Abstract
DNA methylation plays a key role in the silencing of numerous cancer‐related genes, thereby affecting a number of vital cellular processes, including the cell cycle checkpoint, apoptosis, signal transduction, cell adhesion and angiogenesis. Also widely altered in human malignancies is the expression of microRNAs (miRNAs), a class of small noncoding RNAs that act as posttranscriptional regulators of gene expression. Furthermore, emerging evidence now supports the idea that DNA methylation is crucially involved in the dysregulation of miRNAs in cancer. This is in part the result of technological advances that enable more comprehensive analysis of miRNA expression profiles and the epigenome in cancer cells, which has led to the identification of a number of epigenetically regulated miRNAs. As with protein‐coding genes, it appears that miRNA genes involved in regulating cancer‐related pathways are silenced in association with CpG island hypermethylation. In addition, methylation in CpG island shore regions and DNA hypomethylation also appear to contribute to miRNA dysregulation in cancer. Aberrant DNA methylation of miRNA genes is a potentially useful biomarker for detecting cancer and predicting its outcome. Moreover, re‐expression of miRNAs and the replacement of tumor suppressive miRNAs using miRNA mimics or expression vectors could be effective approaches to cancer therapy.
Keywords: MicroRNA, DNA methylation, Epigenetic silencing, Tumor suppressor
Highlights
Dysregulated expression of miRNAs is a common feature in human malignancy.
Epigenetic alterations are important causes of miRNA dysregulation in cancer.
DNA methylation of miRNA genes could be a useful biomarker for cancer diagnosis.
Replacement of tumor suppressive miRNAs could be a cancer treatment strategy.
1. Introduction
microRNAs (miRNAs) are endogenous, small (∼22 nucleotides) noncoding RNAs that mediate post‐transcriptional gene silencing in both plants and animals (Esquela‐Kerscher and Slack, 2006; Esteller, 2011). miRNAs negatively regulate their target genes in one of two ways, depending on the degree of complementarity between themselves and the target messenger RNAs (mRNAs). miRNAs that bind with perfect or nearly perfect complementarity to mRNA sequences induce the RNA‐mediated interference (RNAi) pathway, in which mRNA transcripts are cleaved by a miRNA‐associated RNA‐induced silencing complex (miRISC). This mechanism is mainly observed in plants, though miRNA‐directed mRNA cleavage does occur in animals. Most animal miRNAs are thought to act by binding to imperfectly complementary sites within the 3′ untranslated regions (UTRs) of target mRNAs and inhibiting the initiation of translation via the miRISC complex.
Annotation of their genomic locations suggests most miRNA genes are situated within intergenic regions, though they are also observed within exonic and intronic regions in either sense or antisense orientation. Like genes encoding proteins, miRNA genes are mainly transcribed by RNA polymerase II. They are initially transcribed as large RNA precursors called pri‐miRNAs, which are processed within the nucleus by Drosha, a ribonuclease III enzyme, in collaboration with Pasha (also known as DGCR8). The processing of pri‐miRNAs produces ∼70‐nt‐long molecules called pre‐miRNAs, which fold into imperfect stem‐loop structures. The pre‐miRNAs are exported into the cytoplasm by the nuclear export protein Exportin‐5 (XPO5). Once in the cytoplasm, the pre‐miRNAs are released in a GTP‐dependent manner and processed by another ribonuclease III, Dicer, in complex with the double‐stranded RNA‐binding protein TRBP to produce the functional 22‐nt‐long miRNA.
Several studies have provided evidence that dysregulated miRNA expression contributes to the initiation and progression of human cancers (Croce, 2009; Esquela‐Kerscher and Slack, 2006; Esteller, 2011). Indeed, downregulation of a subset of miRNAs is a commonly observed feature of cancers, suggesting these molecules may act as tumor suppressors. The first report of altered miRNA expression in cancer was related to the frequent chromosomal deletion and downregulated expression of miR‐15 and miR‐16, two miRNAs thought to target the antiapoptotic factor BCL2 in chronic lymphocytic leukemia (Calin et al., 2002). Another example is let‐7, which negatively regulates expression of Ras oncogenes; its downregulation in tumors is thought to contribute to activation of the Ras signaling pathway (Johnson et al., 2005).
Although the mechanism underlying miRNA dysregulation in cancer is not yet fully understood, recent studies have shown that epigenetic mechanisms play important roles in the regulation of miRNA expression. Epigenetic gene silencing due to promoter CpG island hypermethylation is one of the most common mechanisms by which tumor suppressor genes are inactivated during tumorigenesis. To date, approximately half of the classical tumor suppressor genes known to be mutated in familial cancer syndromes have been shown to be inactivated by promoter hypermethylation (Feinberg and Tycko, 2004; Jones and Baylin, 2002). In addition to classical tumor suppressors, increasing numbers of genes related to cell‐cycle control, DNA repair, tumor invasiveness and the response to growth factors are being identified as inactivated by hypermethylation in malignancies (Jones and Baylin, 2007; Suzuki et al., 2008). Moreover, recent advances in microarray and sequencing technologies have enabled comprehensive analysis of the epigenome and miRNA expression in cancer cells, and as a result the list of miRNA genes silenced by methylation in cancer is rapidly growing (Lopez‐Serra and Esteller, 2012) (Figure 1, Table 1). In this review, we will highlight the contribution made by epigenetic alteration of miRNA genes in cancer, and discuss their clinical application as biomarkers and therapeutic targets.
Figure 1.
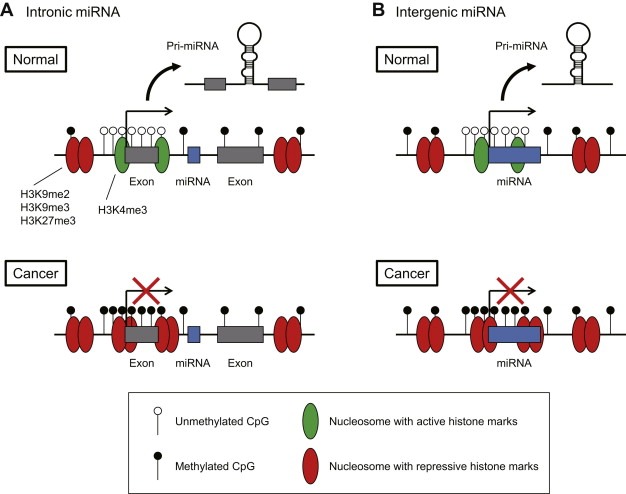
Epigenetically silenced miRNA genes in cancer.
Table 1.
Aberrantly methylated miRNA genes in cancer.
| Gene | Tumor type | Target gene |
|---|---|---|
| miR‐1‐1 | Liver, colon | FOXP1, MET, HDAC4, ANXA2, BDNF |
| miR‐9 family | Multiple types | FGFR1, CDK6, CDX2, E‐cadherin |
| miR‐10b | Stomach | MAPRE1 |
| miR‐34 family | Multiple types | MET, CDK4, CCNE2, C‐MYC, CDK6, E2F3, Notch4 |
| miR‐124 family | Colon, stomach, liver, leukemia, cervix | CDK6, VIM, SMYD3, E2F6, IQGAP1, IGFBP7 |
| miR‐125b | Breast | ETS1 |
| miR‐127 | Prostate, bladder, colon | BCL6 |
| miR‐129‐2 | Endometrium, colon, esophagus, stomach | SOX4 |
| miR‐132 | Pancreas, prostate | TALIN2 |
| miR‐137 | Head and neck, stomach, colon | CDK6, CDC42, LSD1 |
| miR‐143 | Leukemia | MLL‐AF4 |
| miR‐148a | Colon, head & neck, lung, breast, pancreas | TGIF2 |
| miR‐152 | Endometrium, bladder, lung | DNMT1, E2F3, MET, RICTOR |
| miR‐181c | Stomach | NOTCH4, KRAS |
| miR‐193a | Liver, leukemia, bladder | SRSF2, KIT |
| miR‐196b | Stomach | |
| miR‐200 family | Colon, breast, lung, bladder | ZEB1, ZEB2 |
| miR‐203 | Leukemia, liver, MALToma | ABL1, ABCE1, CDK6 |
| miR‐205 | Bladder, lung | ZEB1, ZEB2 |
| miR‐218 | Head and neck | RICTOR |
| miR‐335 | Breast | SOX4, TNC |
| miR‐345 | Colon | BAG3 |
| miR‐375 | Esophagus, melanoma | IGF1R, PDK1 |
| miR‐512 | Stomach | MCL1 |
| miR‐941 | Colon | |
| miR‐1224 | Bladder | |
| miR‐1237 | Colon | |
| miR‐1247 | Colon |
2. Screening methods to identify methylation of miRNA genes
The first evidence of an epigenetic mechanism involved in silencing miRNAs in cancer came from a pharmacological unmasking experiment. Using a miRNA microarray, Saito et al. analyzed the expression profiles of miRNAs in a T24 bladder cancer cell treated with or without the DNA methyltransferase (DNMT) inhibitor 5‐aza‐2′‐deoxycytidine (5‐aza‐dC) and the histone deacetylase (HDAC) inhibitor 4‐phenylbutyric acid (4‐PBA) (Saito et al., 2006). Of 313 miRNA genes evaluated, expression of 17 was upregulated by the drug treatment in T24 cells. Among them, miR‐127 is embedded within a CpG island, and its upregulation was associated not only with DNA demethylation but also with acetylation of histone H3 and trimethylation of histone H3 lysine 4 (H3K4me3), which are marks of active transcription. Experimental evidence confirmed that the proto‐oncogene B‐cell lymphoma 6 (BCL6) is a target of miR‐127, suggesting it can act as a tumor suppressor (Saito et al., 2006). Thereafter, miRNA microarray analysis revealed the epigenetic silencing of miR‐124 family genes in the HCT116 colorectal cancer (CRC) cell line and the same cell line in which both DNMT1 and DNMT3B are genetically disrupted (Lujambio et al., 2007). The silencing of the miR‐34b/c gene was also discovered in the same cells (Toyota et al., 2008), and a number of other methylated miRNA genes have been identified by screening for miRNAs upregulated by epigenetic drug treatment in cancer cells (Hashimoto et al., 2010; Kozaki et al., 2008; Lujambio et al., 2008; Saito et al., 2009).
Methylated miRNA genes have also been identified by analyzing genome‐wide DNA methylation in cancer cells. For instance, through the combined use of methylated CpG island amplification (MCA) and CpG island microarray analysis, the methylation of miR‐9‐1 was identified in pancreatic cancer (Omura et al., 2008). In addition, DNA methylation microarray analysis using the Infinium BeadChip revealed miR‐10b as a target of DNA methylation in gastric cancer. Recently, Yan et al. performed a genome‐wide methylome analysis entailing the deep sequencing of MBD (methylated DNA binding domain)‐isolated DNA in HCT116 cells, and identified a variety of methylated genes, including miR‐941, miR‐1237 and miR‐1247 (Yan et al., 2011).
As with protein‐coding genes, epigenetic regulation of miRNA genes is tightly associated with histone modifications, among which trimethylation of histone H3 lysine 4 (H3K4me3) is a hallmark of active transcription, whereas di‐ or trimethylation of histone H3 lysine 9 (H3K9me2 or H3K9me3) and trimethylation of lysine 27 (H3K27me3) are marks of repression (Figure 1). The combination of chromatin immunoprecipitation (ChIP)‐on‐chip and miRNA microarray analyses in prostate cancer cells revealed that miRNA expression is positively correlated with H3K4me3 and inversely correlated with H3K27me3 in the miRNA promoter regions (Ke et al., 2009). Analysis of histone modification using ChIP‐on‐chip in acute lymphoblastic leukemia (ALL) revealed that the CpG islands of 13 miRNA genes are associated with high H3K9me2 and low H3K4me3, suggesting these miRNAs are epigenetically silenced in ALL (Roman‐Gomez et al., 2009). Subsequent methylation analysis confirmed the hypermethylation of the selected miRNA genes, including the miR‐9 family, miR‐34 family and miR‐124 family genes. Hampering the identification of epigenetically dysregulated miRNA genes in cancer is the limited annotation of the primary transcripts of miRNA genes. Earlier studies have shown that H3K4me3 could be a useful mark with which to identify the active promoter regions of miRNA genes (Marson et al., 2008; Ozsolak et al., 2008). In addition, we recently assessed genome‐wide histone modification by performing deep sequencing (ChIP‐seq) in CRC cells (Suzuki et al., 2011). Using the approach, we identified the putative promoter regions for 174 primary miRNA genes, among which 37 were predicted to be targets of epigenetic silencing in CRC.
The epigenetic silencing of a subset of tumor suppressive miRNAs was discovered through functional screening. Kozaki and colleagues tested the anti‐proliferative effects of a panel of 327 synthetic miRNAs in oral and endometrial cancer cell lines (Tsuruta et al., 2011; Uesugi et al., 2011). Nearly 100 of the 327 miRNAs exerted growth suppressive effects, and approximately half of those were associated with CpG islands. By analyzing the DNA methylation and expression of the candidate miRNA genes, they identified miR‐218 and miR‐152 as targets of DNA methylation in oral and endometrial cancer, respectively.
3. miRNA genes epigenetically silenced in cancer
3.1. miR‐124 family
Epigenetic silencing of miR‐124 family genes was first reported in CRC cells (Lujambio et al., 2007), and is now known to be methylated in various types of cancer. miR‐124 is thought to exert tumor suppressor effects by targeting cyclin‐dependent kinase 6 (CDK6), and epigenetic silencing of miR‐124 leads to CDK6 activation and Rb phosphorylation (Agirre et al., 2009; Lujambio et al., 2007). Within the human genome, three independent loci (miR‐124‐1, miR‐124‐2 and miR‐124‐3) encode the identical mature miR‐124, and all are associated with CpG islands, which may be targets of hypermethylation in cancer (Lujambio et al., 2007). In primary CRC tissues, methylation of miR‐124 family genes is observed in more than 70% of the cases (Lujambio et al., 2007). Methylation of miR‐124 is also reported in hematological malignancies, including approximately half of all cases of ALL (Agirre et al., 2009) and non‐Hodgkin's lymphoma (Wong et al., 2011b). In ALL, moreover, miR‐124 methylation is associated with higher recurrence and mortality rates, and may be an independent prognostic factor for disease‐free and overall survival (Agirre et al., 2009). miR‐124 family genes are also frequently methylated in the gastric mucosa of Helicobacter pylori (H. pylori)‐positive healthy individuals, suggesting their methylation could be induced by chronic inflammation (Ando et al., 2009). Importantly, among H. pylori‐negative individuals, miR‐124 genes were more highly methylated in the noncancerous gastric mucosae of gastric cancer patients than in those of healthy individuals, suggesting miR‐124 methylation may be involved in an epigenetic field defect. In addition, miR‐124 family genes are also frequently methylated in cervical cancer, and ectopic expression of miR‐124 inhibits cell proliferation and migration (Wilting et al., 2010). miR‐124 methylation is also acquired during human papilloma virus‐mediated transformation, suggesting miR‐124 methylation may be a useful marker for detection of cervical cancer and high‐grade precursors.
3.2. miR‐34 family
Members of the miR‐34 gene family (miR‐34a, miR‐34b and miR‐34c) are direct targets of p53, and their ectopic expression in cancer cells induces cell cycle arrest and apoptosis (Bommer et al., 2007; He et al., 2007). Within the human genome, miR‐34a is located on chromosome 1p36, while miR‐34b and miR‐34c are co‐transcribed from a single transcription unit on chromosome 11q23, and both are targets of CpG island hypermethylation in oral, esophageal, gastric, colorectal, pancreatic, breast, lung and renal cancer; malignant mesothelioma; and melanoma (Chen et al., 2012; Kozaki et al., 2008; Kubo et al., 2011; Lodygin et al., 2008; Mazar et al., 2011b; Suzuki et al., 2010; Toyota et al., 2008; Vogt et al., 2011; Wang et al., 2011b). Methylation of miR‐34b/c has also been linked to cancer metastasis (Lujambio et al., 2008) and invasion (Watanabe et al., 2012). Similar to miR‐124, methylation of miR‐34b/c in the gastric mucosa is associated with H. pylori infection, and the noncancerous gastric mucosae of patients with multiple gastric cancers show higher levels of miR‐34b/c methylation than those of patients with a single gastric cancer, indicating its involvement in an epigenetic field defect (Suzuki et al., 2010). In non‐small cell lung cancer, methylation of miR‐34b/c is associated with a high probability of recurrence and poor overall survival (Wang et al., 2011b). In addition, methylation‐associated silencing of miR‐34c was recently shown to promote self‐renewal and epithelial–mesenchymal transition in breast tumor‐initiating cells (Yu et al., 2012). These findings, as well as its contribution to the p53 network, strongly imply that miR‐34 family members act as tumor suppressors in cancer. Introduction of miR‐34b/c into cancer cells leads to the downregulation of candidate target genes, including MET, cyclin‐dependent kinase 4 (CDK4), cyclin E2 (CCNE2) and MYC (Lujambio et al., 2008; Toyota et al., 2008). Likewise, restoration of endogenous miRNA expression through demethylation also downregulates target genes, suggesting miRNAs could be important targets for epigenetic cancer therapy (Toyota et al., 2008).
3.3. miR‐9 family
Methylation of the CpG island in the miR‐9‐1 promoter was first reported in breast and pancreatic cancer (Lehmann et al., 2008; Omura et al., 2008). Shortly thereafter, methylation of miR‐9 family genes (miR‐9‐1, miR‐9‐2 and miR‐9‐3) was also identified in several metastatic cancer cell lines (Lujambio et al., 2008). Consistent with that finding, methylation of miR‐9‐1 is reportedly associated with lymph node metastasis in CRC (Bandres et al., 2009), and methylation of miR‐9‐1 and miR‐9‐3 is correlated with metastatic recurrence of renal cell carcinoma (Hildebrandt et al., 2010). All three miR‐9 family genes are simultaneously methylated in gastric cancer, and ectopic expression of miR‐9 inhibits proliferation, migration and invasion by gastric cancer cells (Tsai et al., 2011a). miR‐9 has been shown to target fibroblast growth factor receptor 1 (FGFR1) and CDK6 in ALL (Rodriguez‐Otero et al., 2011) and caudal‐type homeobox 2 (CDX2) in gastric cancer cells (Rotkrua et al., 2011), suggesting a tumor suppressive function. Interestingly, xenoestrogen, which may increase ones risk of developing breast cancer, can induce methylation‐associated silencing of miR‐9‐3 in breast epithelial cells, indicating that methylation of this miRNA gene could be a hallmark of early breast cancer development (Hsu et al., 2009). In contrast to these findings, however, one recent study showed that miR‐9 is activated by MYC and MYCN in breast cancer, and that miR‐9 promotes metastasis through downregulation of E‐cadherin (CDH1) (Ma et al., 2010). These results are indicative of the functional complexity of miRNAs in cancer cells and suggest that miRNAs may exert opposite effects in different tissues or settings.
3.4. miR‐200 family and miR‐205
The miR‐200 gene family (miR‐200a, miR‐200b, miR‐200c, miR‐141 and miR‐429) and miR‐205 encode key regulators of epithelial–mesenchymal transition (EMT) that act by directly targeting zinc finger E‐box binding homeobox 1 (ZEB1) and ZEB2, which are transcriptional repressors that downregulate CDH1 (Gregory et al., 2008; Korpal et al., 2008; Park et al., 2008). Within the human genome, the miR‐200 family genes are grouped into two polycistronic units, miR‐200b/200a/429 and miR‐200c/141, located on chromosomes 1 and 12, respectively (Davalos et al., 2012). Expression of miR‐200c/141 is regulated by promoter CpG islands in normal mammary epithelial cells and fibroblasts, and their silencing is associated with aberrant methylation in breast and prostate cancer cells (Vrba et al., 2010). Methylation of miR‐200c/141 is tightly correlated with the invasive capacity of breast cancer cells, and induction of EMT by ectopic expression of Twist in immortalized human mammary epithelial cells is accompanied by increased methylation of miR‐200c/141 (Neves et al., 2010). Similarly, in non‐small cell lung cancer, promoter methylation is associated with loss of miR‐200c expression, which is in turn associated with poor differentiation, lymph node metastasis and weaker E‐cadherin expression (Ceppi et al., 2010). Davalos et al. demonstrated that the upstream CpG islands of both units (miR‐200b/200a/429 and miR‐200c/141) are unmethylated in cancer cells with epithelial features, but are both methylated and silenced in transformed cells with mesenchymal characteristics (Davalos et al., 2012). In bladder cancer, both units of the miR‐200 family and miR‐205 are coordinately silenced in association with promoter methylation (Wiklund et al., 2011). Epigenetic silencing of the miR‐200 family and miR‐205 was also observed in carcinogen‐treated lung epithelial cells, suggesting that induction of EMT through miRNA dysregulation occurs early during lung carcinogenesis (Tellez et al., 2011).
3.5. Other miRNA genes
Reduced expression of miR‐1 is reported in many malignancies (Bueno et al., 2008; Nasser et al., 2008; Rao et al., 2010), and methylation of the upstream CpG island and silencing of miR‐1‐1 have been reported in both hepatocellular carcinoma (HCC) (Datta et al., 2008) and CRC (Suzuki et al., 2011). Ectopic expression of miR‐1 in HCC cells inhibits cellular growth through suppression of its target genes, which include MET, forkhead box P1 (FOXP1) and histone deacetylase 4 (HDAC4) (Datta et al., 2008). In CRC cells, miR‐1 suppresses cellular proliferation, motility and invasion by targeting a number of genes, including annexin A2 (ANXA2) and brain‐derived neurotrophic factor (BDNF) (Suzuki et al., 2011), both of which are frequently overexpressed in cancer and are implicated in invasion and metastasis (Diaz et al., 2004; Douma et al., 2004; Emoto et al., 2001). Another recent study also provides evidence that expression of miR‐1 is downregulated in primary CRC, and that miR‐1 suppresses CRC cell proliferation and motility by targeting MET (Reid et al., 2012). Methylation of miR‐1‐1 is observed in approximately 80% of primary CRC tissue samples tested and in 70% of colorectal adenoma tissue samples, suggesting that methylation of miR‐1‐1 is an early event during colorectal tumorigenesis (Suzuki et al., 2011).
miR‐125b is a brain‐enriched miRNA and a good example of an miRNA that has opposite functions in different tumor types; that is, it can exert oncogenic or tumor suppressor effects, depending of the cellular context. Expression of miR‐125b is upregulated in several types of malignancies, including prostate cancer, but it is downregulated in breast cancer, where it acts as a tumor suppressor by directly targeting ETS1 (Zhang et al., 2011a). In breast cancer, the silencing of miR‐125b is associated with methylation of the miR‐125b‐1 promoter, and the weak expression of miR‐125b correlates with a poor prognosis.
Methylation of miR‐129‐2 has been reported in endometrial, esophageal, gastric, and colorectal cancer (Bandres et al., 2009; Chen et al., 2012; Huang et al., 2009; Shen et al., 2010). miR‐129‐2 targets SRY‐related high‐mobility group box 4 (SOX4), an oncogene frequently upregulated in malignancies, and an association between miR‐129‐2 methylation and SOX4 overexpression was found in both endometrial and gastric cancers (Huang et al., 2009; Shen et al., 2010). In endometrial cancer, miR‐129‐2 methylation is associated with microsatellite instability, MLH1 methylation and poor overall survival (Craig et al., 2011). Similarly, downregulated expression of miR‐129 is reportedly associated with poor clinicopathological features in gastric cancer (Tsai et al., 2011b).
Methylation of miR‐137 was first noted in oral cancer (Kozaki et al., 2008) and was subsequently reported in colon (Balaguer et al., 2010; Bandres et al., 2009) and gastric cancer (Chen et al., 2011). miR‐137 methylation is associated with a poorer survival rate among patients with head and neck squamous cell carcinoma (HNSCC) (Langevin et al., 2011), and was detected in oral rinses collected from HNSCC patients, suggesting its utility as a cancer biomarker (Langevin et al., 2010). Within cancer cells, miR‐137 targets CDK6, lysine‐specific demethylase 1 (LSD1) and cell division cycle 42 (CDC42), indicating it to be a tumor suppressor (Balaguer et al., 2010; Kozaki et al., 2008; Liu et al., 2011b), while in normal cells, miR‐137 regulates neuronal differentiation by targeting enhancer of zeste homolog 2 (EZH2) and mindbomb 1 (MIB1) (Smrt et al., 2010; Szulwach et al., 2010).
miR‐145 encodes a well‐known tumor suppressor, expression of which is reduced in a colorectal (Michael et al., 2003), breast (Iorio et al., 2005), ovarian (Iorio et al., 2007) and prostate cancers (Zaman et al., 2010). In breast and colon cancer cells, expression of miR‐145 is induced by p53, and miR‐145 directly targets MYC, suggesting that p53 represses MYC through induction of miR‐145 expression. miR‐145 also suppresses cancer cell proliferation, invasion and metastasis by targeting mucin 1 (MUC1), insulin receptor substrate‐1 (IRS‐1) and Fascin homolog 1 (FSCN1) (Fuse et al., 2011; Sachdeva and Mo, 2010; Shi et al., 2007). In prostate cancer, downregulation of miR‐145 expression is associated with an aggressive phenotype and a poor prognosis, and promoter methylation is associated with its silencing (Zaman et al., 2010). It was also recently shown that both DNA methylation and p53 mutation are major causes of diminished miR‐145 expression in prostate cancer (Suh et al., 2011).
Screening for epigenetically silenced miRNA genes in metastatic cancer cell lines revealed a set of genes that included miR‐148a (Lujambio et al., 2008). Introduction of miR‐148a into methylated cancer cells inhibited cell motility, growth and metastasis. miR‐148a is methylated in colorectal, breast, lung, head and neck cancers, and is associated with lymph node metastasis (Lujambio et al., 2008). Methylation‐induced silencing of miR‐148a is also seen in pancreatic cancer and preneoplastic pancreatic lesions (pancreatic intraepithelial neoplasia; PanIN), suggesting it is an early event in pancreatic carcinogenesis (Hanoun et al., 2010).
Aberrant activation of the proto‐oncogene KIT is observed in various malignancies, including acute myeloid leukemia (AML). Analysis of the 3′ UTR region of KIT suggested several miRNAs may serve as regulators; among those, methylation‐induced silencing of miR‐193a is causally related to KIT overexpression in AML (Gao et al., 2011). Interestingly, in HCC cells, miR‐193a dictates chemoresistance to 5‐fluorouracil by targeting serine/arginine‐rich splicing factor 2 (SRSF2) (Ma et al., 2012). Expression of miR‐193a is regulated by DNA methylation, and 5‐fluorouracil‐sensitive HCC cells show promoter methylation and reduced expression of miR‐196a, suggesting it could be an important prognostic indicator in HCC.
miR‐203 encodes a candidate tumor suppressor and is epigenetically silenced in oral cancer (Kozaki et al., 2008), hematopoietic malignancies (Bueno et al., 2008; Wong et al., 2011a) and HCC (Furuta et al., 2010). miR‐203 directly controls ABL1 expression, and it also targets the BCR‐ABL1 translocation protein induced by Philadelphia chromosome in chronic myelogenous leukemia and B cell ALL in children (Bueno et al., 2008). The CpG island of miR‐203 is specifically methylated in Philadelphia‐positive tumors, as compared to other hematopoietic malignancies, suggesting that epigenetic silencing of miR‐203 enhances the expression of the BCR‐ABL1 oncogene. Epigenetic silencing of miR‐203 has also been shown to activate ABL1 in H. pylori‐associated gastric B‐cell lymphoma of mucosa‐associated lymphoid tissue (MALT lymphoma) (Craig et al., 2011). In HCC cells, miR‐203 suppresses cellular growth and downregulates various target genes, including ATP‐binding cassette, subfamily E, member 1 (ABCE1) and CDK6 (Furuta et al., 2010).
Dysregulated expression of miR‐375 is frequently observed in cancers; for example, it is downregulated in esophageal (Kong et al., 2012), gastric (Tsukamoto et al., 2010), and head and neck cancers (Hui et al., 2011), whereas it is overexpressed in breast cancer (de Souza Rocha Simonini et al., 2010; Giricz et al., 2012). Methylation‐induced silencing of miR‐375 is reported in esophageal squamous cell carcinoma (Kong et al., 2012), HCC and melanoma (Mazar et al., 2011a). In the esophageal cancer cells, miR‐375 suppresses cell proliferation, motility, invasion and metastasis by targeting insulin‐like growth factor 1 receptor (IGF1R) (Kong et al., 2012). In esophageal and gastric cancer, miR‐375 reportedly exerts its tumor suppressive function by downregulating 3‐phosphoinositide‐dependent protein kinase 1 (PDK1), which in turn reduces Akt phosphorylation (Li et al., 2011; Tsukamoto et al., 2010). Interestingly, however, miR‐375 exerts oncogenic effects by targeting RAS dexamethasone‐induced 1 (RASD1), and its overexpression is associated with promoter hypomethylation (see below) (de Souza Rocha Simonini et al., 2010).
4. CpG island shore methylation and miRNA dysregulation
Most of the studies on the epigenetic silencing of miRNA genes in cancer described above have focused on CpG island hypermethylation. By contrast, one recent study demonstrated that CpG island shore methylation is frequently associated with miRNA downregulation in bladder cancer. CpG island shores are regions located within 2 kb of CpG islands, and their methylation strongly affects gene expression (Irizarry et al., 2009). Microarray analysis of miRNA expression coupled with demethylating treatment in bladder cancer cells revealed a number of epigenetically silenced miRNA genes (Dudziec et al., 2011). Interestingly, several miRNA genes are more frequently hypermethylated in the CpG island shore region than the CpG island itself (miR‐9, miR‐149, miR‐210, miR‐212, miR‐328, miR‐503, miR‐1224, miR‐1227 and miR‐1229). Methylation of these genes is associated with tumor grade, stage and prognosis, and the reduced expression of the silenced miRNAs is apparent in urine specimens from bladder cancer patients, suggesting their utility as diagnostic biomarkers.
5. Hypomethylation of miRNA genes in cancer
Although epigenetic dysregulation leads to the silencing of many miRNAs in cancer, several are upregulated though epigenetic mechanisms. The CpG island of let‐7a‐3 is heavily methylated in normal cells but is hypomethylated in lung adenocarcinoma, leading to its elevated expression (Brueckner et al., 2007). In lung cancer cells, let‐7a‐3 exerts oncogenic effects through actions on several genes involved in cell proliferation, adhesion and differentiation. Another study showed that elevated expression of miR‐375 in estrogen receptor α (ERα)‐positive breast cancer cells promotes tumor cell proliferation (de Souza Rocha Simonini et al., 2010). Similarly, overexpression of miR‐375 is caused by loss of repressive histone marks and DNA methylation, which leads to dissociation of the transcriptional repressor CTCF from the miR‐375 promoter, and the binding of ERα to its regulatory region. In addition, whereas expression of miR‐200a and miR‐200b is downregulated in many types of cancer, these genes are overexpressed in pancreatic cancer due to hypomethylation (Li et al., 2010). Moreover, the elevation of miR‐200a and miR‐200b in the serum of pancreatic cancer patients means they could potentially serve as diagnostic biomarkers. miR‐196 family genes (miR‐196a and miR‐196b) are located within the HOX gene cluster and are often overexpressed in tumors, which is indicative of their oncogenic role (Popovic et al., 2009). miR‐196b is embedded within a CpG island, and its overexpression in gastric cancer is associated with its hypomethylation (Tsai et al., 2010). In HCC, expression of miR‐191 is frequently elevated due to hypomethylation of the gene locus (He et al., 2011). miR‐191 directly targets tissue inhibitor of metalloproteinase 3 (TIMP3), and its elevated expression in HCC cells induces transition to mesenchymal‐like cells.
6. Clinical implications of epigenetically silenced miRNAs
A number of studies have demonstrated that the miRNA expression profile is potentially useful for detecting cancers and predicting outcomes. In addition to the altered expression, epigenetically silenced miRNA genes also exhibit aberrant DNA methylation, which could also be a useful marker for cancer diagnosis. For example, as mentioned above, methylation of miR‐124 family genes is an independent prognostic factor for disease‐free and overall survival (Agirre et al., 2009). A more comprehensive analysis of epigenetically silenced miRNA genes in ALL has identified methylation of 13 miRNA genes (miR‐9‐1, miR‐9‐2, miR‐9‐3, miR‐10b, miR‐34b/c, miR124‐1, miR‐124‐2, miR‐124‐3, miR‐132/212, miR‐196b, miR‐203) (Roman‐Gomez et al., 2009). ALLs with at least one methylated miRNA genes, which account for 65% of all ALLs, show significantly poorer disease‐free and overall survival than the unmethylated group, suggesting miRNA gene methylation is an important prognostic factor predictive of disease outcome. Similarly, methylation of multiple miRNA genes is associated with larger tumor size and poorer progression‐free survival in non‐small cell lung cancer (Kitano et al., 2011).
As mentioned above, miR‐34b/c is one of the most frequently methylated miRNA genes among tumors. The CpG island of miR‐34b/c is methylated in more than 90% of primary CRCs, and methylation was detected in 75% of fecal specimens from CRC patients and in 16% of specimens from high‐grade dysplasia patients, suggesting miR‐34b/c methylation could be a useful feces‐based screening marker (Kalimutho et al., 2011). In addition, we recently reported that methylation of a panel of genes, including miR‐34b/c, in mucosal wash fluid collected during colonoscopy could be a useful biomarker for predicting the invasiveness of CRCs (Kamimae et al., 2011).
Given that some epigenetically silenced miRNAs appear to have tumor suppressive potential, restoration of their expression may be an effective strategy for treating cancer. It has been demonstrated experimentally that re‐expression of methylation‐silenced miRNAs through treatment with a demethylating agent leads to downregulation of target oncogenes and suppression of tumor growth (Lujambio et al., 2007; Toyota et al., 2008). In addition, the antitumor effects of miRNA replacement using an oligonucleotide mimic or a miRNA expression vector has been tested in numerous studies (Henry et al., 2011). For example, restoration of miR‐34 family expression in pancreatic cancer cells led to a significant reduction in the fraction of CD44+/CD133+ tumor‐initiating cells and inhibits tumor growth in vitro and in vivo (Ji et al., 2009). In mouse xenograft models of lung cancer, systemic delivery of a miR‐34a mimic using a lipid‐based delivery vehicle inhibited tumor growth (Wiggins et al., 2010). miR‐34a also reportedly inhibits prostate cancer stem cells, and the therapeutic efficacy of its systemic delivery was confirmed in a mouse xenograft model (Liu et al., 2011a). Moreover, using a xenograft model of CRC, polyethylenimine‐mediated delivery of miR‐145 and miR‐33a was shown to downregulate expression of MYC and ERK5, and to suppress tumor growth (Ibrahim et al., 2011).
7. miRNA dysregulation and aberrant DNA methylation
In contrast to the observations described above, several lines of evidence support the idea that dysregulation of miRNA can lead to aberrant DNA methylation in cancer. For instance, using a CRC cell line with hypomorphic DICER (Cummins et al., 2006), we showed that DICER is required to maintain CpG island methylation of several gene promoters in cancer (Ting et al., 2008). To date, several miRNAs able to regulate DNMT genes are reportedly downregulated in cancer. The miR‐29 family (miR‐29a, miR‐29b and miR‐29c), which is downregulated in lung cancer, directly targets DNMT3A and DNMT3B (Fabbri et al., 2007). Ectopic expression of miR‐29 family in lung cancer cells restores expression of methylation‐silenced tumor suppressor genes, including fragile histidine triad (FHIT) and WW domain containing oxidoreductase (WWOX). In CRC, miR‐143 is frequently downregulated, and experimental evidence suggests that miR‐143 targets DNMT3A in CRC cells (Ng et al., 2009). In addition, downregulated expression of miR‐152 in HBV‐related HCC correlates with increased expression of DNMT1 (Huang et al., 2010). Forced expression of miR‐152 in liver cell lines reduces DNMT1 expression and in turn global DNA methylation, whereas inhibition of miR‐152 causes global DNA hypermethylation and increased methylation of the glutathione S‐transferase pi 1 (GSTP1) and CDH1 promoter regions. Similarly, in cholangiocarcinoma cells, DNMT1 is targeted by miR‐148a and miR‐152, and their ectopic expression suppresses DNMT1 and induces expression of the tumor suppressor genes Ras association domain family 1A (RASSF1A) and p16 (Braconi et al., 2010). Recently, miR‐342 was found to be downregulated in CRC cells, and restoration of its expression downregulated DNMT1 and reactivated expression of ADAM metallopeptidase domain 23 (ADAM23), histidine triad nucleotide binding protein 1 (HINT1), RASSF1A and reversion‐inducing‐cysteine‐rich protein with kazal motifs (RECK) through demethylation of their promoter regions (Wang et al., 2011a). And in glioma, miR‐185 targets DNMT1 to regulate the methylation of several gene promoters (Zhang et al., 2011b). Collectively, these results suggest that dysregulation of specific miRNAs may be causally related to aberrant methylation of promoter CpG islands.
8. Concluding remarks
In this review, we highlighted the relationship between epigenetic alteration and dysregulation of miRNAs in cancer. Aberrant DNA methylation and histone modification are major mechanisms underlying miRNA dysregulation in cancer, and methylation of a subset of miRNA genes may be useful biomarkers for detecting cancer and/or predicting clinical outcome. Moreover, replacement of silenced tumor‐suppressive miRNAs in cancer cells could be a promising strategy for cancer treatment. We anticipate that further studies of the cancer epigenome and miRNAs will lead to the discovery of a variety of novel biomarkers and potential therapeutic targets.
Conflict of interest
All authors disclose no conflict of interest.
Acknowledgements
We thank Dr. William Goldman for editing the manuscript. HS is supported by a Grant‐in‐Aid for Scientific Research (C) from the Japan Society for Promotion of Science, a Grant‐in‐Aid for the Third‐term Comprehensive 10‐year Strategy for Cancer Control from the Ministry of Health, Labor, and Welfare, Japan and the A3 Foresight Program from the Japan Society for Promotion of Science.
Suzuki Hiromu, Maruyama Reo, Yamamoto Eiichiro, Kai Masahiro, (2012), DNA methylation and microRNA dysregulation in cancer, Molecular Oncology, 6, doi: 10.1016/j.molonc.2012.07.007.
References
- Agirre, X. , Vilas-Zornoza, A. , Jimenez-Velasco, A. , Martin-Subero, J.I. , Cordeu, L. , Garate, L. , San Jose-Eneriz, E. , Abizanda, G. , Rodriguez-Otero, P. , Fortes, P. , Rifon, J. , Bandres, E. , Calasanz, M.J. , Martin, V. , Heiniger, A. , Torres, A. , Siebert, R. , Roman-Gomez, J. , Prosper, F. , 2009. Epigenetic silencing of the tumor suppressor microRNA Hsa-miR-124a regulates CDK6 expression and confers a poor prognosis in acute lymphoblastic leukemia. Cancer Res. 69, 4443–4453. [DOI] [PubMed] [Google Scholar]
- Ando, T. , Yoshida, T. , Enomoto, S. , Asada, K. , Tatematsu, M. , Ichinose, M. , Sugiyama, T. , Ushijima, T. , 2009. DNA methylation of microRNA genes in gastric mucosae of gastric cancer patients: its possible involvement in the formation of epigenetic field defect. Int. J. Cancer. 124, 2367–2374. [DOI] [PubMed] [Google Scholar]
- Balaguer, F. , Link, A. , Lozano, J.J. , Cuatrecasas, M. , Nagasaka, T. , Boland, C.R. , Goel, A. , 2010. Epigenetic silencing of miR-137 is an early event in colorectal carcinogenesis. Cancer Res. 70, 6609–6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandres, E. , Agirre, X. , Bitarte, N. , Ramirez, N. , Zarate, R. , Roman-Gomez, J. , Prosper, F. , Garcia-Foncillas, J. , 2009. Epigenetic regulation of microRNA expression in colorectal cancer. Int. J. Cancer. 125, 2737–2743. [DOI] [PubMed] [Google Scholar]
- Bommer, G.T. , Gerin, I. , Feng, Y. , Kaczorowski, A.J. , Kuick, R. , Love, R.E. , Zhai, Y. , Giordano, T.J. , Qin, Z.S. , Moore, B.B. , MacDougald, O.A. , Cho, K.R. , Fearon, E.R. , 2007. p53-mediated activation of miRNA34 candidate tumor-suppressor genes. Curr. Biol. 17, 1298–1307. [DOI] [PubMed] [Google Scholar]
- Braconi, C. , Huang, N. , Patel, T. , 2010. MicroRNA-dependent regulation of DNA methyltransferase-1 and tumor suppressor gene expression by interleukin-6 in human malignant cholangiocytes. Hepatology. 51, 881–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brueckner, B. , Stresemann, C. , Kuner, R. , Mund, C. , Musch, T. , Meister, M. , Sultmann, H. , Lyko, F. , 2007. The human let-7a-3 locus contains an epigenetically regulated microRNA gene with oncogenic function. Cancer Res. 67, 1419–1423. [DOI] [PubMed] [Google Scholar]
- Bueno, M.J. , Perez de Castro, I. , Gomez de Cedron, M. , Santos, J. , Calin, G.A. , Cigudosa, J.C. , Croce, C.M. , Fernandez-Piqueras, J. , Malumbres, M. , 2008. Genetic and epigenetic silencing of microRNA-203 enhances ABL1 and BCR-ABL1 oncogene expression. Cancer Cell. 13, 496–506. [DOI] [PubMed] [Google Scholar]
- Calin, G.A. , Dumitru, C.D. , Shimizu, M. , Bichi, R. , Zupo, S. , Noch, E. , Aldler, H. , Rattan, S. , Keating, M. , Rai, K. , Rassenti, L. , Kipps, T. , Negrini, M. , Bullrich, F. , Croce, C.M. , 2002. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. U. S. A. 99, 15524–15529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceppi, P. , Mudduluru, G. , Kumarswamy, R. , Rapa, I. , Scagliotti, G.V. , Papotti, M. , Allgayer, H. , 2010. Loss of miR-200c expression induces an aggressive, invasive, and chemoresistant phenotype in non-small cell lung cancer. Mol. Cancer Res. 8, 1207–1216. [DOI] [PubMed] [Google Scholar]
- Chen, Q. , Chen, X. , Zhang, M. , Fan, Q. , Luo, S. , Cao, X. , 2011. miR-137 is frequently down-regulated in gastric cancer and is a negative regulator of Cdc42. Dig. Dis. Sci. 56, 2009–2016. [DOI] [PubMed] [Google Scholar]
- Chen, X. , Hu, H. , Guan, X. , Xiong, G. , Wang, Y. , Wang, K. , Li, J. , Xu, X. , Yang, K. , Bai, Y. , 2012. CpG island methylation status of miRNAs in esophageal squamous cell carcinoma. Int. J. Cancer. 130, 1607–1613. [DOI] [PubMed] [Google Scholar]
- Craig, V.J. , Cogliatti, S.B. , Rehrauer, H. , Wundisch, T. , Muller, A. , 2011. Epigenetic silencing of microRNA-203 dysregulates ABL1 expression and drives Helicobacter-associated gastric lymphomagenesis. Cancer Res. 71, 3616–3624. [DOI] [PubMed] [Google Scholar]
- Croce, C.M. , 2009. Causes and consequences of microRNA dysregulation in cancer. Nat. Rev. Genet. 10, 704–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummins, J.M. , He, Y. , Leary, R.J. , Pagliarini, R. , Diaz, L.A. , Sjoblom, T. , Barad, O. , Bentwich, Z. , Szafranska, A.E. , Labourier, E. , Raymond, C.K. , Roberts, B.S. , Juhl, H. , Kinzler, K.W. , Vogelstein, B. , Velculescu, V.E. , 2006. The colorectal microRNAome. Proc. Natl. Acad. Sci. U. S. A. 103, 3687–3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta, J. , Kutay, H. , Nasser, M.W. , Nuovo, G.J. , Wang, B. , Majumder, S. , Liu, C.G. , Volinia, S. , Croce, C.M. , Schmittgen, T.D. , Ghoshal, K. , Jacob, S.T. , 2008. Methylation mediated silencing of microRNA-1 gene and its role in hepatocellular carcinogenesis. Cancer Res. 68, 5049–5058. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Davalos, V. , Moutinho, C. , Villanueva, A. , Boque, R. , Silva, P. , Carneiro, F. , Esteller, M. , 2012. Dynamic epigenetic regulation of the microRNA-200 family mediates epithelial and mesenchymal transitions in human tumorigenesis. Oncogene. 31, 2062–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Souza Rocha Simonini, P. , Breiling, A. , Gupta, N. , Malekpour, M. , Youns, M. , Omranipour, R. , Malekpour, F. , Volinia, S. , Croce, C.M. , Najmabadi, H. , Diederichs, S. , Sahin, O. , Mayer, D. , Lyko, F. , Hoheisel, J.D. , Riazalhosseini, Y. , 2010. Epigenetically deregulated microRNA-375 is involved in a positive feedback loop with estrogen receptor alpha in breast cancer cells. Cancer Res. 70, 9175–9184. [DOI] [PubMed] [Google Scholar]
- Diaz, V.M. , Hurtado, M. , Thomson, T.M. , Reventos, J. , Paciucci, R. , 2004. Specific interaction of tissue-type plasminogen activator (t-PA) with annexin II on the membrane of pancreatic cancer cells activates plasminogen and promotes invasion in vitro. Gut. 53, 993–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douma, S. , Van Laar, T. , Zevenhoven, J. , Meuwissen, R. , Van Garderen, E. , Peeper, D.S. , 2004. Suppression of anoikis and induction of metastasis by the neurotrophic receptor TrkB. Nature. 430, 1034–1039. [DOI] [PubMed] [Google Scholar]
- Dudziec, E. , Miah, S. , Choudhry, H.M. , Owen, H.C. , Blizard, S. , Glover, M. , Hamdy, F.C. , Catto, J.W. , 2011. Hypermethylation of CpG islands and shores around specific microRNAs and mirtrons is associated with the phenotype and presence of bladder cancer. Clin. Cancer Res. 17, 1287–1296. [DOI] [PubMed] [Google Scholar]
- Emoto, K. , Yamada, Y. , Sawada, H. , Fujimoto, H. , Ueno, M. , Takayama, T. , Kamada, K. , Naito, A. , Hirao, S. , Nakajima, Y. , 2001. Annexin II overexpression correlates with stromal tenascin-C overexpression: a prognostic marker in colorectal carcinoma. Cancer. 92, 1419–1426. [DOI] [PubMed] [Google Scholar]
- Esquela-Kerscher, A. , Slack, F.J. , 2006. Oncomirs - microRNAs with a role in cancer. Nat. Rev. Cancer. 6, 259–269. [DOI] [PubMed] [Google Scholar]
- Esteller, M. , 2011. Non-coding RNAs in human disease. Nat. Rev. Genet. 12, 861–874. [DOI] [PubMed] [Google Scholar]
- Fabbri, M. , Garzon, R. , Cimmino, A. , Liu, Z. , Zanesi, N. , Callegari, E. , Liu, S. , Alder, H. , Costinean, S. , Fernandez-Cymering, C. , Volinia, S. , Guler, G. , Morrison, C.D. , Chan, K.K. , Marcucci, G. , Calin, G.A. , Huebner, K. , Croce, C.M. , 2007. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc. Natl. Acad. Sci. U. S. A. 104, 15805–15810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg, A.P. , Tycko, B. , 2004. The history of cancer epigenetics. Nat. Rev. Cancer. 4, 143–153. [DOI] [PubMed] [Google Scholar]
- Furuta, M. , Kozaki, K.I. , Tanaka, S. , Arii, S. , Imoto, I. , Inazawa, J. , 2010. miR-124 and miR-203 are epigenetically silenced tumor-suppressive microRNAs in hepatocellular carcinoma. Carcinogenesis. 31, 766–776. [DOI] [PubMed] [Google Scholar]
- Fuse, M. , Nohata, N. , Kojima, S. , Sakamoto, S. , Chiyomaru, T. , Kawakami, K. , Enokida, H. , Nakagawa, M. , Naya, Y. , Ichikawa, T. , Seki, N. , 2011. Restoration of miR-145 expression suppresses cell proliferation, migration and invasion in prostate cancer by targeting FSCN1. Int. J. Oncol. 38, 1093–1101. [DOI] [PubMed] [Google Scholar]
- Gao, X.N. , Lin, J. , Li, Y.H. , Gao, L. , Wang, X.R. , Wang, W. , Kang, H.Y. , Yan, G.T. , Wang, L.L. , Yu, L. , 2011. MicroRNA-193a represses c-kit expression and functions as a methylation-silenced tumor suppressor in acute myeloid leukemia. Oncogene. 30, 3416–3428. [DOI] [PubMed] [Google Scholar]
- Giricz, O. , Reynolds, P.A. , Ramnauth, A. , Liu, C. , Wang, T. , Stead, L. , Childs, G. , Rohan, T. , Shapiro, N. , Fineberg, S. , Kenny, P.A. , Loudig, O. , 2012. Hsa-miR-375 is differentially expressed during breast lobular neoplasia and promotes loss of mammary acinar polarity. J. Pathol. 226, 108–119. [DOI] [PubMed] [Google Scholar]
- Gregory, P.A. , Bert, A.G. , Paterson, E.L. , Barry, S.C. , Tsykin, A. , Farshid, G. , Vadas, M.A. , Khew-Goodall, Y. , Goodall, G.J. , 2008. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat. Cell Biol. 10, 593–601. [DOI] [PubMed] [Google Scholar]
- Hanoun, N. , Delpu, Y. , Suriawinata, A.A. , Bournet, B. , Bureau, C. , Selves, J. , Tsongalis, G.J. , Dufresne, M. , Buscail, L. , Cordelier, P. , Torrisani, J. , 2010. The silencing of microRNA 148a production by DNA hypermethylation is an early event in pancreatic carcinogenesis. Clin. Chem. 56, 1107–1118. [DOI] [PubMed] [Google Scholar]
- Hashimoto, Y. , Akiyama, Y. , Otsubo, T. , Shimada, S. , Yuasa, Y. , 2010. Involvement of epigenetically silenced microRNA-181c in gastric carcinogenesis. Carcinogenesis. 31, 777–784. [DOI] [PubMed] [Google Scholar]
- He, L. , He, X. , Lim, L.P. , de Stanchina, E. , Xuan, Z. , Liang, Y. , Xue, W. , Zender, L. , Magnus, J. , Ridzon, D. , Jackson, A.L. , Linsley, P.S. , Chen, C. , Lowe, S.W. , Cleary, M.A. , Hannon, G.J. , 2007. A microRNA component of the p53 tumour suppressor network. Nature. 447, 1130–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, Y. , Cui, Y. , Wang, W. , Gu, J. , Guo, S. , Ma, K. , Luo, X. , 2011. Hypomethylation of the hsa-miR-191 locus causes high expression of hsa-mir-191 and promotes the epithelial-to-mesenchymal transition in hepatocellular carcinoma. Neoplasia. 13, 841–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry, J.C. , Azevedo-Pouly, A.C. , Schmittgen, T.D. , 2011. MicroRNA replacement therapy for cancer. Pharm. Res. 28, 3030–3042. [DOI] [PubMed] [Google Scholar]
- Hildebrandt, M.A. , Gu, J. , Lin, J. , Ye, Y. , Tan, W. , Tamboli, P. , Wood, C.G. , Wu, X. , 2010. Hsa-miR-9 methylation status is associated with cancer development and metastatic recurrence in patients with clear cell renal cell carcinoma. Oncogene. 29, 5724–5728. [DOI] [PubMed] [Google Scholar]
- Hsu, P.Y. , Deatherage, D.E. , Rodriguez, B.A. , Liyanarachchi, S. , Weng, Y.I. , Zuo, T. , Liu, J. , Cheng, A.S. , Huang, T.H. , 2009. Xenoestrogen-induced epigenetic repression of microRNA-9-3 in breast epithelial cells. Cancer Res. 69, 5936–5945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, J. , Wang, Y. , Guo, Y. , Sun, S. , 2010. Down-regulated microRNA-152 induces aberrant DNA methylation in hepatitis B virus-related hepatocellular carcinoma by targeting DNA methyltransferase 1. Hepatology. 52, 60–70. [DOI] [PubMed] [Google Scholar]
- Huang, Y.W. , Liu, J.C. , Deatherage, D.E. , Luo, J. , Mutch, D.G. , Goodfellow, P.J. , Miller, D.S. , Huang, T.H. , 2009. Epigenetic repression of microRNA-129-2 leads to overexpression of SOX4 oncogene in endometrial cancer. Cancer Res. 69, 9038–9046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui, A.B. , Bruce, J.P. , Alajez, N.M. , Shi, W. , Yue, S. , Perez-Ordonez, B. , Xu, W. , O'Sullivan, B. , Waldron, J. , Cummings, B. , Gullane, P. , Siu, L. , Liu, F.F. , 2011. Significance of dysregulated metadherin and microRNA-375 in head and neck cancer. Clin. Cancer Res. 17, 7539–7550. [DOI] [PubMed] [Google Scholar]
- Ibrahim, A.F. , Weirauch, U. , Thomas, M. , Grunweller, A. , Hartmann, R.K. , Aigner, A. , 2011. MicroRNA replacement therapy for miR-145 and miR-33a is efficacious in a model of colon carcinoma. Cancer Res. 71, 5214–5224. [DOI] [PubMed] [Google Scholar]
- Iorio, M.V. , Ferracin, M. , Liu, C.G. , Veronese, A. , Spizzo, R. , Sabbioni, S. , Magri, E. , Pedriali, M. , Fabbri, M. , Campiglio, M. , Menard, S. , Palazzo, J.P. , Rosenberg, A. , Musiani, P. , Volinia, S. , Nenci, I. , Calin, G.A. , Querzoli, P. , Negrini, M. , Croce, C.M. , 2005. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 65, 7065–7070. [DOI] [PubMed] [Google Scholar]
- Iorio, M.V. , Visone, R. , Di Leva, G. , Donati, V. , Petrocca, F. , Casalini, P. , Taccioli, C. , Volinia, S. , Liu, C.G. , Alder, H. , Calin, G.A. , Menard, S. , Croce, C.M. , 2007. MicroRNA signatures in human ovarian cancer. Cancer Res. 67, 8699–8707. [DOI] [PubMed] [Google Scholar]
- Irizarry, R.A. , Ladd-Acosta, C. , Wen, B. , Wu, Z. , Montano, C. , Onyango, P. , Cui, H. , Gabo, K. , Rongione, M. , Webster, M. , Ji, H. , Potash, J.B. , Sabunciyan, S. , Feinberg, A.P. , 2009. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat. Genet. 41, 178–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji, Q. , Hao, X. , Zhang, M. , Tang, W. , Yang, M. , Li, L. , Xiang, D. , Desano, J.T. , Bommer, G.T. , Fan, D. , Fearon, E.R. , Lawrence, T.S. , Xu, L. , 2009. MicroRNA miR-34 inhibits human pancreatic cancer tumor-initiating cells. PLoS One. 4, e6816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, S.M. , Grosshans, H. , Shingara, J. , Byrom, M. , Jarvis, R. , Cheng, A. , Labourier, E. , Reinert, K.L. , Brown, D. , Slack, F.J. , 2005. RAS is regulated by the let-7 microRNA family. Cell. 120, 635–647. [DOI] [PubMed] [Google Scholar]
- Jones, P.A. , Baylin, S.B. , 2002. The fundamental role of epigenetic events in cancer. Nat. Rev. Genet. 3, 415–428. [DOI] [PubMed] [Google Scholar]
- Jones, P.A. , Baylin, S.B. , 2007. The epigenomics of cancer. Cell. 128, 683–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalimutho, M. , Di Cecilia, S. , Del Vecchio Blanco, G. , Roviello, F. , Sileri, P. , Cretella, M. , Formosa, A. , Corso, G. , Marrelli, D. , Pallone, F. , Federici, G. , Bernardini, S. , 2011. Epigenetically silenced miR-34b/c as a novel faecal-based screening marker for colorectal cancer. Br. J. Cancer. 104, 1770–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamimae, S. , Yamamoto, E. , Yamano, H.O. , Nojima, M. , Suzuki, H. , Ashida, M. , Hatahira, T. , Sato, A. , Kimura, T. , Yoshikawa, K. , Harada, T. , Hayashi, S. , Takamaru, H. , Maruyama, R. , Kai, M. , Nishiwaki, M. , Sugai, T. , Sasaki, Y. , Tokino, T. , Shinomura, Y. , Imai, K. , Toyota, M. , 2011. Epigenetic alteration of DNA in mucosal wash fluid predicts invasiveness of colorectal tumors. Cancer Prev. Res. 4, 674–683. [DOI] [PubMed] [Google Scholar]
- Ke, X.S. , Qu, Y. , Rostad, K. , Li, W.C. , Lin, B. , Halvorsen, O.J. , Haukaas, S.A. , Jonassen, I. , Petersen, K. , Goldfinger, N. , Rotter, V. , Akslen, L.A. , Oyan, A.M. , Kalland, K.H. , 2009. Genome-wide profiling of histone h3 lysine 4 and lysine 27 trimethylation reveals an epigenetic signature in prostate carcinogenesis. PLoS One. 4, e4687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitano, K. , Watanabe, K. , Emoto, N. , Kage, H. , Hamano, E. , Nagase, T. , Sano, A. , Murakawa, T. , Nakajima, J. , Goto, A. , Fukayama, M. , Yatomi, Y. , Ohishi, N. , Takai, D. , 2011. CpG island methylation of microRNAs is associated with tumor size and recurrence of non-small-cell lung cancer. Cancer Sci. 102, 2126–2131. [DOI] [PubMed] [Google Scholar]
- Kong, K.L. , Kwong, D.L. , Chan, T.H. , Law, S.Y. , Chen, L. , Li, Y. , Qin, Y.R. , Guan, X.Y. , 2012. MicroRNA-375 inhibits tumour growth and metastasis in oesophageal squamous cell carcinoma through repressing insulin-like growth factor 1 receptor. Gut. 61, 33–42. [DOI] [PubMed] [Google Scholar]
- Korpal, M. , Lee, E.S. , Hu, G. , Kang, Y. , 2008. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J. Biol. Chem. 283, 14910–14914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozaki, K. , Imoto, I. , Mogi, S. , Omura, K. , Inazawa, J. , 2008. Exploration of tumor-suppressive microRNAs silenced by DNA hypermethylation in oral cancer. Cancer Res. 68, 2094–2105. [DOI] [PubMed] [Google Scholar]
- Kubo, T. , Toyooka, S. , Tsukuda, K. , Sakaguchi, M. , Fukazawa, T. , Soh, J. , Asano, H. , Ueno, T. , Muraoka, T. , Yamamoto, H. , Nasu, Y. , Kishimoto, T. , Pass, H.I. , Matsui, H. , Huh, N.H. , Miyoshi, S. , 2011. Epigenetic silencing of microRNA-34b/c plays an important role in the pathogenesis of malignant pleural mesothelioma. Clin. Cancer Res. 17, 4965–4974. [DOI] [PubMed] [Google Scholar]
- Langevin, S.M. , Stone, R.A. , Bunker, C.H. , Grandis, J.R. , Sobol, R.W. , Taioli, E. , 2010. MicroRNA-137 promoter methylation in oral rinses from patients with squamous cell carcinoma of the head and neck is associated with gender and body mass index. Carcinogenesis. 31, 864–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langevin, S.M. , Stone, R.A. , Bunker, C.H. , Lyons-Weiler, M.A. , Laframboise, W.A. , Kelly, L. , Seethala, R.R. , Grandis, J.R. , Sobol, R.W. , Taioli, E. , 2011. MicroRNA-137 promoter methylation is associated with poorer overall survival in patients with squamous cell carcinoma of the head and neck. Cancer. 117, 1454–1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann, U. , Hasemeier, B. , Christgen, M. , Muller, M. , Romermann, D. , Langer, F. , Kreipe, H. , 2008. Epigenetic inactivation of microRNA gene hsa-mir-9-1 in human breast cancer. J. Pathol. 214, 17–24. [DOI] [PubMed] [Google Scholar]
- Li, A. , Omura, N. , Hong, S.M. , Vincent, A. , Walter, K. , Griffith, M. , Borges, M. , Goggins, M. , 2010. Pancreatic cancers epigenetically silence SIP1 and hypomethylate and overexpress miR-200a/200b in association with elevated circulating miR-200a and miR-200b levels. Cancer Res. 70, 5226–5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X. , Lin, R. , Li, J. , 2011. Epigenetic silencing of microRNA-375 regulates PDK1 expression in esophageal cancer. Dig. Dis. Sci. 56, 2849–2856. [DOI] [PubMed] [Google Scholar]
- Liu, C. , Kelnar, K. , Liu, B. , Chen, X. , Calhoun-Davis, T. , Li, H. , Patrawala, L. , Yan, H. , Jeter, C. , Honorio, S. , Wiggins, J.F. , Bader, A.G. , Fagin, R. , Brown, D. , Tang, D.G. , 2011. The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nat. Med. 17, 211–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, M. , Lang, N. , Qiu, M. , Xu, F. , Li, Q. , Tang, Q. , Chen, J. , Chen, X. , Zhang, S. , Liu, Z. , Zhou, J. , Zhu, Y. , Deng, Y. , Zheng, Y. , Bi, F. , 2011. miR-137 targets Cdc42 expression, induces cell cycle G1 arrest and inhibits invasion in colorectal cancer cells. Int. J. Cancer. 128, 1269–1279. [DOI] [PubMed] [Google Scholar]
- Lodygin, D. , Tarasov, V. , Epanchintsev, A. , Berking, C. , Knyazeva, T. , Korner, H. , Knyazev, P. , Diebold, J. , Hermeking, H. , 2008. Inactivation of miR-34a by aberrant CpG methylation in multiple types of cancer. Cell Cycle. 7, 2591–2600. [DOI] [PubMed] [Google Scholar]
- Lopez-Serra, P. , Esteller, M. , 2012. DNA methylation-associated silencing of tumor-suppressor microRNAs in cancer. Oncogene. 31, 1609–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lujambio, A. , Calin, G.A. , Villanueva, A. , Ropero, S. , Sanchez-Cespedes, M. , Blanco, D. , Montuenga, L.M. , Rossi, S. , Nicoloso, M.S. , Faller, W.J. , Gallagher, W.M. , Eccles, S.A. , Croce, C.M. , Esteller, M. , 2008. A microRNA DNA methylation signature for human cancer metastasis. Proc. Natl. Acad. Sci. U. S. A. 105, 13556–13561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lujambio, A. , Ropero, S. , Ballestar, E. , Fraga, M.F. , Cerrato, C. , Setien, F. , Casado, S. , Suarez-Gauthier, A. , Sanchez-Cespedes, M. , Git, A. , Spiteri, I. , Das, P.P. , Caldas, C. , Miska, E. , Esteller, M. , 2007. Genetic unmasking of an epigenetically silenced microRNA in human cancer cells. Cancer Res. 67, 1424–1429. [DOI] [PubMed] [Google Scholar]
- Ma, K. , He, Y. , Zhang, H. , Fei, Q. , Niu, D. , Wang, D. , Ding, X. , Xu, H. , Chen, X. , Zhu, J. , 2012. DNA methylation-regulated miR-193a-3p dictates resistance of hepatocellular carcinoma to 5-fluorouracil via repression of SRSF2 expression. J. Biol. Chem. 287, 5639–5649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, L. , Young, J. , Prabhala, H. , Pan, E. , Mestdagh, P. , Muth, D. , Teruya-Feldstein, J. , Reinhardt, F. , Onder, T.T. , Valastyan, S. , Westermann, F. , Speleman, F. , Vandesompele, J. , Weinberg, R.A. , 2010. miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat. Cell. Biol. 12, 247–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marson, A. , Levine, S.S. , Cole, M.F. , Frampton, G.M. , Brambrink, T. , Johnstone, S. , Guenther, M.G. , Johnston, W.K. , Wernig, M. , Newman, J. , Calabrese, J.M. , Dennis, L.M. , Volkert, T.L. , Gupta, S. , Love, J. , Hannett, N. , Sharp, P.A. , Bartel, D.P. , Jaenisch, R. , Young, R.A. , 2008. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 134, 521–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazar, J. , DeBlasio, D. , Govindarajan, S.S. , Zhang, S. , Perera, R.J. , 2011. Epigenetic regulation of microRNA-375 and its role in melanoma development in humans. FEBS Lett. 585, 2467–2476. [DOI] [PubMed] [Google Scholar]
- Mazar, J. , Khaitan, D. , DeBlasio, D. , Zhong, C. , Govindarajan, S.S. , Kopanathi, S. , Zhang, S. , Ray, A. , Perera, R.J. , 2011. Epigenetic regulation of microRNA genes and the role of miR-34b in cell invasion and motility in human melanoma. PLoS One. 6, e24922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael, M.Z. , SM, O.C. , van Holst Pellekaan, N.G. , Young, G.P. , James, R.J. , 2003. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol. Cancer Res. 1, 882–891. [PubMed] [Google Scholar]
- Nasser, M.W. , Datta, J. , Nuovo, G. , Kutay, H. , Motiwala, T. , Majumder, S. , Wang, B. , Suster, S. , Jacob, S.T. , Ghoshal, K. , 2008. Down-regulation of micro-RNA-1 (miR-1) in lung cancer. Suppression of tumorigenic property of lung cancer cells and their sensitization to doxorubicin-induced apoptosis by miR-1. J. Biol. Chem. 283, 33394–33405. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Neves, R. , Scheel, C. , Weinhold, S. , Honisch, E. , Iwaniuk, K.M. , Trompeter, H.I. , Niederacher, D. , Wernet, P. , Santourlidis, S. , Uhrberg, M. , 2010. Role of DNA methylation in miR-200c/141 cluster silencing in invasive breast cancer cells. BMC Res. Notes. 3, 219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng, E.K. , Tsang, W.P. , Ng, S.S. , Jin, H.C. , Yu, J. , Li, J.J. , Rocken, C. , Ebert, M.P. , Kwok, T.T. , Sung, J.J. , 2009. MicroRNA-143 targets DNA methyltransferases 3A in colorectal cancer. Br. J. Cancer. 101, 699–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omura, N. , Li, C.P. , Li, A. , Hong, S.M. , Walter, K. , Jimeno, A. , Hidalgo, M. , Goggins, M. , 2008. Genome-wide profiling of methylated promoters in pancreatic adenocarcinoma. Cancer Biol. Ther. 7, 1146–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozsolak, F. , Poling, L.L. , Wang, Z. , Liu, H. , Liu, X.S. , Roeder, R.G. , Zhang, X. , Song, J.S. , Fisher, D.E. , 2008. Chromatin structure analyses identify miRNA promoters. Genes Dev. 22, 3172–3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, S.M. , Gaur, A.B. , Lengyel, E. , Peter, M.E. , 2008. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 22, 894–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popovic, R. , Riesbeck, L.E. , Velu, C.S. , Chaubey, A. , Zhang, J. , Achille, N.J. , Erfurth, F.E. , Eaton, K. , Lu, J. , Grimes, H.L. , Chen, J. , Rowley, J.D. , Zeleznik-Le, N.J. , 2009. Regulation of mir-196b by MLL and its overexpression by MLL fusions contributes to immortalization. Blood. 113, 3314–3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao, P.K. , Missiaglia, E. , Shields, L. , Hyde, G. , Yuan, B. , Shepherd, C.J. , Shipley, J. , Lodish, H.F. , 2010. Distinct roles for miR-1 and miR-133a in the proliferation and differentiation of rhabdomyosarcoma cells. FASEB J. 24, 3427–3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid, J.F. , Sokolova, V. , Zoni, E. , Lampis, A. , Pizzamiglio, S. , Bertan, C. , Zanutto, S. , Perrone, F. , Camerini, T. , Gallino, G. , Verderio, P. , Leo, E. , Pilotti, S. , Gariboldi, M. , Pierotti, M.A. , 2012. miRNA profiling in colorectal cancer highlights miR-1 involvement in MET-dependent proliferation. Mol. Cancer Res. 10, 504–515. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Otero, P. , Roman-Gomez, J. , Vilas-Zornoza, A. , Jose-Eneriz, E.S. , Martin-Palanco, V. , Rifon, J. , Torres, A. , Calasanz, M.J. , Agirre, X. , Prosper, F. , 2011. Deregulation of FGFR1 and CDK6 oncogenic pathways in acute lymphoblastic leukaemia harbouring epigenetic modifications of the MIR9 family. Br. J. Haematol. 155, 73–83. [DOI] [PubMed] [Google Scholar]
- Roman-Gomez, J. , Agirre, X. , Jimenez-Velasco, A. , Arqueros, V. , Vilas-Zornoza, A. , Rodriguez-Otero, P. , Martin-Subero, I. , Garate, L. , Cordeu, L. , San Jose-Eneriz, E. , Martin, V. , Castillejo, J.A. , Bandres, E. , Calasanz, M.J. , Siebert, R. , Heiniger, A. , Torres, A. , Prosper, F. , 2009. Epigenetic regulation of microRNAs in acute lymphoblastic leukemia. J. Clin. Oncol. 27, 1316–1322. [DOI] [PubMed] [Google Scholar]
- Rotkrua, P. , Akiyama, Y. , Hashimoto, Y. , Otsubo, T. , Yuasa, Y. , 2011. MiR-9 downregulates CDX2 expression in gastric cancer cells. Int. J. Cancer. 129, 2611–2620. [DOI] [PubMed] [Google Scholar]
- Sachdeva, M. , Mo, Y.Y. , 2010. MicroRNA-145 suppresses cell invasion and metastasis by directly targeting mucin 1. Cancer Res. 70, 378–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito, Y. , Liang, G. , Egger, G. , Friedman, J.M. , Chuang, J.C. , Coetzee, G.A. , Jones, P.A. , 2006. Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell. 9, 435–443. [DOI] [PubMed] [Google Scholar]
- Saito, Y. , Suzuki, H. , Tsugawa, H. , Nakagawa, I. , Matsuzaki, J. , Kanai, Y. , Hibi, T. , 2009. Chromatin remodeling at Alu repeats by epigenetic treatment activates silenced microRNA-512-5p with downregulation of Mcl-1 in human gastric cancer cells. Oncogene. 28, 2738–2744. [DOI] [PubMed] [Google Scholar]
- Shen, R. , Pan, S. , Qi, S. , Lin, X. , Cheng, S. , 2010. Epigenetic repression of microRNA-129-2 leads to overexpression of SOX4 in gastric cancer. Biochem. Biophys. Res. Commun. 394, 1047–1052. [DOI] [PubMed] [Google Scholar]
- Shi, B. , Sepp-Lorenzino, L. , Prisco, M. , Linsley, P. , deAngelis, T. , Baserga, R. , 2007. Micro RNA 145 targets the insulin receptor substrate-1 and inhibits the growth of colon cancer cells. J. Biol. Chem. 282, 32582–32590. [DOI] [PubMed] [Google Scholar]
- Smrt, R.D. , Szulwach, K.E. , Pfeiffer, R.L. , Li, X. , Guo, W. , Pathania, M. , Teng, Z.Q. , Luo, Y. , Peng, J. , Bordey, A. , Jin, P. , Zhao, X. , 2010. MicroRNA miR-137 regulates neuronal maturation by targeting ubiquitin ligase mind bomb-1. Stem Cells. 28, 1060–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh, S.O. , Chen, Y. , Zaman, M.S. , Hirata, H. , Yamamura, S. , Shahryari, V. , Liu, J. , Tabatabai, Z.L. , Kakar, S. , Deng, G. , Tanaka, Y. , Dahiya, R. , 2011. MicroRNA-145 is regulated by DNA methylation and p53 gene mutation in prostate cancer. Carcinogenesis. 32, 772–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, H. , Takatsuka, S. , Akashi, H. , Yamamoto, E. , Nojima, M. , Maruyama, R. , Kai, M. , Yamano, H.O. , Sasaki, Y. , Tokino, T. , Shinomura, Y. , Imai, K. , Toyota, M. , 2011. Genome-wide profiling of chromatin signatures reveals epigenetic regulation of MicroRNA genes in colorectal cancer. Cancer Res. 71, 5646–5658. [DOI] [PubMed] [Google Scholar]
- Suzuki, H. , Tokino, T. , Shinomura, Y. , Imai, K. , Toyota, M. , 2008. DNA methylation and cancer pathways in gastrointestinal tumors. Pharmacogenomics. 9, 1917–1928. [DOI] [PubMed] [Google Scholar]
- Suzuki, H. , Yamamoto, E. , Nojima, M. , Kai, M. , Yamano, H.O. , Yoshikawa, K. , Kimura, T. , Kudo, T. , Harada, E. , Sugai, T. , Takamaru, H. , Niinuma, T. , Maruyama, R. , Yamamoto, H. , Tokino, T. , Imai, K. , Toyota, M. , Shinomura, Y. , 2010. Methylation-associated silencing of microRNA-34b/c in gastric cancer and its involvement in an epigenetic field defect. Carcinogenesis. 31, 2066–2073. [DOI] [PubMed] [Google Scholar]
- Szulwach, K.E. , Li, X. , Smrt, R.D. , Li, Y. , Luo, Y. , Lin, L. , Santistevan, N.J. , Li, W. , Zhao, X. , Jin, P. , 2010. Cross talk between microRNA and epigenetic regulation in adult neurogenesis. J. Cell Biol. 189, 127–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tellez, C.S. , Juri, D.E. , Do, K. , Bernauer, A.M. , Thomas, C.L. , Damiani, L.A. , Tessema, M. , Leng, S. , Belinsky, S.A. , 2011. EMT and stem cell-like properties associated with miR-205 and miR-200 epigenetic silencing are early manifestations during carcinogen-induced transformation of human lung epithelial cells. Cancer Res. 71, 3087–3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting, A.H. , Suzuki, H. , Cope, L. , Schuebel, K.E. , Lee, B.H. , Toyota, M. , Imai, K. , Shinomura, Y. , Tokino, T. , Baylin, S.B. , 2008. A requirement for DICER to maintain full promoter CpG island hypermethylation in human cancer cells. Cancer Res. 68, 2570–2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyota, M. , Suzuki, H. , Sasaki, Y. , Maruyama, R. , Imai, K. , Shinomura, Y. , Tokino, T. , 2008. Epigenetic silencing of microRNA-34b/c and B-cell translocation gene 4 is associated with CpG island methylation in colorectal cancer. Cancer Res. 68, 4123–4132. [DOI] [PubMed] [Google Scholar]
- Tsai, K.W. , Hu, L.Y. , Wu, C.W. , Li, S.C. , Lai, C.H. , Kao, H.W. , Fang, W.L. , Lin, W.C. , 2010. Epigenetic regulation of miR-196b expression in gastric cancer. Genes Chromosomes Cancer. 49, 969–980. [DOI] [PubMed] [Google Scholar]
- Tsai, K.W. , Liao, Y.L. , Wu, C.W. , Hu, L.Y. , Li, S.C. , Chan, W.C. , Ho, M.R. , Lai, C.H. , Kao, H.W. , Fang, W.L. , Huang, K.H. , Lin, W.C. , 2011. Aberrant hypermethylation of miR-9 genes in gastric cancer. Epigenetics. 6, 1189–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai, K.W. , Wu, C.W. , Hu, L.Y. , Li, S.C. , Liao, Y.L. , Lai, C.H. , Kao, H.W. , Fang, W.L. , Huang, K.H. , Chan, W.C. , Lin, W.C. , 2011. Epigenetic regulation of miR-34b and miR-129 expression in gastric cancer. Int. J. Cancer. 129, 2600–2610. [DOI] [PubMed] [Google Scholar]
- Tsukamoto, Y. , Nakada, C. , Noguchi, T. , Tanigawa, M. , Nguyen, L.T. , Uchida, T. , Hijiya, N. , Matsuura, K. , Fujioka, T. , Seto, M. , Moriyama, M. , 2010. MicroRNA-375 is downregulated in gastric carcinomas and regulates cell survival by targeting PDK1 and 14-3-3zeta. Cancer Res. 70, 2339–2349. [DOI] [PubMed] [Google Scholar]
- Tsuruta, T. , Kozaki, K. , Uesugi, A. , Furuta, M. , Hirasawa, A. , Imoto, I. , Susumu, N. , Aoki, D. , Inazawa, J. , 2011. miR-152 is a tumor suppressor microRNA that is silenced by DNA hypermethylation in endometrial cancer. Cancer Res. 71, 6450–6462. [DOI] [PubMed] [Google Scholar]
- Uesugi, A. , Kozaki, K. , Tsuruta, T. , Furuta, M. , Morita, K. , Imoto, I. , Omura, K. , Inazawa, J. , 2011. The tumor suppressive microRNA miR-218 targets the mTOR component Rictor and inhibits AKT phosphorylation in oral cancer. Cancer Res. 71, 5765–5778. [DOI] [PubMed] [Google Scholar]
- Vogt, M. , Munding, J. , Gruner, M. , Liffers, S.T. , Verdoodt, B. , Hauk, J. , Steinstraesser, L. , Tannapfel, A. , Hermeking, H. , 2011. Frequent concomitant inactivation of miR-34a and miR-34b/c by CpG methylation in colorectal, pancreatic, mammary, ovarian, urothelial, and renal cell carcinomas and soft tissue sarcomas. Virchows Arch. 458, 313–322. [DOI] [PubMed] [Google Scholar]
- Vrba, L. , Jensen, T.J. , Garbe, J.C. , Heimark, R.L. , Cress, A.E. , Dickinson, S. , Stampfer, M.R. , Futscher, B.W. , 2010. Role for DNA methylation in the regulation of miR-200c and miR-141 expression in normal and cancer cells. PLoS One. 5, e8697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H. , Wu, J. , Meng, X. , Ying, X. , Zuo, Y. , Liu, R. , Pan, Z. , Kang, T. , Huang, W. , 2011. MicroRNA-342 inhibits colorectal cancer cell proliferation and invasion by directly targeting DNA methyltransferase 1. Carcinogenesis. 32, 1033–1042. [DOI] [PubMed] [Google Scholar]
- Wang, Z. , Chen, Z. , Gao, Y. , Li, N. , Li, B. , Tan, F. , Tan, X. , Lu, N. , Sun, Y. , Sun, J. , Sun, N. , He, J. , 2011. DNA hypermethylation of microRNA-34b/c has prognostic value for stage non-small cell lung cancer. Cancer Biol. Ther. 11, 490–496. [DOI] [PubMed] [Google Scholar]
- Watanabe, K. , Emoto, N. , Hamano, E. , Sunohara, M. , Kawakami, M. , Kage, H. , Kitano, K. , Nakajima, J. , Goto, A. , Fukayama, M. , Nagase, T. , Yatomi, Y. , Ohishi, N. , Takai, D. , 2012. Genome structure-based screening identified epigenetically silenced microRNA associated with invasiveness in non-small-cell lung cancer. Int. J. Cancer. 130, 2580–2590. [DOI] [PubMed] [Google Scholar]
- Wiggins, J.F. , Ruffino, L. , Kelnar, K. , Omotola, M. , Patrawala, L. , Brown, D. , Bader, A.G. , 2010. Development of a lung cancer therapeutic based on the tumor suppressor microRNA-34. Cancer Res. 70, 5923–5930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiklund, E.D. , Bramsen, J.B. , Hulf, T. , Dyrskjot, L. , Ramanathan, R. , Hansen, T.B. , Villadsen, S.B. , Gao, S. , Ostenfeld, M.S. , Borre, M. , Peter, M.E. , Orntoft, T.F. , Kjems, J. , Clark, S.J. , 2011. Coordinated epigenetic repression of the miR-200 family and miR-205 in invasive bladder cancer. Int. J. Cancer. 128, 1327–1334. [DOI] [PubMed] [Google Scholar]
- Wilting, S.M. , van Boerdonk, R.A. , Henken, F.E. , Meijer, C.J. , Diosdado, B. , Meijer, G.A. , le Sage, C. , Agami, R. , Snijders, P.J. , Steenbergen, R.D. , 2010. Methylation-mediated silencing and tumour suppressive function of hsa-miR-124 in cervical cancer. Mol. Cancer. 9, 167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong, K.Y. , Liang, R. , So, C.C. , Jin, D.Y. , Costello, J.F. , Chim, C.S. , 2011. Epigenetic silencing of MIR203 in multiple myeloma. Br. J. Haematol. 154, 569–578. [DOI] [PubMed] [Google Scholar]
- Wong, K.Y. , So, C.C. , Loong, F. , Chung, L.P. , Lam, W.W. , Liang, R. , Li, G.K. , Jin, D.Y. , Chim, C.S. , 2011. Epigenetic inactivation of the miR-124-1 in haematological malignancies. PLoS One. 6, e19027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, H. , Choi, A.J. , Lee, B.H. , Ting, A.H. , 2011. Identification and functional analysis of epigenetically silenced microRNAs in colorectal cancer cells. PLoS One. 6, e20628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, F. , Jiao, Y. , Zhu, Y. , Wang, Y. , Zhu, J. , Cui, X. , Liu, Y. , He, Y. , Park, E.Y. , Zhang, H. , Lv, X. , Ma, K. , Su, F. , Park, J.H. , Song, E. , 2012. MicroRNA 34c gene down-regulation via DNA methylation promotes self-renewal and epithelial-mesenchymal transition in breast tumor-initiating cells. J. Biol. Chem. 287, 465–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaman, M.S. , Chen, Y. , Deng, G. , Shahryari, V. , Suh, S.O. , Saini, S. , Majid, S. , Liu, J. , Khatri, G. , Tanaka, Y. , Dahiya, R. , 2010. The functional significance of microRNA-145 in prostate cancer. Br. J. Cancer. 103, 256–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Yan, L.X. , Wu, Q.N. , Du, Z.M. , Chen, J. , Liao, D.Z. , Huang, M.Y. , Hou, J.H. , Wu, Q.L. , Zeng, M.S. , Huang, W.L. , Zeng, Y.X. , Shao, J.Y. , 2011. miR-125b is methylated and functions as a tumor suppressor by regulating the ETS1 proto-oncogene in human invasive breast cancer. Cancer Res. 71, 3552–3562. [DOI] [PubMed] [Google Scholar]
- Zhang, Z. , Tang, H. , Wang, Z. , Zhang, B. , Liu, W. , Lu, H. , Xiao, L. , Liu, X. , Wang, R. , Li, X. , Wu, M. , Li, G. , 2011. MiR-185 targets the DNA methyltransferases 1 and regulates global DNA methylation in human glioma. Mol. Cancer. 10, 124 [DOI] [PMC free article] [PubMed] [Google Scholar]


