Abstract
Risk assessment of future breast cancer risk through exposure to sex steroids currently relies on clinical scorings such as mammographic density. Knowledge about the gene expression patterns in existing breast cancer tumors may be used to identify risk factors in the breast tissue of women still free of cancer. The differential effects of estradiol, estradiol together with gestagens, or tibolone on breast cancer‐related gene expression in normal breast tissue samples taken from postmenopausal women may be used to identify gene expression profiles associated with a higher breast cancer risk. Breast tissue samples were taken from 33 healthy postmenopausal women both before and after a six month treatment with either 2mg micronized estradiol [E2], 2mg micronized estradiol and 1mg norethisterone acetate [E2+NETA], 2.5mg tibolone [T] or [no HRT]. Except for [E2], which was only given to women after hysterectomy, the allocation to each of the three groups was randomized. The expression of 102 mRNAs and 46 microRNAs putatively involved in breast cancer was prospectively determined in the biopsies of 6 women receiving [no HRT], 5 women receiving [E2], 5 women receiving [E2+NETA], and 6 receiving [T]. Using epithelial and endothelial markers genes, non‐representative biopsies from 11 women were eliminated. Treatment of postmenopausal women with [E2+NETA] resulted in the highest number of differentially (p<0.05) regulated genes (16.2%) compared to baseline, followed by [E2] (10.1%) and [T] (4.7%). Among genes that were significantly down‐regulated by [E2+NETA] ranked estrogen‐receptor‐1 (ESR1, p=0.019) and androgen receptor (AR, p=0.019), whereas CYP1B1, a gene encoding an estrogen‐metabolizing enzyme, was significantly up‐regulated (p=0.016). Mammary cells triggered by [E2+NETA] and [E2] adjust for steroidogenic up‐regulation through down‐regulation of the estrogen‐receptor pathway. In this prospective study, prolonged administration of [E2+NETA] and to a lesser extent of [E2] but not [T] were associated in otherwise healthy breast tissue with a change in the expression of genes putatively involved in breast cancer. Our data suggest that normal mammary cells triggered by [E2+NETA] adjust for steroidogenic up‐regulation through down‐regulation of the estrogen‐receptor pathway. This feasibility study provides the basis for whole genome analyses to identify novel markers involved in increased breast cancer risk.
Keywords: Hormonal replacement therapy, Menopause, Estradiol, Tibolone, Breast cancer
1. Introduction
Breast cancer risk assessment is an important issue in clinical medicine and various methods such as scoring systems based on epidemiological characteristics (Gail et al., 1989), sex steroid levels in the serum (Key et al., 2002; Missmer et al., 2004; Zeleniuch‐Jacquotte et al., 2004) and mammographic density (Byrne et al., 1995) have been proposed for the early prediction of a higher breast cancer risk in individual postmenopausal women. All these parameters point to the crucial role of sex steroids in creating the environment in the breast which ultimately leads to the manifestation of overt breast cancer disease. The present study was based on the hypothesis that the changes caused by hormonal therapeutics on the expression levels of genes in the mammary tissue of postmenopausal women can be used to identify additional markers for the prediction of incident breast cancer.
Various hormonal treatments have been shown to modify the breast tissue differently and to alter the risk of breast cancer. With mammographic density known to be increased during prolonged administration of hormone replacement therapy (HRT, Greendalem et al., 2003) and to be reduced under tamoxifen (Cuzick et al., 2004), both mammographic density and circulating sex steroid levels have been described as independent markers of an increased risk of breast cancer by HRT (Tamimi et al., 2007). Combined treatments with estrogens and progestagens and to a lesser extent with estrogen‐only preparations enhance mammographic density, whereas tibolone [T] does not affect mammographic density (Lundström et al., 2002; Valdivia et al., 2004). Continued treatment with both estrogens and progestagens increases the incidence of invasive breast cancer in postmenopausal women (Collaborative Group on Hormonal Factors in Breast Cancer, 1997; Schairer et al., 2000; Writing Group for the Women's Health Initiative Investigators, 2002). In contrast, the risk of breast cancer was not significantly influenced in hysterectomized women receiving conjugated estrogens alone (The Women's Health Initiative Steering Committee, 2004) as for elderly women with chronic low circulating estrogen levels at risk of osteoporosis receiving [T] (Cummings et al., 2008), although the recurrence rate was increased by [T] in women previously diagnosed with breast cancer (Kenemans et al., 2009). In case–control studies involving early postmenopausal women, the results with respect to the effect of [T] on breast cancer risk are conflicting (Beral et al., 2003; Banks et al., 2006; Opatrny et al., 2008).
The aim of this prospective study was to explore the effects of three distinctive hormonal preparations, each with a different effect on mammographic density and on circulating sex hormone levels, on the expression of a list of genes putatively known to be involved in breast cancer, such as proliferation and apoptosis markers, breast cancer subtype specific stromal and stem cell markers as well as markers for steroid receptors, steroidogenic and steroid‐metabolizing enzymes (Supplementary Table 1, on line only). This list of genes was extracted from large data bases collected from well characterized breast cancer samples which were used so far to establish prognostic and predictive patterns in already existing breast cancer cases. To evaluate the expression of these genes in relation to increased breast cancer risk of healthy women due to the use of HRT core needle biopsies were taken from the breasts of healthy postmenopausal women before and after six months of hormonal treatment.
2. Subjects and methods
This research project was conceived as a prospective study aiming at identifying differences in the expression profile of genes in core needle biopsies taken from the breasts of postmenopausal women. Except for [E2], which was following established guidelines only given to women after hysterectomy, the allocation to each of the three other treatments, estradiol and norethisterone acetate [E2+NETA], tibolone [T] or [no HRT], was randomized using sealed envelopes. The compliance of the volunteers was controlled by measuring the plasma levels of FSH and SHBG both at the onset and at the end of the intake of the respective hormonal preparations. The serum samples were stored frozen at −70°C and the assay of both FSH and SHBG concentrations was carried out in one single run.
As gene expression profiles detected in the core needle biopsy of a breast tumor have been shown to be representative of the entire tumor (Zanetti‐Dällenbach et al., 2006), we made use of such biopsies taken from the upper outer quadrant of the left breast of healthy postmenopausal women before and after 6 months of each treatment modality.
The inclusion criteria were as follows: healthy postmenopausal women at least three months after natural menopause as identified by secondary amenorrhea in the presence of FSH levels above 30IU/L and estradiol levels below 40pmol/L; normal BMI below 32kg/m2, and having normal prolactin levels. All participants were to be Caucasians. The exclusion criteria were intake of HRT, dihydroepiandrosteron or phyto‐estrogens at least four weeks before recruitment, intake of cardiac medication, any excessive abuse of drugs or alcohol, known existing pathology of the breast, claustrophobia, the presence of metallic implants, such as pacemakers, and an endometrial thickness above 5mm.
The participants were recruited through repeated advertisements in local newspapers. At the beginning of the screening phase, all potential participants were interviewed, thereby identifying their respective health status, reproductive history and presence or absence of climacteric symptoms together with an evaluation of all inclusion and exclusion criteria. Women fulfilling all criteria underwent a series of tests, which included gynecological examination including cervical cytology and transvaginal sonography, mammography, sonography of the breast and whole body Dual Energy X‐ray Absorptiometry (DEXA). The concentrations of the following hormones were measured in serum: TSH, free l‐thyroxine (fT4), estradiol, LH, FSH and prolactin. Thereafter, serum was taken together with breast tissue using a core needle (gauge 14, 22cm length) in local anesthesia. Initially, due to pain concerns, only one sample was taken, but as the procedure proved to be painless, up to three samples were taken. After six months, plasma was taken together with a second set of biopsies following the same protocol as described above. All biopsies were performed in the outer upper quadrant of the left breast and both plasma and biopsies were stored at −70°C until analysis. This study was presented to and approved by the Ethics committee of Basel and all participants signed consent. A flow diagram of cases is presented in Figure 1 and the full study protocol has been published at www.unibas.ch (ID 222544).
Figure 1.
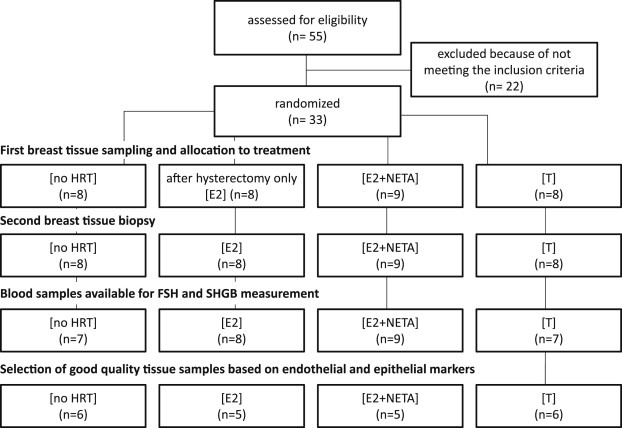
Flow diagram of cases during recruitment and breast tissue sampling.
One hundred and two mRNAs and 46 microRNAs (miRNAs) were selected for this study according to current knowledge of both normal breast physiology and breast cancer biology (Supplementary Table 1, on line only). Aberrant patterns of miRNA expression have been implicated in human disease including ER‐positive and ER‐negative breast cancer (Foekens et al., 2008) and other studies have identified miRNAs regulated by estrogens in human breast cancer cells (Klinge, 2009). The expression of all genes was determined using written standard operating procedures previously established at the Department of Medical Oncology of the Josephine Nefkens Institute and of the Cancer Genomics Centre at the Erasmus University in Rotterdam, The Netherlands, and are described in detail in the Supplementary data section.
3. Results
The recruitment of the volunteers, their randomization and the selection of the tissue samples are summarized in Figure 1. The shortest interval between stop of previous HRT and inclusion into the study was 4 months and 72.7% of all participants reported no previous use of HRT. The volunteers' adherence to the medication was controlled through the expected changes in the serum levels of FSH and SHBG as modified by the respective treatments (Figure 2). The variability of the FSH and SHBG levels in the participants of the [no HRT]‐group was biologically insignificant and corresponds to the natural fluctuations of hormone levels during a six months observation period. The levels of FSH were lower in all individuals of the three groups that indeed underwent hormonal treatment. The serum levels of SHBG dropped significantly in the [T]‐group, whereas they rose in the women treated with [E2+NETA] and [E2]. One individual in the [T]‐group displayed a slight rise in the FSH serum level, whereas another had constant SHBG levels. All changes were as expected and we consider all participants to have been compliant to the treatment.
Figure 2.
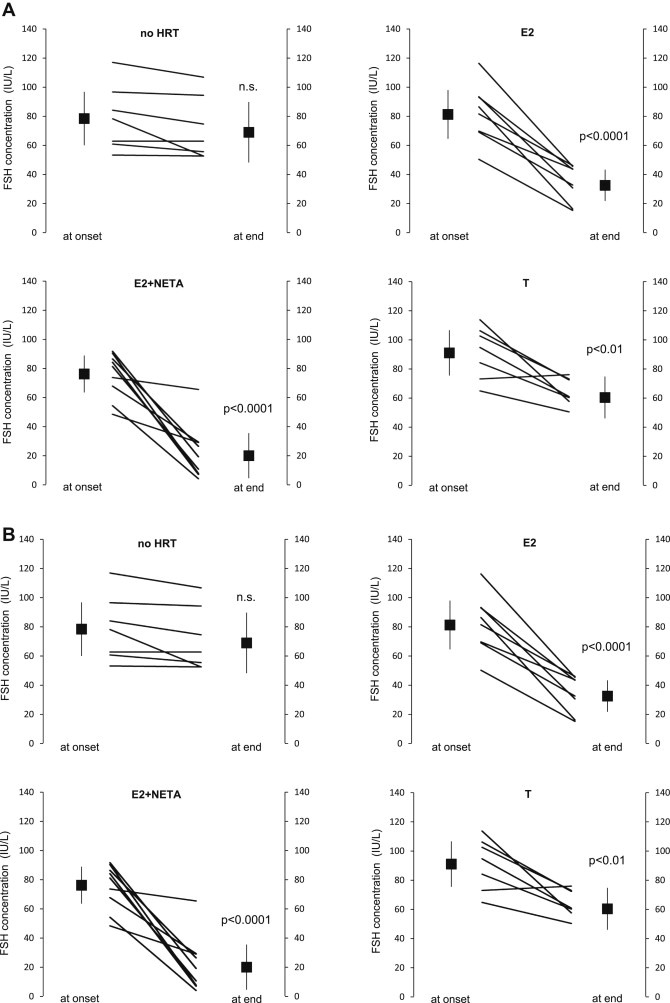
Confirmation of the volunteers' adherence to treatment. A: both the individual and the mean changes in FSH concentration (in IU/L, with the 95% confidence interval) at the onset and at the end of treatment with each of the four treatment modalities are depicted. In one participant the level of FSH rose from 71.4 IU/L to 77.6 IU/L during six month treatment with [T]. This difference was considered to be within the normal variability of the FSH level. B: both the individual and the mean changes in SHBG concentration (in nmol/L, with the 95% confidence interval) at the onset and at the end of treatment with each of the four treatment modalities are depicted. In one individual the level of SHBG failed to drop substantially during the six month intake of [T]. The participant with lack of SHBG change is different from the one with a slight rise in the FSH level (Figure 2A) and both were therefore not considered as being non‐compliant.
Biopsied breast tissue samples were available from both before and after six months treatment of nine women treated with [E2+NETA], eight women treated with [E2], eight women treated with [T] and eight women with [no HRT]. As in most of the 33 recruited women up to three core needle biopsies were taken at each instance, 164 biopsies were available. After RNA isolation, a panel of 3 reference genes (GUSB, HPRT1 and HMBS) was used to evaluate the quality and the quantity of the extracted mRNA. Based on the expression of these genes (<25 Ct, i.e. less than 25 PCR cycles required to pass the fixed fluorescence threshold), we were able to extract good quality (QQ) mRNA from 122 of the 164 individual biopsies (74%) taken from 30 volunteers. Therefore, the RNA of three volunteers was of insufficient quality for this study and these were for that reason excluded from further analysis.
The expression of KRT19, which encodes the epithelial cytokeratin‐19 protein marker, was used to evaluate the relative contribution of epithelial cells in the samples. With the median KRT19 mRNA level after normalization on the reference genes at 13.95 (range: 0.01–54.19), an arbitrary cut‐off was set at 25% of the median (3.49). Thirteen additional samples did not fulfil the criterion of sufficient mRNA KRT19>3.5 (Supplementary Figure 1A, on line only).
In duplicate biopsies, sampled at single time points, we evaluated the concordance of the expression of the selected set of genes and miRNAs. These comparisons demonstrated that only samples were to be included which contained sufficient expression levels of both endothelial and epithelial genes. The number of mismatched samples was thus reduced by excluding samples with low indices of the epithelial marker gene [KRT19] and the endothelial marker genes [CDH5, MCAM, VCAM1 and VWF], with arbitrary cut‐off levels set at 25% of the median value of 13.95 (range: 0.01–54.19, Figure 3A) and 1.29 (range, 0.02–6.13, Supplementary Figure 1B, on line only), respectively. Twenty‐six of the 122 good quality RNA samples did not fulfil the criteria, as they contained insufficient amounts of mRNA of both the epithelial marker KRT19 and the endothelial marker gene set, and were therefore excluded from further analyses. To demonstrate the homogeneity of tissue composition between the four groups after these precautions, one‐way ANOVA was performed on log‐transformed variables followed by Dunnett's post hoc‐test. The three treatment groups were compared with the [no HRT]‐control group. None of the expression levels of 8 genes marking epithelial breast tissue [CEACAM1, KRT5, KRT7, KRT8, KRT18, KRT19, MUC1, EPCAM], none of the expression levels of the 4 genes marking endothelial tissue [CDH5, MCAM, VCAM1 and VWF], none of the 3 genes marking adipose tissue [ADIPOQ, LEP, RETN], nor of the expression levels of 4 genes marking stromal breast tissue [ALCAM, COL1A1, COL1A2, VCAN] were found to be significantly modified by HRT (p>0.1).
Figure 3.
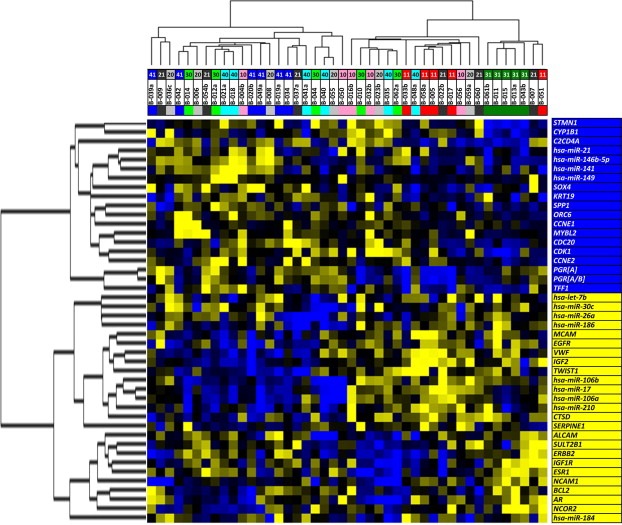
Supervised hierarchical clustering to evaluate similarity among treatment groups. Expression levels of markers expressed differently (p<0.05) among treatment groups. Marker names, shown on the right side of the clustering diagram, were normalized by Spearman rank correlation from −1.0 to 1.0. Blue squares in the cluster diagram indicate a positive relative transcript expression (0–1.0), yellow squares a negative relative expression (−1.0 to 0) and black squares depict a relative expression of zero. Each row depicts a single gene, each column a single case. The dendrogram on top present the relatedness of the profiles of individual cases and the dendrogram on the left the relatedness of the individual genes and miRNAs in this clustering. The longer the dendrogram arm, the greater the difference in between individual cases and genes within a cluster. Cases are color‐ and number‐coded according to the treatment group: (i) gray [20]; [no HRT] before therapy and black [21], [no HRT] after 6 months. (ii) pink [10], [E2] before therapy and red [11], [E2] after 6 months. (iii) light green [30], [E2+NETA] before therapy and dark green [31], [E2+NETA] after 6 months. (iv) light blue [40], [T] before therapy and dark blue [41], [T] after 6 months.
After this series of stringent selections biopsies taken before and after HRT were available from 22 women: 6 women receiving [no HRT], 5 women receiving [E2], 5 women receiving [E2+NETA] and 6 receiving [T]. The demographic characteristics of these subjects are summarized in Table 1. Among all variables, only age at start of therapy (p<0.01) and the age of menarche (p<0.05) were significantly higher in the [E2]‐group compared with the [no HRT]‐group.
Table 1.
Clinical characteristics of the 22 qualified cases at the onset of treatment.
| Group | no HRT | E2+NETA | E2 | Tibolone | p b |
|---|---|---|---|---|---|
| No. of participants | 6 | 5 | 5 | 6 | |
| Age (y)a | 52.5 (51.4–53.6) | 55.4 (51.0–59.8) | 58.2 (55.8–60.6) | 56.0 (52.5.3–60.0) | p <0.01 |
| Age at menarche (y) | 12.7 (11.4–13.9) | 14.0 (13.1–14.9) | 14.4 (13.0–15.8) | 13.5 (12.9–14.1) | p <0.05 |
| Family historyc with breast cancer (no.) | 1 | 1 | 1 | 1 | n.s. |
| TSH (mIU/L) | 2.8 (1.8–3.8) | 2.2 (0.5–3.9) | 2.7 (1.8–3.5) | 2.5 (1.1–4.0) | n.s. |
| fT4 (ng/dL) | 1.2 (1.1–1.4) | 1.1 (1.0–1.2) | 1.2 (0.9–1.4) | 1.2 (1.0–1.3) | n.s. |
| Estradiol (pg/mL) | 8.6 (3.7–13.6) | 8.3 (2.0–14.5) | 9.9 (4.9–14.8) | 8.3 (2.8–13.8) | n.s. |
| FSH (IU/L) | 96.6 (72.1–121.0) | 79.9 (53.4–106.5) | 93.6 (51.6–136.0) | 99.0 (77.6–120.4) | n.s. |
| LH (IU/L) | 42.4 (31.4–53.4) | 39.7 (29.7–49.7) | 44.8 (26.1–63.5) | 39.8 (32.8–46.8) | n.s. |
| Prolactin (mIU/L) | 215 (120–310) | 208 (127–289) | 283 (218–348) | 258 (156–360) | n.s. |
| BMI (kg/m2) | 23.6 (20.1–27.1) | 25.7 (21.9–30.0) | 25,5 (20.4–30.6) | 22.1 (20.7–23.5) | n.s. |
| Lean mass (kg) | 36.7 (29.1–44,3) | 40.8 (36.0–45.5) | 41.4 (38.3–44.6) | 39.0 (34.6–43.4) | n.s. |
| Fat mass (kg) | 22.1 (12.4–31.9) | 24.6 (14.9–3.3) | 28.1 (16.9–39.3) | 18.5 (14.6–22.4) | n.s. |
| Mammographic ACR‐densityd | 2.2 (0.9–3.4) | 1.8 (0.8–2.8) | 2.2 (1.2–3.2) | 2.5 (1.6–3.4) | n.s. |
| Sonographic ACR‐densityd | 1.8 (1.0–2.6) | 1.6 (0.5–2.7) | 1.8 (0.8–2.8) | 2.5 (1.9–3.1) | n.s. |
Median values together with their 95% confidence intervals.
Mann Whitney U unpaired test, value versus [no HRT].
Only close family members were considered (mother or sister). One participant in the [no HRT] group was unable to provide any information about the health status of members of her family.
ACR denominates American College of Radiology.
In order to identify the effects of the four treatments on the expression levels of the selected genes, the data of the 22 biopsies taken at baseline were compared with those after treatment. Forty‐four (31 mRNAs and 13 miRNAs) of the 148 markers showed a significant differential expression in these unpaired Wilcoxon's tests (p<0.05), Table 2. Both [E2+NETA] and [E2] resulted in a significantly higher number of genes differentially expressed before and after treatment (16.2% and 10.1% for [E2+NETA] and [E2], respectively), whereas [T] was associated with the lowest number of differentially expressed markers (4.7%).
Table 2.
Identification of markers differentially expressed among the four treatment groups.
| Markers | Unpaired Wilcoxon test | |||
|---|---|---|---|---|
| no HRT | E2 | E2+NETA | T | |
| n =6 | n =5 | n =5 | n =6 | |
| 22 vs 6 | 22 vs 5 | 22 vs 5 | 22 vs 6 | |
| P (2‐tail) | P (2‐tail) | P (2‐tail) | P (2‐tail) | |
| ALCAM | 5.7E−02 | 4.7E−01 | 3.1E−02 | 2.7E−01 |
| AR | 6.7E−01 | 4.4E−01 | 1.9E−02 | 5.4E−01 |
| BCL2 | 8.4E−01 | 3.3E−01 | 1.6E−02 | 3.9E−01 |
| CCNE1 | 4.7E−02 | 4.2E−02 | 4.0E−01 | 7.6E−01 |
| CCNE2 | 7.6E−01 | 6.0E−01 | 1.9E−02 | 9.8E−01 |
| CDC2 | 4.2E−01 | 4.0E−01 | 1.6E−02 | 6.3E−01 |
| CDC20 | 4.2E−01 | 4.4E−01 | 1.4E−02 | 3.9E−01 |
| CTSD | 2.1E−01 | 5.7E−01 | 3.1E−02 | 2.5E−01 |
| CYP1B1 | 6.3E−01 | 5.1E−01 | 1.6E−02 | 8.9E−01 |
| EGFR | 4.8E−01 | 6.8E−01 | 1.6E−02 | 3.6E−02 |
| ERBB2 | 6.1E−01 | 9.8E−01 | 1.9E−02 | 2.5E−01 |
| ESR1 | 8.0E−01 | 7.8E−01 | 1.9E−02 | 2.3E−01 |
| IGF1R | 8.0E−01 | 9.8E−01 | 1.6E−02 | 5.8E−01 |
| IGF2 | 7.2E−01 | 5.5E−03 | 3.3E−01 | 9.8E−01 |
| KRT19 | 8.0E−01 | 1.6E−02 | 8.5E−01 | 6.5E−01 |
| MCAM | 8.9E−01 | 6.4E−01 | 3.1E−02 | 6.3E−01 |
| MYBL2 | 5.2E−01 | 5.5E−01 | 2.7E−02 | 6.7E−01 |
| NCAM1 | 3.9E−01 | 1.6E−02 | 1.6E−03 | 2.6E−01 |
| NCOR2 | 8.4E−01 | 5.7E−01 | 2.7E−02 | 9.3E−01 |
| NLF1 | 8.4E−01 | 1.8E−01 | 3.0E−03 | 8.9E−01 |
| ORC6L | 8.6E−04 | 2.0E−03 | 1.6E−02 | 4.0E−01 |
| PGRA/B | 5.6E−01 | 1.6E−02 | 8.8E−01 | 7.6E−01 |
| PGRA | 5.4E−01 | 2.7E−02 | 4.7E−01 | 5.6E−01 |
| SERPINE1 | 3.1E−02 | 9.3E−01 | 7.8E−01 | 6.7E−01 |
| SOX4 | 5.6E−01 | 8.6E−02 | 6.4E−01 | 6.1E−03 |
| SPP1 | 2.3E−02 | 9.8E−02 | 9.8E−02 | 7.2E−01 |
| STMN1 | 8.8E−02 | 2.3E−02 | 1.0E−03 | 7.6E−01 |
| SULT2B1 | 2.3E−02 | 6.0E−01 | 2.5E−01 | 1.2E−01 |
| TFF1 | 3.7E−01 | 2.5E−03 | 8.4E−04 | 4.4E−02 |
| TWIST1 | 5.6E−01 | 7.8E−01 | 1.9E−01 | 9.2E−03 |
| VWF | 5.6E−01 | 4.9E−02 | 6.6E−02 | 4.8E−01 |
| hsa‐let‐7b | 9.3E−01 | 9.8E−01 | 1.1E−01 | 2.3E−02 |
| hsa‐miR‐106a | 7.2E−01 | 4.9E−02 | 4.0E−01 | 9.8E−01 |
| hsa‐miR‐106b | 6.3E−01 | 8.3E−01 | 4.9E−02 | 7.2E−01 |
| hsa‐miR‐141 | 5.6E−01 | 3.7E−02 | 7.3E−01 | 6.7E−01 |
| hsa‐miR‐146b‐5p | 2.7E−01 | 1.1E−01 | 3.7E−02 | 1.2E−01 |
| hsa‐miR‐149 | 8.7E−01 | 8.0E−03 | 5.7E−02 | 9.8E−01 |
| hsa‐miR‐17 | 5.2E−01 | 3.7E−02 | 4.7E−01 | 4.2E−01 |
| hsa‐miR‐184 | 6.3E−01 | 2.5E−01 | 9.8E−02 | 2.0E−02 |
| hsa‐miR‐186 | 2.7E−01 | 6.8E−01 | 4.9E−02 | 4.5E−01 |
| hsa‐miR‐21 | 4.1E−02 | 3.7E−01 | 1.8E−01 | 9.3E−01 |
| hsa‐miR‐210 | 8.0E−01 | 4.2E−02 | 9.3E−01 | 5.6E−01 |
| hsa‐miR‐26a | 7.6E−01 | 8.3E−01 | 3.1E−02 | 4.2E−01 |
| hsa‐miR‐30c | 6.7E−01 | 7.3E−01 | 5.7E−02 | 1.3E−02 |
| p <0.05 | 4.1% | 10.1% | 16.2% | 4.7% |
P‐values of markers in the 22 biopsies sampled before treatment compared with the data of biopsies after treatment (Wilcoxon's test). Only the data of genes and miRNAs with p<0.05 in any of the groups before and after treatment are depicted here.
Supervised hierarchical clustering analysis by means of Spearman rank correlation was performed using these 44 genes (p<0.5). Figure 3 shows how this resulted in the identification of two main clusters of cases and 2 main clusters of genes. The cluster on the right side harbors all cases from donors after 6 months in the [E2+NETA]‐group (depicted by green squares) and all cases after 6 months of [E2]‐group (depicted by red squares), with the [E2+NETA]‐group clustering closely together indicating a high level of homology. The individuals receiving [T] (depicted with dark blue squares) are positioned at random together with the controls receiving [no HRT] (depicted by black squares), suggesting that none of the genes examined were much affected by [T].
To authenticate the possible clinical relevance of these markers at the individual level, we corrected our data for multiple comparisons using a false discovery rate of 10% (Benjamini and Hochberg, 1995). After this correction, the expression levels of NCAM1 remained significantly lower and the expression levels of TFF1, STMN1 and C2CD4A [FAM148A, NLF1] significantly higher in the [E2+NETA]‐group after treatment (P FDR adjusted<0.05) (Figure 4).
Figure 4.

Box‐Whisker plots of gene markers significantly differentially expressed after HRT. mRNA expression data of TFF1, STMN1, C2CD4A and NCAM1 in 22 biopsies taken before start of therapy [no HRT] were compared with those of the biopsies trichotomized according the various treatment groups. The box‐plot shows the five statistics (lower whisker is 5% minimum, lower box part is the 25th percentile, solid line in box presents the median, upper box part is 75th percentile, and upper whisker is 95% maximum). Significance levels (*) are relative to the expression levels measured in the control group [no HRT]. PFDR adjusted, 10%<0.05*.
Estrogens may be involved in the development of breast cancer through formation of carcinogenic steroid metabolites and/or induction of proliferation through modulation of nuclear receptor activity (Zhu and Conney, 1998; Rogan et al., 2003; Cavalieri et al., 2006). Among the nuclear receptors AR, ESR1, ESR2 and PGRA and PGRA/B, only [E2+NETA] resulted in a significant decrease in the expression levels of AR (p=0.02) and ESR1 (p=0.01, Student's t‐test on log‐transformed paired variables). Although the expression levels of PGRA were not significantly affected, [E2] significantly up‐regulated those of PGRA/B (p=0.004). Figure 5 shows the individual changes in mRNA levels of these nuclear receptors with respect to the hormonal treatments and demonstrates how an individual reaction to treatment can be detected through breast biopsy and gene expression analysis, e.g. for the prediction of increased breast cancer risk. In addition, an increased cancer risk has been associated with a disruption in the balance in the expression of estrogen‐metabolizing enzymes such as sulfotransferase (Miyoshi et al., 2003), (SULF), steroid sulfatase (Sasano et al., 2009) (SULT) and hydroxysteroid dehydrogenase (Nagasaki et al., 2009). We therefore assessed alterations in expression levels of genes encoding enzymes involved in estrogen metabolism [http://microrna.sanger.ac.uk]. Whereas CYP19A1, HSD17B1, HSD17B2 and SULF1 in combination with hsa‐miR‐150, ‐186, ‐193a‐5p, ‐20a, ‐21, ‐214 contribute to the estrogen‐activating pathways, COMT, CRYZ, CYP1A1, CYP1B1, CYP3A, GSTA1, SULT1E1, SULT2A1 and SULT2B1 in combination with hsa‐miR‐106b, ‐145, ‐26a contribute to the estrogen‐inactivating pathways (Opatrny et al., 2008). Although significant changes were observed in individual genes such as CYP1B1 and hsa‐miR‐21, ‐106b and ‐26a (Table 2), the expression levels of none of these gene panels as a whole were significantly altered after treatment with any of the various treatments (Student's t‐test on log‐transformed paired variables for both panels, p<0.1). Whole genome expression profiling may provide more insight into which pathways are affected by specific types of HRT in particular.
Figure 5.
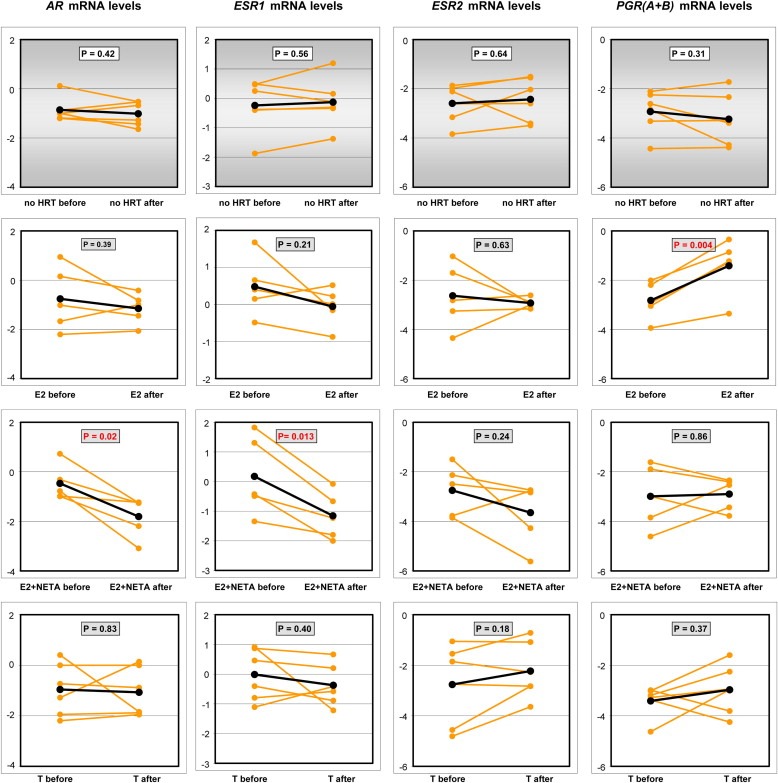
Individual reactions in the change in mRNA levels of nuclear receptors in relation to the hormonal treatment. mRNA expression levels of the nuclear receptors AR, ESR1, ESR2 and PGR in 22 biopsies taken before start of therapy were compared with those measured in biopsies of the same individuals after treatment. T‐test statistics (2‐sided p) are indicated in the graphs.
4. Discussion
Both the early origin of breast cancer and its development into a manifest tumor indicate that these processes depend on a particular endocrine environment. At present, this environment is assessed through personal history, hormonal analyses in the blood and mammographic density. Direct assessment of regulatory pathways at work in the breast and predictive of breast cancer could be more helpful and the analysis of gene expression patterns in women with higher risk of breast cancer may provide the basis for such an approach. Much is known about the gene expression patterns in existing breast cancer tumors. However, no extensive gene expression profiles have yet been published from normal breast tissue of otherwise healthy postmenopausal women with distinctive risks of breast cancer due to the use of HRT.
In this prospective experimental study, we used three commonly used hormonal treatments to establish different endocrine environments, which have previously been shown to have a differential effect on the risk of breast cancer incidence. The relationship between estrogens (both endogenous estrogen levels and estrogen levels after exogenous supply of estrogens) and the risk of postmenopausal breast cancer is well known from epidemiological and interventional studies, but the combined action of estrogens and progestagens has been the most significant clinically (Writing Group for the Women's Health Initiative Investigators, 2002). Although those studies were carried out with conjugated equine estrogens, micronized 17β‐estradiol has been shown to act similarly on breast cancer cell lines (Mueck et al., 2003). In contrast, [T] has been shown not to affect mammographic density and to have the least marked effect on breast cancer metabolism (Lundström et al., 2002; Valdivia et al., 2004). This prospective study was initiated to examine mRNA and miRNA signatures before and after the administration of three different hormonal preparations (Castellano et al., 2009). During this trial none of the participants was diagnosed with breast cancer as evidenced with both mammography and sonography and we found no difference in the incidence of breast cancer among close family members among the four groups. Therefore, we have no indication any of our findings can be explained by the presence of pre‐existent malignant cells in the biopsies.
The present data revealed that, also after correcting for multiple comparisons, [E2+NETA] led to a significantly higher number of disrupted genes as compared to [E2], whereas [T] hardly affected the expression levels. Estrogens are known to increase breast cancer risk through the formation of certain carcinogenic metabolites (Zhu and Conney, 1998; Rogan et al., 2003; Cavalieri et al., 2006). This is also exemplified by the lower incidence risk of breast cancer in women treated with compounds exerting anti‐estrogenic effects such as tamoxifen (Cuzick et al., 2007) and raloxifene (Grady et al., 2008).
In agreement with its known effect on mammographic density, treatment with [E2+NETA] increased the expression of a number of cyclin‐dependent kinases, such as CDK1, CDC20 and CCNE2 (Figure 6C), all strongly involved in cell division and tissue proliferation, and of some stromal cell proliferation markers, such as ALCAM and STMN1 (Table 2), but not of COL1A1, COL1A2 and VCAN. During gestation, both estrogens and progestagens were recently demonstrated to specifically stimulate the expansion of the epithelial compartment of the murine breast through the expansion and differentiation of adult stem cells located there (Asselin‐Labat et al., 2010). This process is mediated through the activation of the nuclear factor‐κB ligand (RANK‐L) (Asselin‐Labat et al., 2010; Joshi et al., 2010; Oakes et al., 2006). Both EGFR and HER2 (encoded by ERBB2 mRNA) are associated with stem cell renewal and activate RANK‐L, but their mRNA expression levels were significantly down‐regulated by treatment with [E2+NETA]. Those findings were described during treatment with another progestagen, medroxyprogesterone acetate, than the one used here. However, this apparently contradictory finding may also be explained by the relative decline of the amount of stem cells in the breast as compared to the rapidly proliferating number of differentiated epithelial cells during prolonged treatment with [E2+NETA]. Of note in this context is that levels of the stem cell marker NCAM1, one of the genes that remained statistically significant after correction for multiple comparisons (Figure 4), were also significantly decreased by treatment with [E2+NETA].
Figure 6.
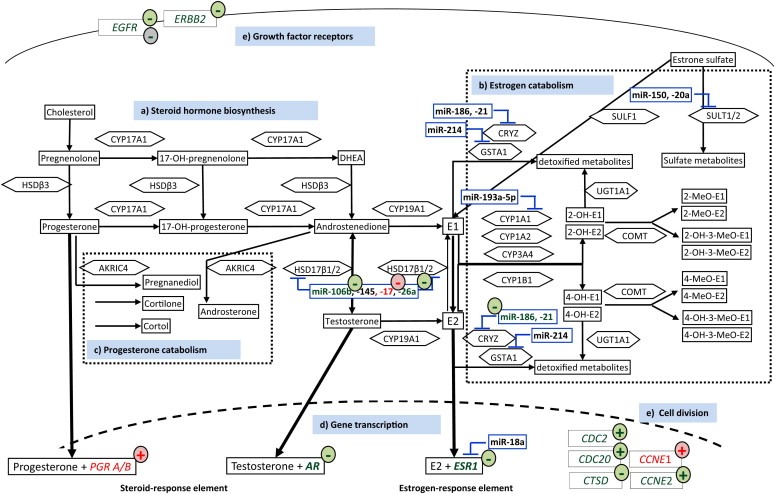
Pathways of steroid hormone synthesis, metabolism and tissue sensitivity to HRT (Kelemen et al., 2008; Pasqualini and Chetrite, 2005; Masson et al., 2010) MiRNA's predicted to target steroidogenic or metabolizing enzymes are encased in blue. Genes measured as either up‐ or down‐regulated in our current study after a specific hormone replacement therapy are marked by colored circles. The effects exerted by [E2+NETA] are depicted in green, the effects exerted by [E2] in red, and the effects exerted by [T] in gray. a) Steroid hormone biosynthesis: Cytochrome P450 17A1 (CYP17A1) catalyzes the conversion of pregnenolone and progesterone to the hormones dehydroepidandrosterone (DHEA) and androstenedione, respectively, which are further metabolized to estrone (E1) and 17β‐estradiol (E2) by CYP19A1 (aromatase). Androgen conversion to estrogen in adipose tissue by CYP19A1 is an important source of bioactive endogenous estrogens among postmenopausal women. Hydroxysteroid dehydrogenase 3ß1 (HSD3ß1) catalyzes the interconversion of pregnenolone and progesterone and of DHEA and androstenedione, whereas HSD17β1 catalyzes the conversion between androstenedione or testosterone and E1 or E2, respectively. The availability of HSD17β1 is regulated by a number of miRNAs, which are inhibited both by [E2+NETA] and [E2]. b) Estrogen metabolism: The ‘sulfatase pathway’ converts stored estrogen sulfates into the bioactive unconjugated E1 and sulfotransferases convert estrogens into the biologically inactive estrogen sulfates. The CYP‐family of enzymes consists of a cluster of enzymes that function in the oxidative metabolic activation and deactivation of compounds including several steroid hormones. E1 and E2 undergo 2‐hydroxylation by the CYP1A1, CYP1A2 and CYP3A4 enzymes and 4‐hydroxylation by CYP1B1. Catecholestrogens are deactivated by catechol‐O‐methyltransferase (COMT). UDP‐glucuronosyltransferase 1A1 (UGT1A1), crystallin zeta, quinone reductase (CRYZ), glutathione S‐transferase alpha 1 (GSTA1) and sulfotransferases (SULT1/2) are detoxifying enzymes that convert endogenous substrates to inactive metabolites. CRYZ is inhibited by miRNA‐186 and ‐21, which are negatively regulated by [E2+NETA], potentially resulting in an accumulation of catechol estrogen quinines, which are potential initiators of breast cancer. c) Progesterone metabolism: Aldo–ketoreductase family 1 member C4 (AKR1C4) catalyzes the conversion of progesterone and androstenedione to their corresponding alcohols. d) Gene transcription: Progesterone, testosterone and E2 bind to their respective nuclear receptor proteins, progesterone receptor (PGR), androgen receptor (AR) and estrogen receptor alpha (ESR1), and activate genes with corresponding responsive elements resulting in gene expression of genes with such responsive elements. The expression of PGRA/B is up‐regulated by [E2], whereas those of ESR1 and of AR are down‐regulated by [E2+NETA]. e) Growth factor receptors and cell division: Two genes encoding membrane bound receptors, EGFR (alternatively symbolized as HER1) and ERBB2 (HER2), both considered being breast cancer oncogenes, are down‐regulated by [E2+NETA], whereas a number of kinases and cell‐division cycle genes, such as CDK1, CDC20 and CCNE2, are up‐regulated by [E2+NETA] or, in the case of CCNE1 by [E2]. In contrast, CTSD, which encodes cathepsin D and which is often used as a breast cancer tumor marker (Cavalieri et al., 2006), is down‐regulated by [E2+NETA]. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
To further examine the impact of HRT on the expression levels of groups of genes and miRNAs, we performed an unsupervised hierarchical clustering to identify gene clusters based on their relative expression in 44 paired samples taken from 22 individuals before and after treatment. Only ANG and SULT2A1 were excluded from this analysis because expression levels of these genes were below the detection limit. Unsupervised two‐dimensional centroid linkage clustering resulted in the identification of 10 groups of markers (mRNAs and miRNAs) with a marker node correlation >0.2 (Supplementary Figure 2, on line only). The observation that hsa‐miR‐10b, ‐145, and ‐342 clustered together with CYP19A1, CYP3A4, CYP1A1 and SULT1E1 and that hsa‐miR‐184 and ‐34b clustered together with HSD17B2 and GSTA1 supports a functional relationship of miRNAs in estrogen metabolism.
Various genes, in particular miRNAs, seem to increase the availability of estradiol and some of its potential oncogenic metabolites through activation of steroidogenic enzymes. For example, the decreased expression of hsa‐miR‐17, ‐26a and ‐106b in the [E2+NETA]‐group (Table 2 and Figure 6B) is expected to up‐regulate their predicted target, 17β‐hydroxysteroid dehydrogenase type 1 (HSD17B1), which in turn catalyzes the conversion of estrone into the biologically more potent estradiol. Indeed, high levels of HSD17B1 have been associated with worse outcome of postmenopausal breast cancer (Gunnarsson et al., 2008). The resulting high levels of estradiol probably explain the significantly increased levels of the estrogen‐inducible TFF1 and STMN1 genes, which we measured in particular breast tissues of women after six month treatment with [E2+NETA] (Figure 4). Among the genes that were significantly down‐regulated by [E2+NETA], ranked estrogen‐receptor‐1 (ESR1, p=0.019) and androgen‐receptor (AR, p=0.019), whereas CYP1B1, a gene encoding an estrogen‐metabolizing enzyme, was significantly up‐regulated (p=0.016).
Our data suggest that normal mammary cells triggered by [E2+NETA] adjust for steroidogenic up‐regulation through down‐regulation of the estrogen‐receptor pathway and are in line with conventional knowledge collected from experimental data with cell lines or with primary cells cultured in vitro.
Our study demonstrates that changes in gene expression patterns that are possibly associated with an increased risk of breast cancer can be observed in healthy breast tissue after a six month administration of both estrogens and progestagens, and to a lesser extent of estrogens alone. This prospective study is the first to demonstrate that a breast cancer‐related gene expression signature can be determined reliably in small healthy breast tissue samples, collected for experimental purposes, and that the expression profile of genes putatively involved in breast cancer correlates with the known effects of different hormonal treatment modalities on mammographic density and breast cancer incidence.
5. Conclusions
The tissue sampling was well tolerated as evidenced by the complete adherence of all participants to the study protocol, which also included a second biopsy after six months. Although only 66.7% (22 out of 33 biopsies) of all breast tissue samples could be used for gene expression analysis, the results of this prospective study clearly demonstrate the feasibility of collecting a sufficient quantity of breast tissue for gene expression profiling from a majority of the healthy volunteers. With our starting hypothesis confirmed, whole genome expression analysis will now be performed in order to enlarge the panel of candidate genes, which may potentially be evaluated for the prediction of a future breast cancer risk. These genes or gene products should be applicable not only for women under HRT, but also for women presenting with high circulating levels of endogenously produced sex steroids. As in this trial plasma was stored frozen both before and at the end of each hormonal treatment, the presence or absence of candidate gene products potentially identifying a higher breast cancer risk may also be determined in peripheral circulation.
We anticipate that the present outline provides a model for further development of objective and quantifiable parameters for the early prediction of increased breast cancer risk.
Conflict of interest
All authors declare no conflict of interest. Helenius J. Kloosterboer was an employee of Organon until August 2006 and has been consultant for companies marketing tibolone after 2006.
Authors' contributions
| Anieta M. Sieuwerts | Study conception, data collection, statistical analyses, results interpretation, manuscript preparation |
| a.sieuwerts@erasmusmc.nl | |
| Giuseppina De Napoli | Data collection and results interpretation |
| DeNapoliG@uhbs.ch | |
| Anne van Galen | Data collection and results interpretation |
| a.vangalen@erasmusmc.nl | |
| Helenius J. Kloosterboer | Study conception, results interpretation |
| helenius@kpnmail.nl | |
| Vanja de Weerd | Technical assistance |
| v.deweerd@erasmusmc.nl | |
| Hong Zhang | Study conception, results interpretation, manuscript preparation |
| hzhang@uhbs.ch | |
| John W.M. Martens | Study conception, results interpretation |
| j.martens@eramusmc.nl | |
| John A. Foekens | Study conception, results interpretation |
| j.foekens@erasmusmc.nl | |
| Christian De Geyter | Study initiation and conception, data collection, results interpretation, manuscript preparation |
| cdegeyter@uhbs.ch |
List of abbreviations
- ACR
American College of Radiology
- AR
androgen receptor
- BMI
body mass index
- DEXA
dual energy X‐ray absorptiometry
- E2
estradiol
- ER
estrogen receptor
- fT4
free l‐thyroxine
- HRT
hormonal replacement therapy
- miRNA
micro RNA
- NETA
norethisterone acetate
- PGR
progesterone receptor
- RANK-L
nuclear factor‐κB ligand
- T
tibolone
Supporting information
Supplementary Fig. 1 Distribution of KRT19 (A) and Endothelial marker set (B) mRNA levels. Evaluation of the distribution of KRT19 and the endothelial marker set mRNA expression is shown for the remaining 122 RNA samples with adequate expression of the reference genes (QQ RNA) used to normalize the data as described in the main body text. Firstly, the median mRNA level of KRT19 was established, after which samples with an expression level less than an arbitrarily cut‐off set at 25% of this median KRT19 level were excluded from further analyses. (A). Next, the median mRNA level of the endothelial marker set‐consisting of CDH5, MCAM, VCAM1 and VWF was established, after which also samples with an expression level less than an arbitrarily cut‐off set at 25% of this median endothelial marker set level were excluded from the final analysis (B).
Supplementary Fig. 2 Unsupervised hierarchical clustering to identify homology in‐between markers. Expression levels of the markers shown on the right site of the figure left were log‐transformed and mean normalized from −1.0 to 1.0. Red squares in the cluster diagram indicate a positive relative transcript expression (0–1.0), green squares a negative relative expression (−1.0 to 0) and black squares depict a relative expression of zero. Each row depicts a single marker (gene or miRNA), each column a single case. The dendrogram on top present the relatedness of the profiles of individual cases and the dendrogram on the left the relatedness of the markers in this clustering. The longer the dendrogram arm, the greater the difference in between individual cases and genes inside a cluster group. Cases are color‐ and number‐coded according therapy group: gray [20]; [no HRT] before therapy; black [21], no HRT after 6 months; pink [10], [E2] before and red [11], [E2] after therapy; light green [30], [E2+NETA] before and dark green [31], E2+NETA after therapy; light blue [40], T before and dark blue [41], [T] after therapy. The 10 clusters of markers with a node correlation >0.2 are visualized by the different colors on the right side of the cluster. To improve readability, the thus identified gene clusters and dendrogram depicting the relatedness of the profiles of individual cases are blown up in the right panel of the figure.
Supplementary data
Supplementary data
Supplementary data
Acknowledgments
The support of Dr. Martin Hund, PhD, and Dr. Isabel Gruber, PhD, is gratefully acknowledged. We are also thankful to the participating postmenopausal women. This work was supported by Organon/Schering‐Plough/Essex Chemie, by the Netherlands Genomic Initiative (NGI)/Netherlands Organisation for Scientific Research (NWO) and by the Repronatal Foundation in Basel, Switzerland. The authors had full responsibility for the design of the study, the collection of the data, the analysis and interpretation of the data, the decision to submit the manuscript for publication and the writing of the manuscript.
Supplementary material 1.
Supplementary data associated with this article can be found, in the on line version, at doi:10.1016/j.molonc.2011.09.003.
Sieuwerts Anieta M., De Napoli Giuseppina, van Galen Anne, Kloosterboer Helenius J., de Weerd Vanja, Zhang Hong, Martens John W.M., Foekens John A. and De Geyter Christian, (2011), Hormone replacement therapy dependent changes in breast cancer‐related gene expression in breast tissue of healthy postmenopausal women, Molecular Oncology, 5, doi: 10.1016/j.molonc.2011.09.003.
None of the authors of this paper have anything to disclose with regard to the content of the manuscript. Helenius Kloosterboer was previously a collaborator of Organon NV, but has meanwhile retired.
Grant Support: This study was financially supported by Organon/Schering‐Plough, by the Netherlands Genomic Initiative (NGI)/Netherlands Organisation for Scientific Research (NWO) and by the Repronatal Foundation.
References
- Asselin-Labat, M.L. , Vaillant, F. , Sheridan, J.M. , Pal, B. , Wu, D. , Simpson, E.R. , Yasuda, H. , Smyth, G.K. , Martin, T.J. , Lindeman, G.F. , Visvader, J.E. , 2010. Control of mammary stem cell function by steroid hormone signalling. Nature. 465, 798–802. [DOI] [PubMed] [Google Scholar]
- Banks, E. , Reeves, G. , Beral, V. , Bull, D. , Crossley, B. , Simmonds, M. , Hilton, E. , Bailey, S. , Barrett, N. , Briers, P. , 2006. Hormone replacement therapy and false positive recall in the Million Women Study: patterns of use, hormonal constituents and consistency of effect. Breast Cancer Res.. 8, R8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini, Y. , Hochberg, Y. , 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Roy. Stat. Soc., Ser. B. 57, 289–300. [Google Scholar]
- Beral, V. , Million Women Study Collaborators, 2003. Breast cancer and hormone-replacement therapy in the Million Women Study. Lancet. 362, 419–427. [DOI] [PubMed] [Google Scholar]
- Byrne, C. , Schairer, C. , Wolfe, J. , Parekh, N. , Salane, M. , Brinton, L.A. , Hoover, R. , Haile, R. , 1995. Mammographic features and breast cancer risk: effects with time, age, and menopause status. J. Natl. Cancer Inst.. 87, 1622–1629. [DOI] [PubMed] [Google Scholar]
- Castellano, L. , Giamas, G. , Jacob, J. , Coombes, R.C. , Lucchesi, W. , Thiruchelvam, P. , Barton, G. , Jiao, L.R. , Wait, R. , Waxman, J. , Hannon, G.J. , Stebbing, J. , 2009. The estrogen receptor-alpha-induced microRNA signature regulates itself and its transcriptional response. Proc. Natl. Acad. Sci. U. S. A.. 106, 15732–15737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalieri, E. , Chakravarti, D. , Guttenplan, J. , Hart, E. , Ingle, J. , Jankowiak, R. , Muti, P. , Rogan, E. , Russo, J. , Santen, R. , Sutter, T. , 2006. Catechol estrogen quinones as initiators of breast and other human cancers: implications for biomarkers of susceptibility and cancer prevention. Biochim. Biophys. Acta. 1766, 63–78. [DOI] [PubMed] [Google Scholar]
- Collaborative Group on Hormonal Factors in Breast Cancer, 1997. Breast cancer and hormone replacement therapy: collaborative reanalysis of data from 51 epidemiological studies of 52705 women with breast cancer and 108411 women without breast cancer. Lancet. 350, 1047–1059. [PubMed] [Google Scholar]
- Cummings, S.R. , Ettinger, B. , Delmas, P.D. , Kenemans, P. , Stathopoulos, V. , Verweij, P. , Mol-Arts, M. , Kloosterboer, L. , Mosca, L. , Christiansen, C. , Bilezikian, J. , Kerzberg, E.M. , Johnson, S. , Zanchetta, J. , Grobbee, D.E. , Seifertm, W. , Eastell, R. , LIFT Trial Investigators, 2008. The effects of tibolone in older postmenopausal women. N. Engl. J. Med.. 359, 697–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuzick, J. , Warwick, J. , Pinney, E. , Warren, R.M. , Duffy, S.W. , 2004. Tamoxifen and breast density in women at increased risk of breast cancer. J. Natl. Cancer Inst.. 96, 621–628. [DOI] [PubMed] [Google Scholar]
- Cuzick, J. , Forbes, J.F. , Sestak, I. , Cawthorn, S. , Hamed, H. , Holli, K. , Howell, A. , International Breast Cancer Intervention Study I Investigators, 2007. Long-term results of tamoxifen prophylaxis for breast cancer – 96-month follow-up of the randomized IBIS-I trial. J. Natl. Cancer Inst.. 99, 272–282. [DOI] [PubMed] [Google Scholar]
- Foekens, J.A. , Sieuwerts, A.M. , Smid, M. , Look, M.P. , de Weerd, V. , Boersma, A.W. , Klijn, J.G. , Wiemer, E.A. , Martens, J.W. , 2008. Four miRNAs associated with aggressiveness of lymph node-negative, estrogen receptor-positive human breast cancer. Proc. Natl. Acad. Sci. U. S. A.. 105, 13021–13026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gail, M.H. , Brinton, L.A. , Byar, D.P. , Corle, D.K. , Green, S.B. , Schairer, C. , Mulvihill, J.J. , 1989. Projecting individualized probabilities of developing breast cancer for white females who are being examined annually. J. Natl. Cancer Inst.. 81, 1879–1886. [DOI] [PubMed] [Google Scholar]
- Grady, D. , Cauley, J.A. , Geiger, M.J. , Kornitzer, M. , Mosca, L. , Collins, P. , Wenger, N.K. , Song, J. , Mershon, J. , Barrett-Connor, E. , Raloxifene Use for The Heart Trial Investigators, 2008. Reduced incidence of invasive breast cancer with raloxifene among women at increased coronary risk. J. Natl. Cancer Inst.. 100, 854–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greendalem, G.A. , Reboussin, B.A. , Slone, S. , Wasilauskas, C. , Pike, M.C. , Ursin, G. , 2003. Postmenopausal hormone therapy and change in mammographic density. J. Natl. Cancer Inst.. 95, 30–37. [DOI] [PubMed] [Google Scholar]
- Gunnarsson, C. , Jerevall, P.L. , Hammar, K. , Olsson, B. , Nordenskjöld, B. , Jansson, A. , Stål, O. , 2008. Amplification of HSD17B1 has prognostic significance in postmenopausal breast cancer. Breast Cancer Res. Treat.. 108, 35–41. [DOI] [PubMed] [Google Scholar]
- Joshi, P.A. , Jackson, H.W. , Beristain, A.G. , Di Grappa, M.A. , Mote, P.A. , Clarke, C.L. , Stingl, J. , Waterhouse, P.D. , Khokha, R. , 2010. Progesterone induces adult mammary stem cell expansion. Nature. 465, 803–807. [DOI] [PubMed] [Google Scholar]
- Kelemen, L.E. , Sellers, T.A. , Vachon, C.M. , 2008. Can genes for mammographic density inform cancer aetiology?. Nat. Rev. Cancer. 8, 812–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenemans, P. , Bundred, N.J. , Foidart, J.M. , Kubista, E. , von Schoultz, B. , Sismondi, P. , Vassilopoulou-Sellin, R. , Yip, C.H. , Egberts, J. , Mol-Arts, M. , Mulder, R. , van Os, S. , Beckmann, M.W. , LIBERATE Study Group, 2009. Safety and efficacy of tibolone in breast-cancer patients with vasomotor symptoms: a double-blind, randomised, non-inferiority trial. Lancet Oncol.. 10, 135–146. [DOI] [PubMed] [Google Scholar]
- Key, T. , Appleby, P. , Barnes, I. , Reeves, G. , Endogenous Hormones and Breast Cancer Collaborative Group, 2002. Endogenous sex hormones and breast cancer in postmenopausal women: reanalysis of nine prospective studies. J. Natl. Cancer Inst.. 94, 606–616. [DOI] [PubMed] [Google Scholar]
- Klinge, C.M. , 2009. Estrogen regulation of MicroRNA expression. Curr. Genomics. 10, 169–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundström, E. , Christow, A. , Kersemaekers, W. , Svane, G. , Azavedo, E. , Söderqvist, G. , Mol-Arts, M. , Barkfeldt, J. , von Schoultz, B. , 2002. Effects of tibolone and continuous combined hormone replacement therapy on mammographic breast density. Am. J. Obstet. Gynecol.. 186, 717–722. [DOI] [PubMed] [Google Scholar]
- Opatrny, L. , Dell'Aniello, S. , Assouline, S. , Suissa, S. , 2008. Hormone replacement therapy use and variations in the risk of breast cancer. BJOG. 115, 169–175. [DOI] [PubMed] [Google Scholar]
- Masson, O. , Bach, A.S. , Derocq, D. , Prébois, C. , Laurent-Matha, V. , Pattingre, S. , Liaudet-Coopman, E. , 2010. Pathophysiological functions of cathepsin D: targeting its catalytic activity versus its protein binding activity?. Biochimie. 92, 1635–1643. [DOI] [PubMed] [Google Scholar]
- Missmer, S.A. , Eliassen, A.H. , Barbieri, R.L. , Hankinson, S.E. , 2004. Endogenous estrogen, androgen, and progesterone concentrations and breast cancer risk among postmenopausal women. J. Natl. Cancer Inst.. 96, 1856–1865. [DOI] [PubMed] [Google Scholar]
- Miyoshi, Y. , Ando, A. , Hasegawa, S. , Ishitobi, M. , Taguchi, T. , Tamaki, Y. , Noguchi, S. , 2003. High expression of steroid sulfatase mRNA predicts poor prognosis in patients with estrogen receptor-positive breast cancer. Clin. Cancer Res.. 9, 2288–2293. [PubMed] [Google Scholar]
- Mueck, A.O. , Seeger, H. , Wallwiener, D. , 2003. Comparison of the proliferative effects of estradiol and conjugated equine estrogens on human breast cancer cells and impact of continuous combined progestogen addition. Climacteric. 6, 221–227. [PubMed] [Google Scholar]
- Nagasaki, S. , Miki, Y. , Akahira, J. , Suzuki, T. , Sasano, H. , 2009. 17beta-hydroxysteroid dehydrogenases in human breast cancer. Ann. N.Y. Acad. Sci.. 1155, 25–32. [DOI] [PubMed] [Google Scholar]
- Oakes, S.R. , Hilton, H.N. , Ormandy, C.J. , 2006. The alveolar switch: coordinating the proliferative cues and cell fate decisions that drive the formation of lobuloalveoli from ductal epithelium. Breast Cancer Res.. 8, 207–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasqualini, J.R. , Chetrite, G.S. , 2005. Recent insight on the control of enzymes involved in estrogen formation and transformation in human breast cancer. J. Steroid Biochem. Mol. Biol.. 93, 221–236. [DOI] [PubMed] [Google Scholar]
- Rogan, E.G. , Badawi, A.F. , Devanesan, P.D. , Meza, J.L. , Edney, J.A. , West, W.W. , Higginbotham, S.M. , Cavalieri, E.L. , 2003. Relative imbalances in estrogen metabolism and conjugation in breast tissue of women with carcinoma: potential biomarkers of susceptibility to cancer. Carcinogenesis. 24, 697–702. [DOI] [PubMed] [Google Scholar]
- Sasano, H. , Nagasaki, S. , Miki, Y. , Suzuki, T. , 2009. New developments in intracrinology of human breast cancer: estrogen sulfatase and sulfotransferase. Ann. N.Y. Acad. Sci.. 1155, 76–79. [DOI] [PubMed] [Google Scholar]
- Schairer, C. , Lubin, J. , Troisi, R. , Sturgeon, S. , Brinton, L. , Hoover, R. , 2000. Menopausal estrogen and estrogen–progestin replacement therapy and breast cancer risk. JAMA. 283, 485–491. [DOI] [PubMed] [Google Scholar]
- Tamimi, R.M. , Byrne, C. , Colditz, G.A. , Hankinson, S.E. , 2007. Endogenous hormone levels, mammographic density, and subsequent risk of breast cancer in postmenopausal women. J. Natl. Cancer Inst.. 99, 1178–1187. [DOI] [PubMed] [Google Scholar]
- The Women's Health Initiative Steering Committee, 2004. Effects of conjugated estrogen on postmenopausal women with hysterectomy: the Women's Health Initiative randomized controlled trial. JAMA. 291, 1701–1712. [DOI] [PubMed] [Google Scholar]
- Valdivia, I. , Campodónico, I. , Tapia, A. , Capetillo, M. , Espinoza, A. , Lavín, P. , 2004. Effects of tibolone and continuous combined hormone therapy on mammographic breast density and breast histochemical markers in postmenopausal women. Fertil. Steril.. 81, 617–623. [DOI] [PubMed] [Google Scholar]
- Writing Group for the Women's Health Initiative Investigators, 2002. Risks and benefits of estrogen plus progestin in healthy postmenopausal women. Principal results from the Women's Health Initiative randomized controlled trial. JAMA. 288, 321–333. [DOI] [PubMed] [Google Scholar]
- Zanetti-Dällenbach, R. , Vuaroqueaux, V. , Wight, E. , Labuhn, M. , Singer, G. , Urban, P. , Eppenberger, U. , Holzgreve, W. , Eppenberger-Castori, S. , 2006. Comparison of gene expression profiles in core biopsies and corresponding surgical breast cancer samples. Breast Cancer Res.. 8, R51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeleniuch-Jacquotte, A. , Shore, R.E. , Koenig, K.L. , Akhmedkhanov, A. , Afanasyeva, Y. , Kato, I. , Kim, M.Y. , Rinaldi, S. , Kaaks, R. , Toniolo, P. , 2004. Postmenopausal levels of oestrogen, androgen, and SHBG and breast cancer: long-term results of a prospective study. Br. J. Cancer. 90, 153–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, B.T. , Conney, A.H. , 1998. Functional role of estrogen metabolism in target cells: review and perspectives. Carcinogenesis. 19, 1–27. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. 1 Distribution of KRT19 (A) and Endothelial marker set (B) mRNA levels. Evaluation of the distribution of KRT19 and the endothelial marker set mRNA expression is shown for the remaining 122 RNA samples with adequate expression of the reference genes (QQ RNA) used to normalize the data as described in the main body text. Firstly, the median mRNA level of KRT19 was established, after which samples with an expression level less than an arbitrarily cut‐off set at 25% of this median KRT19 level were excluded from further analyses. (A). Next, the median mRNA level of the endothelial marker set‐consisting of CDH5, MCAM, VCAM1 and VWF was established, after which also samples with an expression level less than an arbitrarily cut‐off set at 25% of this median endothelial marker set level were excluded from the final analysis (B).
Supplementary Fig. 2 Unsupervised hierarchical clustering to identify homology in‐between markers. Expression levels of the markers shown on the right site of the figure left were log‐transformed and mean normalized from −1.0 to 1.0. Red squares in the cluster diagram indicate a positive relative transcript expression (0–1.0), green squares a negative relative expression (−1.0 to 0) and black squares depict a relative expression of zero. Each row depicts a single marker (gene or miRNA), each column a single case. The dendrogram on top present the relatedness of the profiles of individual cases and the dendrogram on the left the relatedness of the markers in this clustering. The longer the dendrogram arm, the greater the difference in between individual cases and genes inside a cluster group. Cases are color‐ and number‐coded according therapy group: gray [20]; [no HRT] before therapy; black [21], no HRT after 6 months; pink [10], [E2] before and red [11], [E2] after therapy; light green [30], [E2+NETA] before and dark green [31], E2+NETA after therapy; light blue [40], T before and dark blue [41], [T] after therapy. The 10 clusters of markers with a node correlation >0.2 are visualized by the different colors on the right side of the cluster. To improve readability, the thus identified gene clusters and dendrogram depicting the relatedness of the profiles of individual cases are blown up in the right panel of the figure.
Supplementary data
Supplementary data
Supplementary data


