 protein also inhibits Shrub polymerization in a purified system (McMillan et al., 2016). These observations mirror and complement similar cell culture and in vitro studies carried out with the mammalian homologs of Lgd and Shrub, CC2D1A and CHMP4B, respectively, which also show that the DM14-3 repeat of CC2D1A is sufficient for CHMP4B binding (Martinelli et al., 2012).
Crystal structure of the DM14-3 domain
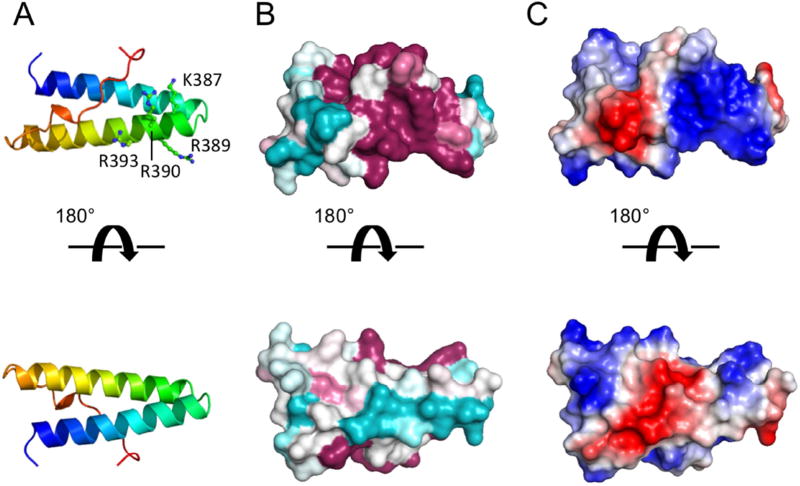
In our effort to uncover the structural basis for binding of Shrub by Lgd, we crystallized the third DM14 domain of Lgd (residues 3596–423) and determined its structure to 1.1 Å resolution by X-ray crystallography (Table 1). The DM14 domain is a short alpha-helical hairpin motif, ~40 Å in length. The 16-residue C-terminal proline-rich tail of the domain curls back into a grooved face of the hairpin, primarily formed by aromatic side-chains that lie between the two helices of the hairpin (Figure 2A, Figure S2A). Near the loop of the hairpin, there is a conserved, positively charged surface patch (Figure 2B,C) created by a cluster of basic residues that includes K387, R389, R390, and R393 of helix two. In the DM14-4 module that fails to bind Shrub (6–106), the residues analogous to K387 and R390 of DM14-3 are Q527 and E530, respectively (Figure S1B), suggesting that the surface of DM14-4 will lack an analogous positively charged surface. Indeed, the DM14 repeats fall into two classes of charge distribution: i) the odd numbered repeats (1 and 3) which have this cluster of basic residues on the second helix, and ii) the even numbered repeats (2 and 4), which do not (see Figure S2B).
Table 1.
Crystallographic data collection and refinement
| Data collection | DM14-3 (SAD) | DM14-3:Shrub (MR) |
|---|---|---|
| Space group | P 41 21 2 | C 1 2 1 |
| Cell dimensions | ||
| a, b, c (Å) | 29.67, 29.67, 139.11 | 120.97, 26.99, 82.84 |
| α, β, γ (°) | 90, 90, 90 | 90, 125.95, 90 |
| Wavelength (Å) | 0.9792 | 0.9792 |
| Resolution (Å) | 50-1.10 (1.12-1.10) | 49-2.00 (2.08-2.00) |
| Rmeas | 7.3 (96.2) | 7.8 (181) |
| CC(1/2) | 99.8 (60.8) | 99.9 (36.5) |
| Completeness (%) | 96.4 (62.3) | 97.4 (93.8) |
| I/ σI | 11.6 (1.13) | 11.4 (0.95) |
| Redundancy | 5.8 (1.6) | 3.5 (3.6) |
|
| ||
| Refinement | ||
|
| ||
| Resolution (Å) | 34.78-1.10 | 49-2.00 |
| No. reflections | 46256 | 51632 |
| Rwork/Rfree | 0.170/0.178 | 0.240/0.269 |
| No. atoms | ||
| Protein | 1032 | 1408 |
| Water | 118 | 68 |
| B-factors | ||
| Protein | 14.0 | 62 |
| Water | 28.4 | 56 |
| Rms deviations | ||
| Bond lengths (Å) | 0.006 | 0.001 |
| Bond angles (°) | 0.897 | 0.37 |
| Ramachandran (%) | ||
| Favored | 100 | 98.3 |
| Allowed | 0 | 1.7 |
| Outlier | 0 | 0 |
| Rotamer Outliers (%) | 0 | 0.67 |
| Clashscore | 0 | 2.46 |
Figure 2.
Structure of the third DM14 domain (residues 359–423) of Lgd. A, Cartoon representation, colored on a sliding scale from N-terminus (blue) to C-terminus (red). B, Surface representation, colored by conservation on a sliding scale from cyan (least conserved) to maroon (most conserved). C, Surface representation colored by electrostatic potential from negative (red) to positive (blue). See also Figure S2.
To investigate potential functional differences between the “odd” and “even” repeats, we tested for phenotypic rescue in lgd null and shrub sensitized genetic backgrounds (Figure S2C–F). Lgd molecules with deletion of the “even” DM14-2 and DM14-4 repeats were capable of full phenotypic rescue of both the lgd null and sensitized null mutant backgrounds (Figure S2C–D), similar to the rescue observed with full-length Lgd (Figure 1E–F). In contrast, deletion of the “odd” repeats resulted in failure to rescue either of the two backgrounds (Figure S2E–F).
Structural basis for interaction between DM14-3 and Shrub
Prior attempts to crystallize a stable DM14-Shrub complex (Martinelli et al., 2012; McMillan et al., 2016) have resulted in structures containing only the Shrub component. To circumvent the difficulty of preventing DM14-3 dissociation during crystallization, we generated a single fusion protein that contains, from N- to C-terminus, the DM14-3 module of Lgd (359–422), an 11-residue glycine linker, and a truncated form of Shrub (17–130). Point mutations R59E and R70E were also installed in Shrub to further suppress polymerization (McMillan et al., 2016). The fusion protein crystallized, and we were able to solve its X-ray structure to 2.0 Å resolution by molecular replacement using the isolated DM14-3 (this work) and Shrub (PDB 5J45) structures as search models. In the structure representing the complex, the helical hairpins of Lgd and Shrub are oriented nearly perpendicularly to each other, with a crossing angle of ~110 ° (Figure 3A, left). The polyglycine linker is not observed in the structure, strongly suggesting that the orientation of the DM14 domain with respect to Shrub has not been constrained by the linker. In the complex, the electron density for the first helix of the DM14 domain is weak by comparison to the rest of the structure, resulting in high overall crystallographic R-factors after refinement (Table 1), and it is possible that this helix can adopt a second conformation with low occupancy. Notwithstanding this nuance about the first helix, however, a 2FO-FC map contoured at 1.5σ (Figure S3) clearly shows strong density for the residues at the DM14-Shrub interface described below.
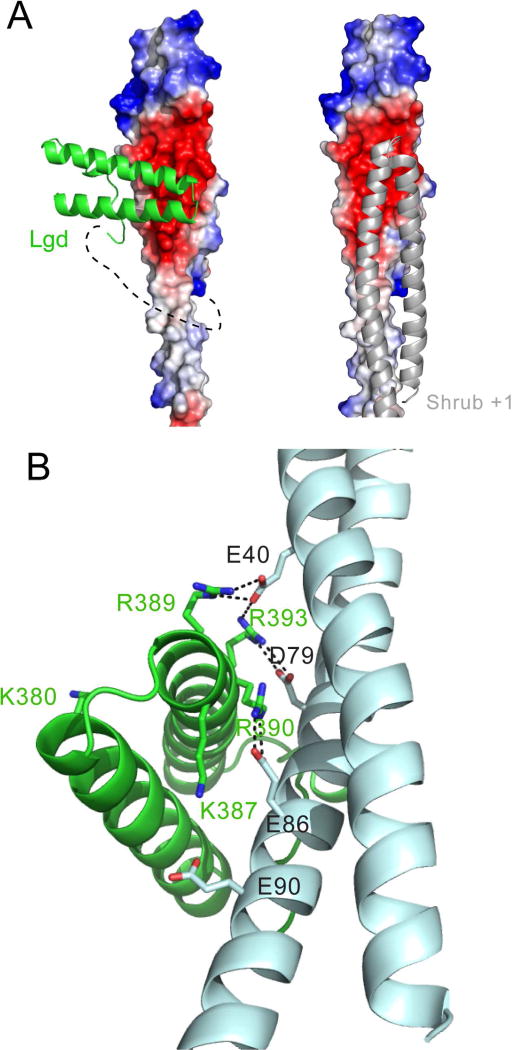
Figure 3.
Crystal structure of the Lgd-Shrub fusion protein. A, Left panel: Lgd-Shrub complex. DM14-3 is shown as a cartoon (green), and Shrub as an electrostatic surface on a sliding scale from negative (red) to positive (blue). The polyglycine linker connecting Lgd and Shrub, illustrated here with a dotted line, is not visible in the structure. Right panel: crystallographic model of a Shrub homopolymer (based on PDB entry 5J45). B, Key residues at the Lgd:Shrub binding interface. Lgd is shown as a cartoon in green. Shrub is shown as a cartoon in cyan. Charged residues at the interface are shown as sticks with salt bridges illustrated by dotted lines. See also Figure S3.
The character of the interaction is predominantly electrostatic, as the basic face of DM14-3 directly contacts the acidic face of Shrub. In a crystallographic model of the Shrub homopolymer (derived from pdb 5J45), this acidic patch also serves as the acceptor for the basic face of the +1 Shrub protomer (Figure 3A, right) and is required for in vitro polymerization (McMillan et al., 2016). The C-terminal proline-rich tail of DM14-3 runs parallel to the first helix of Shrub and makes additional van der Waals interactions with Shrub. In total, 576 Å2 are buried at the Lgd-Shrub interface.
There is a network of electrostatic interactions at the Lgd-Shrub interface (Figure 3B), in which a series of arginine side chains of the DM14-3 domain of Lgd lie on the same face of consecutive turns of the second helix, engaging the negatively charged surface of Shrub. R389 and R393 of Lgd make electrostatic contacts with E40 of Shrub, R393 of Lgd forms a salt-bridge with D79 of Shrub, and R390 of Lgd interacts with E86 of Shrub. K387 of Lgd and E90 of Shrub also contribute to the local surface electrostatic potential of the two proteins, but are not within hydrogen bonding distance in the crystal structure. Notably, all three acidic residues of Shrub (E40, D79, and E86) that form salt bridges with Lgd also form a salt bridge with a neighbouring Shrub protomer in the crystallographic homopolymer (PDB ID code 5J45).
Mutation of the Lgd-Shrub interface blocks binding and cellular function
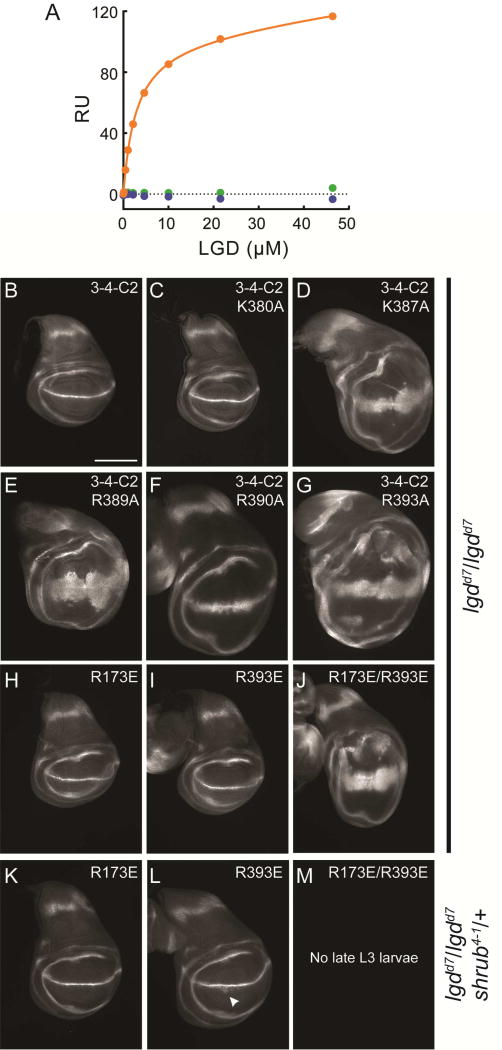
The interface responsible for the interaction of Lgd with Shrub in the X-ray structure was interrogated using site-directed mutagenesis of charged residues at the contact interface on each protein. In a binding assay using SPR, an R393A mutation in Lgd abolished detectable binding to Shrub (Figure 4A). On the Shrub side of the interface, detectable binding was eliminated by an E86R mutation (Figure 4A), also consistent with the crystallographic model.
Figure 4.
Analysis of the Lgd-Shrub interface. A, Interaction between DM14 domain three of Lgd (residues 359–423) and Shrub (6–106) measured using SPR. Interaction between wildtype molecules is shown in orange. Interaction between wildtype Shrub and Lgd R393A is shown in blue. Interaction between Shrub E86R and wildtype Lgd is shown in green. B-G, Truncated Lgd (3-4-C2 construct) molecules with the listed mutations were transgenically expressed in the indicated genetic backgrounds. Fly wing discs were stained for Wg expression as a surrogate for MVB dysfunction. H-M, Full-length Lgd molecules (1-2-3-4-C2) with the listed mutations were transgenically expressed in the indicated genetic backgrounds. Fly wing discs were stained for Wg expression as a surrogate for MVB dysfunction. Scale bar: 200 µm. See also Table S1.
The Lgd-Shrub interface was also probed in the in vivo assay of ESCRT function described above. In order to avoid disrupting the interface required for Shrub self-association into the ESCRT-III polymer, we focused our analysis on the effects of point mutations in the DM14-3 module of Lgd. Single alanine mutations were generated at five basic residues of the DM14-3 module, in the context of the truncated Lgd protein containing the DM14-3 repeat through the C-terminus (3-4-C2). Four of these mutations were at the putative electrostatic interface (K387A, R389A, R390A, R393A), while an additional control K380A mutation was introduced onto the face of DM14-3 that points away from the interface (Figure 3B). Single alanine substitutions of each interface residue disrupted phenotypic rescue of the lgdd7/lgdd7 background (Figure 4D–G), whereas the K380A control mutation had no effect (Figure 4C).
Mutation of the electrostatic interface was also probed in the context of the full-length Lgd molecule. Prior experiments suggested the potential for partial functional redundancy between the first and third DM14 repeat (Figures 1 and S2). Notably, DM14-1 shares sequence identity with the DM14-3 repeat at the residues equivalent to K387, R389, R390, and R393 (K167, R169, R170 and R173, Figure S1B). Full-length Lgd constructs were thus generated with glutamate mutations at equivalent positions in DM14-1 (R173E) or DM14-3 (R393E), and tested for phenotypic rescue. Transgenic expression of full-length Lgd with the R173E mutation in the first repeat rescued the lgd and sensitized null background (Figure 4H, K); Lgd with the R393E also rescued the lgd mutant background (Figure 4I), but provided less complete rescue of the sensitized null background (Figure 4L). Full-length Lgd with mutations in both DM14-1 and DM14-3 (R173E/R393E), however, did not rescue the lgd null phenotype (Figure 4J). Moreover, this construct failed to rescue the early L3 lethality of the sensitized null background, indicating that it lacks detectable Lgd function in this phenotypic assay (Figure 4M). Overall, these results are consistent with partial functional redundancy between the electrostatically similar DM14-1 and DM14-3 repeats, but suggest that DM14-3 may have a more dominant role in Lgd function.
DISCUSSION
Dynamic assembly of the ESCRT-III polymer at the membrane induces budding and fission. Strict control of this process is required, and many cellular components have been shown to direct Shrub to the membrane, and subsequently trigger assembly of the polymer (McCullough et al., 2013; Tang et al., 2016a). In the absence of this activation trigger, Shrub is maintained in a monomeric state within the cytosol. Conversion from monomer to fission-active polymer is thought to require a major structural rearrangement from an auto-inhibited helical bundle to an elongated helical hairpin. In both forms, a negatively charged face of Shrub is surface exposed; a second, positive face is available upon remodelling and provides a complementary electrostatic surface for stacking of the protomers (McMillan et al., 2016; Tang et al., 2016b).
Our structural data show that Lgd binds the negatively changed face of Shrub in a manner that overlaps with the surface required for polymer assembly. Moreover, a single interaction-competent DM14 repeat is sufficient for rescue of the lgd null phenotype in vivo. The binding mode between Shrub and Lgd appears to be conserved in humans, as CC2D1A/Lgd2 and CC2D1B/Lgd1 can rescue lgd mutants in a shrb dependent manner (Drusenheimer et al., 2015) and charge reversal of the acidic face of CHMP4B, the mammalian homolog of Shrub, also disrupts interaction with human Lgd2/CC2D1A (Martinelli et al., 2012). In yeast, where the ESCRT pathway has been extensively defined, an Lgd ortholog appears to be lacking, suggesting that Lgd serves a regulatory role, as opposed to an obligate function in polymer assembly and ESCRT activity.
Genetic studies indicate that Lgd is a positive regulator of ESCRT function in vivo. Shrub heterozygosity, which is normally tolerated in the fly, is synthetically lethal when combined with Lgd loss of function (Troost et al., 2012). Since Lgd inhibits polymerization in vitro, it is not immediately clear how Shrub activity is instead potentiated by Lgd in the cell. Recent findings challenge the presumption that Snf7 proteins exclusively favor a closed form in the cytosol by showing that solubilized Snf7 is conformationally dynamic (Tang et al., 2016b). In addition, studies with the C. elegans ortholog of Shrub, Vps32, which exists in an oligomer-monomer equilibrium in solution and does not require additional factors for polymerization, raise the possibility of non-productive polymerization of shrb orthologs in the cytosol (Shen et al., 2014). Lgd, which should be competent for binding to both “open” and “closed” forms, could confer increased fidelity of polymerization by suppressing premature polymerization of the “open” form, and thus prevent depletion of monomeric Shrub subunits by reducing non-productive cytosolic polymerization. A second, not mutually exclusive possibility, is that Lgd actively aids in delivery of monomeric Shrub to sites of membrane remodelling, or increases the effective concentration of Shrub at these sites to stimulate productive polymerization. Further work will be required to determine whether Lgd plays an active role in assembling Shrub polymers or has a more passive role in maintenance of its monomeric building blocks and prevention of premature, non-productive polymerization.
The DM14 domain is a unique feature of the Lgd protein family. The third DM14 repeat of both the fly and human Lgd proteins independently binds Shrub/CHMP4B as an isolated protein module. In flies, the DM14-3 repeat even suffices in lieu of all four DM14 domains in partially suppressing lgd loss of function phenotypes. The mechanistic role of the remaining DM14 modules, however, is less clear. DM14-1 and DM14-3 are most similar at the level of sequence and electrostatic character, suggesting that DM14-1 may function similarly to DM14-3 by acting as a partner for Shrub monomers, with the requirement for its contribution revealed under more stringent conditions, such as shrub heterozygosity. The “even” numbered DM14 units are highly divergent in sequence from DM14-1 and DM14-3, and do not appear to be necessary or sufficient for interaction with Shrub. If these DM14 repeats are indeed functionally divergent, how do they contribute to the biological activity of Lgd? One possibility is that they bind to other Snf7 domain containing proteins, generating “2 + 2” complexes or assemblies that promote ESCRT activity. Alternatively, the additional DM14 units may have evolved new functions and protein interaction partners that are not core components of the ESCRT complex. Indeed, protein partners outside of the ESCRT pathway, such as phosphodiesterase 4D, have been identified (Al-Tawashi and Gehring, 2013). The structure and mechanism elucidated here for the DM14-3 subunit should provide a roadmap to our understanding of these additional interactions and cellular functions.
METHODS
Protein Production
Shrub (residues 6–106) and Lgd (359–423, 359–560, 496–560) constructs were cloned into a pETHSUL vector using BamHI and XhoI restriction sites. Point mutants were produced by site-directed mutagenesis. The Lgd-Shrub fusion protein was produced as a synthetic DNA encoding residues 359–422 of Lgd, an 11-residue glycine linker, and residues 17–130 of Shrub, with Shrub mutations R59E and R70E to further inhibit polymerization. Constructs were transformed into BL21(DE3) cells and expressed by IPTG induction at 18°C for 1 6 hr. Lgd and the Lgd-Shrub fusion protein were purified by affinity chromatography using HisPur Cobalt (Thermo Scientific) followed by cleavage of the His6-SUMO tag using Ulp1 peptidase. Untagged protein was then purified by size exclusion chromatography in 10 mM HEPES 7.4, 150 mM NaCl, 1 mM TCEP. Shrub protein was purified in 1M NaCl, as previously described, to maintain monomeric form (McMillan et al., 2016). For SPR experiments, the Sumo fusion was not cleaved from Shrub prior to purification.
Surface Plasmon Resonance
Sumo-Shrub was amine-coupled to CM5 sensor chips at a 500 RU density (GE Healthcare) following pre-concentration of 0.01 mg/ml protein in 50 mM acetate buffer pH 4.8, and 500 mM NaCl. The experimental running buffer was 10 mM HEPES 7.4, 150 mM NaCl, 1 mM TCEP, and 0.01% w/v Tween-20. Between analyte injections, the chip surface was regenerated with 10 mM HEPES 7.4, 2 M NaCl. Data were fit with Prism software using a one-site saturation with linear non-specific binding model.
Crystallization and data collection
DM14-3 (Lgd 359–423) was labeled with selenomethionine during growth in minimal media, and purified in 10 mM HEPES, 100 mM NaCl, and 1 mM TCEP. DM14-3 was crystallized in 96 well sitting drop plates using 30% PEG2000 MME, Tris pH 8.0, at 20 C. Crystals were cryopreserved by dipping in 40% PEG2000 MME, Tris pH 8.0 prior to plunging in liquid nitrogen. Lgd-Shrub fusion protein was crystallized in 96 well sitting drop plates using 18% PEG 3350, 0.1 M sodium citrate pH 5.5, at 4C. Crystals were cryopreserved by dipping in 20% glycerol, 25% PEG 3350, 0.1 M sodium citrate pH 5.4 before plungung into liquid nitrogen. Data were obtained at the Advanced Photon Source, Beam-line 24-ID-C (NE-CAT).
Structure Determination
Diffraction images were indexed and integrated using XDS (Kabsch, 2010). Initial phases for the DM14-3 structure were deduced by single wavelength anomalous diffraction in Phenix (Adams et al., 2010). The Lgd-Shrub fusion protein was solved by molecular replacement using the isolated DM14-3 structure and PDB 5J45 (Shrub) coordinates as search models. Refinement was performed in Phenix and Buster with manual building/review in COOT (Emsley et al., 2010).
Structure analysis and molecular graphics
Sequence alignments were performed using Clustal Omega (Sievers and Higgins, 2014). Homology models were generated using the Phyre2 server (Kelley et al., 2015) with energy minimization in Phenix. Buried surface area calculations were performed using PDBePISA (Krissinel and Henrick, 2007). Surface conservation scores were generated with the ConSurf server (Ashkenazy et al., 2016) using maximum likelihood calculation. Structure figures were produced using Pymol (Schrödinger).
Drosophila genetics
The following fly stocks were used: lgdd7 (Klein, 2003), shrub4-1 (Sweeney et al., 2006). The Lgd rescue constructs are based on constructs described previously (Troost et al., 2012). All DM14 deletion constructs include the intact N-terminus (amino acid 1–141) and the C2 domain with the C-terminus (amino acid 558–816). They were modified by side-directed mutagenesis or Gibson assembly. The constructs were inserted into the genomic attP landing site 86Fb (Bischof et al., 2007) by BestGene.
Wing imaginal discs were dissected from late L3 larvae of the particular genotypes. Antibody staining was performed according to standard prcedures. Antibodies used: Wingless (4D4, Developmental Studies Hybridoma Bank), Alexa fluorophore-conjugated secondary antibodies (Invitrogen). Images were obtained with a Zeiss Axio Imager Z1 with a Zeiss Apotome.
Supplementary Material
Highlights.
Lgd modulation of Shrub suppresses ligand-independent Notch activity
X-ray structure of Lgd DM14 domain three shows a compact helical hairpin
DM14 domain three of Lgd binds Shrub and masks its homopolymerization interface
Mutation of the Shrub-binding interface disrupts Lgd function
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft (DFG) through SFB 1208, project B01, to TK and by NIH award 5 R01 CA092433 to SCB. This work is based upon research conducted at the Northeastern Collaborative Access Team beamlines, which are funded by the National Institute of General Medical Sciences from the National Institutes of Health (P41 GM103403). This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357. We are grateful to S. Tannebaum for excellent technical assistance and L. Korfmacher and V. Strotmann for help during some of the experiments.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
Conceptualization, B.J.M., S.C.B. and T.K.; Methodology, B.J.M., S.C.B., and T.K.; Investigation, B.J.M., C.T., A.D., T.C.M.S., T.K.; Writing – B.J.M., C.T., S.C.B. and T.K..; Funding Acquisition, S.C.B. and T.K.; Resources, S.C.B. and T.K..; Supervision, B.J.M., S.C.B., and T.K.
ACCESSION NUMBERS
Coordinates and X-ray data sets have been deposited in the Protein Data Bank (PDB) under accession numbers 5VNY (DM14 domain alone) and 5VO5.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Tawashi A, Gehring C. Phosphodiesterase activity is regulated by CC2D1A that is implicated in non-syndromic intellectual disability. Cell Commun Signal. 2013;11:47. doi: 10.1186/1478-811X-11-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashkenazy H, Abadi S, Martz E, Chay O, Mayrose I, Pupko T, Ben-Tal N. ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 2016;44:W344–350. doi: 10.1093/nar/gkw408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basel-Vanagaite L, Attia R, Yahav M, Ferland RJ, Anteki L, Walsh CA, Olender T, Straussberg R, Magal N, Taub E, et al. The CC2D1A, a member of a new gene family with C2 domains, is involved in autosomal recessive non-syndromic mental retardation. J Med Genet. 2006;43:203–210. doi: 10.1136/jmg.2005.035709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof J, Maeda RK, Hediger M, Karch F, Basler K. An optimized transgenesis system for Drosophila using germ-line-specific fC31 integrases. PNAS. 2007;104:3312–3317. doi: 10.1073/pnas.0611511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson LA, Hurley JH. In vitro reconstitution of the ordered assembly of the endosomal sorting complex required for transport at membrane-bound HIV-1 Gag clusters. Proc Natl Acad Sci U S A. 2012;109:16928–16933. doi: 10.1073/pnas.1211759109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton JG, Martin-Serrano J. Parallels between cytokinesis and retroviral budding: a role for the ESCRT machinery. Science. 2007;316:1908–1912. doi: 10.1126/science.1143422. [DOI] [PubMed] [Google Scholar]
- Drusenheimer N, Migdal B, Jäckel S, Tveriakhina L, Scheider K, Schulz K, Gröper J, Köhrer K, Klein T. The Mammalian Orthologs of Drosophila Lgd, CC2D1A and CC2D1B, Function in the Endocytic Pathway, but Their Individual Loss of Function Does Not Affect Notch Signalling. PLoS Genet. 2015;11 doi: 10.1371/journal.pgen.1005749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher CM, Knoblich J. The conserved c2 domain protein lethal (2) giant discs regulates protein trafficking in Drosophila. Dev Cell. 2006;11:641–653. doi: 10.1016/j.devcel.2006.09.014. [DOI] [PubMed] [Google Scholar]
- Hurley JH. ESCRTs are everywhere. EMBO J. 2015;34:2398–2407. doi: 10.15252/embj.201592484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaekel R, Klein T. The Drosophila Notch Inhibitor and tumor Suppressor Gene lethal (2) giant discs Encodes a conserved Regulator of Endosomal Trafficking. Dev Cell. 2006;11:655–669. doi: 10.1016/j.devcel.2006.09.019. [DOI] [PubMed] [Google Scholar]
- Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein T. The tumour suppressor gene l(2)giant discs is required to restrict the activity of Notch to the dorsoventral boundary during Drosophila wing development. Dev Biol. 2003;255:313–333. doi: 10.1016/s0012-1606(02)00052-0. [DOI] [PubMed] [Google Scholar]
- Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- Manzini MC, Xiong L, Shaheen R, Tambunan DE, Di Costanzo S, Mitisalis V, Tischfield DJ, Cinquino A, Ghaziuddin M, Christian M, et al. CC2D1A regulates human intellectual and social function as well as NF-κB signaling homeostasis. Cell Rep. 2014;8:647–655. doi: 10.1016/j.celrep.2014.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinelli N, Hartlieb B, Usami Y, Sabin C, Dordor A, Miguet N, Avilov SV, Ribeiro EA, Jr, Göttlinger H, Weissenhorn W. CC2D1A Is a Regulator of ESCRT-III CHMP4B. J Mol Biol. 2012;419:75–88. doi: 10.1016/j.jmb.2012.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matias NR, Mathieu J, Huynh JR. Abscission is regulated by the ESCRT-III protein shrub in Drosophila germline stem cells. PLoS Genet. 2015;11:e1004653. doi: 10.1371/journal.pgen.1004653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullough J, Clippinger AK, Talledge N, Skowyra ML, Saunders MG, Naismith TV, Colf LA, Afonine P, Arthur C, Sundquist WI, et al. Structure and membrane remodeling activity of ESCRT-III helical polymers. Science. 2015;350:1548–1551. doi: 10.1126/science.aad8305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullough J, Colf LA, Sundquist WI. Membrane fission reactions of the mammalian ESCRT pathway. Annu Rev Biochem. 2013;82:663–692. doi: 10.1146/annurev-biochem-072909-101058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan BJ, Tibbe C, Jeon H, Drabek AA, Klein T, Blacklow SC. Electrostatic Interactions between Elongated Monomers Drive Filamentation of Drosophila Shrub, a Metazoan ESCRT-III Protein. Cell Rep. 2016;16:1211–1217. doi: 10.1016/j.celrep.2016.06.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morawa K, Schneider M, Klein T. Lgd regulates the activity of the BMP/Dpp signalling pathway during Drosophila oogenesis. Development. 2015;142:1325–1335. doi: 10.1242/dev.112961. [DOI] [PubMed] [Google Scholar]
- Morita E, Sandrin V, Alam SL, Eckert DM, Gygi SP, Sundquist WI. Identification of human MVB12 proteins as ESCRT-I subunits that function in HIV budding. Cell Host Microbe. 2007;2:41–53. doi: 10.1016/j.chom.2007.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita E, Sandrin V, McCullough J, Katsuyama A, Baci Hamilton I, Sundquist WI. ESCRT-III protein requirements for HIV-1 budding. Cell Host Microbe. 2011;9:235–242. doi: 10.1016/j.chom.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saksena S, Wahlman J, Teis D, Johnson AE, Emr SD. Functional Reconstitution of ESCRT-III Assembly and Disassembly. Cell. 2009;136:97–109. doi: 10.1016/j.cell.2008.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider M, Troost T, Grawe F, Martinez-Arias A, Klein T. Activation of Notch in lgd mutant cells requires the fusion of late endosomes with the lysosome. J Cell Sci. 2013;126:645–656. doi: 10.1242/jcs.116590. [DOI] [PubMed] [Google Scholar]
- Schuh AL, Hanna M, Quinney K, Wang L, Sarkeshik A, Yates JR, 3rd, Audhya A. The VPS-20 subunit of the endosomal sorting complex ESCRT-III exhibits an open conformation in the absence of upstream activation. Biochem J. 2015;466:625–637. doi: 10.1042/BJ20141202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen QT, Schuh AL, Zheng Y, Quinney K, Wang L, Hanna M, Mitchell JC, Otegui MS, Ahlquist P, Cui Q, et al. Structural analysis and modeling reveals new mechanisms governing ESCRT-III spiral filament assembly. J Cell Biol. 2014;206:763–777. doi: 10.1083/jcb.201403108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F, Higgins DG. Clustal Omega, accurate alignment of very large numbers of sequences. Methods Mol Biol. 2014;1079:105–116. doi: 10.1007/978-1-62703-646-7_6. [DOI] [PubMed] [Google Scholar]
- Sweeney NT, Brennan JE, Jan YN, Gao FB. The coiled-coil protein shrub controls neuronal morphogenesis in Drosophila. Curr Biol. 2006;16:1006–1011. doi: 10.1016/j.cub.2006.03.067. [DOI] [PubMed] [Google Scholar]
- Tang S, Buchkovich NJ, Henne WM, Banjade S, Kim YJ, Emr SD. ESCRT-III activation by parallel action of ESCRT-I/II and ESCRT-0/Bro1 during MVB biogenesis. Elife. 2016a;5 doi: 10.7554/eLife.15507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang S, Henne WM, Borbat PP, Buchkovich NJ, Freed JH, Mao Y, Fromme JC, Emr SD. Structural basis for activation, assembly and membrane binding of ESCRT-III Snf7 filaments. eLife. 2016b;4 doi: 10.7554/eLife.12548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troost T, Jaeckel S, Ohlenhard N, Klein T. The tumour suppressor Lethal (2) giant discs is required for the function of the ESCRT-III component Shrub/CHMP4. J Cell Sci. 2012;125:763–776. doi: 10.1242/jcs.097261. [DOI] [PubMed] [Google Scholar]
- Usami Y, Popov S, Weiss ER, Vriesema-Magnuson C, Calistri A, Gottlinger HG. Regulation of CHMP4/ESCRT-III function in human immunodeficiency virus type 1 budding by CC2D1A. J Virol. 2012;86:3746–3756. doi: 10.1128/JVI.06539-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollert T, Wunder C, Lippincott-Schwartz J, Hurley JH. Membrane scission by the ESCRT-III complex. Nature. 2009;458:172–177. doi: 10.1038/nature07836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M, Raingo J, Chen ZJ, Kavalali ET. Cc2d1a, a C2 domain containing protein linked to nonsyndromic mental retardation, controls functional maturation of central synapses. J Neurophysiol. 2011;105:1506–1515. doi: 10.1152/jn.00950.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.