Abstract
Cancer represents a complex group of heterogeneous diseases. While many cancers share fundamental biological processes (hallmarks of cancer) necessary for their development and progression, cancers also distinguish themselves by their dependence on distinct oncogenic pathways. Over the last decade, targeted therapies have been introduced to the clinic with variable success. In truth, single targeted therapies may be successful in only a subset of malignancies but insufficient to address malignancies that often rely on multiple pathways, thus evading single targeted agents. Investigators have recently identified potentially functional components of the human genome that were previously thought to have no biological function. This discovery has added to the already established complexity of gene regulation in the pathogenesis of cancer. Non‐coding RNAs represent key regulators of gene expression. Improved knowledge of their biogenesis and function may in turn lead to a better understanding of the heterogeneity of malignancies and eventually be leveraged as diagnostic, prognostic and therapeutic targets. MicroRNAs (miRNAs or miRs) for example, have the capacity for the regulation of multiple genes and thus redirection or reprogramming of biological pathways. However, several other members of the non‐coding RNA family may be of equal biological relevance. In this review, we provide a perspective on emerging concepts in the clinical application of miRNA and other non‐coding RNAs as biomarkers in cancer with an eye on the eventual integration of both miRNA and other non‐coding RNA biology into our understanding of cancer pathogenesis and treatment.
Keywords: MicroRNA, Non-coding RNA, Epigenetics
Highlights
Non‐coding RNAs (ncRNAs) are important in both gene regulation and biological processes fundamental to cancer.
ncRNAs are divided into short (<200 nt) and long non‐coding RNAs (300 nt–100 kb).
Circulating miRNA and targeted in vivo delivery are emerging concepts in the study of non‐coding RNAs.
1. Introduction
With the development of sophisticated platforms for interrogating the human genome, in the last several years, investigators have identified regions of the human genome (greater than 90%) that were previously thought to be non‐functional and non‐coding (thus termed non‐coding RNAs) (Dermitzakis et al., 2005). These families of non‐coding RNAs (ncRNAs) have an increasingly important role in gene regulation and biological processes fundamental to both normal development and disease. These paradigm shifting observations have served as the basis for a rapidly expanding field of investigation in the biology of non‐coding RNAs and elucidating their role alongside protein‐coding genes in cancer initiation and progression. microRNAs (miRNAs or miRs) represent perhaps the best studied of this group of non‐coding RNAs(Grimson et al., 2007; Lee et al., 1993). Most recent estimates suggest that there are over 1000 human miRNAs and that up to 60% of the human genome may be under the regulation of miRNAs (Friedman et al., 2009). miRNAs regulate a large number of essential biological functions that are critical to normal development with deregulation of these same miRNAs later in life contributing to the development of diseases such as cancer. In cancer, miRNA expression and function may be both tissue and cell specific with miRNAs serving as tumor suppressors, oncogenes or in some cases both (Calin et al., 2002). The traditional dogma has been that these small, 18–25 nt, molecules can cause either mRNA degradation or inhibit translation (Calin and Croce, 2006; Croce, 2009). However, there are lines of investigation to suggest that miRNAs may induce gene expression as well. One identified mechanism is through effects on nonsense‐mediated decay that regulates both aberrant and normal miRNAs (Bruno, Molecular Cell, 2011).
One particularly appealing characteristic of miRNAs lies in their capacity for the regulation of multiple genes and thus their ability to redirect or reprogram biological pathways(Volinia et al., 2010). This has in part fueled the move towards understanding the role for miRNAs from a “systems biology” perspective as well as the development of miRNA‐based therapeutics. However, this same appealing characteristic makes the investigation of miRNA biology particularly challenging. Strategies to prioritize miRNA/target candidates and organ directed therapies have yet to be fully realized. As a result, the translation of miRNA biology to everyday clinical practice remains a challenge. Nevertheless, progress is being made. For example, miRNA expression patterns are being tested and validated as part of clinical decision algorithms in clinical trials. After numerous studies of global miRNA profiling and reductionist approaches focused on single miRNA/single target mRNA relationships, investigators have now transitioned to the next chapter of miRNA investigation by focusing on a few areas: 1)the integration of miRNA biology into a more global picture of the reprogramming of biological pathways 2) the development of non‐invasive assays for miRNA detection and novel delivery systems for miRNA‐based therapeutics and 3) further validation of the functional role for other non‐coding RNAs in cancer biology.
It is important to note that miRNAs represent only one of a family of non‐coding RNAs many of whose function is just beginning to be uncovered (Figure 1). Other components of the human genome include tRNAs, rRNAs, small nuclear RNAs (snRNAs) and long non‐coding RNAs (lncRNAs) to name a few (Gibb et al., 2011). Among lncRNAs, long intervening non‐coding RNAs (lincRNAs) and ultraconserved regions (UCRs) appear to be globally deregulated in cancer and carry regulatory function (Calin et al., 2007). Our understanding for these molecules, however, is in its infancy with the majority of studies being confined to the identification of abnormal expression patterns in cancer. It is clear that lncRNAs are likely to play just as important of a role in both normal development and tumorigenesis. Furthermore, studies suggest that there may be interactions between both short and long non‐coding RNAs thus further complicating the picture. We are just beginning to uncover the mechanisms by which these family members function both independently and in concert to promote or prevent tumor development.
Figure 1.
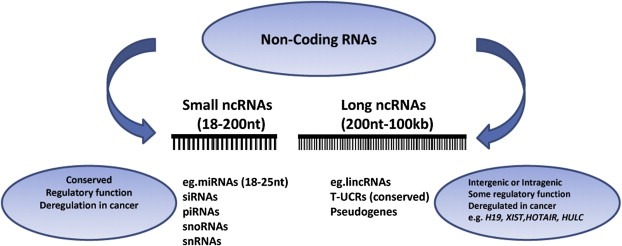
Overview of non‐coding RNAs: miRNAs (microRNAs), siRNAs (small interfering RNAs), piRNAs (piwi interacting RNAs), snoRNAs (small nucleolar RNAs), snRNA (small nuclear RNAs), lincRNAs (long intergenic RNAs), tUCRs (transcribed ultraconserved regions).
2. miRNA deregulation and disease
First described in 2002 as being altered in Chronic Lymphocytic Leukemia (CLL), miRNAs are now known to be globally altered in both solid and hematological malignancies (Calin et al., 2002).The mechanisms for miRNA deregulation are several‐fold including their frequent location to fragile chromosomal sites and cancer‐associated genomic regions (CAGRs), impaired processing, environmental exposures (eg. cigarette smoke), oncogene or tumor suppressor targeting and sequence variants of both miRNA and target mRNA (Chang et al., 2011; Izzotti et al., 2010). Each of these areas is the subject of intense investigation. There is increasing evidence that miRNAs are susceptible to epigenetic modifications (Saito and Jones, 2006). Several individual miRNAs including miR‐126, miR‐145, miR‐127 and miR‐29 have been shown be at least partially under the regulation of methylation. A recent comprehensive review of 45 studies determined that out of 122 epigenetically regulated miRNAs, 55 were associated with more than one malignancy while 67 appeared to be cancer specific (Kunej et al., 2011). The investigators further identified chromosomes, 1, 7, 11, 14 and 19 as the most common locations for epigenetically modified miRNAs.
miRNA biogenesis represents a complex, multi‐step process that is dependent upon a series of enzymatic and protein reactions which have previously been described (Lopez‐Serra and Esteller, 2011). Investigators have focused primarily on deficiencies in essential components of the miRNA processing machinery (Dicer, Drosha, Ago) and their link to tumor initiation and progression (Chiosea et al., 2007; Kumar et al., 2007). However, there are several other mechanisms that may compromise miRNA processing and function. For example, defective export of pre‐miRNAs is another mechanism by which miRNA production may be impaired (Melo et al., 2010). A mutated form of the Exportin 5 gene (XPO5) within a subset of cancer cell lines harboring microsatellite instability was localized to the nucleus thus interfering with both pre‐miRNA processing and function. Interestingly, in vitro re‐expression of the wild type form caused an up‐regulation of miRNAs with known tumor suppressive functions.
In addition to the contributions of chromosomal aberrations, epigenetic modifications and exogenous stimuli to miRNA deregulation, which have been described elsewhere, nucleotide polymorphisms within miRNA may also regulate both miRNA processing and targeting. There are several such examples in the literature. For example, single nucleotide polymorphisms (SNPs) in the pri‐miR‐16 led to a decrease in miRNA processing. This mutation resulted in with the development of CLL in vivo (Calin et al., 2005). Pre‐miR‐196a2 SNPs have been associated with risk for the development of solid tumors including esophageal, lung, gastric and breast cancers (Chu et al., 2011). Target mRNA polymorphisms can also compromise miRNA/mRNA binding and thus activity. The Let‐7/KRAS relationship is well established including successful directed targeting in murine models of lung cancer (Trang et al., 2010). More recently, studies have demonstrated that single nucleotide polymorphisms (SNPs) in the 3′UTR of KRAS have functional relevance by interfering with Let‐7 binding and regulation. These findings have subsequently been expanded to population based studies linking KRAS 3′UTR variants to cancer risk. For example, Paranjape et al. examined 2 separate groups of breast cancer patients (415 patients and 457 controls in group 1 and 690 patients and 360 controls in group 2) for a previously described KRAS 3′UTR variant (Paranjape et al., 2011). They identified an association between KRAS 3″UTR variants and premenopausal patients with tripe negative breast cancer (OR 2.3).
3. Emerging concepts in clinical applications for miRNAs
Given the fundamental role for miRNAs in tumor development and their global deregulation, they are ideally suited as either surrogate or therapeutic biomarkers in cancer. Global patterns of miRNA expression have been used for both diagnostic and prognostic purposes as well to guide therapeutic decisions. For example, both elevated miR‐155 and low let7a‐2 expression correlated with poor survival in lung cancer (Yanaihara et al., 2006). Elevated levels of miR‐21 correlated with poor survival in breast cancer (Yan et al., 2008) and increased miR‐21 correlated with poor survival in colorectal carcinoma (Schetter et al., 2008). The number of such studies correlating individual or panels of miRNAs with clinical outcome is constantly growing with variable reproducibility. Therefore, the question of how such biomarkers should inform clinical decision making remains a matter of debate. Two rapidly changing areas of investigation are that of circulating miRNAs and targeted delivery systems. The non‐invasive detection or “liquid biopsy” of cancers has been particularly elusive for the majority of cancers. Such assays would be beneficial in further risk stratifying at risk populations, provide less invasive means for diagnosis among patients deemed to be high risk for more invasive procedures and could potentially be used as biomarkers for both treatment response and surveillance. The development of an in vivo miRNA delivery system that translates clinically is a challenging task. However, initiatives to develop such on‐target miRNAs while minimizing toxicity are ongoing.
3.1. Circulating miRNA
Circulating tumors cells (CTCs), circulating DNA and gene expression are all being investigated with varying degrees of success (Lianidou and Markou, 2011; Taniguchi et al., 2011). In the last few years a multitude of studies have shown that miRNAs are detectable in the peripheral circulation in cancer as well as cardiac and rheumatological diseases (Mitchell et al., 2008; Hunter et al., 2008). Investigators have identified miRNA signatures in serum, plasma, peripheral blood mononuclear cells (PBMCs) and whole blood that appear to distinguish patients with cancers such as prostate (miR‐141), lung (miR‐21, miR‐210 and miR‐486‐5 p), breast (miR‐195 and let‐7a) and lymphoma from patients without disease (Heneghan et al., 2010; Lawrie, 2007; Mitchell et al., 2008; Shen et al., 2011). Circulating miRNA signatures have also been integrated into clinical trials. One recent study by Boeri et al. demonstrated the value of circulating miRNAs in patients enrolled in a longitudinal lung cancer CT‐screening trial (Boeri et al., 2011). Patterns of miRNA expression were used to both distinguish patients with and without lung cancer as well as used as a predictive tool for those at risk for the development of lung cancer. Investigators have yet to reach consensus on the ideal platform for circulating miRNA detection but novel nanotechnology based approaches are on the horizon (Wang et al., 2011).
Despite these findings, the actual functional implications for circulating miRNA, their location and the optimal methods of detection have yet to be clarified. The recent detection of a subset of stable miRNAs within circulating exosomes has generated a new area linking miRNA to cell–cell communication and function (Hunter et al., 2008). Importantly, exosomes represent only one type of microvesicle along with apoptotic bodies and ectosomes (Lee et al., 2011) each of which undergo a distinct process of biogenesis. Exosomes are membrane bound particles (30–100 nm) that originate from endosomes in a variety of host cell types including platelets, macrophages and tumor cells (Rani et al., 2011; Yang et al., 2011a). Depending on their host cell of origin and initiating event, exosomes may carry miRNAs, lipids, mRNA or proteins leading to varying functions (Valadi et al., 2007). Investigators have known for quite some time that microvesicles could carry molecules implicated in carcinogenesis (Hawari et al., 2004; Kim et al., 2002). However, the concept of microvesicles as carriers for miRNA is still a relatively new and evolving concept. Microvesicle packaged miRNA may be released from several malignancies including renal cell (Grange et al., 2011), hepatocellular (Kogure et al., 2011) and glioblastoma (Skog et al., 2008). Some very interesting observations suggest that microvesicles may not be the only source for circulating miRNA. Arroyo et al recently detected a high percentage of circulating protein bound miRNAs (Arroyo et al., 2011). Further investigation revealed that these miRNAs co‐purified with the Ago2 ribonucleoprotein complex, an essential component of the RNA‐induced silencing complex (RISC). Clearly, this increases the complexity of identifying the ideal circulating miRNA signatures in disease and requires further validation.
3.2. miRNA delivery systems
The development of miRNA‐based therapeutics is primarily based on either selective inhibition of oncogenic miRNAs or binding or selective over‐expression of tumor suppressive miRNAs. Both in vivo and in vitro approaches to augmenting or silencing miRNA have been met with reasonable success. Organ specific transgenic and knockout models are invaluable for understanding the underlying mechanistic role for a given miRNA. For example, targeted over‐expression of the oncomiR miR‐21 increased tumorigenesis in a Kras‐driven murine model while targeted deletion of miR‐21 reduced lung tumorigenesis (Hatley et al., 2010). While such approaches are necessary to uncover molecular mechanisms of miRNA effect, they may prove difficult to translate to human application. Systemic delivery of a liver‐specific DNA‐LNA miR‐122 anti‐miRNA (SPC3649) as therapy for chronic Hepatitis C infection represents the most successful application of miRNA targeting to achieve clinical application in a model of chronic HCV infection (Lanford et al., 2010).
In order to better translate targeted miRNA‐based therapeutics investigators are conducting complementary in vivo studies using various platforms for miRNA delivery in models of cancer. However, issues of organ specificity, off target toxicity and delivery stability still exist. There are several approaches to miRNA targeting including antagomiRs that are modified with a cholesterol conjugated 2′‐O‐methyl in order to maintain stability while minimizing degradation (Garzon et al., 2010). In contrast, Locked Nucleic Acids (LNAs) have a methylene bridge that functionally locks ribose conformation (Garzon et al., 2010). This change results in increased binding affinity and stability. Anti‐miRNA Oligonucleotides (AMOs) represent one of the simplest forms of miRNA targeting by forming direct complementarity and thus inhibit specific miRNA.
Given the redundancies that exist between miRNA seed sequences and target mRNA, individual targeting of miRNA may be insufficient to achieve physiological effect. MiRNA sponges represent one approach to targeting a family of miRNAs as opposed to single miRNA targeting with antisense oligonucleotides (Ebert and Sharp, 2010). Sponges function by using multiple complementary 3′UTR mRNA sites for a specific miRNA. This approach has been successfully applied in vivo for several cancers. Sponges targeting miR‐9 and miR‐31 (Ma et al., 2010; Valastyan et al., 2009) have been used to reduce and induce tumor development in murine models of breast cancer. Another more recently adopted approach involves using locked nucleic acid (LNA) modified phosphothioate oligonucleotides that are complementary to common seed sequences for several miRNAs (Garzon et al., 2010). This allows for targeted interference of clusters of miRNAs. Obad et al demonstrated this approach in an elegant study for miR‐155, 21 and let‐7 both in vitro and in vivo (Obad et al., 2011).
An emerging area of investigation involves the development of nanotechnology based approaches to miRNA delivery. Nanoparticle based formulations have been used primarily for in vitro delivery of siRNAs. However, an increasing number of studies are using similar technologies for miRNA delivery to improve stability and targeted organ delivery. Trang et al demonstrated that systemic delivery of miRNA mimics packaged in a neutral emulsion could be used to reduce tumor burden in both orthotopic and autochronous murine models of lung cancer (Trang et al., 2011). We recently used a liposomal packaged miRNA formulation to achieve short‐term lung specific delivery (Wu et al., 2011). Similarly, liposome‐polycation‐hyaluronic acid (LPH) particles as a carrier for miRNA modified with a tumor targeting monoclonal antibody (GC4 single‐chain variable fragment) have been used to target lung metastases in a murine model of metastatic melanoma (Chen et al., 2010).
4. LNCRNAs, UCRS, snoRNAs: new or old players in cancer biology?
The advent of platforms for genome wide analyses including deep sequencing has allowed for the identification of an increasing number of other previously unrecognized ncRNAs. ncRNAs are essentially divided into short (less than 200 nt) and long non‐coding RNAs (300 nt −100 kb) (lncRNAs) (Gibb et al., 2011). While the majority of studies have focused on miRNAs, lncRNAs are likely to be of equal importance to fundamental biology.
4.1. Long non‐coding RNAs
Long non‐coding RNAs (lncRNAs) range in length between 300 nucleotides to 100 kb (Gibb et al., 2011). Investigators have determined that these non‐coding RNAs also have a role in the regulation of biological processes such as imprinting, cell cycle regulation, splicing, differentiation and transcription. Two of the first described lncRNAs H19 (imprinting) and XIST (X inactive specific transcript) were actually identified prior to the discovery of miRNAs but have not been investigated to the extent of miRNAs (Brown et al., 1991; Hung and Chang, 2010). Since their initial discovery, lncRNAs and their deregulation has been described across both solid and hematological malignancies. Similar to miRNAs, lncRNAs have both cell and tissue specificity and regulatory function. Several studies now demonstrate their functional role in tumor progression.
4.1.1. Hox antisense intergenic RNA (HOTAIR)
HOTAIR is a 2.2 kb, long intergenic non‐coding RNA (lincRNA) localized to the HOXC locus (12q13.3) (Rinn et al., 2007). A recent, thorough examination of the HOTAIR sequence across both mammals and non‐mammalian vertebrates revealed that HOTAIR is poorly conserved across species and only exists in mammals (Gupta et al., 2010). However, studies suggest that variability may also exist in HOTAIR function between mammals (Brannan et al., 1990). Co‐expressed with HOXC genes, HOTAIR binds to the Polycomb protein termed Polycomb Repressor Complex 2 (PRC2) (Tsai et al., 2010). Polycomb group proteins (PcG) are critical to the maintenance of differentiation during embryogenesis through gene silencing (Tsai et al., 2010). PRC2 is a histone H3 lysine 27 (H3K27) methylase silencing complex. By recruiting PRC2, HOTAIR targets PRC2 to regulate in trans HOXD gene expression. Specifically, the functions of HOTAIR are dependent upon binding to sub‐complexes of PRC2 (Schorderet and Duboule, 2011). The murine homolog for HOTAIR (termed mHOTAIR) has variable sequence homology and function compared to human HOTAIR. Interestingly, while HOXc mutant null mice displayed very little mHOTAIR, there were little differences in target HOXD expression or PRC2 targets (Schorderet and Duboule, 2011). The underlying mechanisms for this discrepancy between human and murine functions of HOTAIR are under investigation. Gupta et al., measured HOTAIR levels in a 132 breast cancer tumors and identified an association between high HOTAIR and the presence of both metastases and death (Gupta et al., 2010). They also confirmed that HOTAIR over‐expression lead to an alteration in genome wide PRC2 occupancy and that the HOTAIR mediated effects on invasive capacity were PRC2 dependent. Since that study, others have followed suit in examining the role for HOTAIR in other solid tumors. Kogo et al, recently showed that HOTAIR levels were decreased in colorectal carcinoma tissues compared to adjacent uninvolved tissues (Kogo et al., 2011). Upon further stratifying cases into “high” and “low” HOTAIR expressing tumors, the investigators detected an association between high levels and both liver metastases and poor survival. These findings were complemented by in vitro studies demonstrating that HOTAIR gain of function promoted tumor cell invasion and tissue analysis further validating the link between HOTAIR expression and PRC2 based genes. An independent study also showed that high HOTAIR expression was an independent prognostic factor in patients with hepatocellular carcinoma (Yang et al., 2011b).
4.1.2. MALAT‐1
First identified in (NSCLC) non‐small cell lung cancer, metastasis associated in lung adenocarcinoma transcript‐1 (MALAT‐1) is a non‐coding RNA that has been implicated in several malignancies (Ji et al., 2003). MALAT‐1 is an intergenic transcript (7 kb) located on chromosome 11 (Clark and Mattick, 2011). In 2003, Ji et al. first noted that expression patterns of the non‐coding MALAT‐1 transcript correlated with metastases and prognosis in early stage NSCLC. MALAT‐1 expression also correlated with chemotherapeutic response in osteosarcoma (Fellenberg et al., 2007) and recurrence of hepatocellular carcinoma in patients undergoing liver transplantation (Lai et al., 2011). As would be expected, in vitro studies show that MALT‐1 regulates migratory, invasive and survival capacity in cancer cells (Lai et al., 2011; Tano et al., 2010).
4.1.3. HULC
Highly up‐regulated in liver cancer (HULC) is a lncRNA first identified in 2007 (Panzitt et al., 2007). Using HCC‐specific cDNA libraries by subtractive suppressive hybridization followed by an HCC‐specific cDNA microarray, Panzitt identified an EST within the most increased cluster of genes. Termed HULC, this lncRNA was found to be evolutionary conserved in primates. HULC may also function as a biomarker. For example, elevated levels of HULC in focal nodular hyperplasia, a benign liver tumor, suggests that HULC deregulation may contribute to early tumorigenesis. Colorectal carcinomas that metastasize to the liver also exhibited increased HULC (Matouk et al., 2009). The regulatory mechanisms for HULC up‐regulation in HCC remain relatively unknown. However, Wang et al., identified a PKA dependent interaction between phosphoCREB and the HULC promoter (Wang et al., 2010). An intriguing aspect of lncRNA involves the observations that lnRNAs and sncRNAs such as miRNAs may functionally interact in order to promote tumorigenesis. Wang et al., determined that HULC binds to miR‐372, which then leads to de‐repression of PRKACB, phosphorylation of CREB and subsequent induction of HULC (Wang et al., 2010).
4.1.4. SPRY4‐IT1
Another lnCRNA termed SPRY4‐IT1 is over‐expressed in melanoma and localized to the cellular cytoplasm. Gain of function of this lnCRNAs attenuated cellular growth while increasing death suggesting a tumor suppressive role (Khaitan et al., 2011).
4.2. Transcribed ultraconserved regions (tUCRS)
In 2004, Berjerano et al. first described in the human genome the presence of 481 segments that were larger than 200 bp and harbored 100% identity with mouse and rat (Bejerano et al., 2004). Termed ultraconserved regions (UCRs), these strictly conserved RNAs carry minimal to no variation within the human population and may be classified according to location (intergenic, intronic, exonic, partly exonic and exon containing). Given this conservation, UCRs are likely to be of importance to both ontogeny and phylogeny and have functionality in basic biology. Calin et al. first analyzed these 481 genomic segments within both normal and cancer tissues and determined that transcripts of these UCRs termed ultraconserved genes (UCGs) harbored both tissue specificity as well as distinct patterns of expression in solid and hematological malignancies (Calin et al., 2007). For example, a five UCR signature (uc.269A(N), uc.160(N), uc.215(N), uc.346A(P), and uc.348(N)) distinguished two phenotypically distinct groups of CLL. More recent studies have confirmed that much like miRNAs, UCRs tended to be localized to CAGRs.
Up until recently, the mechanisms for the regulation of UCR expression and function have remained a mystery. However, investigators are now starting to identify these mechanisms. Not surprisingly, UCRs are susceptible to many of the same regulatory mechanisms as miRNAs. Wojcik et al. identified both germline and somatic sequence variations and polymorphisms within 28 UCRs in the blood from a cohort of CLL patients and colorectal carcinoma tissues compared to normal (Wojcik et al., 2010). Furthermore, sequence variations in UCRs such as uc.276 have functional interactions with both miR‐125a and miR‐638 depending on its orientation. Yang et al. further supported this observation by identifying a rare G allele SNP (rs2056166) that was associated with breast cancer risk (Yang et al., 2008). There is now evidence to suggest that epigenetic modifications regulate UCRs (Lujambio et al., 2010). Global profiling for UCRs in a HCT‐116 colorectal cancer cell line both prior to and following treatment with the demethylating agent (5‐aza‐2deoxycytidine) led to the identification of 6 UCRs that were up‐regulated upon treatment and had evidence of an upstream CpG island. (uc.392 antisense, uc.469 antisense, uc.282 antisense, uc.160, uc.346 and uc.283 antisense). Three of these UCRs (uc.160, uc.346 and uc.283 antisense) were then further validated both in vitro and in several solid tumors as silenced by methylation.
4.3. Small nucleolar RNAs (snoRNAs)
Small nucleolar RNAs (snoRNAs) are non‐protein coding (∼70 nt) TNAs that have traditionally been utilized as endogenous controls for miRNA quantification. However, recent investigation that these RNAs may be deregulated in cancers, thus they may contribute to tumorigenesis. SnoRNAs such as RNU44, 48 and 43 direct 2′‐O′‐ribose methylation in ribosome biogenesis. Gee et al conducted a high throughput assessment for snoRNAs in a cohort breast and head and neck patients (Gee et al., 2011). They determined that not only did the snoRNAs vary between samples but that specific snoRNAs such as RNU44 correlated with molecular subtype and survival. Another study demonstrated a link between expression patterns of snoRNAs and a host gene Growth arrest specific 5 (GAS5) and breast cancer (Mourtada‐Maarabouni et al., 2009). The mechanisms for snoRNAs in tumor development remain largely unknown. However, the finding of deregulated snoRNAs in cancer should give one pause when considering suitable endogenous controls for non‐coding RNA analysis.
5. Conclusions and future directions
The discovery of functional non‐coding regions of the human genome that were previously thought to be non‐functional has had a significant impact on our understanding of both normal embryogenesis and the development of human disease. miRNAs have emerged as suitable diagnostic and prognostic biomarkers with the capacity for guiding treatment decisions in the clinic. Three particularly exciting and emerging areas of investigation include 1) the non‐invasive detection of miRNAs as biomarkers with the recognition that exosome packaged miRNAs may function as conduits for cell–cell communication and 2) the continued development of novel modalities for in vivo miRNA delivery in cancer and 3) the emergence of other functional ncRNAs. While the majority of investigation has been focused on miRNAs, it is now clear that other non‐coding RNAs may function to direct genetic programming thus contributing to cancer initiation and progression. Furthermore, there is likely to be a functional interdependence of these classes of short and long ncRNAs. We anticipate that such a complex yet elegant orchestration of non‐coding components of the human genome will lead to an improved understanding of the molecular underpinnings of cancer and eventually to the development of novel biomarkers and targeted therapies in cancer.
Nana-Sinkam S. Patrick and Croce Carlo M., (2011), Non‐coding RNAs in cancer initiation and progression and as novel biomarkers, Molecular Oncology, 5, doi: 10.1016/j.molonc.2011.10.003.
Contributor Information
S. Patrick Nana-Sinkam, Email: Patrick.Nana-Sinkam@osumc.du.
Carlo M. Croce, Email: Carlo.Croce@osumc.edu
References
- Arroyo, J.D. , Chevillet, J.R. , Kroh, E.M. , Ruf, I.K. , Pritchard, C.C. , Gibson, D.F. , Mitchell, P.S. , Bennett, C.F. , Pogosova-Agadjanyan, E.L. , Stirewalt, D.L. , Tait, J.F. , Tewari, M. , 2011. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl. Acad. Sci. U. S. A. 108, 5003–5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejerano, G. , Pheasant, M. , Makunin, I. , Stephen, S. , Kent, W.J. , Mattick, J.S. , Haussler, D. , 2004. Ultraconserved elements in the human genome. Science. 304, 1321–1325. [DOI] [PubMed] [Google Scholar]
- Boeri, M. , Verri, C. , Conte, D. , Roz, L. , Modena, P. , Facchinetti, F. , Calabro, E. , Croce, C.M. , Pastorino, U. , Sozzi, G. , 2011. MicroRNA signatures in tissues and plasma predict development and prognosis of computed tomography detected lung cancer. Proc. Natl. Acad. Sci. U. S. A. 108, 3713–3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brannan, C.I. , Dees, E.C. , Ingram, R.S. , Tilghman, S.M. , 1990. The product of the H19 gene may function as an RNA. Mol. Cell Biol.. 10, 28–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, C.J. , Ballabio, A. , Rupert, J.L. , Lafreniere, R.G. , Grompe, M. , Tonlorenzi, R. , Willard, H.F. , 1991. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature. 349, 38–44. [DOI] [PubMed] [Google Scholar]
- Calin, G.A. , Croce, C.M. , 2006. MicroRNA signatures in human cancers. Nat. Rev. Cancer. 6, 857–866. [DOI] [PubMed] [Google Scholar]
- Calin, G.A. , Dumitru, C.D. , Shimizu, M. , Bichi, R. , Zupo, S. , Noch, E. , Aldler, H. , Rattan, S. , Keating, M. , Rai, K. , Rassenti, L. , Kipps, T. , Negrini, M. , Bullrich, F. , Croce, C.M. , 2002. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. U. S. A. 99, 15524–15529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin, G.A. , Ferracin, M. , Cimmino, A. , Di Leva, G. , Shimizu, M. , Wojcik, S.E. , Iorio, M.V. , Visone, R. , Sever, N.I. , Fabbri, M. , Iuliano, R. , Palumbo, T. , Pichiorri, F. , Roldo, C. , Garzon, R. , Sevignani, C. , Rassenti, L. , Alder, H. , Volinia, S. , Liu, C.G. , Kipps, T.J. , Negrini, M. , Croce, C.M. , 2005. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N. Engl. J. Med.. 353, 1793–1801. [DOI] [PubMed] [Google Scholar]
- Calin, G.A. , Liu, C.G. , Ferracin, M. , Hyslop, T. , Spizzo, R. , Sevignani, C. , Fabbri, M. , Cimmino, A. , Lee, E.J. , Wojcik, S.E. , Shimizu, M. , Tili, E. , Rossi, S. , Taccioli, C. , Pichiorri, F. , Liu, X. , Zupo, S. , Herlea, V. , Gramantieri, L. , Lanza, G. , Alder, H. , Rassenti, L. , Volinia, S. , Schmittgen, T.D. , Kipps, T.J. , Negrini, M. , Croce, C.M. , 2007. Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer Cell. 12, 215–229. [DOI] [PubMed] [Google Scholar]
- Chang, S. , Wang, R.H. , Akagi, K. , Kim, K.A. , Martin, B.K. , Cavallone, L. , Haines, D.C. , Basik, M. , Mai, P. , Poggi, E. , Isaacs, C. , Looi, L.M. , Mun, K.S. , Greene, M.H. , Byers, S.W. , Teo, S.H. , Deng, C.X. , Sharan, S.K. , 2011. Tumor suppressor BRCA1 epigenetically controls oncogenic microRNA-155. Nat. Med.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y. , Zhu, X. , Zhang, X. , Liu, B. , Huang, L. , 2010. Nanoparticles modified with tumor-targeting scFv deliver siRNA and miRNA for cancer therapy. Mol. Ther.. 18, 1650–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiosea, S. , Jelezcova, E. , Chandran, U. , Luo, J. , Mantha, G. , Sobol, R.W. , Dacic, S. , 2007. Overexpression of dicer in precursor lesions of lung adenocarcinoma. Cancer Res.. 67, 2345–2350. [DOI] [PubMed] [Google Scholar]
- Chu, H. , Wang, M. , Shi, D. , Ma, L. , Zhang, Z. , Tong, N. , Huo, X. , Wang, W. , Luo, D. , Gao, Y. , Zhang, Z. , 2011. Hsa-miR-196a2 Rs11614913 polymorphism contributes to cancer susceptibility: evidence from 15 case-control studies. PLoS. One. 6, e18108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, M.B. , Mattick, J.S. , 2011. Long noncoding RNAs in cell biology. Semin. Cell Dev. Biol.. 22, 366–376. [DOI] [PubMed] [Google Scholar]
- Croce, C.M. , 2009. Causes and consequences of microRNA dysregulation in cancer. Nat. Rev. Genet.. 10, 704–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dermitzakis, E.T. , Reymond, A. , Antonarakis, S.E. , 2005. Conserved non-genic sequences - an unexpected feature of mammalian genomes. Nat. Rev. Genet.. 6, 151–157. [DOI] [PubMed] [Google Scholar]
- Ebert, M.S. , Sharp, P.A. , 2010. Emerging roles for natural microRNA sponges. Curr. Biol.. 20, R858–R861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellenberg, J. , Bernd, L. , Delling, G. , Witte, D. , Zahlten-Hinguranage, A. , 2007. Prognostic significance of drug-regulated genes in high-grade osteosarcoma. Mod. Pathol.. 20, 1085–1094. [DOI] [PubMed] [Google Scholar]
- Friedman, R.C. , Farh, K.K. , Burge, C.B. , Bartel, D.P. , 2009. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res.. 19, 92–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzon, R. , Marcucci, G. , Croce, C.M. , 2010. Targeting microRNAs in cancer: rationale, strategies and challenges. Nat. Rev. Drug Discov.. 9, 775–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gee, H.E. , Buffa, F.M. , Camps, C. , Ramachandran, A. , Leek, R. , Taylor, M. , Patil, M. , Sheldon, H. , Betts, G. , Homer, J. , West, C. , Ragoussis, J. , Harris, A.L. , 2011. The small-nucleolar RNAs commonly used for microRNA normalisation correlate with tumour pathology and prognosis. Br. J. Cancer. 104, 1168–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibb, E.A. , Brown, C.J. , Lam, W.L. , 2011. The functional role of long non-coding RNA in human carcinomas. Mol. Cancer. 10, 38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grange, C. , Tapparo, M. , Collino, F. , Vitillo, L. , Damasco, C. , Deregibus, M.C. , Tetta, C. , Bussolati, B. , Camussi, G. , 2011. Microvesicles released from human renal cancer stem cells stimulate angiogenesis and formation of lung premetastatic niche. Cancer Res.. 71, 5346–5356. [DOI] [PubMed] [Google Scholar]
- Grimson, A. , Farh, K.K. , Johnston, W.K. , Garrett-Engele, P. , Lim, L.P. , Bartel, D.P. , 2007. MicroRNA targeting specificity in mammals: determinants beyond seed pairing 1. Mol. Cell. 27, 91–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta, R.A. , Shah, N. , Wang, K.C. , Kim, J. , Horlings, H.M. , Wong, D.J. , Tsai, M.C. , Hung, T. , Argani, P. , Rinn, J.L. , Wang, Y. , Brzoska, P. , Kong, B. , Li, R. , West, R.B. , van de Vijver, M.J. , Sukumar, S. , Chang, H.Y. , 2010. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 464, 1071–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatley, M.E. , Patrick, D.M. , Garcia, M.R. , Richardson, J.A. , Bassel-Duby, R. , van Rooij, E. , Olson, E.N. , 2010. Modulation of K-Ras-dependent lung tumorigenesis by MicroRNA-21. Cancer Cell. 18, 282–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawari, F.I. , Rouhani, F.N. , Cui, X. , Yu, Z.X. , Buckley, C. , Kaler, M. , Levine, S.J. , 2004. Release of full-length 55-kDa TNF receptor 1 in exosome-like vesicles: a mechanism for generation of soluble cytokine receptors. Proc. Natl. Acad. Sci. U. S. A. 101, 1297–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heneghan, H.M. , Miller, N. , Kelly, R. , Newell, J. , Kerin, M.J. , 2010. Systemic miRNA-195 differentiates breast cancer from other malignancies and is a potential biomarker for detecting noninvasive and early stage disease. Oncologist. 15, 673–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung, T. , Chang, H.Y. , 2010. Long noncoding RNA in genome regulation: prospects and mechanisms. RNA. Biol.. 7, 582–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter, M.P. , Ismail, N. , Zhang, X. , Aguda, B.D. , Lee, E.J. , Yu, L. , Xiao, T. , Schafer, J. , Lee, M.L. , Schmittgen, T.D. , Nana-Sinkam, S.P. , Jarjoura, D. , Marsh, C.B. , 2008. Detection of microRNA expression in human peripheral blood microvesicles. PLoS. ONE. 3, e3694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izzotti, A. , Larghero, P. , Longobardi, M. , Cartiglia, C. , Camoirano, A. , Steele, V.E. , De Flora, S. , 2010. Dose-responsiveness and persistence of microRNA expression alterations induced by cigarette smoke in mouse lung. Mutat. Res.. [DOI] [PubMed] [Google Scholar]
- Ji, P. , Diederichs, S. , Wang, W. , Boing, S. , Metzger, R. , Schneider, P.M. , Tidow, N. , Brandt, B. , Buerger, H. , Bulk, E. , Thomas, M. , Berdel, W.E. , Serve, H. , Muller-Tidow, C. , 2003. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 22, 8031–8041. [DOI] [PubMed] [Google Scholar]
- Khaitan, D. , Dinger, M.E. , Mazar, J. , Crawford, J. , Smith, M.A. , Mattick, J.S. , Perera, R.J. , 2011. The melanoma-upregulated long noncoding RNA SPRY4-IT1 modulates apoptosis and invasion. Cancer Res.. 71, 3852–3862. [DOI] [PubMed] [Google Scholar]
- Kim, C.W. , Lee, H.M. , Lee, T.H. , Kang, C. , Kleinman, H.K. , Gho, Y.S. , 2002. Extracellular membrane vesicles from tumor cells promote angiogenesis via sphingomyelin. Cancer Res.. 62, 6312–6317. [PubMed] [Google Scholar]
- Kogo, R. , Shimamura, T. , Mimori, K. , Kawahara, K. , Imoto, S. , Sudo, T. , Tanaka, F. , Shibata, K. , Suzuki, A. , Komune, S. , Miyano, S. , Mori, M. , 2011. Long non-coding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res.. [DOI] [PubMed] [Google Scholar]
- Kogure, T. , Lin, W.L. , Yan, I.K. , Braconi, C. , Patel, T. , 2011. Intercellular nanovesicle-mediated microRNA transfer: a mechanism of environmental modulation of hepatocellular cancer cell growth. Hepatology. 54, 1237–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, M.S. , Lu, J. , Mercer, K.L. , Golub, T.R. , Jacks, T. , 2007. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat. Genet.. 39, 673–677. [DOI] [PubMed] [Google Scholar]
- Kunej, T. , Godnic, I. , Ferdin, J. , Horvat, S. , Dovc, P. , Calin, G.A. , 2011. Epigenetic regulation of microRNAs in cancer: an integrated review of literature. Mutat. Res.. [DOI] [PubMed] [Google Scholar]
- Lai, M.C. , Yang, Z. , Zhou, L. , Zhu, Q.Q. , Xie, H.Y. , Zhang, F. , Wu, L.M. , Chen, L.M. , Zheng, S.S. , 2011. Long non-coding RNA MALAT-1 overexpression predicts tumor recurrence of hepatocellular carcinoma after liver transplantation. Med. Oncol.. [DOI] [PubMed] [Google Scholar]
- Lanford, R.E. , Hildebrandt-Eriksen, E.S. , Petri, A. , Persson, R. , Lindow, M. , Munk, M.E. , Kauppinen, S. , Orum, H. , 2010. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 327, 198–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrie, C.H. , 2007. MicroRNA expression in lymphoma. Expert Opin. Biol. Ther.. 7, 1363–1374. [DOI] [PubMed] [Google Scholar]
- Lee, R.C. , Feinbaum, R.L. , Ambros, V. , 1993. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 75, 843–854. [DOI] [PubMed] [Google Scholar]
- Lee, T.H. , D'Asti, E. , Magnus, N. , Al Nedawi, K. , Meehan, B. , Rak, J. , 2011. Microvesicles as mediators of intercellular communication in cancer-the emerging science of cellular ‘debris’. Semin. Immunopathol.. 33, 455–467. [DOI] [PubMed] [Google Scholar]
- Lianidou, E.S. , Markou, A. , 2011. Circulating tumor cells in breast cancer: detection systems, molecular characterization, and future challenges. Clin. Chem.. 57, 1242–1255. [DOI] [PubMed] [Google Scholar]
- Lopez-Serra, P. , Esteller, M. , 2011. DNA methylation-associated silencing of tumor-suppressor microRNAs in cancer. Oncogene. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lujambio, A. , Portela, A. , Liz, J. , Melo, S.A. , Rossi, S. , Spizzo, R. , Croce, C.M. , Calin, G.A. , Esteller, M. , 2010. CpG island hypermethylation-associated silencing of non-coding RNAs transcribed from ultraconserved regions in human cancer. Oncogene. 29, 6390–6401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, L. , Young, J. , Prabhala, H. , Pan, E. , Mestdagh, P. , Muth, D. , Teruya-Feldstein, J. , Reinhardt, F. , Onder, T.T. , Valastyan, S. , Westermann, F. , Speleman, F. , Vandesompele, J. , Weinberg, R.A. , 2010. miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat. Cell Biol.. 12, 247–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matouk, I.J. , Abbasi, I. , Hochberg, A. , Galun, E. , Dweik, H. , Akkawi, M. , 2009. Highly upregulated in liver cancer noncoding RNA is overexpressed in hepatic colorectal metastasis. Eur. J. Gastroenterol. Hepatol.. 21, 688–692. [DOI] [PubMed] [Google Scholar]
- Melo, S.A. , Moutinho, C. , Ropero, S. , Calin, G.A. , Rossi, S. , Spizzo, R. , Fernandez, A.F. , Davalos, V. , Villanueva, A. , Montoya, G. , Yamamoto, H. , Schwartz, S. , Esteller, M. , 2010. A genetic defect in exportin-5 traps precursor microRNAs in the nucleus of cancer cells. Cancer Cell. 18, 303–315. [DOI] [PubMed] [Google Scholar]
- Mitchell, P.S. , Parkin, R.K. , Kroh, E.M. , Fritz, B.R. , Wyman, S.K. , Pogosova-Agadjanyan, E.L. , Peterson, A. , Noteboom, J. , O'Briant, K.C. , Allen, A. , Lin, D.W. , Urban, N. , Drescher, C.W. , Knudsen, B.S. , Stirewalt, D.L. , Gentleman, R. , Vessella, R.L. , Nelson, P.S. , Martin, D.B. , Tewari, M. , 2008. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. U. S. A. 105, 10513–10518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourtada-Maarabouni, M. , Pickard, M.R. , Hedge, V.L. , Farzaneh, F. , Williams, G.T. , 2009. GAS5, a non-protein-coding RNA, controls apoptosis and is downregulated in breast cancer. Oncogene. 28, 195–208. [DOI] [PubMed] [Google Scholar]
- Obad, S. , dos Santos, C.O. , Petri, A. , Heidenblad, M. , Broom, O. , Ruse, C. , Fu, C. , Lindow, M. , Stenvang, J. , Straarup, E.M. , Hansen, H.F. , Koch, T. , Pappin, D. , Hannon, G.J. , Kauppinen, S. , 2011. Silencing of microRNA families by seed-targeting tiny LNAs. Nat. Genet.. 43, 371–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panzitt, K. , Tschernatsch, M.M. , Guelly, C. , Moustafa, T. , Stradner, M. , Strohmaier, H.M. , Buck, C.R. , Denk, H. , Schroeder, R. , Trauner, M. , Zatloukal, K. , 2007. Characterization of HULC, a novel gene with striking up-regulation in hepatocellular carcinoma, as noncoding RNA. Gastroenterology. 132, 330–342. [DOI] [PubMed] [Google Scholar]
- Paranjape, T. , Heneghan, H. , Lindner, R. , Keane, F.K. , Hoffman, A. , Hollestelle, A. , Dorairaj, J. , Geyda, K. , Pelletier, C. , Nallur, S. , Martens, J.W. , Hooning, M.J. , Kerin, M. , Zelterman, D. , Zhu, Y. , Tuck, D. , Harris, L. , Miller, N. , Slack, F. , Weidhaas, J. , 2011. A 3’-untranslated region KRAS variant and triple-negative breast cancer: a case-control and genetic analysis. Lancet Oncol.. 12, 377–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rani, S. , O'Brien, K. , Kelleher, F.C. , Corcoran, C. , Germano, S. , Radomski, M.W. , Crown, J. , O'Driscoll, L. , 2011. Isolation of exosomes for subsequent mRNA, MicroRNA, and protein profiling. Methods Mol. Biol.. 784, 181–195. [DOI] [PubMed] [Google Scholar]
- Rinn, J.L. , Kertesz, M. , Wang, J.K. , Squazzo, S.L. , Xu, X. , Brugmann, S.A. , Goodnough, L.H. , Helms, J.A. , Farnham, P.J. , Segal, E. , Chang, H.Y. , 2007. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 129, 1311–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito, Y. , Jones, P.A. , 2006. Epigenetic activation of tumor suppressor microRNAs in human cancer cells. Cell Cycle. 5, 2220–2222. [DOI] [PubMed] [Google Scholar]
- Schetter, A.J. , Leung, S.Y. , Sohn, J.J. , Zanetti, K.A. , Bowman, E.D. , Yanaihara, N. , Yuen, S.T. , Chan, T.L. , Kwong, D.L. , Au, G.K. , Liu, C.G. , Calin, G.A. , Croce, C.M. , Harris, C.C. , 2008. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 299, 425–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schorderet, P. , Duboule, D. , 2011. Structural and functional differences in the long non-coding RNA hotair in mouse and human. PLoS. Genet.. 7, e1002071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, J. , Liu, Z. , Todd, N.W. , Zhang, H. , Liao, J. , Yu, L. , Guarnera, M.A. , Li, R. , Cai, L. , Zhan, M. , Jiang, F. , 2011. Diagnosis of lung cancer in individuals with solitary pulmonary nodules by plasma microRNA biomarkers. BMC. Cancer. 11, 374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skog, J. , Wurdinger, T. , van Rijn, S. , Meijer, D.H. , Gainche, L. , Sena-Esteves, M. , Curry, W.T. , Carter, B.S. , Krichevsky, A.M. , Breakefield, X.O. , 2008. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol.. 10, 1470–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi, K. , Uchida, J. , Nishino, K. , Kumagai, T. , Okuyama, T. , Okami, J. , Higashiyama, M. , Kodama, K. , Imamura, F. , Kato, K. , 2011. Quantitative detection of EGFR mutations in circulating tumor DNA derived from lung adenocarcinomas. Clin. Cancer Res.. [DOI] [PubMed] [Google Scholar]
- Tano, K. , Mizuno, R. , Okada, T. , Rakwal, R. , Shibato, J. , Masuo, Y. , Ijiri, K. , Akimitsu, N. , 2010. MALAT-1 enhances cell motility of lung adenocarcinoma cells by influencing the expression of motility-related genes. FEBS Lett.. 584, 4575–4580. [DOI] [PubMed] [Google Scholar]
- Trang, P. , Medina, P.P. , Wiggins, J.F. , Ruffino, L. , Kelnar, K. , Omotola, M. , Homer, R. , Brown, D. , Bader, A.G. , Weidhaas, J.B. , Slack, F.J. , 2010. Regression of murine lung tumors by the let-7 microRNA. Oncogene. 29, 1580–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trang, P. , Wiggins, J.F. , Daige, C.L. , Cho, C. , Omotola, M. , Brown, D. , Weidhaas, J.B. , Bader, A.G. , Slack, F.J. , 2011. Systemic delivery of tumor suppressor microRNA mimics using a neutral lipid emulsion inhibits lung tumors in mice. Mol. Ther.. 19, 1116–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai, M.C. , Manor, O. , Wan, Y. , Mosammaparast, N. , Wang, J.K. , Lan, F. , Shi, Y. , Segal, E. , Chang, H.Y. , 2010. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 329, 689–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valadi, H. , Ekstrom, K. , Bossios, A. , Sjostrand, M. , Lee, J.J. , Lotvall, J.O. , 2007. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol.. 9, 654–659. [DOI] [PubMed] [Google Scholar]
- Valastyan, S. , Reinhardt, F. , Benaich, N. , Calogrias, D. , Szasz, A.M. , Wang, Z.C. , Brock, J.E. , Richardson, A.L. , Weinberg, R.A. , 2009. A pleiotropically acting microRNA, miR-31, inhibits breast cancer metastasis. Cell. 137, 1032–1046. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Volinia, S. , Galasso, M. , Costinean, S. , Tagliavini, L. , Gamberoni, G. , Drusco, A. , Marchesini, J. , Mascellani, N. , Sana, M.E. , Abu, J.R. , Desponts, C. , Teitell, M. , Baffa, R. , Aqeilan, R. , Iorio, M.V. , Taccioli, C. , Garzon, R. , Di Leva, G. , Fabbri, M. , Catozzi, M. , Previati, M. , Ambs, S. , Palumbo, T. , Garofalo, M. , Veronese, A. , Bottoni, A. , Gasparini, P. , Harris, C.C. , Visone, R. , Pekarsky, Y. , de la, C.A. , Bloomston, M. , Dillhoff, M. , Rassenti, L.Z. , Kipps, T.J. , Huebner, K. , Pichiorri, F. , Lenze, D. , Cairo, S. , Buendia, M.A. , Pineau, P. , Dejean, A. , Zanesi, N. , Rossi, S. , Calin, G.A. , Liu, C.G. , Palatini, J. , Negrini, M. , Vecchione, A. , Rosenberg, A. , Croce, C.M. , 2010. Reprogramming of miRNA networks in cancer and leukemia. Genome Res.. 20, 589–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J. , Liu, X. , Wu, H. , Ni, P. , Gu, Z. , Qiao, Y. , Chen, N. , Sun, F. , Fan, Q. , 2010. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res.. 38, 5366–5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y. , Zheng, D. , Tan, Q. , Wang, M.X. , Gu, L.Q. , 2011. Nanopore-based detection of circulating microRNAs in lung cancer patients. Nat. Nanotechnol.. 6, 668–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojcik, S.E. , Rossi, S. , Shimizu, M. , Nicoloso, M.S. , Cimmino, A. , Alder, H. , Herlea, V. , Rassenti, L.Z. , Rai, K.R. , Kipps, T.J. , Keating, M.J. , Croce, C.M. , Calin, G.A. , 2010. Non-codingRNA sequence variations in human chronic lymphocytic leukemia and colorectal cancer. Carcinogenesis. 31, 208–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, Y. , Crawford, M. , Yu, B. , Mao, Y. , Nana-Sinkam, S.P. , Lee, L.J. , 2011. MicroRNA delivery by cationic lipoplexes for lung cancer therapy. Mol. Pharm.. 8, 1381–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, L.X. , Huang, X.F. , Shao, Q. , Huang, M.Y. , Deng, L. , Wu, Q.L. , Zeng, Y.X. , Shao, J.Y. , 2008. MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. RNA. 14, 2348–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanaihara, N. , Caplen, N. , Bowman, E. , Seike, M. , Kumamoto, K. , Yi, M. , Stephens, R.M. , Okamoto, A. , Yokota, J. , Tanaka, T. , Calin, G.A. , Liu, C.G. , Croce, C.M. , Harris, C.C. , 2006. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 9, 189–198. [DOI] [PubMed] [Google Scholar]
- Yang, M. , Chen, J. , Su, F. , Yu, B. , Su, F. , Lin, L. , Liu, Y. , Huang, J.D. , Song, E. , 2011. Microvesicles secreted by macrophages shuttle invasion-potentiating microRNAs into breast cancer cells. Mol. Cancer. 10, 117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, R. , Frank, B. , Hemminki, K. , Bartram, C.R. , Wappenschmidt, B. , Sutter, C. , Kiechle, M. , Bugert, P. , Schmutzler, R.K. , Arnold, N. , Weber, B.H. , Niederacher, D. , Meindl, A. , Burwinkel, B. , 2008. SNPs in ultraconserved elements and familial breast cancer risk. Carcinogenesis. 29, 351–355. [DOI] [PubMed] [Google Scholar]
- Yang, Z. , Zhou, L. , Wu, L.M. , Lai, M.C. , Xie, H.Y. , Zhang, F. , Zheng, S.S. , 2011. Overexpression of long non-coding RNA HOTAIR predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation. Ann. Surg. Oncol.. 18, 1243–1250. [DOI] [PubMed] [Google Scholar]


