Abstract
The integrity of the human genome is constantly threatened by genotoxic agents that cause DNA damage. Inefficient or inaccurate repair of DNA lesions triggers genome instability and can lead to cancer development or even cell death. Cells counteract the adverse effects of DNA lesions by activating the DNA damage response (DDR), which entails a coordinated series of events that regulates cell cycle progression and repair of DNA lesions. Efficient DNA repair in living cells is complicated by the packaging of genomic DNA into a condensed, often inaccessible structure called chromatin. Cells utilize post‐translational histone modifications and ATP‐dependent chromatin remodeling to modulate chromatin structure and increase the accessibility of the repair machinery to lesions embedded in chromatin. Here we review and discuss our current knowledge and recent advances on DNA damage‐induced chromatin changes and their implications for the mammalian DNA damage response, genome stability and carcinogenesis. Exploiting our improving understanding of how modulators of chromatin structure orchestrate the DDR may provide new avenues to improve cancer management.
Keywords: Chromatin, Cancer, DNA damage response
Highlights
Efficient DNA repair is complicated by the packaging of genomic DNA into chromatin.
Chromatin structure is modulated by histone modification and chromatin remodeling.
Chromatin modifying enzymes increase the accessibility of DNA lesions in chromatin.
DNA damage‐induced chromatin changes promote DNA repair and genomic stability.
Targeting chromatin modifying enzymes may provide new avenues for cancer therapies.
1. Introduction
Maintaining genomic stability is of vital importance to protect cells against the harmful consequences of DNA damage, which, if not properly dealt with, can lead to genomic instabilty, cancer or cell death (Negrini et al., 2010; Jackson and Bartek, 2009). Exposure to solar ultra‐violet (UV) light triggers the formation of tens of thousands of DNA lesions per cell per hour in the form of DNA intra‐strand cross‐links in genomic DNA (Jackson and Bartek, 2009). These lesions are either cytotoxic and can trigger cell death or accelerated ageing, or they are mutagenic and can lead to the onset of skin cancer (Mitchell et al., 2003). Genomic integrity is also threatened by the formation of chromosomal double‐strand DNA breaks (DSBs), which arise after exposure of cells to ionizing radiation or chemotherapeutical agents, or through the collapse of replication forks when replication blocks at DNA intra‐strand cross‐links induced by solar UV light (Hoeijmakers, 2001). To protect against the adverse effects of these DNA lesions, cells activate a coordinated series of events that regulate cell cycle progression and repair of these lesions (Hoeijmakers, 2001; Polo and Jackson, 2011; Bekker‐Jensen and Mailand, 2011; Ciccia and Elledge, 2010). This cellular response to DNA damage is highly relevant to counteract tumor development, which is underscored by the fact that several cancer‐prone human disorders, including Xeroderma pigmentosum, Nijmegen breakage syndrome and Ataxia telangiectasia, are caused by defects in DNA repair genes (reviewed in Ciccia and Elledge, 2010).
Cells utilize an impressive arsenal of DNA repair enzymes to remove a variety of lesions from their genome. No single repair pathway can cope with all kinds of lesions and cells have evolved several different repair pathways that are highly conserved throughout the kingdoms of life (Hoeijmakers, 2001; Polo and Jackson, 2011). Damages that affect only one of the two DNA strands are removed by excision repair, in which the injury and a flanking DNA region is removed and the resulting gap is re‐synthesized using the complementary DNA strand as a template (Giglia‐Mari et al., 2010). Oxidative lesions (usually of endogenous origin) are mainly removed by base excision repair (BER), whereas nucleotide excision repair (NER) removes a variety of helix‐distorting lesions including UV‐induced damage and bulky adducts from the genome (Giglia‐Mari et al., 2010; de Laat et al., 1999). Damages that affect both DNA strand, such as double‐strand breaks (DSB) are acted upon by homologous recombination (HR) which, following resection of the DNA ends, utilizes the sister‐chromatid that is present after DNA replication as a template to direct the error‐free repair of DSBs. Alternatively, cells can fuse the broken ends together in the absence of a second copy by a mechanism termed non‐homologous end‐joining (NHEJ). NHEJ repairs DSBs in an error‐free or error‐prone manner and is the dominant repair pathway in mammalian cells (Polo and Jackson, 2011; Wyman and Kanaar, 2006).
DNA repair is tightly interwoven with cell cycle progression and unrepaired DNA lesions in the mammalian genome trigger signaling pathways that delay or impede the cell cycle before DNA replication (G1/S arrest) or cell division (G2/M arrest) occurs (Polo and Jackson, 2011; Warmerdam and Kanaar, 2010). DNA damage‐induced signaling critically depends on the activity of the ATM and ATR kinases, which coordinate the assembly of repair and checkpoint proteins at damaged sites (Warmerdam and Kanaar, 2010; Huen and Chen, 2010). While ATM is activated by DSBs (Lee and Paull, 2005), activation of ATR is brought about by single‐stranded DNA regions, which arise due to stalled replication forks, or due to the processing of DSBs or UV‐induced DNA lesions into ssDNA repair intermediates (Shiotani and Zou, 2009; Zou and Elledge, 2003; Vrouwe et al., 2011). Ultimately, these signaling pathways, through activation of the CHK1 and CHK2 kinases, tumor suppressor protein p53 and many other factors, regulate the activity of key cell cycle regulators and DNA repair enzymes to tightly coordinate DNA repair with cell cycle progression (Warmerdam and Kanaar, 2010; Huen and Chen, 2010).
Genetic, biochemical and cellular studies have uncovered much of the molecular mechanisms underlying the signaling and repair of DNA lesions in mammalian cells (Polo and Jackson, 2011; Ciccia and Elledge, 2010; Giglia‐Mari et al., 2010). Efficient repair of DNA damage, however, is complicated by the fact that genomic DNA is packaged, through histone and non‐histone proteins, into a condensed structure called chromatin. The DNA repair machinery has to overcome this physical barrier to gain access to damaged DNA and repair DNA lesions (Misteli and Soutoglou, 2009; Dinant et al., 2008; van Attikum and Gasser, 2009). Post‐translational modifications of chromatin as well as ATP‐dependent chromatin remodeling factors help to overcome this barrier and facilitate access to damaged DNA by altering chromatin structure at sites of DNA damage. It is becoming clear that modulating chromatin structure is essential for the regulation of DNA damage‐induced signaling and repair pathways (Misteli and Soutoglou, 2009; van Attikum and Gasser, 2009). Here we review and discuss our current knowledge and recent advances on DNA damage‐induced chromatin changes and their implications for the mammalian DNA damage response, genome stability and carcinogenesis.
2. Histone modifications
Numerous covalent modifications of core histones that are tightly linked to gene activity and DNA repair have been identified. These modifications include the acetylation, methylation, phosphorylation and ubiquitylation of several different histone tails (Jenuwein and Allis, 2001; Turner, 2002). In the following section, we discuss how these histone modifications orchestrate the mammalian DDR in the context of chromatin.
2.1. Histone phosphorylation
Phosphorylation plays a central role in the DNA damage response (DDR) and several hundreds of proteins are phosphorylated in response to DNA damage by the ATM, ATR and DNA‐PK kinases (Matsuoka et al., 2007). Phosphorylation of DDR proteins often provides a means for phospho‐specific interactions and many DDR factors harbor phospho‐binding domains such as BRCT (breast cancer C‐terminal) or FHA (forkhead‐associated) domains (Mohammad and Yaffe, 2009).
Histones are also subject to DNA damage‐induced phosphorylation of which the archetypical example is the phosphorylation of histone variant H2AX at S139 (γH2AX). This phosphorylation is a prerequisite for the binding of MDC1 (through its BRCT domain) and the subsequent accumulation of many different DDR factors at DNA lesions (Figure 2), which can be visualized by the formation of structures known as ionizing radiation‐induced foci (IRIF) (Stucki et al., 2005). H2AX‐deficient mice exhibit genomic instability and increased risk of developing tumors (Celeste et al., 2002), underscoring the importance of this modification. Phosphorylation of H2AX has several distinct roles in the DDR. Firstly, γH2AX is required for checkpoint activation in response to ionizing radiation (IR). Cells lacking H2AX manifest a G2/M checkpoint defect after exposure to low doses of IR (Fernandez‐Capetillo et al., 2002). Secondly, γH2AX is required for the accumulation of the ubiquitin ligases RNF8 and RNF168, which, through extensive ubiquitylation (discussed below), mediate the subsequent recruitment of BRCA1 and 53BP1 into IRIF (Figure 2) (Doil et al., 2009; Huen et al., 2007; Kolas et al., 2007; Mailand et al., 2007; Stewart et al., 2009; Wang and Elledge, 2007). The RNF8–RNF168 pathway, through 53BP1 recruitment, mediates the phosphorylation and subsequent dissociation of the heterochromatin‐associated KAP1 protein (Ziv et al., 2006; Goodarzi et al., 2008; Noon et al., 2010; Lee et al., 2010b). This pathway promotes repair of heterochromatic DSBs (∼15% of all DSBs) by either NHEJ in G1 (Goodarzi et al., 2008) or HR in G2 (Beucher et al., 2009). Thirdly, the interaction between MDC1 and γH2AX is required for homologous recombination (HR), a function which is independent of the γH2AX‐dependent accumulation of BRCA1 and 53BP1 in IRIF (Xie et al., 2007). Exactly how γH2AX‐bound MDC1 promotes HR is currently unclear. Fourthly, γH2AX has an essential role in generating DNA ends which can be efficiently joined by NHEJ during V(D)J recombinant in lymphocytes. This is achieved by preventing CtIP‐dependent DNA end resection, which would lead to aberrant end‐joining, and by favoring processing of DNA ends by the Artemis nuclease, which is essential for efficient joining during V(D)J recombination (Zha et al., 2010; Helmink et al., 2010). This role of γH2AX was only recently recognized due to functional redundancy with the XRCC4‐like factor XLF (Zha et al., 2010). Interestingly, the binding of MDC1 to γH2AX is modulated by another residue in the same histone tail. The phosphorylation of H2AX at T142 by the tyrosine kinase WSTF and its dephosphorylation by EYA1/3 regulates the binding of MDC1 to phosphorylated S139 (Xiao et al., 2009; Cook et al., 2009). H2AX is constitutively phosphorylated at T142 and this acts as a switch by regulating the recruitment of repair factors (when dephosphorylated) or apoptotic factors (when it remains phosphorylated), providing a regulatory mechanism to activate the DDR or trigger cell death (Cook et al., 2009).
Figure 2.
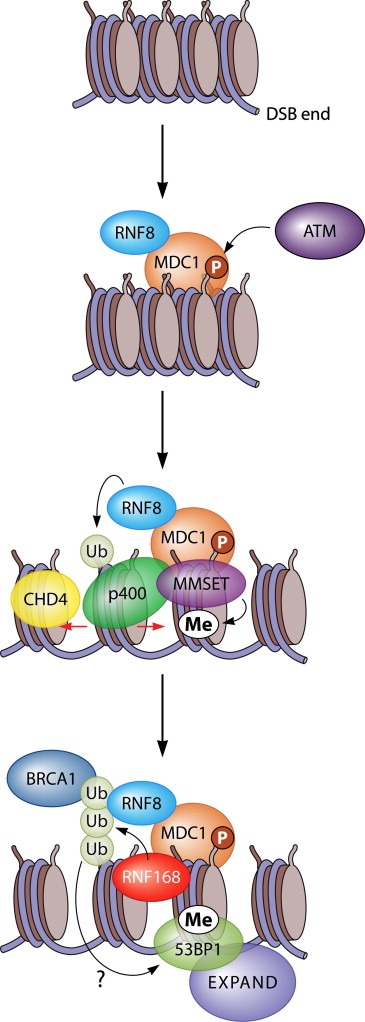
The RNF8‐ and RNF168‐dependent ubiquitin signaling pathway at DSBs is stimulated by CHD4‐ and p400‐mediated chromatin remodeling. ATM phosphorylates H2AX (to form γH2AX) in DSB‐flanking chromatin to trigger MDC1 assembly and subsequent binding of ubiquitin ligase RNF8. The ensuing ubiquitylation by RNF8 (here indicated as mono‐ubiquitylation) and the subsequent downstream events are enhanced by the chromatin remodeling activities of p400 and CHD4, which presumably create a more open and accessible chromatin state that is amendable for ubiquitylation. While p400 is recruited to DSBs in an MDC1‐dependent manner, the mechanistic basis of CHD4 stimulation of the RNF8 pathways is not clear. Upon binding, MDC1 also mediates recruitment of the MMSET methyltransferase, which mediates the de novo formation of di‐methylated H4K20 at DSBs. What follows is the association of RNF168 with RNF8‐catalyzed ubiquitin, poly‐ubiquitin chain conjugation (here indicated as H2A poly‐ubiquitylation) by RNF168, and subsequently efficient recruitment of BRCA1 through its ubiquitin‐binding partner RAP80. Likewise, RNF8‐ and RNF168‐mediated ubiquitylation, together with MMSET‐induced H4K20 methylation, synergistically promote efficient 53BP1 assembly, which in turn triggers the recruitment of the EXPAND1 chromatin modulator.
Other histones are also subject to phosphorylation and dephosphorylation in response to DNA damage. For instance, H2B is phosphorylated at S14 in a manner that requires prior induction of γH2AX (Fernandez‐Capetillo et al., 2004) However, its function is currently not clear. H3 is constitutively phosphorylated on T11 by the CHK1 kinase, which triggers H3K9 acetylation (by histone acetyltransferase GCN5) and transcriptional activation. The induction of DNA damage by UV irradiation triggers the dissociation of CHK1 from chromatin, followed by the dephosphorylation of H3T11P by phosphatase PP1γ, which results in the DNA damage‐induced repression of genes such as cyclin B1 and Cdk1 (Shimada et al., 2008, 2010).
2.2. Histone acetylation
Histone acetylation is a well‐known modulator of higher‐order chromatin structure and often functions to promote the accessibility of proteins to chromatin. It is becoming increasingly clear that the acetylation and deacetylation of histone tails are also tightly controlled at sites of DSBs, which is underscored by the finding that several histone acetyltransferases (HATs), as well as histone deacetylases (HDACs), assemble and operate at DSBs.
For example, H2AX is acetylated at K5 by the HAT TIP60 in response to IR. This acetylation is required for the mono‐ and poly‐ubiquitylation of H2AX at K119, which is regulated by TIP60 and UBC13 (see also section on ubiquitylation) (Ikura et al., 2007). Both acetylation and ubiquitylation of H2AX trigger the DNA damage‐induced dissociation of H2AX from nucleosomes, an event that occurs independently of S139 phosphorylation (Ikura et al., 2007). On the other hand, H2AX is also constitutively acetylated at K36 by the p300/CBP HATs (Jiang et al., 2010a). This mark is required for cells to survive exposure to ionizing radiation. It operates independently of γH2AX (Jiang et al., 2010a), yet through an unknown mechanism.
Several examples of DNA damage‐induced acetylation and deacetylation of core histones have also been described. For example, The nucleosome acetyltransferase of H4 (NuA4) complex, which contains the TIP60 and TRRAP subunits (Doyon and Cote, 2004), mediates acetylation of H4 in response to DSBs, which is required for efficient recruitment of repair proteins 53BP1, BRCA1, MDC1 and RAD51 and HR (Murr et al., 2006). The need for TIP60‐TRRAP recruitment and HR could be partially overcome by forced chromatin relaxation using various drugs, suggesting that the function of TIP60‐TRRAP in this context is to promote access of HR proteins to DNA that is wrapped in chromatin. Besides HR, histone acetylation also significantly stimulates NHEJ through the actions of the homologous HATs p300 and CBP, which are both recruited to DSBs, where they mediate acetylation of several sites on H3 and H4 (Ogiwara et al., 2011). Indeed, HAT inhibitor treatment and depletion of p300/CBP renders cells highly sensitive to IR, suppresses efficient recruitment of the KU complex to DSBs, and significantly impairs DSB repair by NHEJ (Ogiwara et al., 2011).
Another example of histone acetylation involved in promoting chromatin accessibility is the acetylation of H3K14, which increases after IR in a manner that depends on HMGN1. This acetylation indirectly regulates the activation of ATM kinase (Kim et al., 2009a). In agreement, loss of HMGN1 not only reduces IR‐induced phosphorylation of ATM substrates, but also renders human cells sensitive to IR and UV, and increases tumorigenesis in mice (Birger et al., 2005). The need for HMGN1 in ATM activation can by bypassed by forced chromatin relaxation using histone deacetylase inhibitors (Kim et al., 2009a), suggesting that HMNG indeed regulates the DDR by promoting chromatin structural changes at sites of DNA lesions.
The acetylation (and deacetylation; see below) of H4K16 plays a key role in the regulation of the DDR and the acetylation of this mark is directly linked to the unfolding of higher‐order chromatin structure (Shogren‐Knaak et al., 2006). The steady‐state levels of acetylated H4K16 increase in response to DSBs, an event that requires the activity of the HAT MOF1 (Li et al., 2010; Miller et al., 2010). While deletion of MOF1 does not affect γH2AX, it severely impairs the recruitment of MDC1 as well as that of BRCA1 and 53BP1 (Figure 1) (Li et al., 2010; Sharma et al., 2010). Furthermore, loss of MOF1 leads to defects in DNA repair, resulting in a persistent DNA damage‐induced G2/M arrest and the formation chromosome aberrations (Li et al., 2010; Sharma et al., 2010). The binding of MDC1 to γH2AX requires H4K16 acetylation and an acidic pocket on H2AX, suggesting that the interaction between the H2AX‐H4 tails in the same nucleosome or between different nucleosomes is required to establish a chromatin configuration required for MDC1 to associate with γH2AX (Li et al., 2010). MOF1 was also found to interact with DNA‐PK and to promote NHEJ and HR (Sharma et al., 2010).
Figure 1.
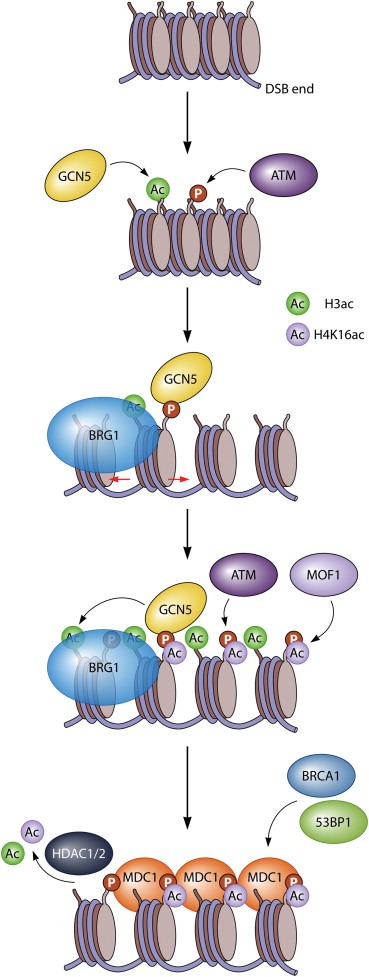
Histone acetylation acts synergistically with SWI/SNF chromatin remodeling to mediate efficient H2AX phosphorylation and assembly of MDC1. DSB‐flanking chromatin is modified by ATM and GCN5, which phosphorylate H2AX (to form γH2AX), and acetylate H3 (at low levels), respectively. The bromodomain‐containing SWI/SNF ATPase BRG1 binds to acetylated histones and presumably increases the accessibility of DSB‐flanking chromatin. GCN5 binds to γH2AX leading to further acetylation, while acetylation‐dependent BRG1 recruitment, in turn, further stimulates ATM‐mediated H2AX phosphorylation. This creates a chromatin micro‐environment that is rich in γH2AX and acetylated H3. γH2AX, together with MOF1‐mediated acetylation of H4K16, mediates efficient recruitment of MDC1, which in turn facilitates recruitment of BRCA1 and 53BP1. Reversal of histone acetylation, particularly H4K16ac and H3K56ac, occurs by HDAC1 and HDAC2 and promotes DSB repair by NHEJ.
In addition to acetylation, also deacetylation plays an important role in the DDR. The steady‐state levels of H3K9, H3K56 and H4K16 acetylation reversibly decrease following DNA damage induction (Tjeertes et al., 2009), which in case of H3K56 and H4K16 deacetylation is mediated by HDAC1 and HDAC2 (Figure 1) (Miller et al., 2010). An HDAC‐mediated decrease of H4K16ac occurs rapidly following DSB induction (Miller et al., 2010), while the H4K16ac levels increase by the activity of MOF1 at later time‐points after DNA damage (Li et al., 2010; Miller et al., 2010). Recruitment of HDAC1/2 – subunits of the Nucleosome Remodeling and Histone Deacatylase (NuRD) complex – is mediated by the CHD4 ATPase subunit of this complex, which is partly recruited by PARP (Polo et al., 2010; Chou et al., 2010). To what extent HDAC1/2 activity and H3K56 hypo‐acetylation depends on PARP or chromatin remodeling driven by CHD4 remains to be seen. Histone deacetylation by HDAC1 and HDAC2 regulates efficient disassembly of repair factors Artemis and KU and, thus, plays an important role in promoting DSB repair by NHEJ (Miller et al., 2010). However, the underlying mechanism is currently not understood. In contrast to the reported deacetylation of H3K56 (Miller et al., 2010; Tjeertes et al., 2009), an increase in H3K56 levels in response to DNA damage has also been reported (Das et al., 2009; Vempati et al., 2010). It may be that H3K56ac, like H4K16ac, is a reversible and highly dynamic mark during the DDR. In human cells, the HATs p300 and CBP were found to mediate H3K56 acetylation and histones bearing this mark are incorporated at sites of DNA repair (Das et al., 2009; Vempati et al., 2010). Interestingly, increased levels of H3K56 acetylation were found to correlate with tumorigenicity, suggesting a link between this mark and cancer development (Das et al., 2009). Whether this reflects increased acetylation or defects in deacteylation is currently not clear.
The response to UV‐induced DNA lesions also involves histone acetylation and several studies have demonstrated that histones H3 and H4 are hyper‐acetylated in human cells following UV irradiation to stimulate NER (Brand et al., 2001; Ramanathan and Smerdon, 1986). For example, UV‐induced DNA lesions trigger acetylation of H3K9 and H4K16 (Guo et al., 2010). While UV damage‐induced H3K9ac requires the activity of GCN5 in the TFTC and STAGA complexes (Brand et al., 2001; Guo et al., 2010; Martinez et al., 2001; Rubbi and Milner, 2003), it is not clear how increased levels of H4K16ac are brought about, but this may involve MOF1 as reported for DSBs (Li et al., 2010; Sharma et al., 2010). Consistent with a role for histone acetylation in NER, depletion of GCN5 significantly impairs the recruitment of repair proteins and DNA repair by NER (Guo et al., 2010). An increase in H4 acetylation, which depends on the tumor suppressors p53 and p33ING2, was also reported upon UV irradiation (Wang et al., 2006a). The underlying mechanism however remains unclear as p33ING2 and p53 are not recruited to UV‐induced DNA lesions (Rubbi and Milner, 2003; Wang et al., 2006a). Finally, the repair factor CSB recruits acetyltransferase p300 to transcription‐coupled repair complexes, suggesting that histone acetylation plays a role in this repair pathway as well (Fousteri et al., 2006). Future research will undoubtedly unravel in more detail how this chromatin modification contributes to NER.
2.3. Histone ubiquitylation
The conjugation of ubiquitin to a histone was the first example of a post‐translational histone modification (Goldknopf et al., 1975). Around 10–15% of all H2A molecules in the human cell nucleus is ubiquitylated at K119 (uH2A) at any given time, while the steady‐state levels of ubiquitylated H2B (at K120; uH2B) is considerably lower (∼1%) (Jason et al., 2002). While uH2A is linked to polycomb‐mediated transcriptional repression (Wang et al., 2004), uH2B, which is located on the opposite face of the nucleosome, is primarily linked to transcriptional activation (Kim et al., 2009b). Histones H3 and H4 are also ubiquitylated although at very low levels (∼0.1%) (Wang et al., 2006b). Histone ubiquitylation is emerging as a key regulatory modification in the DDR, which is underscored by the plethora of ubiquitin ligases and ubiquitin‐specific proteases that are directly recruited to DSBs (Messick and Greenberg, 2009).
The archetypical example of DNA damage‐induced ubiquitin conjugation is the ubiquitylation of histones H2A and H2AX at K119 (uH2A), which is mediated by the RNF8 and RNF168 ubiquitin ligases together with the E2‐conjugating enzyme UBC13 (Huen et al., 2007; Mailand et al., 2007; Bergink et al., 2006; Marteijn et al., 2009; Pinato et al., 2009). RNF8 is recruited through phospho‐specific interactions with MDC1 and histone ubiquitylation by RNF8, in concert with ubiquitylation mediated by RNF168, is required for efficient recruitment of BRCA1 and 53BP1 in response to DSBs (Doil et al., 2009; Huen et al., 2007; Kolas et al., 2007; Mailand et al., 2007; Stewart et al., 2009) and UV‐induced DNA lesions (Figure 2) (Marteijn et al., 2009). The BRCA1 complex involves the ubiquitin interacting motif (UIM)‐containing RAP80 subunit that binds specifically to Lys63‐linked ubiquitin chains catalyzed by RNF8 and RNF168, providing a molecular basis for the ubiquitin‐dependent targeting of the BRCA1 complex to DSBs (Doil et al., 2009; Sato et al., 2009; Sims and Cohen, 2009; Wang et al., 2007a). Induction of uH2A in response to DSBs is dependent on ATM/DNA‐PK and γH2AX (Mailand et al., 2007), while UV‐induced uH2A is mediated through ATR and MDC1, but does not require H2AX (Bergink et al., 2006; Marteijn et al., 2009). Histone ubiquitylation by RNF8 and RNF168 was recently implicated in transcriptional silencing at DSBs (Shanbhag et al., 2010). Several ubiquitin‐specific proteases, including BRCC36, USP3, and USP28 are required for an efficient DDR (Nicassio et al., 2007; Shao et al., 2009; Zhang et al., 2006), suggesting that the steady‐state levels of DNA damage‐induced ubiquitylation substrates is tightly regulated. It is interesting to note that while most of these deubiquitylating enzymes directly reverse RNF8‐ and RNF168‐mediated ubiquitylation at DSBs (Doil et al., 2009; Nicassio et al., 2007; Shao et al., 2009), the OTUB1 deubiquitylase inhibits RNF168‐mediated ubiquitylation by the non‐catalytic inhibition of the E2 conjugase UBC13 (Nakada et al., 2010), underscoring the multi‐layered nature of DDR regulation.
In addition to RNF8 and RNF168, a protein complex containing the canonical H2A ubiquitin ligase, RNF2, was recently also implicated in DNA damage‐induced H2A ubiquitylation (Chou et al., 2010; Ismail et al., 2010; Ginjala et al., 2011). The polycomb‐repressive complex 1 (PRC1), which contains RNF2 and the proto‐oncogene BMI1, is directly recruited to DSBs by the FHA/BRCT domain of NBS1 (Ismail et al., 2010) and possibly partly regulated by PARP (Chou et al., 2010) although this was not found in another study (Ginjala et al., 2011). While the knock‐down of RNF8 was shown to significantly reduce H2A mono‐ubiquitylation (Mailand et al., 2007), it was also reported that RNF8 depletion significantly affects γH2AX di‐ubiquitylation, while a significant level of mono‐ubiquitylation remained (Huen et al., 2007). The PRC1 complex is recruited to DSBs independently of PRC2 and carries out mono‐ubiquitylation of γH2AX. In agreement, depletion of PRC1 in RNF8‐deficient cells resulted in loss of the residual γH2AX mono‐ubiquitylation (Ismail et al., 2010), suggesting a two‐step mechanism in which RNF2‐mediated mono‐ubiquitylation of H2AX occurs prior to RNF8‐mediated di‐ubiquitylation. Similarly, depletion of BMI1 leads to impaired DSB‐induced mono‐ubiquitylation of H2A, but not poly‐ubiquitin conjugation at DSBs (Ginjala et al., 2011). Loss of PRC1 components delays, but not abolishes, the association of BRCA1, RAP80 and 53BP1 to DSBs (Ismail et al., 2010; Ginjala et al., 2011). In this model, RNF8‐mediated di‐ubiquitylation of γH2AX requires prior mono‐ubiquitylation by either RNF8 itself (which seems to be insufficient) or PRC1, which may explain the delayed recruitment of BRCA1 and 53BP1 due to loss of PRC1 recruitment. Given that several other ubiquitin ligases, such as RAD18, HERC2 and BRCA1 also accumulate at DSBs (Wang et al., 2007a; Bekker‐Jensen et al., 2010; Huang et al., 2009), it cannot be excluded, however, that these or perhaps other unidentified ubiquitin ligases may also contribute to ubiquitin‐dependent signaling at DSB.
To what extent the RNF8–RNF168 pathway and the PRC1 response overlap is currently unclear (Ismail et al., 2010; Ginjala et al., 2011). While one study suggested that the RNF8 and the RNF2/BMI1 pathways are mechanistically distinct and do not affect each other (Ismail et al., 2010), another study found that BMI1 recruitment is dependent on γH2AX and RNF8, suggesting that BMI1 acts downstream of RNF8 (Ginjala et al., 2011). To complicate things further, H2AX di‐ or poly‐ubiquitylation (but not mono‐ubiquitylation) at K119 was found to be mediated by TIP60 together with UBC13. In this study, both H2AX acetylation and ubiquitylation were found to be independent of H2AX phosphorylation, while another study found H2AX di‐ubiquitylation mediated by RNF8‐UBC13 to be dependent on the phosphorylation status of H2AX (Huen et al., 2007). Thus, while the role of RNF8 in DSB‐induced H2A ubiquitylation is firmly established, understanding exactly how BMI1/RNF2 and TIP60 are involved in the regulation of this modification will require further studies.
Histone H2A was shown to be ubiquitylated following exposure to UV light and this not only requires ATR and NER, but also several other DDR factors (Bergink et al., 2006). The CUL4A‐DDB2 complex was found to increase the levels of ubiquitylated H2A following UV irradiation after these levels are initially decreased (Kapetanaki et al., 2006). However, another report did not observe DDB2‐dependent ubiquitylation of H2A (Wang et al., 2006a). Furthermore, H2A ubiquitylation following UV exposure was also shown to depend on the H3–H4 chaperone CAF1. It was proposed that H2A is ubiquitylated in the cytoplasm and that this uH2A is incorporated at sites of UV‐induced DNA damage following DNA repair (Zhu et al., 2009). It was unclear however whether and how UV‐induced uH2A would be translocated into the nucleus and targeted to UV lesions. However, a recent study solved this issue by demonstrating that the RNF8 ubiquitin ligase is recruited to UV‐induced DNA lesions to mediate H2A ubiquitylation in concert with UBC13 (Marteijn et al., 2009). In parallel to the response to DSBs, RNF8 recruitment to UV‐induced lesions requires a phospho‐specific interaction with MDC1 and is required for the accumulation of BRCA1 and 53BP1 to photo‐lesions (Marteijn et al., 2009). Interestingly, depletion of RNF2 also impairs UV‐induced H2A ubiquitylation (Bergink et al., 2006; Marteijn et al., 2009). How RNF2 affects uH2A formation in response to UV remains unclear. It may be that its depletion leads to a decrease in nuclear ubiquitin levels. Alternatively, RNF2 may be directly involved in the H2A ubiquitylation process at UV‐induced lesions, by analogy to its role in the formation of DSB‐associated uH2A (Ismail et al., 2010; Ginjala et al., 2011).
In addition to H2A ubiquitylation, also H2B ubiquitylation (at K120) was recently recognized to play a role in the DDR (Moyal et al., 2011; Nakamura et al., 2011). During transcription, H2B ubiquitylation (uH2B) is carried out by the RNF20/RNF40 complex (Pavri et al., 2006). Likewise, RNF20/40‐induced uH2B is directly involved in the DDR and promotes DNA repair by both HR and NHEJ (Moyal et al., 2011; Nakamura et al., 2011). RNF20/40 is directly recruited to DSBs and damage‐induced uH2B requires ATM‐mediated phosphorylation of RNF20/40 (Moyal et al., 2011). The recruitment mechanism of RNF20/40 to DSBs remains to be established as recruitment required neither ATM nor NBS1 (Moyal et al., 2011; Nakamura et al., 2011). At present, it seems that the MDC1–RNF8–uH2A pathway is not affected by, nor does it affect, the RNF20/40–uH2B pathway, suggesting that ATM‐dependent uH2A and uH2B pathways may be mechanistically distinct. Interfering with RNF20/40‐mediated uH2B leads to reduced accumulation of NHEJ (XRCC4 and KU) and HR proteins (BRCA1/2 and RAD51), reduced DNA end resection, defects in the repair of DSBs and, accordingly, delayed disappearance of γH2AX (Moyal et al., 2011; Nakamura et al., 2011). As a result, depletion of RNF20/40 renders cells sensitive to IR and radiometric drugs (Moyal et al., 2011; Nakamura et al., 2011; Chernikova et al., 2010). Significant crosstalk has been reported between uH2B and histone methylation at H3K4 and H3K79 during transcription (Kim et al., 2009b). While one study reported no difference in H3K4me or K79me following DNA damage (Moyal et al., 2011), another study reported elevated H3K4me at DSBs (Nakamura et al., 2011). Depletion of RNF20 triggers transformation and tumorigenesis in cultured cells, and cancers were found to often have decreased levels of RNF20, identifying this gene as a potential tumor suppressor. As depletion of RNF20 results in lower levels of p53 and 53BP1 expression (Shema et al., 2008), it may be that not only its function in the DDR (Moyal et al., 2011; Nakamura et al., 2011), but also its role in the regulation of gene expression contributes to carcinogenesis.
Also H3 and H4 ubiquitylation is induced in response to UV irradiation, which is mediated by the CUL4A‐DDB1–DDB2 complex (Wang et al., 2006a). The DDB2 subunit of this complex has high affinity to photo‐lesions and, thus, targets the activity of the CUL4A ubiquitin ligase complex to UV‐damaged chromatin (Groisman et al., 2003). The DDB2‐CUL4A‐mediated ubiquitylation of H3 and H4 following UV irradiation results in the destabilization of nucleosomes and may contribute to enhanced recruitment of NER factors to DNA lesions (Wang et al., 2006a).
Although not induced by DNA damage, H4 is constitutively ubiquitylated at K91 by BBAP, a protein which is over‐expressed in chemotherapy‐resistant lymphomas. Disruption of BBAP results in impaired mono‐ and di‐methylation of H4K20, due to lower chromatin‐bound levels of mono‐methylase PR‐SET7 (see also below), which, in turn, results in compromised accumulation of 53BP1 at DSBs (Yan et al., 2009).
2.4. Methylation
The methylation of histones plays an import role in the DDR and histone lysine residues are found in vivo in mono‐, di‐, and tri‐methylated states, which are brought about by the activity of various histone methyltransferases. Several repair factors harbor methyl‐binding pockets such as the chromodomain (CD) and tudor domain (Taverna et al., 2007), which target such proteins to methylated histone tails.
The tri‐methylation of H3K9 is a hallmark of condensed heterochromatin, which is considered inhibitory to DNA repair. The heterochromatin 1 proteins (HP1α, β, γ) directly bind to H3K9me3 through their CDs, which is thought to contribute to heterochromatin maintenance (Cheutin et al., 2003). HP1β was found to dissociate from H3K9me3 in response to DSBs as a result of its casein kinase 2‐mediated phosphorylation (Ayoub et al., 2008). Dissociation of HP1β was suggested to promote induction of γH2AX in response to DNA damage (Ayoub et al., 2008). Compatible with such as scenario, the CD of TIP60 interacts directly with H3K9me3 and this interaction is stimulated by depletion of HP1β (Sun et al., 2009). Recruitment of TIP60 is mediated by the MRN complex, but its HAT activity requires binding to H3K9 methylation. Indeed, genetic deletion of Suv3‐9H1/H2 (the enzymes that mediate H3K9me3) impairs activation of TIP60 (Sun et al., 2009). The need to re‐organize heterochromatin to promote efficient DSB repair is underscored by the finding that ∼15% of the DSBs that are associated with heterochromatin require ATM‐dependent phosphorylation of KAP1 to be efficiently repaired (Goodarzi et al., 2008). Depletion of the three HP1 isoforms, however, overcomes the need for ATM signaling to repair heterochromatin‐associated DSBs (Goodarzi et al., 2008), possibly due to a more accessible organization of heterochromatin in cells lacking HP1 proteins. Although it may appear that the binding of HP1 proteins is inhibitory to repair, all three isoforms of HP1 are actually recruited to DSBs, oxidative DNA lesions and UV‐induced DNA lesions independently of their ability to bind to H3K9me3, suggesting additional roles in the DDR (Dinant and Luijsterburg, 2009; Luijsterburg et al., 2009; Zarebski et al., 2009). Genetic deletion studies in nematodes show that the Caenorhabditis elegans HP1 isoforms (HPL1 and HPL2) have redundant roles in the UV‐DDR as deletion of both isoforms lead to extreme sensitivity to UV. In contrast, loss of HPL1 confers IR resistance to nematodes, while loss of HPL2 results in extreme IR sensitivity (Luijsterburg et al., 2009). In agreement with differential roles for HP1 proteins in the DDR, human cells over‐expressing HP1α and HP1β, but not HP1γ, display higher levels of chromosomal aberrations, show increased sensitivity to IR and promote tumorigenicity upon injection into mice in a CD‐dependent manner (Sharma et al., 2003). In addition, over‐expression of HP1α has been found in several carcinomas (De Koning et al., 2009). However, reduced levels of H3K9me2 and H3K9me3 have been linked to several human cancers (Wang et al., 2007b; Cloos et al., 2006), and deletion of HP1β, but not HP1α, in mice lead to genomic instability (Aucott et al., 2008). Interestingly, genetic ablation of HP1β in mice has a more profound impact on genomic instability than loss of the enzymes that mediate H3K9me3, suggesting that HP1 proteins have additional roles in the DDR that do not require their methyl‐binding properties (Aucott et al., 2008; Billur et al., 2010; Peters et al., 2001). Compatible with such a scenario, recent evidence suggests a role for HP1α in promoting HR, possibly by stimulating DNA end resection and the subsequent recruitment of RAD51 (Baldeyron et al., 2011). Although more research is required to fully understand HP1 function in the DDR, it is becoming clear that proper regulation of HP1 and H3K9me levels are crucial to trigger the DDR and prevent carcinogenesis.
Another example of histone methylation that occurs during the DDR is the di‐methylation of H3K36, which is induced following DNA damage in a manner that requires the activity of the Metnase/SETMAR methyltransferase. H3K36me2 promotes the association of NBS1 and the KU complex and directly stimulates repair by NHEJ (Fnu et al., 2010) through an unknown mechanism.
The variable domain of antigen receptors is generated by the assembly of V, D and J gene segments in B lymphocytes, which requires the formation of DSBs by the RAG1–RAG2 complex followed by joining of the DNA ends by NHEJ. The methylation of H3 at K4 plays a key role in this process, which is brought about by the direct binding of the RAG2 PHD finger to H3K4me3 (Matthews et al., 2007). Reduced H3K4me3 levels and impaired binding of RAG2 to this modifications result in impaired V(D)J recombination and is linked to human immunodeficiency syndromes (Matthews et al., 2007). H3K4me3 is not only essential to target the RAG complex to V, D and J gene segments, but is also crucial to stimulate the catalytic activity of the RAG complex (Shimazaki et al., 2009), which is mediated by the methyl‐binding‐dependent alleviation of auto‐inhibition by the RAG complex (Grundy et al., 2010).
The 53BP1 protein, which harbors tandem tudor domains, is probably the most well‐known example of a DNA repair factor that binds to a methylated histone lysine residue (Taverna et al., 2007). The 53BP1 tudor domains were initially reported to bind to H3K79me2, the formation of which is mediated by Dot1L. Depletion of this enzyme was found to impair 53BP1 recruitment to DSBs (Huyen et al., 2004). However, subsequent studies showed that 53BP1 has a rather low affinity to H3K79me2 (KD ∼2 mM). Moreover, depletion of Dot1L by siRNAs or genetic deletion of Dot1L did not affect 53BP1 accumulation after IR (Botuyan et al., 2006), suggesting that the contribution of H3K79me2 to 53BP1 recruitment is limited. The 53BP1 tudors bind considerably better to H4K20me1 (KD ∼50 μM), but have the highest affinity for H4K20me2 (KD ∼20 μM). They do not bind to H4K20me3 in vitro. Accordingly, the crystal structure of the 53BP1 tudor domains revealed a binding pocket that accommodates H4K20me2, but excludes H4K20me3 (Botuyan et al., 2006). The methyltransferase MMSET was recently shown to induce de novo H4K20me2 at sites of DSBs, and this event appeared to be essential for the recruitment of 53BP1 in vivo (Pei et al., 2011). The activity of MMSET is targeted to DSBs by direct phospho‐specific interactions with MDC1 (Pei et al., 2011), but does not require RNF8 or RNF168 (Figure 2). Consistent with a role for MMSET in DNA damage‐induced di‐methylation of H4K20 (Pei et al., 2011), genetic deletion of the canonical H4K20 di‐ and tri‐methyltransferases, Suv4‐20H1 and H2, did not abolish 53BP1 recruitment to DSBs (Schotta et al., 2008). However, depletion of the H4K20 mono‐methylase PR‐Set7 did abolish 53BP1 recruitment (Botuyan et al., 2006; Oda et al., 2010), likely due to the fact that H4K20me2 induction by MMSET requires prior mono‐methylation by PR‐SET7. The 53BP1–H4K20me interaction plays a role in suppressing HR and promoting XRCC4‐dependent NHEJ (Xie et al., 2007). Notably, the NHEJ function of 53BP1 seems largely independent of H2AX (Xie et al., 2007), which may explain the transient recruitment of 53BP1 in H2AX‐deficient cells (Celeste et al., 2003). The accumulation of 53BP1 in IRIF does depend on γH2AX and the MDC1–RNF8–RNF168 pathway (Doil et al., 2009; Mailand et al., 2007) and this pathway mediates efficient 53BP1‐dependent repair through local phosphorylation of KAP1 in heterochromatin (Noon et al., 2010), which is required for ∼15% of the DSBs that are associated with dense chromatin and are repaired in an ATM‐dependent manner (Goodarzi et al., 2008). This pathway involves the 53BP1‐dependent recruitment of the MRN complex (through the BRCT domains of 53BP1) to locally enrich ATM kinase activity, which explains the requirement for 53BP1 to phosphorylate KAP1 at heterochromatic DSBs (Lee et al., 2010b). In this context, 53BP1 recruitment depends on ubiquitylation by RNF8–RNF168 through an unknown mechanism since no ubiquitin‐binding domains have been identified in 53BP1. Whether this pathway involves binding to H4K20me2 and how this is mediated through ubiquitylation by RNF8–RNF168 remains to be established. It is interesting to note that recent evidence linked chromatin remodeling activity to 53BP1. The PWWP domain‐containing protein EXPAND1 interacts with 53BP1 through its BRCT domain and is stably associated with chromatin in undamaged cells (Huen et al., 2010). EXPAND1 accumulation in IRIF depends on H2AX, MDC1, RNF8 and 53BP1 (Figure 2). Interestingly, EXPAND1 promotes increased chromatin accessibility and depletion of EXPAND1 leads to a delay in disappearance of γH2AX and sensitivity to IR (Huen et al., 2010). However, loss of EXPAND1 did not seem to affect the ATM–KAP1 pathway to facilitate heterochromatic DSBs, which does require 53BP1 (Goodarzi et al., 2008; Noon et al., 2010). Elucidating the precise role of EXPAND1 in the DDR will require further studies.
3. Chromatin remodeling
A number of large multi‐protein complexes (200 kDa–2 MDa) can enzymatically modulate chromatin structure at the expense of ATP hydrolysis (Varga‐Weisz and Becker, 2006; Becker and Horz, 2002). These nucleosome remodeling factors contain an ATPase of the SWI2/SNF2 (Switch/Sucrose non‐fermentable) family, as well as several targeting and regulatory subunits (Varga‐Weisz and Becker, 2006). It is becoming increasingly clear that ATP‐dependent chromatin remodeling factors play an essential role in the DDR. In the following section we discuss the involvement of chromatin remodeling factors in the DDR and outline how these factors promote access for DNA repair proteins to DNA lesions buried in chromatin (1, 2).
Table 1.
Chromatin remodeling factors in the DDR.
| Chromatin remodeling complex | Recruitment by | Role in DDR | Reference |
|---|---|---|---|
| BRM and BRG1 (SWI/SNF) | H3 acetylation (by GCN5 or p300/CBP), recognized by the bromodomains of BRG1 | Spreading of γH2AX and subsequent IRIF formation, accumulation of KU and repair by NHEJ | (Ogiwara et al., 2011; Lee et al., 2010a) |
| BRIT‐BRM/BRG1 (SWI/SNF) | BRIT1 recruits SWI/SNF complex through phosphorylated BAF170 to DSBs | Accumulation of NHEJ, HR and signaling proteins and repair by NHEJ and HR. | (Peng et al., 2009; Rai et al., 2006) |
| BRG1 (SWI/SNF) | Unknown | Required for CPD repair by NER (mechanism is currently unclear) | (Gong et al., 2008; Zhao et al., 2009; Zhang et al., 2009) |
| SNF2H (ISWI) | RNF20‐RNF40, and possibly ACF1 | Accumulation of HR proteins, repair by HR and NHEJ | (Nakamura et al., 2011; Lan et al., 2010) |
| INO80 | ARP5 and ARP8 to DSBs and DDB1 to UV damage | Involved in repair by HR (YY1 subunit binds to holidays junctions), stimulates repair of UV damage by NER through enhanced recruitment of XPC and XPA | (Kashiwaba et al., 2010; Kitayama et al., 2009; Wu et al., 2007; Jiang et al., 2010b) |
| p400 (NuA4 complex) | MDC1 | Regulates nucleosome destabilization in concert with TIP60‐mediated acetylation, stimulates RNF8‐mediated ubiquitylation and BRCA1, 53BP1 recruitment to DSBs | (Xu et al., 2010) |
| ALC1 (CHD1‐like protein) | PARP (the macrodomain of ALC1 recognizes PAR) | Stimulates repair of ssDNA breaks and DSB, interacts with KU, XRCC1 and APLF (requires poly‐ADP‐ribosylation (PAR)). APLF stimulates ligation during NHEJ, but whether this requires ALC1 is unclear | (Ahel et al., 2009; Gottschalk et al., 2009; Rulten et al., 2011) |
| CHD2 | Unknown | Unknown, but loss of CHD2 leads to higher and persistent levels of γH2AX in mice | (Nagarajan et al., 2009) |
| CHD4 | PARP and possibly components of the RNF8 pathway | Prevents accumulation of spontaneous DNA damage, cell cycle progression through p53 deacetylation, mediates recruitment of HDACs to promote NHEJ, stimulates RNF8‐mediated ubiquitylation and RNF168 and BRCA1 recruitment to DSBs | (Polo et al., 2010; Chou et al., 2010; Larsen et al., 2010; Smeenk et al., 2010; Pegoraro et al., 2009) |
| EXPAND | 53BP1 (requires its BRCT domain) | Increased chromatin accessibility and survival after IR, although the underlying mechanism are currently unclear | (Huen et al., 2010) |
Table 2.
Histone modification in the DDR.
| Histone modification | Catalyzed by | Role in DDR | Reference |
|---|---|---|---|
| H2AX S139ph (γH2AX) | ATM, ATR, DNA‐PKcs | Checkpoint activation, accumulation of numerous proteins in IRIF, HR, NHEJ during V(D)J recombination | (Fernandez‐Capetillo et al., 2002; Doil et al., 2009; Huen et al., 2007; Kolas et al., 2007; Mailand et al., 2007; Stewart et al., 2009; Wang and Elledge, 2007; Xie et al., 2007; Zha et al., 2010; Helmink et al., 2010) |
| H2AX T142ph | WSTF (reversed by EYA1/3) | Switch between DNA repair and apoptosis | (Xiao et al., 2009; Cook et al., 2009) |
| H2B S14ph | Unknown | Unknown | (Fernandez‐Capetillo et al., 2004) |
| H3 T11ph | CHK1 (reversed by PP1γ) | Constitutive mark that is lost after UV exposure resulting in gene repression | (Shimada et al., 2008, 2010) |
| H2AX K5ac | TIP60 | Required for mono‐ and poly‐ub and subsequent dissociation of H2AX from chromatin | (Ikura et al., 2007) |
| H2AX K36ac | p300/CBP | Promotes IR survival independently of γH2AX | (Jiang et al., 2010a) |
| H4 K5ac, K8ac, K12ac, K16ac | TIP60‐TRRAP (NuA4 complex) | Recruitment of MDC1, BRCA1. 53BP1, RAD51 and repair by HR | (Murr et al., 2006) |
| H3 K18ac, H4 K5ac, K8ac, K12ac, K16ac | p300/CBP | Recruitment of SWI/SNF complex, KU accumulation and repair by NHEJ | (Ogiwara et al., 2011) |
| H3 K9ac, K14ac, K18ac, K23ac | GCN5 | Recruitment of SWI/SNF complex through binding to H3K14ac and spreading of γH2AX | (Lee et al., 2010a) |
| H3 K14ac | Requires HMGN | Regulates activity of ATM kinase | (Kim et al., 2009b) |
| H4 K16ac | MOF1 (reversed by HDAC1/2) | Levels transiently decrease (reversal by HDACs) after IR and then increase (by MOF1). Decrease promotes retention of NHEJ factors, while subsequent increase regulates MDC1, BRCA1 and 53BP1 accumulation, and repair by HR and NHEJ. | (Li et al., 2010; Miller et al., 2010; Sharma et al., 2010) |
| H3 K56ac | p300/CBP (reversed by HDAC1/2) | Levels decrease (reversal by HDACs) after IR, which promotes NHEJ. Conversely, incorporation of new H3K56ac‐bearing histones at repair sites has also been reported | (Miller et al., 2010; Tjeertes et al., 2009; Das et al., 2009; Vempati et al., 2010) |
| H3 K9ac | GCN5 in TFTC and STAGA complexes | Increased following UV exposure, stimulates XPC recruitment and repair by NER | (Brand et al., 2001; Guo et al., 2010; Martinez et al., 2001) |
| H2A K119ub | RNF8 (mono/di‐ub), RNF168 (poly‐ub) possibly assisted by RNF2 (mono‐ub) (reversed by USP3 and BRCC36, and inhibited by OTUB1) | Accumulation of BRCA1, RAD18 and 53BP1 after IR and UV, checkpoint activation and repair of DSBs in heterochromatin, transcriptional silencing at DSBs | (Doil et al., 2009; Huen et al., 2007; Kolas et al., 2007; Mailand et al., 2007; Stewart et al., 2009; Wang and Elledge, 2007; Noon et al., 2010; Bergink et al., 2006; Marteijn et al., 2009; Shanbhag et al., 2010; Nicassio et al., 2007; Shao et al., 2009; Ginjala et al., 2011; Bekker‐Jensen et al., 2010; Huang et al., 2009) |
| H2AX K119ub | RNF2 (mono‐ub), RNF8 (di‐ub) and TIP60‐UBC13 (di‐ and poly‐ub) | Presumably similar to ubi H2A at K119 | (Huen et al., 2007; Ikura et al., 2007; Ismail et al., 2010) |
| H2B K120ub | RNF20‐RNF40 | Accumulation of KU, XRCC4, BRCA1 and RAD51, stimulates repair by NHEJ and HR | (Moyal et al., 2011; Nakamura et al., 2011) |
| H3 and H4 ub (residues unknown) | DDB2‐DDB1‐CUL4A | Destabilize nucleosomes after UV exposure to stimulate NER | (Wang et al., 2006a) |
| H4 K91ub | BBAP | Constitutive mark required for induction of H4K20me and 53BP1 recruitment to DSBs | (Yan et al., 2009) |
| H3 K9me3 | Suv3‐9H1/Suv3‐9H2 (recognized by chromodomains of HP1 and TIP60 proteins) | DNA damage‐induced phosphorylation of HP1β leads to its dissociation from H3K9me and stimulates TIP60 activation. HP1 proteins are also recruited to DSBs (independently of H3K9me) to promote repair | (Ayoub et al., 2008; Sun et al., 2009; Dinant and Luijsterburg, 2009; Luijsterburg et al., 2009; Zarebski et al., 2009; Sharma et al., 2003; Baldeyron et al., 2011) |
| H3 K36me2 | Metnase/SETMAR | Promotes accumulation of NBS1 and KU and stimulates repair by NHEJ | (Fnu et al., 2010) |
| H3 K4me3 | SET1 (recognized by the PHD finger of RAG2) | Targets and stimulates the RAG complex involved in V(D)J recombination | (Matthews et al., 2007; Shimazaki et al., 2009; Grundy et al., 2010) |
| H4 K20me2 | Suv4‐20H1/Suv4‐20H2 or MMSET | Mediated by MMSET which is recruited to DSBs by MDC1. Required for recruitment of 53BP1 through its tandem tudor domain. The 53BP1‐H4K20me‐interaction suppresses HR, but promotes NHEJ independently of γH2AX | (Xie et al., 2007; Botuyan et al., 2006; Pei et al., 2011) |
3.1. SWI/SNF
Mammalian chromatin remodeling complexes of the SWI/SNF family contain either the BRM or BRG1 ATPase, which slide and eject nucleosomes (Clapier and Cairns, 2009). A role for SWI/SNF complexes in DSB repair has recently been established. BRM and BRG1 are recruited to DSBs, where they directly interact with γH2AX‐containing nucleosomes (Lee et al., 2010a). This interaction involves binding of the bromodomain of BRG1 to H3 acetylation, which induces additional H3 acetylation (at K9, K14, K18, and K23) in γH2AX‐containing nucleosomes, but not in bulk chromatin, through recruitment of the acetyltransferase GCN5 (Figure 1). This feedback mechanism promotes efficient induction of γH2AX after IR, perhaps due to increased accessibility of the ATM kinase (Lee et al., 2010a). Depletion of BRG1 renders cells sensitive to IR, impairs induction of γH2AX and leads to defects in DSB repair (Park et al., 2006). In analogy to this mechanism, BRM and BRG1 are also required for efficient NHEJ in a manner that depends on the acetyltransferase activity of the CBP and p300 enzymes (Ogiwara et al., 2011). Indeed, depletion of these HATs impairs the recruitment of BRM, and depletion of BRM, in turn, impairs the efficient recruitment of the KU complex at DSBs and leads to considerable defects in repair by NHEJ (Ogiwara et al., 2011). Thus, histone acetylation by CBP/p300 mediates recruitment of SWI/SNF, which is required for KU binding and efficient NHEJ.
A SWI/SNF complex containing BRG1–BRM–BAF170–BAF155–SNF5 is recruited to DSBs by BRIT1, which accumulates in IRIF (Rai et al., 2006; Peng et al., 2009). Several human cancers display decreased levels of BRIT1 and their tumorigenicity is inversely correlated with BRIT1 levels (Rai et al., 2006). The interaction between BRIT1 and the SWI/SNF complex is regulated by the ATM/ATR‐dependent phosphorylation of BAF170, which is essential for proper DSB repair through NHEJ and HR (Peng et al., 2009). In agreement, depletion of BRIT1 impairs the accumulation of several repair factors including KU70, RPA, RAD51, NBS1, MDC1, and BRCA1 (Rai et al., 2006; Peng et al., 2009; Lin et al., 2005). Surprisingly, while γH2AX formation seems not to be affected by BRIT1 depletion (Rai et al., 2006), accumulation of BRM/BRG1 is mainly BRIT1‐dependent (Peng et al., 2009), which seems to be at odds with the finding that γH2AX induction is impaired by depletion of BRM/BRG1 (Lee et al., 2010a; Park et al., 2006). Further studies will be needed to address whether the BRG1‐dependent feedback loop, which induces γH2AX spreading, and the BRIT‐dependent recruitment of BRG1, which promotes the recruitment of numerous repair factors, constitute mechanistically distinct pathways.
SWI/SNF ATPases are often inactivated in human cancers suggesting that they may function as tumor suppressor proteins (Varela et al., 2011). In addition, their loss in mammalian cells confers high sensitivity to UV irradiation, suggesting a possible role for SWI/SNF in the repair of UV‐induced DNA damage. Indeed, several studies agree that loss of BRM/BRG1 selectively leads to impaired CPD (but not 6‐4 PP) repair following UV irradiation. However, the underlying mechanism is at present unclear mainly due to conflicting reports. The repair defect has been attributed to an indirect apoptotic response that is suppressed by SWI/SNF (Gong et al., 2008), or impaired accumulation of XPG/PCNA, but not of XPC and DDB2, at UV‐induced DNA lesions (Zhao et al., 2009). This latter observation was not in agreement with another report, which suggested that loss of SWI/SNF impairs recruitment of XPC to DNA lesions (Zhang et al., 2009). Given that XPC and XPG are essential for 6‐4PP and CPD repair, scenarios in which BRG1 is required for the recruitment of XPG and/or XPC seem incompatible with efficient removal of 6‐4PP following BRG1 depletion. Future studies are required to solve this riddle and elucidate how SWI/SNF affects NER.
Screens for UV sensitivity in C. elegans revealed that loss of the nematode orthologues of BRM, SNF5 and BAF155 result in UV sensitivity (Lans et al., 2010). Loss of SNF5 – especially when combined with loss of p53 – is linked to malignant tumor formation (Isakoff et al., 2005; Versteege et al., 1998), which does not result from genomic instability, but rather from transcriptional changes in genes that regulate cell cycle progression (Isakoff et al., 2005; Klochendler‐Yeivin et al., 2006; McKenna et al., 2008). For instance, loss of SNF5 activates genes regulated by polycomb factor EZH2 (required for H3K27me), and inactivation of EZH2 blocks tumor formation driven by loss of SNF5 (Wilson et al., 2010). Whether SNF5 is directly involved in the DDR is currently unclear. Conflicting results concerning the DNA damage sensitivity of SNF5‐deficient cells have been reported (Klochendler‐Yeivin et al., 2006; McKenna et al., 2008). Recent studies suggested a role for SNF5 in UV‐induced γH2AX formation, which was suggested to be due to defective ATM, but not ATR recruitment (Ray et al., 2009). The significance of these findings awaits further studies as the formation of γH2AX as well as phospho‐ATM following UV irradiation was previously shown to be strictly ATR‐dependent and did not require ATM activity (Vrouwe et al., 2011; Marti et al., 2006; Hanasoge and Ljungman, 2007; O'Driscoll et al., 2003; Stiff et al., 2006). Like BRG1 depletion, SNF5 knock‐down was shown to selectively affect CPD repair following UV irradiation, which was attributed to loss of p21 and GAD45a expression (Gong et al., 2008).
3.2. ISWI
Mammalian ISWI remodeling complexes, such as CHRAC, WICH and NURF, contain either the SNF2H or SNF2L ATPase, which can mediate nucleosome sliding and histone displacement (Clapier and Cairns, 2009). Micro‐irradiation of mammalian cells recently revealed that SNF2H and SNF2L are directly recruited to DSBs (Erdel et al., 2010). Indeed, SNF2H and ACF1, a subunit of the CHRAC complex (Poot et al., 2000), are also recruited to DSBs, in a manner depending on the SLIDE domain of SNF2H, which is required for the interaction with ACF1 (Lan et al., 2010). Depletion of ACF1 or SNF2H renders human cells sensitive to DSBs and knock‐down of ACF1, SNF2H and the other CHRAC subunits CHRAC15 and CHRAC17 impairs both NHEJ and HR. Repair by NHEJ is stimulated by direct and DNA damage‐independent interactions between ACF1 and KU70/80, which are required for KU recruitment to DSBs (Figure 3). In the presence of DSBs, ACF1 recruits other components of CHRAC complex, and the ATPase activity of SNF2H promotes efficient NHEJ (Lan et al., 2010). Whether CHRAC activity is required to recruit other NHEJ proteins remains to be established. SNF2H also promotes HR and its depletion impairs RPA, BRCA1 and RAD51 foci formation and inhibits HR (Nakamura et al., 2011). Although it was suggested that SNF2H promotes HR as a downstream effector of RNF20/40‐mediated H2B ubiquitylation (Nakamura et al., 2011), this proposed mechanism remains to be tested experimentally. Simultaneous depletion of SNF2H and RNF20 indeed impairs HR to the same extent as depletion of either one of the proteins (Nakamura et al., 2011), but whether SNF2H recruitment depends on uH2B is currently not clear.
Figure 3.
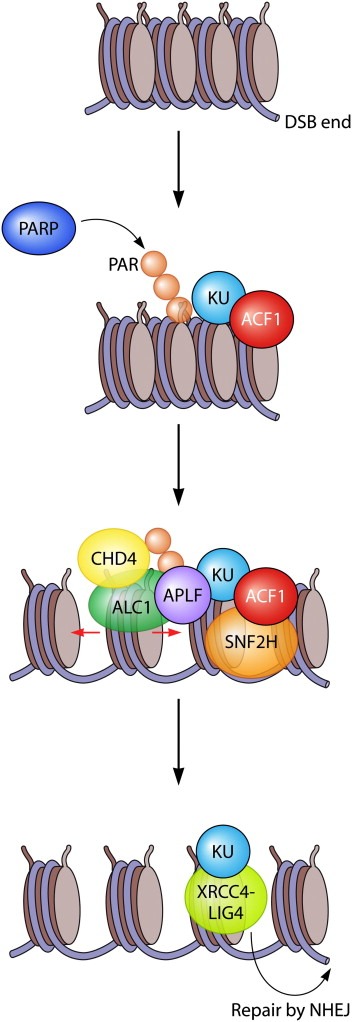
PARP‐dependent recruitment of chromatin remodelers ALC1 and CHD4 and ACF1‐dependent recruitment of chromatin remodeler SNF2H promotes efficient repair by NHEJ. Single‐stranded breaks and/or DSBs are recognized by PARP and chromatin surrounding these sites is subsequently poly‐ADP‐ribosylated. The KU complex binds to broken DNA ends in an ACF1‐dependent manner. Whether ACF1 recruitment also requires KU is not clear. ACF1 recruits the SNF2H remodeling complex to promote NHEJ and HR. Poly‐ADP‐ribosylation mediates recruitment of remodeling factor ALC1 through its macrodomain. ALC1 interacts with KU and APLF in an ADP‐ribose‐dependent manner and as such may stimulate single‐stranded break repair and (most likely NHEJ‐dependent) DSB repair. The CHD4 ATPase is also recruited in a PARP‐dependent manner and this may lead to the assembly of other NuRD components, including HDAC1 and HDAC2, to promote NHEJ. The recruitment of APLF by PARP3 promotes XRCC4/LIG4‐dependent ligation during NHEJ. Whether this requires ALC1‐mediated chromatin remodeling remains unanswered.
A recent screen in C. elegans revealed that loss of the nematode orthologues of SNF2H results in UV sensitivity (Lans et al., 2010). Knock‐down of ACF1 or SNF2H in human cells indeed led to a mild sensitivity to UV light (Lan et al., 2010). Repair by NER in di‐nucleosomes is enhanced by ACF1 in vitro (Ura et al., 2001), and ACF1 appears to be recruited to UV‐induced DNA lesions in vivo (Luijsterburg et al., 2009). Whether SNF2H is recruited to photo‐lesions and stimulates NER in vivo awaits further studies.
3.3. INO80
Mammalian INO80 chromatin remodeling factors contain either the INO80 ATPase or related SWR1‐like factors such as the p400 ATPase. A defining feature among INO80‐like remodelers is a long insertion in the middle of the conserved ATPase domain (Clapier and Cairns, 2009).
The polycomb transcription factor YY1 forms a complex with several INO80 components, including the INO80 ATPase and TIP49A/B, as well as the actin‐related proteins ARP4, ARP5, and ARP8. The YY1–INO80 complex has been shown to play an important role in the mammalian DDR. INO80 is recruited to sites of laser‐generated DSBs in a yH2AX‐ and YY1‐independent, but ARP8‐ and ARP5‐dependent manner (Kashiwaba et al., 2010; Kitayama et al., 2009). Loss of YY1 leads to chromosome aberrations and depletion of YY1 or INO80 conferred a defect in HR repair (Wu et al., 2007). In vitro experiments indeed suggest that YY1 binds to recombination intermediates (holiday‐junctions), which may be important for its role in HR (Wu et al., 2007). In addition to the YY1 complex, INO80 was also found in a complex containing DNA‐PK, suggesting it may have additional roles in NHEJ (Wu et al., 2007).
INO80 has also been implicated in DNA repair by NER. Indeed, loss of INO80 or ARP5 leads to a marked reduction in the removal of UV‐induced 6‐4PPs and CPDs (Jiang et al., 2010b). INO80 is recruited to DNA lesions independently of XPC and XPA, suggesting an early role in NER. Consistent with this, cells deficient in INO80 have a markedly reduced recruitment of NER factors XPC and XPA to DNA lesions. INO80 interacts with the DDB1 subunit of UV‐DDB (Jiang et al.,2010b), and this interaction may play a role in its recruitment to lesions. Whether INO80 is recruited by DDB1–DDB2 awaits analysis of its assembly at photo‐lesions in DDB2‐deficient XP‐E cells. In agreement with a role in NER, depletion of YY1, INO80 or TIP49B renders cells sensitive to UV irradiation and impairs removal of UV‐induced DNA lesions (Wu et al., 2007).
The TIP60 acetyltransferase is part of the nucleosome acetyltransferase of H4 (NuA4) complex together with TRRAP and the INO80‐related p400 ATPase (Doyon and Cote, 2004). Expression of a catalytically inactive, dominant‐negative version of p400 renders cells sensitive to IR and leads to increased levels of chromosome aberrations. Furthermore, p400 is essential for bleomycin‐induced nucleosome destabilization, which also requires the HAT activity of TIP60 (Xu et al., 2010). The p400‐mediated nucleosome destabilization requires H2AX phosphorylation (by either ATM or DNA‐PKcs) and MDC1 (Xu et al., 2010). Interestingly, while p400 is not required for RNF8 recruitment to DSBs, it is essential for efficient DNA damage‐induced ubiquitin conjugation, as well as BRCA1 and 53BP1 accumulation at DSBs (Figure 2) (Xu et al., 2010). Together, these results suggest that MDC1 targets the chromatin‐remodeling activity of p400 to DSBs, which results in a chromatin structure that is amendable to RNF8‐dependent ubiquitylation and BRCA1 and 53BP1 recruitment (Xu et al., 2010). Although these results seem to imply that the NuA4 complex is recruited to DSBs by MDC1, these results seem inconsistent with the findings that MDC1 recruitment is impaired in cells deficient in NuA4 subunit TRRAP (Murr et al., 2006). Based on the finding that KAP1 phosphorylation, which is part of an ATM‐dependent pathway to regulate access to DNA lesions in heterochromatin (Ziv et al., 2006; Goodarzi et al., 2008; Noon et al., 2010), was not involved in p400‐dependent chromatin changes, it was suggested that the ATM–KAP1 pathway may promote access to lesions in heterochromatin, while the TIP60/p400 pathway may regulate access to lesions in euchromatin (Xu and Price, 2011). Although this is an intriguing scenario, it seems at present incompatible with the finding that the HAT activity of TIP60 requires its binding to H3K9me3 (Sun et al., 2009), a histone modification found exclusively in heterochromatin.
3.4. CHD
Mammalian CHD family members contain two tandemly arranged CDs on the N‐terminus of their ATPase, which are involved in binding methylated histone tails as well as DNA. The CHD remodeling complexes, including the NuRD complex, can slide or eject histones and have both activatory and inhibitory roles in transcription regulation (Clapier and Cairns, 2009; Hall and Georgel, 2007). Several CHD family members have been implicated in the DDR.
The CHD1‐like protein ALC1, whose gene is often amplified in liver cancer, is a SNF2‐like chromatin remodeling factor that contains a macrodomain, which binds ADP‐ribose. ALC1 was recently found to accumulate at sites of DNA damage in a Poly(ADP‐ribose) polymerase (PARP)‐ and macrodomain‐dependent manner (Figure 3) (Ahel et al., 2009; Gottschalk et al., 2009). The ALC1 ATPase activity requires the N‐terminal tail of H4 for chromatin remodeling and is stimulated by poly(ADP ribosyl)ation (Ahel et al., 2009). ALC1 interacts with the KU complex, XRCC1 and APLF in a PARP‐dependent manner (Ahel et al., 2009), suggesting roles in single‐ and double‐strand break repair. Indeed, APLF was shown to enhance DNA ligation during NHEJ by promoting the retention of XRCC4‐DNA Ligase IV complex after its recruitment to DNA breaks in a PARP3‐dependent manner (Figure 3) (Rulten et al., 2011). Accordingly, depletion of APLF or ALC1 renders cells sensitive to single‐ and double‐strand breaks (Ahel et al., 2009; Mehrotra et al., 2011; Bekker‐Jensen et al., 2007). Recent work showed that APLF is a histone chaperone that may deposit or exchange histones, such as macroH2A1.1, at sites of DNA repair (Mehrotra et al., 2011; Timinszky et al., 2009). Whether it cooperates with ALC1 to fulfill this function during NHEJ is not clear.
Mice heterozygous for a truncated allele of CHD2 showed a significant susceptibility to developing lymphomas (Nagarajan et al., 2009). At the cellular levels, CHD2 homozygote mutant cells showed increased accumulation of spontaneous DNA damage, higher and persistent levels of DNA damage‐induced γH2AX, and increased sensitivity to DNA damage (Nagarajan et al., 2009). However, the underlying molecular mechanisms and the exact role of CHD2 in the DDR remain to be solved.
The CHD4 ATPase has recently been implicated in the DDR through several, distinct mechanisms (Polo et al., 2010; Chou et al., 2010; Larsen et al., 2010; Smeenk et al., 2010; Pegoraro et al., 2009). Firstly, loss of CHD4 and other NuRD components leads to chromatin structural defects (such as reduced H3K9me3), increased accumulation of spontaneous DNA damage (Pegoraro et al., 2009), and perturbed cell cycle progression (G1 arrest) caused by p53/p21 induction, the latter of which is likely due to a loss of p53 deacetylation by the NuRD complex (Polo et al., 2010; Larsen et al., 2010; Smeenk et al., 2010). Secondly, CHD4 was shown to be recruited to DSBs in a PARP‐dependent manner (Figure 3) (Polo et al., 2010; Chou et al., 2010). Recruitment of CHD4 is required for the accumulation of HDAC1 and HDAC2 at DSBs (Polo et al., 2010), which stimulate NHEJ (Miller et al., 2010). It is tempting to speculate that PARP‐dependent recruitment of the NuRD complex stimulates repair by NHEJ, but whether this involves chromatin remodeling by CHD4 or only the HDAC1 activity of the NuRD complex remains to be established. Thirdly, depletion of CHD4 leads to a marked reduction in DNA damage‐induced ubiquitin conjugation and impaired recruitment of RNF168 and BRCA1 to DSBs (Larsen et al., 2010; Smeenk et al., 2010) in a manner depending on the ATPase activity of CHD4 (Figure 2) (Smeenk et al., 2010). These finding are consistent with a model in which CHD4 creates a local chromatin environment that is amendable to chromatin ubiquitylation, which, in turn, promotes the recruitment of RNF168 and BRCA1 (Larsen et al., 2010; Smeenk et al., 2010). However, the mechanistic basis for CHD4‐dependent stimulation of histone ubiquitylation is currently not understood. Thus, the NuRD complex acts through distinct mechanisms and operates genome‐wide as well as directly at DSBs to maintain genome stability. Its importance in the DDR is underscored by the fact that it protects cells against the cytotoxic effects of IR (Miller et al., 2010; Polo et al., 2010; Chou et al., 2010; Larsen et al., 2010; Smeenk et al., 2010).
4. Conclusions and future directions
In the recent years, great progress has been made in identifying new ATP‐dependent chromatin remodeling factors and histone modifications that play a role in the orchestration of DDR. Based on these recent findings, it is anticipated that many more new chromatin modulators in the DDR await identification. Intuitively, it may be expected that once an accessible chromatin micro‐environment has been establish early in the DDR, such an environment is also amendable to the later stages of the DDR. However, a picture is now emerging in which the various steps in the DDR are tightly regulated by structural chromatin changes mediated by distinct chromatin remodeling factors and histone modifications. It is feasible that the distinct stages in the DDR require a specific chromatin configuration that can then be acted upon by repair factors. Such a scenario necessitates the continual re‐configuration of chromatin throughout the different stages of the DDR and could explain why so many different ATP‐dependent chromatin remodeling factors and histone modifications have emerged as regulators of distinct events during the DDR.
A major challenge in this field will be to unravel how the combinatorial action of numerous chromatin modulators shapes the chromatin landscape during the DDR, which will require an interdisciplinary approach using cell biological, biochemical and biophysical techniques. Understanding the precise interplay between DNA repair pathways, chromatin modulators and histone modifications will require further development of technical advances in super‐resolution microscopy, quantitative proteomics approaches and genetic screening methodologies (Meerbrey et al., 2011; Huang et al., 2010; Walther and Mann, 2010).
Many if not all cancer cells have defects in one or more aspects of the DDR and, as a result, this may make such cells more vulnerable to cancer therapies that aim at targeting tumor‐related DDR defects (Jackson and Bartek, 2009). In this respect, targeting chromatin modulators in the DDR may provide new avenues to improve cancer therapies. For example, inhibitors of PARP are non‐toxic to normal cells but highly toxic to HR‐defective cells, such as BRCA1‐ or BRCA2‐deficient tumor cells (see also the contribution of T. Helleday, in this issue) (Bryant et al., 2005; Farmer et al., 2005). Several studies have implicated a role for chromatin modulators in HR (e.g. TIP60, BRIT1, INO80 and SNF2H (Murr et al., 2006; Nakamura et al., 2011; Peng et al., 2009; Wu et al., 2007)), suggesting that tumors in which the genes encoding such factors are mutated or epigenetically silenced may be subject to treatment with PARP inhibitors. Likewise, histone deacetylase (HDAC) inhibitors are promising drugs that selectively kill cancer cells (Ropero and Esteller, 2007). Although the toxicity of HDAC inhibitors may be caused by changes in the expression level of certain proteins, recent results have also directly implicated histone deacetylation by HDACs in repair by NHEJ (Miller et al., 2010). Accordingly, HDAC inhibitors selectively sensitize tumor cells to radiomimetic drugs and IR (Munshi et al., 2005). Given the strong links between histone lysine methylation and the DDR, it is interesting to note that a few pharmacological inhibitors of histone methylation have now also been developed (Tan et al., 2007; Kubicek et al., 2007). However, further identification and characterization of selective histone methylation inhibitors as anti‐cancer drugs is currently in its infancies.
In summary, improving our understanding of the interplay between DNA repair pathways and chromatin modulators will not only lead to more comprehensive insights into the molecular mechanism of the mammalian DDR in normal cells, but will undoubtedly also provide new avenues for improving cancer therapies.
Abbreviations and acronyms
- ADP
Adenosine diphosphate
- 53BP1
p53‐binding protein 1
- 6‐4PP
pyrimidine‐pyrimidone (6‐4) photoproducts
- ACF1
ATP‐utilizing chromatin assembly and remodeling factor
- ALC1
amplified in liver cancer 1
- APLF
aprataxin‐PNK‐like factor
- ARP
actin‐related protein
- ATM
ataxia telangiectasia mutated
- ATR
ataxia telangiectasia and Rad3‐related
- BER
base excision repair
- BRCA
breast cancer susceptibility gene
- BRCT
breast cancer C‐terminal
- BRG1
brahma‐related gene 1
- BRIT1
BRCT‐repeat inhibitor of hTERT expression
- BRM
brahma
- CBP
Creb‐binding protein
- CD
chromodomain
- CHD
chromodomain helicase DNA‐binding protein
- CHK
checkpoint kinase 1
- CHRAC
chromatin accessibility complex
- CPD
cyclobutane pyrimidine dimers
- CtIP
C‐terminal binding protein interacting protein
- DDR
DNA damage response
- DNA‐PK
DNA‐dependent protein kinase
- DSB
DNA double‐strand break
- FHA
forkhead‐associated
- GCN5
general control nonderepressible 5
- HAT
histone acetyltransferase
- HDAC
histone deacetylase
- HMG
high mobility group
- HP1
heterochromatin protein 1
- HR
homologous recombination
- INO80
inositol requiring 80
- IR
ionizing radiation
- ISWI
imitation switch
- MDC1
mediator of DNA damage checkpoint protein 1
- MMSET
multiple‐myeloma‐related WHSC1
- MOF
males absent on the first
- NBS1
Nijmegen breakage syndrome
- NER
nucleotide excision repair
- NHEJ
non‐homologous end‐joining
- NuA4
nucleosome acetyltransferase of H4
- NuRD
nucleosome remodeling and histone deacetylation
- PARP
poly(ADP‐ribosyl) polymerase
- PRC
polycomb‐recessive comlplex
- RAG
recombination activating gene 1
- RNF
really interesting new gene
- RPA
replication protein A
- SWI/SNF
switch/sucrose non‐fermentable
- TIP60
TAT‐interactive protein 60
- UBC
ubiquitin conjugase
- USP
ubiquitin‐specific protease
- UV
ultra‐violet
- XP
xeroderma pigmentosum
- XRCC
X‐ray cross‐complementation group
- YY1
Yin Yang 1
Acknowledgement
This work was supported by the Netherlands Organization for Scientific Research (NWO Rubicon grant to MSL; NWO‐VIDI grant to HVA), the European Molecular Biology Organization (EMBO long‐term fellowship to MSL) and the Human Frontiers Science Program (HFSP‐CDA grant to HVA).
Luijsterburg Martijn S. and van Attikum Haico, (2011), Chromatin and the DNA damage response: The cancer connection, Molecular Oncology, 5, doi: 10.1016/j.molonc.2011.06.001.
References
- Ahel, D. , Horejsi, Z. , Wiechens, N. , 2009. Poly(ADP-ribose)-dependent regulation of DNA repair by the chromatin remodeling enzyme ALC1. Science. 325, (5945) 1240–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Attikum, H. , Gasser, S.M. , 2009. Crosstalk between histone modifications during the DNA damage response. Trends Cell Biol. [DOI] [PubMed] [Google Scholar]
- Aucott, R. , Bullwinkel, J. , Yu, Y. , 2008. HP1-beta is required for development of the cerebral neocortex and neuromuscular junctions. J. Cell Biol.. 183, (4) 597–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayoub, N. , Jeyasekharan, A.D. , Bernal, J.A. , Venkitaraman, A.R. , 2008. HP1-beta mobilization promotes chromatin changes that initiate the DNA damage response. Nature. 453, (7195) 682–686. [DOI] [PubMed] [Google Scholar]
- Baldeyron, C. , Soria, G. , Roche, D. , Cook, A.J. , Almouzni, G. , 2011. HP1{alpha} recruitment to DNA damage by p150CAF-1 promotes homologous recombination repair. J. Cell Biol.. 193, (1) 81–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker, P.B. , Horz, W. , 2002. ATP-dependent nucleosome remodeling. Annu. Rev. Biochem.. 71, 247–273. [DOI] [PubMed] [Google Scholar]
- Bekker-Jensen, S. , Mailand, N. , 2011. Assembly and function of DNA double-strand break repair foci in mammalian cells. DNA Repair (Amst). 9, (12) 1219–1228. [DOI] [PubMed] [Google Scholar]
- Bekker-Jensen, S. , Fugger, K. , Danielsen, J.R. , 2007. Human Xip1 (C2orf13) is a novel regulator of cellular responses to DNA strand breaks. J. Biol. Chem.. 282, (27) 19638–19643. [DOI] [PubMed] [Google Scholar]
- Bekker-Jensen, S. , Rendtlew Danielsen, J. , Fugger, K. , 2010. HERC2 coordinates ubiquitin-dependent assembly of DNA repair factors on damaged chromosomes. Nat. Cell Biol.. 12, (1) 80–86. 81–12 [DOI] [PubMed] [Google Scholar]
- Bergink, S. , Salomons, F.A. , Hoogstraten, D. , 2006. DNA damage triggers nucleotide excision repair-dependent monoubiquitylation of histone H2A. Genes Dev.. 20, (10) 1343–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beucher, A. , Birraux, J. , Tchouandong, L. , 2009. ATM and Artemis promote homologous recombination of radiation-induced DNA double-strand breaks in G2. EMBO J.. 28, (21) 3413–3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billur, M. , Bartunik, H.D. , Singh, P.B. , 2010. The essential function of HP1 beta: a case of the tail wagging the dog?. Trends Biochem. Sci.. 35, (2) 115–123. [DOI] [PubMed] [Google Scholar]
- Birger, Y. , Catez, F. , Furusawa, T. , 2005. Increased tumorigenicity and sensitivity to ionizing radiation upon loss of chromosomal protein HMGN1. Cancer Res.. 65, (15) 6711–6718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botuyan, M.V. , Lee, J. , Ward, I.M. , 2006. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell. 127, (7) 1361–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand, M. , Moggs, J.G. , Oulad-Abdelghani, M. , 2001. UV-damaged DNA-binding protein in the TFTC complex links DNA damage recognition to nucleosome acetylation. EMBO J.. 20, (12) 3187–3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant, H.E. , Schultz, N. , Thomas, H.D. , 2005. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 434, (7035) 913–917. [DOI] [PubMed] [Google Scholar]
- Celeste, A. , Petersen, S. , Romanienko, P.J. , 2002. Genomic instability in mice lacking histone H2AX. Science. 296, (5569) 922–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celeste, A. , Fernandez-Capetillo, O. , Kruhlak, M.J. , 2003. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nat. Cell Biol.. 5, (7) 675–679. [DOI] [PubMed] [Google Scholar]
- Chernikova, S.B. , Dorth, J.A. , Razorenova, O.V. , Game, J.C. , Brown, J.M. , 2010. Deficiency in Bre1 impairs homologous recombination repair and cell cycle checkpoint response to radiation damage in mammalian cells. Radiat. Res.. 174, (5) 558–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheutin, T. , McNairn, A.J. , Jenuwein, T. , Gilbert, D.M. , Singh, P.B. , Misteli, T. , 2003. Maintenance of stable heterochromatin domains by dynamic HP1 binding. Science. 299, (5607) 721–725. [DOI] [PubMed] [Google Scholar]
- Chou, D.M. , Adamson, B. , Dephoure, N.E. , 2010. A chromatin localization screen reveals poly (ADP ribose)-regulated recruitment of the repressive polycomb and NuRD complexes to sites of DNA damage. Proc. Natl. Acad. Sci. U S A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia, A. , Elledge, S.J. , 2010. The DNA damage response: making it safe to play with knives. Mol. Cell. 40, (2) 179–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapier, C.R. , Cairns, B.R. , 2009. The biology of chromatin remodeling complexes. Annu. Rev. Biochem.. 78, 273–304. [DOI] [PubMed] [Google Scholar]
- Cloos, P.A. , Christensen, J. , Agger, K. , 2006. The putative oncogene GASC1 demethylates tri- and dimethylated lysine 9 on histone H3. Nature. 442, (7100) 307–311. [DOI] [PubMed] [Google Scholar]
- Cook, P.J. , Ju, B.G. , Telese, F. , Wang, X. , Glass, C.K. , Rosenfeld, M.G. , 2009. Tyrosine dephosphorylation of H2AX modulates apoptosis and survival decisions. Nature. 458, (7238) 591–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das, C. , Lucia, M.S. , Hansen, K.C. , Tyler, J.K. , 2009. CBP/p300-mediated acetylation of histone H3 on lysine 56. Nature. 459, (7243) 113–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Koning, L. , Savignoni, A. , Boumendil, C. , 2009. Heterochromatin protein 1alpha: a hallmark of cell proliferation relevant to clinical oncology. EMBO Mol. Med.. 1, (3) 178–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinant, C. , Luijsterburg, M.S. , 2009. The emerging role of HP1 in the DNA damage response. Mol. Cell Biol.. 29, (24) 6335–6340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinant, C. , Houtsmuller, A.B. , Vermeulen, W. , 2008. Chromatin structure and DNA damage repair. Epigenetics Chromatin. 1, (1) 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doil, C. , Mailand, N. , Bekker-Jensen, S. , 2009. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell. 136, (3) 435–446. [DOI] [PubMed] [Google Scholar]
- Doyon, Y. , Cote, J. , 2004. The highly conserved and multifunctional NuA4 HAT complex. Curr. Opin. Genet. Dev.. 14, (2) 147–154. [DOI] [PubMed] [Google Scholar]
- Erdel, F. , Schubert, T. , Marth, C. , Langst, G. , Rippe, K. , 2010. Human ISWI chromatin-remodeling complexes sample nucleosomes via transient binding reactions and become immobilized at active sites. Proc. Natl. Acad. Sci. U S A. 107, (46) 19873–19878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer, H. , McCabe, N. , Lord, C.J. , 2005. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 434, (7035) 917–921. [DOI] [PubMed] [Google Scholar]
- Fernandez-Capetillo, O. , Chen, H.T. , Celeste, A. , 2002. DNA damage-induced G2-M checkpoint activation by histone H2AX and 53BP1. Nat. Cell Biol.. 4, (12) 993–997. [DOI] [PubMed] [Google Scholar]
- Fernandez-Capetillo, O. , Allis, C.D. , Nussenzweig, A. , 2004. Phosphorylation of histone H2B at DNA double-strand breaks. J. Exp. Med.. 199, (12) 1671–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fnu, S. , Williamson, E.A. , De Haro, L.P. , 2010. Methylation of histone H3 lysine 36 enhances DNA repair by nonhomologous end-joining. Proc. Natl. Acad. Sci. U S A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fousteri, M. , Vermeulen, W. , van Zeeland, A.A. , Mullenders, L.H. , 2006. Cockayne syndrome A and B proteins differentially regulate recruitment of chromatin remodeling and repair factors to stalled RNA polymerase II in vivo. Mol. Cell. 23, (4) 471–482. [DOI] [PubMed] [Google Scholar]
- Giglia-Mari, G. , Zotter, A. , Vermeulen, W. , 2010. DNA damage response. Cold Spring Harb Perspect. Biol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginjala, V. , Nacerddine, K. , Kulkarni, A. , 2011. BMI1 is recruited to DNA breaks and contributes to DNA damage induced H2A ubiquitination and repair. Mol. Cell Biol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldknopf, I.L. , Taylor, C.W. , Baum, R.M. , 1975. Isolation and characterization of protein A24, a “histone-like” non-histone chromosomal protein. J. Biol. Chem.. 250, (18) 7182–7187. [PubMed] [Google Scholar]
- Gong, F. , Fahy, D. , Liu, H. , Wang, W. , Smerdon, M.J. , 2008. Role of the mammalian SWI/SNF chromatin remodeling complex in the cellular response to UV damage. Cell Cycle. 7, (8) 1067–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodarzi, A.A. , Noon, A.T. , Deckbar, D. , 2008. ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Mol. Cell. 31, (2) 167–177. [DOI] [PubMed] [Google Scholar]
- Gottschalk, A.J. , Timinszky, G. , Kong, S.E. , 2009. Poly(ADP-ribosyl)ation directs recruitment and activation of an ATP-dependent chromatin remodeler. Proc. Natl. Acad. Sci. U S A. 106, (33) 13770–13774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groisman, R. , Polanowska, J. , Kuraoka, I. , 2003. The ubiquitin ligase activity in the DDB2 and CSA complexes is differentially regulated by the COP9 signalosome in response to DNA damage. Cell. 113, (3) 357–367. [DOI] [PubMed] [Google Scholar]
- Grundy, G.J. , Yang, W. , Gellert, M. , 2010. Autoinhibition of DNA cleavage mediated by RAG1 and RAG2 is overcome by an epigenetic signal in V(D)J recombination. Proc. Natl. Acad. Sci. U S A. 107, (52) 22487–22492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, R. , Chen, J. , Mitchell, D.L. , Johnson, D.G. , 2010. GCN5 and E2F1 stimulate nucleotide excision repair by promoting H3K9 acetylation at sites of damage. Nucleic Acids Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, J.A. , Georgel, P.T. , 2007. CHD proteins: a diverse family with strong ties. Biochem. Cell Biol.. 85, (4) 463–476. [DOI] [PubMed] [Google Scholar]
- Hanasoge, S. , Ljungman, M. , 2007. H2AX phosphorylation after UV irradiation is triggered by DNA repair intermediates and is mediated by the ATR kinase. Carcinogenesis. 28, (11) 2298–2304. [DOI] [PubMed] [Google Scholar]
- Helmink, B.A. , Tubbs, A.T. , Dorsett, Y. , 2010. H2AX prevents CtIP-mediated DNA end resection and aberrant repair in G1-phase lymphocytes. Nature. 469, (7329) 245–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeijmakers, J.H. , 2001. Genome maintenance mechanisms for preventing cancer. Nature. 411, (6835) 366–374. [DOI] [PubMed] [Google Scholar]
- Huang, J. , Huen, M.S. , Kim, H. , 2009. RAD18 transmits DNA damage signalling to elicit homologous recombination repair. Nat. Cell Biol.. 11, (5) 592–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, B. , Babcock, H. , Zhuang, X. , 2010. Breaking the diffraction barrier: super-resolution imaging of cells. Cell. 143, (7) 1047–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huen, M.S. , Chen, J. , 2010. Assembly of checkpoint and repair machineries at DNA damage sites. Trends Biochem. Sci.. 35, (2) 101–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huen, M.S. , Grant, R. , Manke, I. , 2007. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell. 131, (5) 901–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huen, M.S. , Huang, J. , Leung, J.W. , 2010. Regulation of chromatin architecture by the PWWP domain-containing DNA damage-responsive factor EXPAND1/MUM1. Mol. Cell. 37, (6) 854–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huyen, Y. , Zgheib, O. , Ditullio, R.A. , 2004. Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature. 432, (7015) 406–411. [DOI] [PubMed] [Google Scholar]
- Ikura, T. , Tashiro, S. , Kakino, A. , 2007. DNA damage-dependent acetylation and ubiquitination of H2AX enhances chromatin dynamics. Mol. Cell Biol.. 27, (20) 7028–7040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isakoff, M.S. , Sansam, C.G. , Tamayo, P. , 2005. Inactivation of the Snf5 tumor suppressor stimulates cell cycle progression and cooperates with p53 loss in oncogenic transformation. Proc. Natl. Acad. Sci. U S A. 102, (49) 17745–17750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail, I.H. , Andrin, C. , McDonald, D. , Hendzel, M.J. , 2010. BMI1-mediated histone ubiquitylation promotes DNA double-strand break repair. J. Cell Biol.. 191, (1) 45–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, S.P. , Bartek, J. , 2009. The DNA-damage response in human biology and disease. Nature. 461, (7267) 1071–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jason, L.J. , Moore, S.C. , Lewis, J.D. , Lindsey, G. , Ausio, J. , 2002. Histone ubiquitination: a tagging tail unfolds?. Bioessays. 24, (2) 166–174. [DOI] [PubMed] [Google Scholar]
- Jenuwein, T. , Allis, C.D. , 2001. Translating the histone code. Science. 293, (5532) 1074–1080. [DOI] [PubMed] [Google Scholar]
- Jiang, X. , Xu, Y. , Price, B.D. , 2010. Acetylation of H2AX on lysine 36 plays a key role in the DNA double-strand break repair pathway. FEBS Lett.. 584, (13) 2926–2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, Y. , Wang, X. , Bao, S. , 2010. INO80 chromatin remodeling complex promotes the removal of UV lesions by the nucleotide excision repair pathway. Proc. Natl. Acad. Sci. U S A. 107, (40) 17274–17279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapetanaki, M.G. , Guerrero-Santoro, J. , Bisi, D.C. , Hsieh, C.L. , Rapic-Otrin, V. , Levine, A.S. , 2006. The DDB1-CUL4ADDB2 ubiquitin ligase is deficient in xeroderma pigmentosum group E and targets histone H2A at UV-damaged DNA sites. Proc. Natl. Acad. Sci. U S A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashiwaba, S. , Kitahashi, K. , Watanabe, T. , Onoda, F. , Ohtsu, M. , Murakami, Y. , 2010. The mammalian INO80 complex is recruited to DNA damage sites in an ARP8 dependent manner. Biochem. Biophys. Res. Commun.. 402, (4) 619–625. [DOI] [PubMed] [Google Scholar]
- Kim, J. , Guermah, M. , McGinty, R.K. , 2009. RAD6-Mediated transcription-coupled H2B ubiquitylation directly stimulates H3K4 methylation in human cells. Cell. 137, (3) 459–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, Y.C. , Gerlitz, G. , Furusawa, T. , 2009. Activation of ATM depends on chromatin interactions occurring before induction of DNA damage. Nat. Cell Biol.. 11, (1) 92–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitayama, K. , Kamo, M. , Oma, Y. , 2009. The human actin-related protein hArp5: nucleo-cytoplasmic shuttling and involvement in DNA repair. Exp. Cell Res.. 315, (2) 206–217. [DOI] [PubMed] [Google Scholar]
- Klochendler-Yeivin, A. , Picarsky, E. , Yaniv, M. , 2006. Increased DNA damage sensitivity and apoptosis in cells lacking the Snf5/Ini1 subunit of the SWI/SNF chromatin remodeling complex. Mol. Cell Biol.. 26, (7) 2661–2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolas, N.K. , Chapman, J.R. , Nakada, S. , 2007. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science. 318, (5856) 1637–1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubicek, S. , O'Sullivan, R.J. , August, E.M. , 2007. Reversal of H3K9me2 by a small-molecule inhibitor for the G9a histone methyltransferase. Mol. Cell. 25, (3) 473–481. [DOI] [PubMed] [Google Scholar]
- de Laat, W.L. , Jaspers, N.G. , Hoeijmakers, J.H. , 1999. Molecular mechanism of nucleotide excision repair. Genes Dev.. 13, (7) 768–785. [DOI] [PubMed] [Google Scholar]
- Lan, L. , Ui, A. , Nakajima, S. , 2010. The ACF1 complex is required for DNA double-strand break repair in human cells. Mol. Cell. 40, (6) 976–987. [DOI] [PubMed] [Google Scholar]
- Lans, H. , Marteijn, J.A. , Schumacher, B. , Hoeijmakers, J.H. , Jansen, G. , Vermeulen, W. , 2010. Involvement of global genome repair, transcription coupled repair, and chromatin remodeling in UV DNA damage response changes during development. PLoS Genet.. 6, e1000941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen, D.H. , Poinsignon, C. , Gudjonsson, T. , 2010. The chromatin-remodeling factor CHD4 coordinates signaling and repair after DNA damage. J. Cell Biol.. 190, (5) 731–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J.H. , Paull, T.T. , 2005. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science. 308, (5721) 551–554. [DOI] [PubMed] [Google Scholar]
- Lee, H.S. , Park, J.H. , Kim, S.J. , Kwon, S.J. , Kwon, J. , 2010. A cooperative activation loop among SWI/SNF, gamma-H2AX and H3 acetylation for DNA double-strand break repair. EMBO J.. 29, (8) 1434–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J.H. , Goodarzi, A.A. , Jeggo, P.A. , Paull, T.T. , 2010. 53BP1 promotes ATM activity through direct interactions with the MRN complex. EMBO J.. 29, (3) 574–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X. , Corsa, C.A. , Pan, P.W. , 2010. MOF and H4 K16 acetylation play important roles in DNA damage repair by modulating recruitment of DNA damage repair protein Mdc1. Mol. Cell Biol.. 30, (22) 5335–5347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, S.Y. , Rai, R. , Li, K. , Xu, Z.X. , Elledge, S.J. , 2005. BRIT1/MCPH1 is a DNA damage responsive protein that regulates the Brca1-Chk1 pathway, implicating checkpoint dysfunction in microcephaly. Proc. Natl. Acad. Sci. U S A. 102, (42) 15105–15109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luijsterburg, M.S. , Dinant, C. , Lans, H. , 2009. Heterochromatin protein 1 is recruited to various types of DNA damage. J. Cell Biol.. 185, (4) 577–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailand, N. , Bekker-Jensen, S. , Faustrup, H. , 2007. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell. 131, (5) 887–900. [DOI] [PubMed] [Google Scholar]
- Marteijn, J.A. , Bekker-Jensen, S. , Mailand, N. , 2009. Nucleotide excision repair-induced H2A ubiquitination is dependent on MDC1 and RNF8 and reveals a universal DNA damage response. J. Cell Biol.. 186, (6) 835–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marti, T.M. , Hefner, E. , Feeney, L. , Natale, V. , Cleaver, J.E. , 2006. H2AX phosphorylation within the G1 phase after UV irradiation depends on nucleotide excision repair and not DNA double-strand breaks. Proc. Natl. Acad. Sci. U S A. 103, (26) 9891–9896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez, E. , Palhan, V.B. , Tjernberg, A. , 2001. Human STAGA complex is a chromatin-acetylating transcription coactivator that interacts with pre-mRNA splicing and DNA damage-binding factors in vivo. Mol. Cell Biol.. 21, (20) 6782–6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka, S. , Ballif, B.A. , Smogorzewska, A. , 2007. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 316, (5828) 1160–1166. [DOI] [PubMed] [Google Scholar]
- Matthews, A.G. , Kuo, A.J. , Ramon-Maiques, S. , 2007. RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination. Nature. 450, (7172) 1106–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna, E.S. , Sansam, C.G. , Cho, Y.J. , 2008. Loss of the epigenetic tumor suppressor SNF5 leads to cancer without genomic instability. Mol. Cell Biol.. 28, (20) 6223–6233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meerbrey, K.L. , Hu, G. , Kessler, J.D. , 2011. The pINDUCER lentiviral toolkit for inducible RNA interference in vitro and in vivo. Proc. Natl. Acad. Sci. U S A. 108, (9) 3665–3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrotra, P.V. , Ahel, D. , Ryan, D.P. , 2011. DNA repair factor APLF is a histone chaperone. Mol. Cell. 41, (1) 46–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messick, T.E. , Greenberg, R.A. , 2009. The ubiquitin landscape at DNA double-strand breaks. J. Cell Biol.. 187, (3) 319–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, K.M. , Tjeertes, J.V. , Coates, J. , 2010. Human HDAC1 and HDAC2 function in the DNA-damage response to promote DNA nonhomologous end-joining. Nat. Struct. Mol. Biol.. 17, (9) 1144–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli, T. , Soutoglou, E. , 2009. The emerging role of nuclear architecture in DNA repair and genome maintenance. Nat. Rev. Mol. Cell Biol.. 10, (4) 243–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell, J.R. , Hoeijmakers, J.H. , Niedernhofer, L.J. , 2003. Divide and conquer: nucleotide excision repair battles cancer and ageing. Curr. Opin. Cell Biol.. 15, (2) 232–240. [DOI] [PubMed] [Google Scholar]
- Mohammad, D.H. , Yaffe, M.B. , 2009. 14-3-3 proteins, FHA domains and BRCT domains in the DNA damage response. DNA Repair (Amst). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyal, L. , Lerenthal, Y. , Gana-Weisz, M. , 2011. Requirement of ATM-dependent monoubiquitylation of histone H2B for timely repair of DNA double-strand breaks. Mol. Cell. 41, (5) 529–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munshi, A. , Kurland, J.F. , Nishikawa, T. , 2005. Histone deacetylase inhibitors radiosensitize human melanoma cells by suppressing DNA repair activity. Clin. Cancer Res.. 11, (13) 4912–4922. [DOI] [PubMed] [Google Scholar]
- Murr, R. , Loizou, J.I. , Yang, Y.G. , 2006. Histone acetylation by Trrap-Tip60 modulates loading of repair proteins and repair of DNA double-strand breaks. Nat. Cell Biol.. 8, (1) 91–99. [DOI] [PubMed] [Google Scholar]
- Nagarajan, P. , Onami, T.M. , Rajagopalan, S. , Kania, S. , Donnell, R. , Venkatachalam, S. , 2009. Role of chromodomain helicase DNA-binding protein 2 in DNA damage response signaling and tumorigenesis. Oncogene. 28, (8) 1053–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakada, S. , Tai, I. , Panier, S. , 2010. Non-canonical inhibition of DNA damage-dependent ubiquitination by OTUB1. Nature. 466, (7309) 941–946. [DOI] [PubMed] [Google Scholar]
- Nakamura, K. , Kato, A. , Kobayashi, J. , 2011. Regulation of homologous recombination by RNF20-dependent H2B ubiquitination. Mol. Cell. 41, (5) 515–528. [DOI] [PubMed] [Google Scholar]
- Negrini, S. , Gorgoulis, V.G. , Halazonetis, T.D. , 2010. Genomic instability – An evolving hallmark of cancer. Nat. Rev. Mol. Cell Biol.. 11, (3) 220–228. [DOI] [PubMed] [Google Scholar]
- Nicassio, F. , Corrado, N. , Vissers, J.H. , 2007. Human USP3 is a chromatin modifier required for S phase progression and genome stability. Curr. Biol.. 17, (22) 1972–1977. [DOI] [PubMed] [Google Scholar]
- Noon, A.T. , Shibata, A. , Rief, N. , 2010. 53BP1-dependent robust localized KAP-1 phosphorylation is essential for heterochromatic DNA double-strand break repair. Nat. Cell Biol.. 12, (2) 177–184. [DOI] [PubMed] [Google Scholar]
- Oda, H. , Hubner, M.R. , Beck, D.B. , 2010. Regulation of the histone H4 monomethylase PR-Set7 by CRL4(Cdt2)-mediated PCNA-dependent degradation during DNA damage. Mol. Cell. 40, (3) 364–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Driscoll, M. , Ruiz-Perez, V.L. , Woods, C.G. , Jeggo, P.A. , Goodship, J.A. , 2003. A splicing mutation affecting expression of ataxia-telangiectasia and Rad3-related protein (ATR) results in Seckel syndrome. Nat. Genet.. 33, (4) 497–501. [DOI] [PubMed] [Google Scholar]
- Ogiwara, H. , Ui, A. , Otsuka, A. , 2011. Histone acetylation by CBP and p300 at double-strand break sites facilitates SWI/SNF chromatin remodeling and the recruitment of non-homologous end joining factors. Oncogene. [DOI] [PubMed] [Google Scholar]
- Park, J.H. , Park, E.J. , Lee, H.S. , 2006. Mammalian SWI/SNF complexes facilitate DNA double-strand break repair by promoting gamma-H2AX induction. EMBO J.. 25, (17) 3986–3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavri, R. , Zhu, B. , Li, G. , 2006. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell. 125, (4) 703–717. [DOI] [PubMed] [Google Scholar]
- Pegoraro, G. , Kubben, N. , Wickert, U. , Gohler, H. , Hoffmann, K. , Misteli, T. , 2009. Ageing-related chromatin defects through loss of the NURD complex. Nat. Cell Biol.. 11, (10) 1261–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei, H. , Zhang, L. , Luo, K. , 2011. MMSET regulates histone H4K20 methylation and 53BP1 accumulation at DNA damage sites. Nature. 470, (7332) 124–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, G. , Yim, E.K. , Dai, H. , 2009. BRIT1/MCPH1 links chromatin remodelling to DNA damage response. Nat. Cell Biol.. 11, (7) 865–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters, A.H. , O'Carroll, D. , Scherthan, H. , 2001. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell. 107, (3) 323–337. [DOI] [PubMed] [Google Scholar]
- Pinato, S. , Scandiuzzi, C. , Arnaudo, N. , Citterio, E. , Gaudino, G. , Penengo, L. , 2009. RNF168, a new RING finger, MIU-containing protein that modifies chromatin by ubiquitination of histones H2A and H2AX. BMC Mol. Biol.. 10, 55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo, S.E. , Jackson, S.P. , 2011. Dynamics of DNA damage response proteins at DNA breaks: a focus on protein modifications. Genes Dev.. 25, (5) 409–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo, S.E. , Kaidi, A. , Baskcomb, L. , Galanty, Y. , Jackson, S.P. , 2010. Regulation of DNA-damage responses and cell-cycle progression by the chromatin remodelling factor CHD4. EMBO J.. 29, (18) 3130–3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poot, R.A. , Dellaire, G. , Hulsmann, B.B. , 2000. HuCHRAC, a human ISWI chromatin remodelling complex contains hACF1 and two novel histone-fold proteins. EMBO J.. 19, (13) 3377–3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai, R. , Dai, H. , Multani, A.S. , 2006. BRIT1 regulates early DNA damage response, chromosomal integrity, and cancer. Cancer Cell. 10, (2) 145–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanathan, B. , Smerdon, M.J. , 1986. Changes in nuclear protein acetylation in UV-damaged human cells. Carcinogenesis. 7, (7) 1087–1094. [DOI] [PubMed] [Google Scholar]
- Ray, A. , Mir, S.N. , Wani, G. , 2009. Human SNF5/INI1, a component of the human SWI/SNF chromatin remodeling complex, promotes nucleotide excision repair by influencing ATM recruitment and downstream H2AX phosphorylation. Mol. Cell Biol.. 29, (23) 6206–6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ropero, S. , Esteller, M. , 2007. The role of histone deacetylases (HDACs) in human cancer. Mol. Oncol.. 1, (1) 19–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubbi, C.P. , Milner, J. , 2003. p53 is a chromatin accessibility factor for nucleotide excision repair of DNA damage. EMBO J.. 22, (4) 975–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rulten, S.L. , Fisher, A.E. , Robert, I. , 2011. PARP-3 and APLF function together to accelerate nonhomologous end-joining. Mol. Cell. 41, (1) 33–45. [DOI] [PubMed] [Google Scholar]
- Sato, Y. , Yoshikawa, A. , Mimura, H. , Yamashita, M. , Yamagata, A. , Fukai, S. , 2009. Structural basis for specific recognition of Lys 63-linked polyubiquitin chains by tandem UIMs of RAP80. EMBO J.. 28, (16) 2461–2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schotta, G. , Sengupta, R. , Kubicek, S. , 2008. A chromatin-wide transition to H4K20 monomethylation impairs genome integrity and programmed DNA rearrangements in the mouse. Genes Dev.. 22, (15) 2048–2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanbhag, N.M. , Rafalska-Metcalf, I.U. , Balane-Bolivar, C. , Janicki, S.M. , Greenberg, R.A. , 2010. ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks. Cell. 141, (6) 970–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao, G. , Lilli, D.R. , Patterson-Fortin, J. , Coleman, K.A. , Morrissey, D.E. , Greenberg, R.A. , 2009. The Rap80-BRCC36 de-ubiquitinating enzyme complex antagonizes RNF8-Ubc13-dependent ubiquitination events at DNA double strand breaks. Proc. Natl. Acad. Sci. U S A. 106, (9) 3166–3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma, G.G. , Hwang, K.K. , Pandita, R.K. , 2003. Human heterochromatin protein 1 isoforms HP1(Hsalpha) and HP1(Hsbeta) interfere with hTERT-telomere interactions and correlate with changes in cell growth and response to ionizing radiation. Mol. Cell Biol.. 23, (22) 8363–8376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma, G.G. , So, S. , Gupta, A. , 2010. MOF and histone H4 acetylation at lysine 16 are critical for DNA damage response and double-strand break repair. Mol. Cell Biol.. 30, (14) 3582–3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shema, E. , Tirosh, I. , Aylon, Y. , 2008. The histone H2B-specific ubiquitin ligase RNF20/hBRE1 acts as a putative tumor suppressor through selective regulation of gene expression. Genes Dev.. 22, (19) 2664–2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada, M. , Niida, H. , Zineldeen, D.H. , 2008. Chk1 is a histone H3 threonine 11 kinase that regulates DNA damage-induced transcriptional repression. Cell. 132, (2) 221–232. [DOI] [PubMed] [Google Scholar]
- Shimada, M. , Haruta, M. , Niida, H. , Sawamoto, K. , Nakanishi, M. , 2010. Protein phosphatase 1gamma is responsible for dephosphorylation of histone H3 at Thr 11 after DNA damage. EMBO Rep.. 11, (11) 883–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimazaki, N. , Tsai, A.G. , Lieber, M.R. , 2009. H3K4me3 stimulates the V(D)J RAG complex for both nicking and hairpinning in trans in addition to tethering in cis: implications for translocations. Mol. Cell. 34, (5) 535–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiotani, B. , Zou, L. , 2009. Single-stranded DNA orchestrates an ATM-to-ATR switch at DNA breaks. Mol. Cell. 33, (5) 547–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shogren-Knaak, M. , Ishii, H. , Sun, J.M. , Pazin, M.J. , Davie, J.R. , Peterson, C.L. , 2006. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 311, (5762) 844–847. [DOI] [PubMed] [Google Scholar]
- Sims, J.J. , Cohen, R.E. , 2009. Linkage-specific avidity defines the lysine 63-linked polyubiquitin-binding preference of rap80. Mol. Cell. 33, (6) 775–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeenk, G. , Wiegant, W.W. , Vrolijk, H. , Solari, A.P. , Pastink, A. , van Attikum, H. , 2010. The NuRD chromatin-remodeling complex regulates signaling and repair of DNA damage. J. Cell Biol.. 190, (5) 741–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart, G.S. , Panier, S. , Townsend, K. , 2009. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell. 136, (3) 420–434. [DOI] [PubMed] [Google Scholar]
- Stiff, T. , Walker, S.A. , Cerosaletti, K. , 2006. ATR-dependent phosphorylation and activation of ATM in response to UV treatment or replication fork stalling. EMBO J.. 25, (24) 5775–5782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stucki, M. , Clapperton, J.A. , Mohammad, D. , Yaffe, M.B. , Smerdon, S.J. , Jackson, S.P. , 2005. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell. 123, (7) 1213–1226. [DOI] [PubMed] [Google Scholar]
- Sun, Y. , Jiang, X. , Xu, Y. , 2009. Histone H3 methylation links DNA damage detection to activation of the tumour suppressor Tip60. Nat. Cell Biol.. 11, (11) 1376–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, J. , Yang, X. , Zhuang, L. , 2007. Pharmacologic disruption of polycomb-repressive complex 2-mediated gene repression selectively induces apoptosis in cancer cells. Genes Dev.. 21, (9) 1050–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taverna, S.D. , Li, H. , Ruthenburg, A.J. , Allis, C.D. , Patel, D.J. , 2007. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat. Struct. Mol. Biol.. 14, (11) 1025–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timinszky, G. , Till, S. , Hassa, P.O. , 2009. A macrodomain-containing histone rearranges chromatin upon sensing PARP1 activation. Nat. Struct. Mol. Biol.. 16, (9) 923–929. [DOI] [PubMed] [Google Scholar]
- Tjeertes, J.V. , Miller, K.M. , Jackson, S.P. , 2009. Screen for DNA-damage-responsive histone modifications identifies H3K9Ac and H3K56Ac in human cells. EMBO J.. 28, (13) 1878–1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, B.M. , 2002. Cellular memory and the histone code. Cell. 111, (3) 285–291. [DOI] [PubMed] [Google Scholar]
- Ura, K. , Araki, M. , Saeki, H. , 2001. ATP-dependent chromatin remodeling facilitates nucleotide excision repair of UV-induced DNA lesions in synthetic dinucleosomes. EMBO J.. 20, (8) 2004–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varela, I. , Tarpey, P. , Raine, K. , 2011. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature. 469, (7331) 539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga-Weisz, P.D. , Becker, P.B. , 2006. Regulation of higher-order chromatin structures by nucleosome-remodelling factors. Curr. Opin. Genet. Dev.. 16, (2) 151–156. [DOI] [PubMed] [Google Scholar]
- Vempati, R.K. , Jayani, R.S. , Notani, D. , Sengupta, A. , Galande, S. , Haldar, D. , 2010. p300-mediated acetylation of histone H3 lysine 56 functions in DNA damage response in mammals. J. Biol. Chem.. 285, (37) 28553–28564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versteege, I. , Sevenet, N. , Lange, J. , 1998. Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature. 394, (6689) 203–206. [DOI] [PubMed] [Google Scholar]
- Vrouwe, M.G. , Pines, A. , Overmeer, R.M. , Hanada, K. , Mullenders, L.H. , 2011. UV-induced photolesions elicit ATR-kinase-dependent signaling in non-cycling cells through nucleotide excision repair-dependent and -independent pathways. J. Cell Sci.. 124, (3) 435–446. [DOI] [PubMed] [Google Scholar]
- Walther, T.C. , Mann, M. , 2010. Mass spectrometry-based proteomics in cell biology. J. Cell Biol.. 190, (4) 491–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, B. , Elledge, S.J. , 2007. Ubc13/Rnf8 ubiquitin ligases control foci formation of the Rap80/Abraxas/Brca1/Brcc36 complex in response to DNA damage. Proc. Natl. Acad. Sci. U S A. 104, (52) 20759–20763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H. , Wang, L. , Erdjument-Bromage, H. , 2004. Role of histone H2A ubiquitination in polycomb silencing. Nature. 431, (7010) 873–878. [DOI] [PubMed] [Google Scholar]
- Wang, H. , Zhai, L. , Xu, J. , 2006. Histone H3 and H4 ubiquitylation by the CUL4-DDB-ROC1 ubiquitin ligase facilitates cellular response to DNA damage. Mol. Cell. 22, (3) 383–394. [DOI] [PubMed] [Google Scholar]
- Wang, J. , Chin, M.Y. , Li, G. , 2006. The novel tumor suppressor p33ING2 enhances nucleotide excision repair via inducement of histone H4 acetylation and chromatin relaxation. Cancer Res.. 66, (4) 1906–1911. [DOI] [PubMed] [Google Scholar]
- Wang, B. , Matsuoka, S. , Ballif, B.A. , 2007. Abraxas and RAP80 form a BRCA1 protein complex required for the DNA damage response. Science. 316, (5828) 1194–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, G.G. , Allis, C.D. , Chi, P. , 2007. Chromatin remodeling and cancer, Part I: covalent histone modifications. Trends Mol. Med.. 13, (9) 363–372. [DOI] [PubMed] [Google Scholar]
- Warmerdam, D.O. , Kanaar, R. , 2010. Dealing with DNA damage: relationships between checkpoint and repair pathways. Mutat. Res.. 704, (1–3) 2–11. [DOI] [PubMed] [Google Scholar]
- Wilson, B.G. , Wang, X. , Shen, X. , 2010. Epigenetic antagonism between polycomb and SWI/SNF complexes during oncogenic transformation. Cancer Cell. 18, (4) 316–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, S. , Shi, Y. , Mulligan, P. , 2007. A YY1-INO80 complex regulates genomic stability through homologous recombination-based repair. Nat. Struct. Mol. Biol.. 14, (12) 1165–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyman, C. , Kanaar, R. , 2006. DNA double-strand break repair: all's well that ends well. Annu. Rev. Genet.. 40, 363–383. [DOI] [PubMed] [Google Scholar]
- Xiao, A. , Li, H. , Shechter, D. , 2009. WSTF regulates the H2A.X DNA damage response via a novel tyrosine kinase activity. Nature. 457, (7225) 57–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, A. , Hartlerode, A. , Stucki, M. , 2007. Distinct roles of chromatin-associated proteins MDC1 and 53BP1 in mammalian double-strand break repair. Mol. Cell. 28, (6) 1045–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Y. , Price, B.D. , 2011. Chromatin dynamics and the repair of DNA double strand breaks. Cell Cycle. 10, (2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Y. , Sun, Y. , Jiang, X. , 2010. The p400 ATPase regulates nucleosome stability and chromatin ubiquitination during DNA repair. J. Cell Biol.. 191, (1) 31–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, Q. , Dutt, S. , Xu, R. , 2009. BBAP monoubiquitylates histone H4 at lysine 91 and selectively modulates the DNA damage response. Mol. Cell. 36, (1) 110–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarebski, M. , Wiernasz, E. , Dobrucki, J.W. , 2009. Recruitment of heterochromatin protein 1 to DNA repair sites. Cytometry A. 75, (7) 619–625. [DOI] [PubMed] [Google Scholar]
- Zha, S. , Guo, C. , Boboila, C. , 2010. ATM damage response and XLF repair factor are functionally redundant in joining DNA breaks. Nature. 469, (7329) 250–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, D. , Zaugg, K. , Mak, T.W. , Elledge, S.J. , 2006. A role for the deubiquitinating enzyme USP28 in control of the DNA-damage response. Cell. 126, (3) 529–542. [DOI] [PubMed] [Google Scholar]
- Zhang, L. , Zhang, Q. , Jones, K. , Patel, M. , Gong, F. , 2009. The chromatin remodeling factor BRG1 stimulates nucleotide excision repair by facilitating recruitment of XPC to sites of DNA damage. Cell Cycle. 8, (23) 3953–3959. [DOI] [PubMed] [Google Scholar]
- Zhao, Q. , Wang, Q.E. , Ray, A. , 2009. Modulation of nucleotide excision repair by mammalian SWI/SNF chromatin-remodeling complex. J. Biol. Chem.. 284, (44) 30424–30432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, Q. , Wani, G. , Arab, H.H. , El-Mahdy, M.A. , Ray, A. , Wani, A.A. , 2009. Chromatin restoration following nucleotide excision repair involves the incorporation of ubiquitinated H2A at damaged genomic sites. DNA Repair (Amst). 8, (2) 262–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziv, Y. , Bielopolski, D. , Galanty, Y. , 2006. Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM- and KAP-1 dependent pathway. Nat. Cell Biol.. 8, (8) 870–876. [DOI] [PubMed] [Google Scholar]
- Zou, L. , Elledge, S.J. , 2003. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 300, (5625) 1542–1548. [DOI] [PubMed] [Google Scholar]


