Abstract
Current hypotheses suggest that tumors originate from cells that carry out a process of “malignant reprogramming” driven by genetic and epigenetic alterations. Multiples studies reported the existence of stem‐cell‐like cells that acquire the ability to self‐renew and are able to generate the bulk of more differentiated cells that form the tumor. This population of cancer cells, called cancer stem cells (CSC), is responsible for sustaining the tumor growth and, under determined conditions, can disseminate and migrate to give rise to secondary tumors or metastases to distant organs. Furthermore, CSCs have shown to be more resistant to anti‐tumor treatments than the non‐stem cancer cells, suggesting that surviving CSCs could be responsible for tumor relapse after therapy. These important properties have raised the interest in understanding the mechanisms that govern the generation and maintenance of this special population of cells, considered to lie behind the on/off switches of gene expression patterns. In this review, we summarize the most relevant epigenetic alterations, from DNA methylation and histone modifications to the recently discovered miRNAs that contribute to the regulation of cancer stem cell features in tumor progression, metastasis and response to chemotherapy.
Keywords: Cancer stem cells, Epigenetics, DNA methylation, Histone modifications, microRNAs
1. Introduction
During the last decades great advances have been made in basic research on cancer, in identifying the genetic changes and chromosomal alterations responsible for cell transformation, tumor initiation and progression (Fearon and Vogelstein, 1990; Vogelstein and Kinzler, 2004). Recent studies of epigenetic changes in cancer demonstrate the relevance that these have in cancer etiology (Jones and Baylin, 2007; Esteller, 2008). A great body of evidence supports that both processes are responsible for tumor development. Indeed, alterations in DNA methylation, histone modifications, polycomb, miRNAs and chromatin remodeling complex function are mechanisms that directly contribute to tumorigenesis.
Several epigenetic mechanisms are connected and work synergistically in order to regulate the expression of specific genes. DNA methylation is an important regulatory pathway that is altered in carcinogenesis. DNA methyltransferases are the enzymes responsible on the deposition of methyl groups on cytosines. DNA methylation patterns are maintained through cell division cycles by DNA methyltransferase 1 (DNMT1), which recognizes semi‐methylated DNA (Eden et al., 2003), while DNMT3a and DNMT3b mediate de novo DNA methylation (Okano et al., 1999). Methyl‐cytosines are recognized and bound by methyl‐binding proteins, inducing transcriptional repression by recruiting transcriptional co‐repressors (Klose and Bird, 2006). In mammals, nearly all methylation occurs at CpG sites, especially in areas of repetitive sequences. On the contrary, CpGs‐enriched regions (CpG islands) close to 5′‐end of genes appear protected from such modification, suggesting that DNA methylation of promoter regions is a regulatory mechanism of gene expression (Bird, 2002). The cancer epigenome is characterized by global DNA hypomethylation and gene specific hypermethylation (Esteller, 2008; Feinberg et al., 2006). Different studies indicated that DNA hypomethylation occurs at early stages in cancer development, contributing to chromosomal instability and tumor progression (Esteller, 2008; Holm et al., 2005). DNA hypomethylation also leads to the specific activation of key genes involved in tumorigenesis, such as R‐Ras, Cyclin D2, MASPIN, melanoma‐associated antigen (MAGE) (Akiyama et al., 2003; De Smet et al., 1996; Nakamura and Takenaga, 1998; Nishigaki et al., 2005) and loss of imprinting (LOI) genes (Cui et al., 2003; Ogawa et al., 1993). On the other hand, the silencing of tumor‐suppressor genes, such as retinoblastoma 1 (RB1), CDKN2A (p16), von Hippel–Lindau tumor suppressor (VHL), MutL protein homolog 1 (MLH1) and BRCA1 (Tsai and Baylin, 2011), as well as APC and Wnt‐signaling genes in colorectal carcinomas are associated to promoter DNA hypermethylation and chromatin hypoacetylation (Hiltunen et al., 1997; Suzuki et al., 2004).
Alterations in the balance among many of the histone marks lead to deregulated gene transcription and are related to cancer (Fraga et al., 2005; Seligson et al., 2005). The nucleosomes, the basic chromatin units, are composed of DNA wrapped around octamers of the core histones H2A, H2B, H3 and H4. The amino‐terminal tails of histones are subjected to a variety of post‐translational modifications (reviewed in Torres‐Padilla et al., 2007) and, together with the linker histone H1, can compact the nucleosomal DNA forming high‐order structures. The more studied histone modifications are the methylation of lysine (K) residues mostly on H3, which, dependent on the lysine residue, can be either activating or repressive, and the acetylation of K residues on histones H3 and H4, which are more abundant in transcriptionally permissive euchromatin. The H3K4me3 is mediated by the Trithorax group of proteins (TrxG) and marks nucleosomes found in the promoter regions of actively transcribed genes (Santos‐Rosa et al., 2002; Bernstein et al., 2002). H3K27me3 marks are mediated by the Polycomb repressive complex 2 (PRC2), composed of Polycomb group proteins (PcGs). PcGs were initially identified as homeotic regulators which establish epigenetic patterns during development, imprinting and X‐inactivation (Sparmann and van Lohuizen, 2006). Genes marked by H3K27me3 are usually methylated and silenced. The catalytic subunit of PRC2, Enhancer of zeste homolog 2 (EZH2) (Otte and Kwaks, 2003; Ringrose and Paro, 2004), initiates the silencing process through H3K27 methylation (Sun et al., 2002; Lee et al., 2006; Vire et al., 2006). This mark allows the recruitment of PRC1 and other co‐repressors onto chromatin, resulting in the heterochromatinization of the region through formation of higher‐order chromatin structures spanning around the starting sites of the modification (Zhao et al., 2006). High levels of PRC2 components are present in embryonic stem cells (ESCs), which decline quickly upon the onset of differentiation, while the expression and function of some of the components of the PRC1 and PRC2 complexes have been found altered in cancer (Tsang and Cheng, 2011). Concerning histone acetylation, H3K4ac and H3K9ac are the more known marks correlating with accessible euchromatin and transcriptionally active regions. Histone acetylation is catalyzed by histone acetyltransferases (HATs) and removed by histone deacetylases (HDACs) (Lee and Workman, 2007). Modulation of the histone acetylation program interferes with the differentiation process, and therefore, is not surprising that drugs targeting HDACS are already being used to enhance differentiation or reprogramming events. In addition, the PRC2‐mediated transcriptional repression of genes implicates histone deacetylation. EZH2 is physically and functionally linked to HDAC1 and HDAC2, which are frequently found overexpressed in various types of cancer (Halkidou et al., 2004; Song et al., 2005). Furthermore, HATs such as G9a, and aberrant fusion proteins formed through chromosomal translocations of HAT and HAT‐related genes (MOZ, MORF, CBP and p300) (Yang, 2004) or chromosomal translocations of MLL, have been related to cancer (Krivtsov and Armstrong, 2007). Together, these studies show the complex interplay between HMT, DNMTs, HATs and HDACs epigenetic pathways that contribute to aberrant gene expression in cancer cells and illustrate how aberrant DNA methylation can be initiated and maintained in cancer.
Additionally, ATP‐dependent chromatin‐remodeling factors were found significantly altered in cancer. These enzymes are represented by several subunits, encoded by 30 genes in mammals, and divided in four main families on the basis of the sequence and the structure of the ATPase subunit: SWI/SNF (switch/sucrose nonfermentable), ISWI (imitation switch), CDH (chromodomain helicase DNA‐binding) and INO80 (inositol‐requiring 80). All those subunits interact with each other in numerous combinations, creating assemblies such as BAF (SWI/SNF), NuRD, ISWI, CDH1 and Tip60‐p400 complexes (reviewed in Ho and Crabtree, 2010). In mammals, Brm (Brahma) and Brg1 ATPases together with other 12 subunits compose the major histone remodeling complex, BAF (homologous to the SWI/SNF yeast complex). The selective disruption of chromatin remodelers interferes with ESCs proliferation and differentiation and alterations in the function of this family of chromatin‐modifying complexes were associated with cancer development (Roberts and Orkin, 2004; Reisman et al., 2009). Thus, the BAF47 and BAF250A subunits are inactivated in different types of tumors (Biegel et al., 2000; Grand et al., 1999; Jones et al., 2010) and loss of heterozygosity of Brm and Brg1, is found in primary lung cancers (Reisman et al., 2003). At the present the epigenetic changes caused by these mutations have yet to be defined.
miRNAs are small non‐coding RNAs (19–25 nucleotides long) that regulate mRNAs at post‐transcriptional level. These non‐coding RNAs are involved in the control of ESCs pluripotency (Gangaraju and Lin, 2009) and aberrant expression of some of them is related to tumor growth and metastasis. Recently, it was found that the expression of miRNAs may also be regulated by promoter DNA methylation, adding a new level of complexity to the epigenetic regulation of tumorigenesis (Lujambio et al., 2007; Bandres et al., 2009).
However, most of the extensive characterization of epigenetic alterations related above and associated to malignant transformation and cancer has been obtained from whole cells populations forming the tumor or cancer cell lines. Considering that tumors present cell heterogeneity, the identification of specific epigenetic mutations and the characterization of their effects in the subset of cancer cells responsible to maintain the tumor growth, the cancer stem cells (CSCs, see below), become a key issue. In this review we will focus on the recent data regarding epigenetic modifications in CSCs and in the bulk of tumor cells (non stem cancer cells‐NSCCs) with a less self‐renewal potential, and their impact on tumor initiation, progression and chemotherapy response.
2. Relevance of stem‐like cells in cancer
Tumors have long been viewed as a caricature of normal tissue development, where tumor cellular heterogeneity could represent abortive attempts by a subset of undifferentiated tumor cells to undergo functional differentiation (Pierce and Speers, 1988). In this regard, some tumors show a pathological architecture and hierarchical organization reminiscent to that present in the origin tissue (Schepers et al., 2012). The CSC model suggests that a subset of cancer cells, which presents similar self‐renewal and multipotency properties to adult stem cells‐responsible for tissue regeneration and homeostasis is capable to sustain the tumor growth in contrast to the bulk of tumor cells with a more differentiated phenotype (NSCCs) (Visvader, 2011). CSCs were initially described in myeloid leukemia. The use of cell surface markers that identify hematopoietic stem cells enabled the isolation of tumor cells expressing these markers. In contrast to the NSCCs, CSCs were able to regenerate all the cell types present in the parental tumor after serial transplantation into immunodeficient mice (Bonnet and Dick, 1997; Lapidot et al., 1994). During last years, CSCs were identified in human solid tumors, such as brain, breast, prostate, head and neck, pancreas, liver, ovary, melanoma, skin and colon cancer, by using different stem cell surface markers, including CD133, CD44, EpCAM, CD24, Lgr5, by the expression of ALDH1 or by the ability to extrude DNA dye (side population cells) (Visvader, 2011; Zhang et al., 2011). However, some reports indicated that cells identified by some of these markers were heterogeneous populations of cells enriched in or containing CSCs and the phenotypic features of these CSCs might change during tumor progression (reviewed in Baccelli and Trumpp, 2012). Therefore, it is necessary to define this subset of tumor cells using more functional assays, which relied on the ability to regenerate a tumor with identical features to the parental tumor upon transplantation into immunodeficient mice. However, this approach, that imperfectly recapitulates the in vivo situation found in patients, has some limitations associated to isolation of tumor cells, injection in heterotopic sites and the possible mis‐interpretation of tumor initiating capability of the isolated cells depending on the immunodeficiency of mice used in the xenograft assays (Quintana et al., 2008). This raised the debate about the existence and role of the CSCs in the physiological tumor progression. Recently, lineage tracing experiments performed in mouse models demonstrated that adult stem cells suffering specific mutation are the cells in the origin of tumors in skin, colon and brain (Alcantara Llaguno et al., 2009; Lapouge et al., 2011; Schepers et al., 2012; White et al., 2011). Although in some tumor types these cells of origin were demonstrated to act as CSCs (Chen et al., 2012; Driessens et al., 2012; Schepers et al., 2012), some reports indicated that CSCs can be also originated from more committed progenitors (Cozzio et al., 2003; Krivtsov et al., 2006; Somervaille and Cleary, 2006) that acquire stemness‐related features as a consequence of accumulation of genetic or epigenetic alterations. Thus, progeny of mutated stem cells could acquire CSCs features and function.
In order to understand the relationship between CSCs and normal stem cells, the transcriptional program in human intestinal stem cells and colorectal carcinoma was compared and the results showed certain resemblance of gene expression signature between both populations of cells (Schepers et al., 2012). In addition, an adult stem cell transcriptional profile was activated in diverse human epithelial cancers, which correlated with recurrent disease, metastasis and death (Merlos‐Suarez et al., 2012; Wong et al., 2008). Furthermore, ESCs‐gene expression signature, defined in part by some miRNAs, PRC2‐regulated genes and the transcription factors Oct4, Nanog, Sox2, Klf4, is more frequently found in poorly differentiated tumors with poor clinical outcome (Chiou et al., 2010; Eberle et al., 2010; Schoenhals et al., 2009). The relevance of the Oct4 and Nanog in cancer is also supported by the notion that ectopic expression of these transcription factors in lung and ovarian cancer cells increased the percentage of cells with CSC properties, enhanced drug resistance and promoted epithelial‐to‐mesenchymal transition (EMT) (Chiou et al., 2010; Kong et al., 2010; Siu et al., in press).
The relevance of CSCs to clinical cancer rely on the observation that CSC population are more resistant to anti‐tumor therapies than NSCCs (Dallas et al., 2009; Dylla et al., 2008; Eramo et al., 2006; Hermann et al., 2007; Ma et al., 2008; Todaro et al., 2007). Indeed, the frequently used anti‐tumor therapy would eliminate preferentially NSCCs, being the surviving CSCs responsible for the tumor relapse. Using a combination of experimental approaches that selectively eliminate glioblastoma CSCs, and conventional anti‐tumor therapy that suppress the bulk of dividing cells, it was recently demonstrated that glioblastoma growth was dramatically impeded in vivo, increasing the survival of tumor carrying mice (Chen et al., 2012).
In addition, several lines of evidence indicate that cell initiating metastasis could be founded within subpopulations of CSCs. Thus, CSCs possess some of the features characteristics that make them likely candidates for metastasis initiating activities, such as the tumor‐initiating capacity, necessary for the generation of secondary tumors in distant organs and the expression of EMT markers (Mani et al., 2008), which is associated with the tumor cell ability to migrate.
The frequency of CSCs is variable in different tumors and it may depend on the cell type of origin, tumor microenvironment, accumulated somatic mutations and stage of malignant progression reached by the tumor (Gupta et al., 2009; Vermeulen et al., 2010). Tumor niche is composed of diverse immune and stromal cells, blood vessels and matrix glycoproteins, which provide a highly specialized microenvironment for cancer cells (Bissell and Hines, 2011; Hanahan and Coussens, 2012; Shiao et al., 2011). Contact and communication between these elements are critical for stem cell self‐renewal and multipotency. CSCs are frequently enriched in regions around tumor vessels and necrosis (Calabrese et al., 2007), the latter associated with restricted oxygen/hypoxia. Furthermore, hypoxia that maintains the undifferentiated states of embryonic, hematopoietic, mesenchymal and neural stem cells, influence proliferation and cell‐fate commitment of cancer cells (Mohyeldin et al., 2010) through hypoxia‐inducible factors‐1 (HIF‐1 and HIF being the latter preferentially expressed in CSCs (Bar et al., 2010; Heddleston et al., 2009; Li and Rich, 2010; Seidel et al., 2010). Hence, the signals provided by the niche could be important to regulate the properties of CSCs as well the dynamic interplay between CSCs and NSCCs during tumor progression. The CSC concept has, therefore, evolved to model the complex and highly dynamic processes of tumorigenesis, tumor relapse and metastasis. Alterations in adult stem/progenitors cell homeostasis induced by genetic and epigenetics defaults could reprogram these cells to acquire more advantageous features in response to the tumor microenvironment requirements, thus, leading to CSCs generation.
3. Epigenetic regulation of CSC properties
Multiples studies have been focused on decoding the genetic and epigenetic mechanisms responsible for the acquisition of stemness features and CSC genesis. Epigenetic mechanisms are involved in the regulation of the embryonic and adult stem cell transcriptional program, controlling self‐renewal and differentiation processes. Multiple observations indicate that the establishment and maintenance of CSC features can be orchestrated by a similar way, switching CSC markers on and off to generate heterogeneous population of cells with distinct phenotypes and features. Genetic and epigenetic changes would provide survival advantages in CSC subpopulation and contribute to tumor initiation capability and tumor progression. The relevance of the DNA methylation in CSC regulation and tumor growth was illustrated in leukemia stem cells. The abrogation of DNA methyltransferase Dnmt1 expression blocked the leukemia development. Furthermore, haploinsufficiency of Dnmt1 resulted in tumor suppressor gene derepression, reduced bivalent chromatine marks, impaired CSC self‐renewal and delayed leukemogenesis (Trowbridge et al., 2012). Although promoter hypermethylation of some tumor suppressor genes, that drive oncogenesis at early stages, was already present in the CSCs and was preserved in NSCC subpopulation, the promoter methylation status of some CSC markers can show differences in both tumor cell populations. Indeed, the methylation of CD133 promoter was heterogeneous between CD133+ and CD133− subpopulations isolated from brain (Gopisetty et al., in press; Yi et al., 2008) and epithelial ovarian cancer (Baba et al., 2009). Interestingly, the methylation of the CD133 promoter appeared during the differentiation of CD133+ CSCs to CD133− NSCCs cells, correlating with the decreased expression of this surface glycoprotein (Baba et al., 2009; Gopisetty et al., in press). Likewise, CD44, CD133 and Mushasi‐1 promoters presented a hypomethylated status which was associated with high expression of these CSC markers in triple‐negative breast tumors (Kagara et al., 2012). This suggests that aberrant DNA methylation in tumors is dynamic and contributes to the transition between active and repressive state of gene transcription.
There is evidence that PcG complexes target similar sets of CpG‐containing genes in ESCs as in cancer cells (Ohm et al., 2007; Schlesinger et al., 2007; Widschwendter et al., 2007). These targeted genes can be responsible for the CSC phenotype emerging during tumorigenesis (Schlesinger et al., 2007; Widschwendter et al., 2007). An aberrant methylation may, then, help to abnormally lock in the activation of stem cell pathways and contribute to the self‐renewing ability of CSCs during tumor progression. The induced expression of EZH2 in hematopoietic stem cells promoted myeloid expansion in a knock‐in mouse model, indicating a stem cell‐specific EZH2 oncogenic role in myeloid disorders (Herrera‐Merchan et al., 2012). In addition, several studies demonstrated that the upregulation of EZH2 expression in some tumors contribute to the maintenance of a reversible and undifferentiated stem‐like phenotype in cancer cells (Burdach et al., 2009; Chang et al., 2011b) and the expansion of breast CSCs (Chang et al., 2011b). Furthermore, the pharmacological inhibition and downregulation of EHZ2 inhibits CSC self‐renewal in vitro, reduced the expression of CSC markers and block the in vivo tumor‐initiating capacity in different tumor types (Bao et al., 2012; Crea et al., 2012b; Rizzo et al., 2011; Suva et al., 2009). Similarly, BMI1, a subunit of PRC1 complex previously implicated in leukemogenesis, is upregulated by the ESC transcription factor Sall4, through increase of H3K4me3 and H3K79me2 marks on the BMI1 promoter, and this mechanism can regulate self‐renewal in normal and leukemic stem cells (Yang et al., 2007). BMI1 is also expressed in CD133+ cells in human glioblastomas and its knockdown resulted in the inhibition of the clonogenic potential, as well the ability to induce brain tumor formation in vivo (Abdouh et al., 2009).
Methylation of H3 at lysine 4 (H3K4) is frequently associated with active promoters. LSD1/KDM1 is a histone demethylase that suppresses gene expression by converting H3K4me2 to H3K4me and unmethylated H3K4. Inhibitors of LSD1 inhibited specifically the proliferation of pluripotent cancer cells from teratocarcinoma, embryonic carcinoma and seminoma but not from normal somatic cells or non‐pluripotent cancer cells (Wang et al., 2011). Interestingly, a new mechanism that mediate tumor hypoxic responses was recently reported, which links microenvironmental regulation of epigenetic modifying proteins to cancer cellular heterogeneity. The histone methyltransferase mixed‐lineage leukemia 1 (MLL1) is induced by hypoxia in glioblastoma and CSCs were found to express higher levels of this enzyme than matched NSCC. Downregulation of MLL1 induced the repression of HIF2α protein and target genes concomitantly with the reduction of CSC self‐renewal and tumorigenicity (Heddleston et al., 2012).
Several miRNAs implicated in development cooperate with PcGs complexes and DNA methylation to regulate the balance between self‐renewal and differentiation in CSCs (Esquela‐Kerscher and Slack, 2006; Volinia et al., 2006). Let‐7 is one of the most consistently and significantly reduced miRNAs in different types of cancers and frequently linked to tumor malignant progression (Johnson et al., 2005; Viswanathan et al., 2009). Similarly to that described in ESCs, let‐7/Lin28 loop plays a critical role in the breast CSC maintenance (Viswanathan et al., 2009; Yang et al., 2010). Breast CSCs isolated from human tumors expressed reduced levels of let‐7 compared to NSCCs. Interestingly, increased expression of let‐7 enhance differentiation, leading to reduced CSC self‐renewal and ability to develop tumor and metastasis in immunodeficient mice (Yu et al., 2007). Similarly, a reduced expression of let‐7 family that leads to overexpression of EZH2 was found in aggressive human prostate cancer and re‐expression of let‐7 decreased EZH2 expression and repressed CSC self‐renewal (Kong et al., 2012).
Contrary to let‐7, the expression of miR‐200 family members was unaffected during transformation, but it became specifically downregulated in breast CSCs in comparison to NSCCs. Indeed, miR‐200b and miR‐200c overexpression strongly inhibited the proliferation of CSCs and their ability to form tumors in vivo (Iliopoulos et al., 2010; Lo et al., 2011; Shimono et al., 2009), and this effect was mediated by targeting different subunits of PcGs complexes. MiR‐200c repressed the expression of BMI1 (Lo et al., 2011; Shimono et al., 2009), while loss of miR‐200b increased Suz12 expression and H3K27 methylation. Interestingly, ectopic expression of Suz12 in transformed cells was able to generate CSCs (Iliopoulos et al., 2010). In addition, miR‐200c levels are regulated by a complicated loop comprising of Bmi1 and ZEB1 (Wellner et al., 2009). These findings reveled that miR‐200 family members play an important role regulation the CSC formation and function, implicating PcG complexes in this process.
In addition, miR‐34a modulates CSC function in tumor growth and metastasis. The expression of miR‐34a is regulated by p53 and miR‐34a induces apoptosis, cell‐cycle arrest or senescence when is introduced in cancer cells (Bommer et al., 2007; Chang et al., 2007; He et al., 2007; Raver‐Shapira et al., 2007; Tarasov et al., 2007). Recently, it was demonstrated that miR‐34a, which is under‐expressed in CSCs, negatively regulates the tumor initiating capacity of prostate (Liu et al., 2011), pancreatic (Ji et al., 2009b) and breast (Yu et al., 2012) CSCs. Interestingly, systemically delivered miR‐34a inhibited prostate cancer metastasis and extended survival of tumor‐bearing mice (Liu et al., 2011). In addition, miR‐34a targets CD44 and CD44 knockdown phenocopied miR‐34a over‐expression in inhibiting prostate cancer regeneration and metastasis, indicating the relevance of CD44 marker in CSC function (Liu et al., 2011). Similarly to the axis p53‐miR‐34a regulation, TA‐p73 and p63, homologous to p53, may also upregulate the expression of tumor suppressor miRNAs, such as let‐7, miR‐15/16a, miR‐145, miR‐29, miR‐30 and miR‐146a (Boominathan, 2010), highlighting the relevance of tumor suppressors genes in the regulation of miRNAs expression. Although some cancers maintain wild‐type human p53, it was found that miR‐380‐5p, which is highly expressed neuroblastomas with poor outcome, represses p53 expression via a conserved sequence in the p53 3′‐untranslated region (3′‐UTR). MiR‐380 over‐expression cooperates with activated HRas oncoprotein to transform primary cells, blocks oncogene‐induced senescence and forms tumors in mice (Swarbrick et al., 2010). Differential miRNA expression profiling of CD133+ CSCs and CD133− NSCCs from human hepatocellular carcinoma identified a high expression of miR‐130b in CSCs. Finally, miR‐130b that targets TP53INP1 (Ma et al., 2010), miR‐181 that targets regulators of differentiation such as CDX2, GATA6 and NLK (Ji et al., 2009a), and miR‐371‐3 (Cairo et al., 2010), are over‐expressed in CSCs compared to NSCCs in different tumor types, resulting in enhanced self‐renewal and tumorigenicity in vivo. Recent data suggest that Myc up‐regulates the miR‐371‐3 cluster with concomitant down‐regulation of the miR‐100/let‐7a‐2/miR‐125b‐1 cluster, contributing to the aggressive phenotype of liver tumors originating from hepatic progenitors cells (Cairo et al., 2010).
4. Deregulation of pathways controlling CSC self‐renewal by epigenetic alterations
Some of the most characterized signaling pathways controlling self‐renewal and differentiation in adult stem cells, such as Wnt/β‐catenin, Hedgehog, Notch and TGF‐β/BMP pathways are frequently modulated in cancer by epigenetic mechanisms.
4.1. Wnt/β‐catenin signaling pathway
The canonical Wnt signaling pathway, which through β‐catenin modulates the expression of specific target genes, is an important regulator of stem cells and CSCs and is aberrantly activated during the development of various human cancers (Clevers, 2006; Fodde and Brabletz, 2007; Jin et al., 2011; Vermeulen et al., 2010). Gain‐of‐function mutations of the CTNNB1 gene (encoding β‐catenin) and loss‐of‐function mutations of APC and AXIN genes were identified as the main mechanisms associated to Wnt signaling dysfunction in cancers (Barker and Clevers, 2006; Lindvall et al., 2007; Polakis, 2000). A number of genes involved in the Wnt/β‐catenin signaling are methylated and silenced in breast cancer, including the Wnt inhibitors WIF1, SFRP1‐5 and DKK1, as well APC and SRY‐box containing gene 17 (SOX17) (Klarmann et al., 2008; Suzuki et al., 2004; Zhang et al., 2008). Recent studies indicate that Wnt/β‐catenin pathway can be also regulated by histone modifications in cancer. Genome wide profiling studies revealed an enrichment of EZH2 and associated H3K27me3 on Wnt genes in Drosophila and mammalian cells (Bracken et al., 2006; Kirmizis et al., 2004). In support to this notion, over‐expression of EZH2 in mammary gland induces β‐catenin nuclear accumulation and activation of Wnt pathway and causes intraductal epithelial hyperplasia (Li et al., 2009a). Additionally, the transcriptional repression of DACT3, a Wnt antagonist, was associated with bivalent H3K27me3 and H3K4me3 histone modifications (Jiang et al., 2008) and Dkk‐1 repressed expression was induced by decreased H4K16Ac and increased H3K27me3, and by the recruitment of SirT1, EZH2, Suz12 and BMI1 to its promoter (Hussain et al., 2009). MiRNAs have also been implicated in the regulation of different players of the Wnt/β‐catenin pathways (Inui et al., 2010). MiR‐200a, miR‐1826 and miR‐320 directly targets β‐catenin mRNA and a direct correlation was found between decreased levels of these and the upregulation of β‐catenin and tumor growth in different cancer types (Hirata et al., 2012; Saydam et al., 2009; Sun et al., 2012). In addition, β‐catenin/Lef1 transactivates the miR‐371‐373 cluster involved in CSC self‐renewal, and in turn, these miRNAs modulate the Wnt/β‐catenin signaling by targeting DKK1 inhibitor (Zhou et al., 2012). MiR‐15a and miR‐16‐1 clusters, down‐regulated in prostate cancer, target CCND1 (encoding cyclin D1) and WNT3A expression and reconstitution assays of miR‐15a and miR‐16‐1 expression resulted in impaired tumor growth in vivo (Bonci et al., 2008). MiR‐135a and miR‐135b target APC and suppress its expression, inducing β‐catenin signaling (Nagel et al., 2008). Interestingly, a significant up‐regulation of these miRNAs was found in colorectal adenomas and carcinomas, which correlated with low APC mRNA levels (Nagel et al., 2008). On the other hand, Wnt/β‐catenin signaling can regulate also tumor growth, cancer cell migration and invasion by negatively regulating the miR‐122a expression in liver cancer (Wang et al., 2009a).
4.2. Hedgehog signaling pathway
In mammals, Hedgehog (Hh) signaling pathway controls the proliferation of stem and progenitor cells in different tissues and alterations in this pathway has been related to tumor development (Ingham and Placzek, 2006; Jiang and Hui, 2008; Pasca di Magliano and Hebrok, 2003). Binding of Hh to its receptor Patched (Ptch‐1) activates the transmembrane protein Smoothened (Smo), which subsequently activates the Gli family of transcription factors, leading to activation of target genes. Gli1 expression and function are regulated at different levels by epigenetic mechanisms. Gli1 function is downregulated by the chromatin remodeling protein SNF5, through its interaction with Gli1‐regulated promoters and recently SNF5 was found to be inactivated in human malignant rhabdoid tumors, coincidental with the activation of the Hh‐Gli1 pathway (Jagani et al., 2010). Gli1 and Gli2 are acetylated proteins and their HDAC‐mediated deacetylation promotes Hh pathway transcriptional activation. A positive auto‐regulatory loop was described, where Hh activation induced upregulation of HDAC1. This mechanism is turned off by HDAC1 degradation through an E3 ubiquitin ligase complex, formed by Cullin3 and REN, which is lost in human medulloblastoma (Canettieri et al., 2010). In addition, Gli1 expression is downregulated by miR‐324‐5p, and loss of miR‐324‐5p leads to tumor formation (Ferretti et al., 2008). A recurrent amplification of the miR‐17/92 cluster proto‐oncogene was found in 6% of pediatric medulloblastomas, which are characterized by present Sonic hedgehog (Shh) signaling activation compared with other subgroups of medulloblastoma (Northcott et al., 2009). Shh treatment of primary cerebellar granule neural progenitors resulted in increased miR‐17/92 expression, indicating a functional collaboration between the miR‐17/92 cluster and the Shh signaling pathway in the development of medulloblastomas (Northcott et al., 2009; Uziel et al., 2009).
4.3. BMP and TGF‐β signaling pathways
Bone morphogenetic proteins (BMPs) regulate a wide variety of biological processes that range from proliferation and differentiation to apoptosis, depending on developmental stage. Deregulation of the molecular effectors of BMP signaling may contribute to cancer (Fukuda and Taga, 2005; Plikus et al., 2008; Rendl et al., 2008). BMP2/4 induces differentiation of neural stem cells and glioblastoma CSCs. This signaling is impaired by the EZH2‐dependent epigenetic silencing of BMP receptor 1B (BMPR1B). Ectopic over‐expression of BMPR1B or demethylation of its promoter restores differentiation capabilities of BMP signaling, leading to loss of tumorigenicity (Lee et al., 2008). Furthermore, BMP‐6 has been identified as an inhibitor of breast cancer EMT by rescuing E‐cadherin expression (see below). Current data suggest that this process is mediated by the BMP‐6‐induced transcriptional inhibition of miR‐21, which is over‐expressed in aggressive breast cancers (Du et al., 2009). However, increased expression of miR‐21 also positively correlated with TGF‐β1 and it was found that TGF‐β upregulates the expression of miR‐21, facilitating tumor progression (Qian et al., 2009). TGF‐β acts as a tumor suppressor in tumor initiation and early steps of tumor progression and inactivation of TGF‐β tumor suppressor pathway is a main step in the development of a variety of human tumors. However, at late stage it induces tumor growth, EMT and metastasis (Majumdar et al., 2012). MiR‐106b‐25 and miR‐17‐92 clusters were described as key‐modulators of TGF‐β signaling (Petrocca et al., 2008). By functioning both upstream and downstream of pSMAD2, miR‐17‐92 activation triggers downregulation of multiple key effectors along the TGF‐β signaling cascade, as well as direct inhibition of TGF‐β‐responsive genes, which regulate tumor growth, migration and CSC function in various cancers (Dews et al., 2010; Ernst et al., 2010; Mestdagh et al., 2010; Tili et al., 2010). In addition, the elevated expression of miR‐181b/d correlated with upregulation of TGF‐β and its downstream mediators SMAD 2, 3 and 4 in a mouse model of hepatocellular carcinoma. In turn, miR‐181b was augmented upon exposure of hepatic cells to TGF‐β, showing the involvement of TGF‐β signaling pathway in miR‐181b expression. In turn, miR‐181b enhanced matrix metallopeptidases MMP2 and MMP9 activity and promoted growth, migration and invasion of hepatocellular carcinoma (Wang et al., 2010). In addition, TGF‐β can induce specific miRNA expression, such as miR‐23a/27a/24 cluster, which is up‐regulated in hepatocellular carcinoma (Huang et al., 2008) and miR‐155, induced in breast cancer (Kong et al., 2008), to escape from tumor‐suppressive response in developing tumors. Indeed, TGF‐β increased miR‐155 expression through Smad4 function and the knockdown of miR‐155 suppressed TGF‐β‐induced EMT, as well as cell migration and invasion. These data suggest that miR‐155 may play an important role in TGF‐β‐induced EMT and cell migration.
4.4. Notch signaling pathway
Notch signaling, a highly conserved regulatory signaling network, is crucial for the correct development and growth of numerous organs and tissues. When subverted, it can induce tumorigenesis at times, serving as an oncogene and at others, behaving as a tumor suppressor (Katoh, 2007; Miele et al., 2006; Weng et al., 2006). Notch is a transmembrane receptor, whose intracellular part is cleaved off upon binding of a specific ligand, producing the Notch intracellular domain (NICD). NICD is translocated to the nucleus, where it targets the DNA binding protein RBP‐Jkappa. In the absence of Notch, RBP‐Jkappa represses Notch target genes by recruiting a co‐repressor complex. Epigenetic modifications affecting Notch pathway at different levels (receptor, ligands or downstream effectors) have been associated with cancer development and progression. Indeed, Notch ligand Jagged 2 is over‐expressed in multiple myeloma, coincidental with an aberrantly acetylated JAGGED 2 promoter region. This acetylation is regulated by the recruitment of HDACs to promoter regions through interaction with nuclear co‐repressors such as SMRT. Hence, reduced levels of SMRT decreased the HDAC recruitment to the JAGGED 2 promoter region, leading to increase transcriptional expression and upregulation of Notch signaling in myeloma cell lines (Ghoshal et al., 2009). These findings match with the effects of the HDAC inhibitor valproic acid, inducing the expression of Notch genes and leading to an increase of the invasiveness of non‐invasive osteosarcoma cell lines (Hughes, 2009). Notch pathway is also regulated by miRNAs, and this process has been related to the promotion of tumor growth and invasion in different cancer types. In this regard, miR‐200c and miR‐141 directly inhibited Jagged 1, impeding proliferation of human metastatic prostate cancer cells (Vallejo et al., 2011). In addition, miR‐34a inhibited invasiveness through to downregulation of the expression of JAGGED 1 and JAGGED 2 in glioma cells (Li et al., 2009b; Pang et al., 2010). Enhanced expression of miR‐34a as well as the inhibition of Notch signaling suppressed the invasiveness in these cells and in osteosarcoma cells (Hughes, 2009; Pang et al., 2010). In addition, the involvement of miR‐34 by negatively regulating the Notch1 and Notch2 expression in self‐renewal/differentiation was recently demonstrated in pancreatic CSCs. These CSCs showed high levels of both receptors, coincidental to decreased expression of miR‐34 (Ji et al., 2008). The expression of Notch1 is also regulated by miR‐326 in glioma cells following a feedback loop, where Notch1 downregulates miR‐326 expression and this miRNA inhibit Notch proteins and activity (Kefas et al., 2009). Additionally, the expression of other Notch receptors such as Notch4 and Notch3 are respectively targets of miR‐181c, which in turn, exhibits a DNA methylation‐dependent silencing expression in some gastric carcinomas (Hashimoto et al., 2010). Other epigenetic mechanisms that activate Notch signaling were related to the role of miR‐146a, that targets Numb (Kuang et al., 2009), a negative regulator of Notch signaling found in a large proportion of breast carcinomas (Pece et al., 2004; Stylianou et al., 2006), and miR‐199b‐5p, that targets the transcription factor Hes‐1, inhibiting medulloblastoma cell growth. Moreover, over‐expression of miR‐199b‐5p decreased the CSCs (CD133+) and also blocked expression of several cancer stem‐cell genes (Garzia et al., 2009).
Together, these findings support the notion that a complex network of signaling pathways, responsible for controlling self‐renewal and differentiation fate in normal stem cells, can get deregulated due to the aberrant function of multiples epigenetic mechanisms during the course of tumorigenesis, inducing the proliferation and self‐renewal of CSCs. Interestingly some of these epigenetic alterations are also described in normal ESCs, indicating that the re‐acquisition of these may have a deep impact in CSC features essential to promote tumor progression and metastasis.
5. Epigenetic regulation of metastasis and chemotherapy response
Tumor cells dissemination and metastasis has been related to epithelial‐to‐mesenchymal transition (EMT), a trans‐differentiation program that converts adherent epithelial cells into individual migratory cells (Polyak and Weinberg, 2009). EMT and the reverse process, termed the mesenchymal‐to‐epithelial transition (MET), play central roles in embryogenesis (Perez‐Pomares and Munoz‐Chapuli, 2002; Thiery and Sleeman, 2006), as well as in cancer invasion and metastasis (Guarino et al., 2007; Polyak and Weinberg, 2009). EMT process in cancer involves disruption of normal epithelial integrity, loss of morphological features of polarized epithelia, including cell–cell adhesion, planar and apical–basal polarity and lack of motility, and the acquisition instead of mesenchymal features, such as motility, invasiveness and a heightened resistance to apoptosis (Polyak and Weinberg, 2009). This process is triggered by a diverse set of stimuli including growth factor signaling, tumor–stromal cell interactions and hypoxia, whose crosstalk can lead to reprogramming of epithelial cells to mesenchymal state. Currently, it is considered that disseminated cancer cells require self‐renewal capability, similar to that exhibited by stem cells, in order to produce macroscopic metastases. A direct link between the EMT and the gain of stem cell properties was previously demonstrated (Mani et al., 2008). Indeed, an increase in the proportion of immortalized human mammary epithelial cells exhibiting stem cell markers and an increased ability to form mammospheres, was observed after EMT induction. Furthermore, stem cells isolated from mouse and human mammary glands or mammary carcinomas expressed EMT markers (Mani et al., 2008). Hence, signaling pathways involved in the regulation of stem cell function and niche–stem cell interactions may play a role in triggering EMT, potentially by establishing and maintaining stem cell‐like characteristics. TGF‐β induces EMT through multiple signaling mechanisms (Massague, 2008; Yang and Weinberg, 2008) and by influencing the activities of other EMT‐inducing signal transduction pathways, including Notch, Wnt and integrin signaling (Polyak and Weinberg, 2009).
EMT is characterized by the loss of E‐cadherin expression, which emerges as a critical step (Dohadwala et al., 2006; Dumont et al., 2008; Gibbons et al., 2009). This results in the liberation of β‐catenin, which is normally sequestered by the cytoplasmic tail of E‐cadherin. The resulting free β‐catenin may then migrate to the nucleus and induce expression of EMT transcription factors (Polyak and Weinberg, 2009; Vincan and Barker, 2008). However, the activation of β‐catenin signaling, although necessary in some cells, may not be sufficient to orchestrate all the program leading to EMT, as some β‐catenin activated cancer cells do not necessary exhibit EMT markers. Loss of E‐cadherin function can be induced by mutations in its encoding gene (CDH1), as that identified in hereditary diffuse gastric cancer and in lobular breast carcinomas (Berx et al., 1996; Dunbier and Guilford, 2001). However, other mechanisms are implicated in the regulation of E‐cadherin expression, as direct inhibition by zinc finger transcriptional repressors ZEB1, ZEB2, Snail1, and Twist1 (Cano et al., 2000; Comijn et al., 2001; Eger et al., 2005; Yang et al., 2004) and epigenetic mechanisms as CpG hypermethylation and alterations of histone modifications (Lombaerts et al., 2006; Peinado et al., 2004). Methylation of CpG islands in the repressed E‐cadherin promoter result in recruitment of HDAC and histone deacetylation, which is essential for the silencing of E‐cadherin gene (Koizume et al., 2002; Wang and Shang, in press). E‐cadherin repression is also mediated by EZH2 and PRC2 complex, which is recruited to E‐cadherin promoter via Snail (Cao et al., 2008; Herranz et al., 2008). Indeed, EZH2 silencing results in inhibition of invasion and migration in different cancer cells. Together with BMI1, is essential for the anchorage‐independent growth of metastatic cancer cells (Crea et al., 2012a). In addition, the treatment of cancer cells with DNMT inhibitors increase invasiveness, tumorigenicity and metastatic capability, concomitantly with the upregulation of EMT‐associated genes (Ateeq et al., 2008; Guo et al., 2002). In this regard, research of global epigenetic changes associated to EMT identified promoter hypomethylation of a set of genes, which are highly expressed in breast CSCs (Bloushtain‐Qimron et al., 2008). In fact, the over‐expression of one of these factors, FOXC1, induced a complete EMT program in breast cancer cell lines and increased invasion and motility features (Bloushtain‐Qimron et al., 2008). On the other hand, global reduction in the heterochromatin mark H3K9Me2 and an increase in H3K4Me3 and H3K36Me3 was reported during the induction of EMT by TGF‐β. These changes depended largely on LSD1, and loss of LSD1 function had marked effects on EMT‐driven cell migration and chemoresistance (McDonald et al., 2012).
During the last years multiples miRNAs have been reported as modulators of transcription factors involved in EMT and metastasis. The miR‐200 family members (i.e miR‐200a, miR‐200b, miR‐200c, miR‐141), miR‐429, and miR‐205 are key determinants in the regulation of EMT and cancer cell invasion and migration by directly targeting ZEB1 and ZEB2 factors (Gregory et al., 2008; Korpal et al., 2008; Park et al., 2008). Loss of miR‐200 results in increased ZEB1 and ZEB2 levels, leading to the repression of E‐cadherin and EMT induction. However, a reciprocal feedback loop, in which ZEB1 and ZEB2 bind to promoter regions of the miR‐200 family to repress its transcription, was described (Bracken et al., 2008; Burk et al., 2008). MiR‐200 is also involved in the modulation of Sox2, Klf4, BMI1 and Suz12, previously described to regulate stemness in cancer cells (Gregory et al., 2008; Liu et al., 2012a; Shimono et al., 2009). However, other mechanisms may be involved in the regulation of miR‐200. Thus, the exposure of immortalized human bronchial epithelial cells (HBECs) to tobacco carcinogens induced a persistent and irreversible dedifferentiation program marked by EMT and the emergence of CSCs. EMT induction was initially driven by chromatin remodeling through H3K27me3 enrichment and later by DNA methylation to silence the expression of miR‐200c and miR‐205 (Tellez et al., 2011). Furthermore, p53 has shown to have a role in regulating both EMT and EMT‐associated stem cell properties by directly binding to the miR‐200c promoter and activating its expression. Loss of p53 in mammary epithelial cells leads to decreased expression of miR‐200c and activation of the EMT program and increased mammary stem cell population. Contrariwise, the enhanced p53 expression, induced by ectopic expression or by etoposide treatment, was able to reverse TGF‐β induced mesenchymal phenotype to an epithelial phenotype, blocking the E‐cadherin repression mediated by TGF‐β (Chang et al., 2011a). The relevance of the miR‐200 family is highlighted by the correlation between reduced levels of miR‐200c and an increased expression of EMT and stemness markers in a cohort of high grade breast tumors (Chang et al., 2011a). A link between DCAMKL‐1 (a microtubule‐associated kinase considered a stem cell marker) (May et al., 2008; May et al., 2010) which promotes pancreas tumorigenesis and EMT was established. Knockdown of DCAMKL‐1 in pancreatic cancer cells resulted in downregulation of Snail, Slug, and Twist and induction of miR‐200a (Sureban et al., 2011). Recent studies indicated that miR‐34a/c expression is significantly decreased in metastases and human primary tumors with lymph node metastases (Yang et al., in press) and the expression of miR‐34a was shown to be regulated by ZEB1 and by methylation of its promoter (Ahn et al., 2012; Yu et al., 2012).
More miRNAs have been related to EMT and metastasis, such as miR‐495. MiR‐495 is highly up‐regulated in breast CSCs and directly targets E‐cadherin and REDD1, promoting tumorigenesis and cell invasion under hypoxia conditions (Hwang‐Verslues et al., 2011). MiR‐21 and miR‐31 were prominently elevated under the synergistic actions of TGF‐β/TNF‐α in colon cancer cells. Consistent with this, over‐expression of either miR‐21 or miR‐31 significantly enhanced the effect of TGF‐β inducing EMT and invasiveness by directly targeting the expression of Tiam1, a guanidine exchange factor of the Rac GTPase. Therefore, miR‐21 and miR‐31 are downstream effectors of TGF‐β in facilitating invasion and metastasis of colon carcinoma cells (Cottonham et al., 2010). Accordingly, an increased expression of miR‐21 was observed in gastric and lung tumors correlating with poor prognosis (Liu et al., in press‐b; Xu et al., 2012). Further studies indicated that miR‐21 promotes invasion and metastasis by modulating PTEN expression through AKT and ERK1/2 pathways (Han et al., 2012; Liu et al., in press‐b).
Emerging evidences suggest that resistance to chemotherapy in tumors is associated with CSC features and with the acquisition of an EMT‐like phenotype (Shah et al., 2007; Wang et al., 2009b). Indeed, several pancreatic cancer cell lines with high expression of the E‐cadherin and low expression of the ZEB1 showed to be sensitive to three conventional chemotherapeutic agents (gemcitabine, 5 fluorouracil and cisplatin). By contrast, pancreatic cancer cell lines that showed EMT characteristics were resistant to these drugs (Arumugam et al., 2009). Furthermore, pancreatic cancer cells selected to be resistant to gemcitabine acquired EMT features, as well as increased cell migration and invasion capabilities (Arumugam et al., 2009). Interestingly, the reversion of the EMT phenotype by downregulation of Notch signaling, which resulted in decreased expression of vimentin, ZEB1, Snail1 and Slug and nuclear factor κB (NFκB), lead to decrease of resistance to chemotherapy (Shah et al., 2007). The above evidence suggests that the epigenetic modifications involved in the acquisition of these properties (EMT and CSC features) could have an impact in the response to chemotherapy. Hence, miR‐200 family is downregulated in pancreatic cancer cells resistant to gemcitabine, and the re‐expression of the miR‐200 resulted in resistant cells becoming sensitive to the drug (Li et al., 2009c). High levels of miR‐21 has been related to resistance to chemotherapy and poor survival in pancreatic cancer (Ali et al., 2010; Giovannetti et al., 2010; Hwang et al., 2010; Moriyama et al., 2009). Many members of the let7 family are also found downregulated in EMT‐type cells resistant to gemcitabine (Li et al., 2009c). Inhibition of miR‐221 lead to arrest of the cell cycle, induction of apoptosis and sensitization of pancreatic cancer cells to the effects of gemcitabine (Park et al., 2009). In addition, an upregulation of miR‐221/222 was reveled in breast cancer cell resistant to the fulvestrant. Ectopic expression of these miRNAs increase the resistance to this drug in breast cell lines and resulted in deregulation of multiple oncogenic signaling pathways previously associated with drug resistance. Activation of β‐catenin by miR‐221/222 contributed to fulvestrant resistance, whereas TGF‐β‐mediated growth inhibition was repressed by the two miRNAs (Rao et al., 2011).
ABCG2 is a ubiquitous ATP‐binding cassette transmembrane protein that plays a role in stem cell biology and clinical drug resistance. The increased expression of ABCG2 and the consequent acquisition of chemoresistance were associated with several chromatin modifications in the ABCG2 promoter, such as increased acetylated H3, H3K4me3 and H3 serine 10 phosphorylation and decreased HDAC1. Indeed, only those cells exhibiting permissive histone mark and lack of H3K9me3 repressive marks allowed recruitment of RNA polymerase II and Brg1 to the ABCG2 promoter, resulting in increased ABCG2 expression, suggesting that chromatin remodeling may impact the response to chemotherapy (To et al., 2008). Moreover, ABCG2 protein expression was shown to be regulated by miR‐328 and miR‐519c in breast cancer cells, as indicated by the downregulated expression of this transporter after ectopic expression of miR‐328‐ or 519c (Li et al., 2011). Recently it was reported that miR‐200c target ABCG2, ABCG5 and MDR1 and ectopic expression of this miRNA or downregulation of BMI1 reduced drug resistance and melanoma xenograft growth and metastasis in vivo (Liu et al., 2012a).
Altogether, these observations suggest that modulating the expression of miRNAs responsible for EMT, CSC phenotype and chemoresistance would improve the response to therapy. Hence, doxorubicin treatment of breast tumors growing in immunodeficient mice caused a significant regression of the tumor, but relapse of the disease was observed. Treatment of either miR‐200 or siRNA against Suz12 had only a very slight effect on tumor growth, presumably because these treatments did not affect NSCCs. Strikingly, the combinations of doxorubicin with either miR‐200b or Suz12 depletion caused even stronger regression of tumor growth, and relapse was prevented (Iliopoulos et al., 2010). Furthermore, the direct inhibition of miR‐21 or the reduction of its expression after treatment with SP600125, an inhibitor of the miR‐21‐regulator AP‐1, increased topotecan sensitivity of cancer cells (Misawa et al., 2010). These promising findings will provide a new strategy for cancer therapy by impairing the CSC resistance to chemotherapy.
6. Concluding remarks
In this review, we described the complex circuit of epigenetic mechanisms that contributes to the acquisition and maintenance of self‐renewal and stemness features by a population of cancer cells, resulting in the generation of cancer stem cells, as it is summarized in Figure 1. Indeed, histones modifiers and remodelers, DNA methyltransferases and miRNAs, appear severely deregulated in tumor cells, while a small subpopulation within the tumor emerge with stem cell‐like features. Multiple studies demonstrated that this population of cells conserves/acquire the expression of stem cell markers and in most of the cases, a hierarchical organization similar to that presented in the tissue of origin. Interestingly, the re‐expression of pluripotency‐associated genes and adult stem cell gene expression signature, as well as the upregulation of EMT‐ and metastasis‐related genes correlate with poor prognosis and with resistance to chemotherapy. Furthermore, several signaling pathways involved in the homeostasis of adult and sometimes embryonic stem cells are altered in cancer by epigenetic mechanisms, providing to CSC signals to self‐renewal, maintain an undifferentiated status and survive in the tumor microenvironment. In this regard, interaction with the tumor niche can provide signals to induce cancer cells migration and metastasis. However, further studies will be necessary to reveal specific signals that regulate, through epigenetic mechanisms, EMT and metastasis. Interestingly, the epigenetic origins of some of the aberrant signals that operate in tumor progression facilitate their reversion by specific inhibitors. In consequence, targeting the stemness‐like properties of this special population of cancer cells with agents that modify their epigenetic landscape can contribute to the sensitization of CSCs to chemotherapy, impeding tumor relapse. This is a rapidly emerging field in oncology and might represent a promising strategy for cancer therapy in the future.
Figure 1.
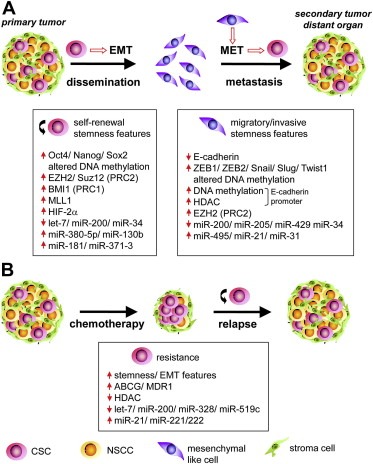
Summary representing the most prominent alterations of epigenetic pathways that regulate CSC/EMT features (A) and resistance to chemotherapy (B).
Muñoz Purificación, Iliou Maria S., Esteller Manel, (2012), Epigenetic alterations involved in cancer stem cell reprogramming, Molecular Oncology, 6, doi: 10.1016/j.molonc.2012.10.006.
References
- Abdouh, M. , Facchino, S. , Chatoo, W. , Balasingam, V. , Ferreira, J. , Bernier, G. , 2009. BMI1 sustains human glioblastoma multiforme stem cell renewal. J. Neurosci. 29, 8884–8896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn, Y.H. , Gibbons, D.L. , Chakravarti, D. , Creighton, C.J. , Rizvi, Z.H. , Adams, H.P. , Pertsemlidis, A. , Gregory, P.A. , Wright, J.A. , Goodall, G.J. , 2012. ZEB1 drives prometastatic actin cytoskeletal remodeling by downregulating miR-34a expression. J. Clin. Invest. 122, 3170–3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama, Y. , Maesawa, C. , Ogasawara, S. , Terashima, M. , Masuda, T. , 2003. Cell-type-specific repression of the maspin gene is disrupted frequently by demethylation at the promoter region in gastric intestinal metaplasia and cancer cells. Am. J. Pathol. 163, 1911–1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcantara Llaguno, S. , Chen, J. , Kwon, C.H. , Jackson, E.L. , Li, Y. , Burns, D.K. , Alvarez-Buylla, A. , Parada, L.F. , 2009. Malignant astrocytomas originate from neural stem/progenitor cells in a somatic tumor suppressor mouse model. Cancer Cell. 15, 45–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali, S. , Ahmad, A. , Banerjee, S. , Padhye, S. , Dominiak, K. , Schaffert, J.M. , Wang, Z. , Philip, P.A. , Sarkar, F.H. , 2010. Gemcitabine sensitivity can be induced in pancreatic cancer cells through modulation of miR-200 and miR-21 expression by curcumin or its analogue CDF. Cancer Res. 70, 3606–3617. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Arumugam, T. , Ramachandran, V. , Fournier, K.F. , Wang, H. , Marquis, L. , Abbruzzese, J.L. , Gallick, G.E. , Logsdon, C.D. , McConkey, D.J. , Choi, W. , 2009. Epithelial to mesenchymal transition contributes to drug resistance in pancreatic cancer. Cancer Res. 69, 5820–5828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ateeq, B. , Unterberger, A. , Szyf, M. , Rabbani, S.A. , 2008. Pharmacological inhibition of DNA methylation induces proinvasive and prometastatic genes in vitro and in vivo. Neoplasia. 10, 266–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba, T. , Convery, P.A. , Matsumura, N. , Whitaker, R.S. , Kondoh, E. , Perry, T. , Huang, Z. , Bentley, R.C. , Mori, S. , Fujii, S. , 2009. Epigenetic regulation of CD133 and tumorigenicity of CD133+ ovarian cancer cells. Oncogene. 28, 209–218. [DOI] [PubMed] [Google Scholar]
- Baccelli, I. , Trumpp, A. , 2012. The evolving concept of cancer and metastasis stem cells. J. Cell. Biol. 198, 281–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandres, E. , Agirre, X. , Bitarte, N. , Ramirez, N. , Zarate, R. , Roman-Gomez, J. , Prosper, F. , Garcia-Foncillas, J. , 2009. Epigenetic regulation of microRNA expression in colorectal cancer. Int. J. Cancer. 125, 2737–2743. [DOI] [PubMed] [Google Scholar]
- Bao, B. , Ali, S. , Banerjee, S. , Wang, Z. , Logna, F. , Azmi, A.S. , Kong, D. , Ahmad, A. , Li, Y. , Padhye, S. , Sarkar, F.H. , 2012. Curcumin analogue CDF inhibits pancreatic tumor growth by switching on suppressor microRNAs and attenuating EZH2 expression. Cancer Res. 72, 335–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar, E.E. , Lin, A. , Mahairaki, V. , Matsui, W. , Eberhart, C.G. , 2010. Hypoxia increases the expression of stem-cell markers and promotes clonogenicity in glioblastoma neurospheres. Am. J. Pathol. 177, 1491–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker, N. , Clevers, H. , 2006. Mining the Wnt pathway for cancer therapeutics. Nat. Rev. Drug Discov. 5, 997–1014. [DOI] [PubMed] [Google Scholar]
- Bernstein, B.E. , Humphrey, E.L. , Erlich, R.L. , Schneider, R. , Bouman, P. , Liu, J.S. , 2002. Proc. Natl. Acad. Sci. U.S.A. 99, (13) 8695–8700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berx, G. , Cleton-Jansen, A.M. , Strumane, K. , de Leeuw, W.J. , Nollet, F. , van Roy, F. , Cornelisse, C. , 1996. E-cadherin is inactivated in a majority of invasive human lobular breast cancers by truncation mutations throughout its extracellular domain. Oncogene. 13, 1919–1925. [PubMed] [Google Scholar]
- Biegel, J.A. , Fogelgren, B. , Zhou, J.Y. , James, C.D. , Janss, A.J. , Allen, J.C. , Zagzag, D. , Raffel, C. , Rorke, L.B. , 2000. Mutations of the INI1 rhabdoid tumor suppressor gene in medulloblastomas and primitive neuroectodermal tumors of the central nervous system. Clin. Cancer Res. 6, 2759–2763. [PubMed] [Google Scholar]
- Bird, A. , 2002. DNA methylation patterns and epigenetic memory. Genes Dev. 16, (1) 6–21. [DOI] [PubMed] [Google Scholar]
- Bissell, M.J. , Hines, W.C. , 2011. Why don't we get more cancer? A proposed role of the microenvironment in restraining cancer progression. Nat. Med. 17, 320–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloushtain-Qimron, N. , Yao, J. , Snyder, E.L. , Shipitsin, M. , Campbell, L.L. , Mani, S.A. , Hu, M. , Chen, H. , Ustyansky, V. , Antosiewicz, J.E. , 2008. Cell type-specific DNA methylation patterns in the human breast. Proc. Natl. Acad. Sci. U.S.A. 105, 14076–14081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bommer, G.T. , Gerin, I. , Feng, Y. , Kaczorowski, A.J. , Kuick, R. , Love, R.E. , Zhai, Y. , Giordano, T.J. , Qin, Z.S. , Moore, B.B. , 2007. p53-mediated activation of miRNA34 candidate tumor-suppressor genes. Curr. Biol. 17, 1298–1307. [DOI] [PubMed] [Google Scholar]
- Bonci, D. , Coppola, V. , Musumeci, M. , Addario, A. , Giuffrida, R. , Memeo, L. , D'Urso, L. , Pagliuca, A. , Biffoni, M. , Labbaye, C. , 2008. The miR-15a-miR-16-1 cluster controls prostate cancer by targeting multiple oncogenic activities. Nat. Med. 14, 1271–1277. [DOI] [PubMed] [Google Scholar]
- Bonnet, D. , Dick, J.E. , 1997. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat. Med. 3, 730–737. [DOI] [PubMed] [Google Scholar]
- Boominathan, L. , 2010. The guardians of the genome (p53, TA-p73, and TA-p63) are regulators of tumor suppressor miRNAs network. Cancer Metastasis Rev. 29, 613–639. [DOI] [PubMed] [Google Scholar]
- Bracken, A.P. , Dietrich, N. , Pasini, D. , Hansen, K.H. , Helin, K. , 2006. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev. 20, 1123–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken, C.P. , Gregory, P.A. , Kolesnikoff, N. , Bert, A.G. , Wang, J. , Shannon, M.F. , Goodall, G.J. , 2008. A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial-mesenchymal transition. Cancer Res. 68, 7846–7854. [DOI] [PubMed] [Google Scholar]
- Burdach, S. , Plehm, S. , Unland, R. , Dirksen, U. , Borkhardt, A. , Staege, M.S. , Muller-Tidow, C. , Richter, G.H. , 2009. Epigenetic maintenance of stemness and malignancy in peripheral neuroectodermal tumors by EZH2. Cell Cycle. 8, 1991–1996. [DOI] [PubMed] [Google Scholar]
- Burk, U. , Schubert, J. , Wellner, U. , Schmalhofer, O. , Vincan, E. , Spaderna, S. , Brabletz, T. , 2008. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 9, 582–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairo, S. , Wang, Y. , de Reynies, A. , Duroure, K. , Dahan, J. , Redon, M.J. , Fabre, M. , McClelland, M. , Wang, X.W. , Croce, C.M. , Buendia, M.A. , 2010. Stem cell-like micro-RNA signature driven by Myc in aggressive liver cancer. Proc. Natl. Acad. Sci. U.S.A. 107, 20471–20476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabrese, C. , Poppleton, H. , Kocak, M. , Hogg, T.L. , Fuller, C. , Hamner, B. , Oh, E.Y. , Gaber, M.W. , Finklestein, D. , Allen, M. , 2007. A perivascular niche for brain tumor stem cells. Cancer Cell. 11, 69–82. [DOI] [PubMed] [Google Scholar]
- Canettieri, G. , Di Marcotullio, L. , Greco, A. , Coni, S. , Antonucci, L. , Infante, P. , Pietrosanti, L. , De Smaele, E. , Ferretti, E. , Miele, E. , 2010. Histone deacetylase and Cullin3-REN(KCTD11) ubiquitin ligase interplay regulates Hedgehog signalling through Gli acetylation. Nat. Cell. Biol. 12, 132–142. [DOI] [PubMed] [Google Scholar]
- Cano, A. , Perez-Moreno, M.A. , Rodrigo, I. , Locascio, A. , Blanco, M.J. , del Barrio, M.G. , Portillo, F. , Nieto, M.A. , 2000. The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat. Cell. Biol. 2, 76–83. [DOI] [PubMed] [Google Scholar]
- Cao, Q. , Yu, J. , Dhanasekaran, S.M. , Kim, J.H. , Mani, R.S. , Tomlins, S.A. , Mehra, R. , Laxman, B. , Cao, X. , Yu, J. , 2008. Repression of E-cadherin by the polycomb group protein EZH2 in cancer. Oncogene. 27, 7274–7284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers, H. , 2006. Wnt/beta-catenin signaling in development and disease. Cell. 127, 469–480. [DOI] [PubMed] [Google Scholar]
- Comijn, J. , Berx, G. , Vermassen, P. , Verschueren, K. , van Grunsven, L. , Bruyneel, E. , Mareel, M. , Huylebroeck, D. , van Roy, F. , 2001. The two-handed E box binding zinc finger protein SIP1 downregulates E-cadherin and induces invasion. Mol. Cell. 7, 1267–1278. [DOI] [PubMed] [Google Scholar]
- Cottonham, C.L. , Kaneko, S. , Xu, L. , 2010. miR-21 and miR-31 converge on TIAM1 to regulate migration and invasion of colon carcinoma cells. J. Biol. Chem. 285, 35293–35302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cozzio, A. , Passegue, E. , Ayton, P.M. , Karsunky, H. , Cleary, M.L. , Weissman, I.L. , 2003. Similar MLL-associated leukemias arising from self-renewing stem cells and short-lived myeloid progenitors. Genes Dev. 17, 3029–3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crea, F. , Fornaro, L. , Bocci, G. , Sun, L. , Farrar, W.L. , Falcone, A. , Danesi, R. , 2012. EZH2 inhibition: targeting the crossroad of tumor invasion and angiogenesis. Cancer Metastasis Rev. 31, (3–4) 753–761. [DOI] [PubMed] [Google Scholar]
- Crea, F. , Hurt, E.M. , Mathews, L.A. , Cabarcas, S.M. , Sun, L. , Marquez, V.E. , Danesi, R. , Farrar, W.L. , 2012. Pharmacologic disruption of Polycomb Repressive Complex 2 inhibits tumorigenicity and tumor progression in prostate cancer. Mol. Cancer. 10, 40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui, H. , Cruz-Correa, M. , Giardiello, F.M. , Hutcheon, D.F. , Kafonek, D.R. , Brandenburg, S. , Wu, Y. , He, X. , Powe, N.R. , Feinberg, A.P. , 2003. Loss of IGF2 imprinting: a potential marker of colorectal cancer risk. Science. 299, 1753–1755. [DOI] [PubMed] [Google Scholar]
- Chang, C.J. , Chao, C.H. , Xia, W. , Yang, J.Y. , Xiong, Y. , Li, C.W. , Yu, W.H. , Rehman, S.K. , Hsu, J.L. , Lee, H.H. , 2011. p53 regulates epithelial-mesenchymal transition and stem cell properties through modulating miRNAs. Nat. Cell. Biol. 13, 317–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, C.J. , Yang, J.Y. , Xia, W. , Chen, C.T. , Xie, X. , Chao, C.H. , Woodward, W.A. , Hsu, J.M. , Hortobagyi, G.N. , Hung, M.C. , 2011. EZH2 promotes expansion of breast tumor initiating cells through activation of RAF1-beta-catenin signaling. Cancer Cell. 19, 86–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, T.C. , Wentzel, E.A. , Kent, O.A. , Ramachandran, K. , Mullendore, M. , Lee, K.H. , Feldmann, G. , Yamakuchi, M. , Ferlito, M. , Lowenstein, C.J. , 2007. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol. Cell. 26, 745–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J. , Li, Y. , Yu, T.S. , McKay, R.M. , Burns, D.K. , Kernie, S.G. , Parada, L.F. , 2012. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 488, 522–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou, S.H. , Wang, M.L. , Chou, Y.T. , Chen, C.J. , Hong, C.F. , Hsieh, W.J. , Chang, H.T. , Chen, Y.S. , Lin, T.W. , Hsu, H.S. , Wu, C.W. , 2010. Coexpression of Oct4 and Nanog enhances malignancy in lung adenocarcinoma by inducing cancer stem cell-like properties and epithelial-mesenchymal transdifferentiation. Cancer Res. 70, 10433–10444. [DOI] [PubMed] [Google Scholar]
- Dallas, N.A. , Xia, L. , Fan, F. , Gray, M.J. , Gaur, P. , van Buren, G. , Samuel, S. , Kim, M.P. , Lim, S.J. , Ellis, L.M. , 2009. Chemoresistant colorectal cancer cells, the cancer stem cell phenotype, and increased sensitivity to insulin-like growth factor-I receptor inhibition. Cancer Res. 69, 1951–1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet, C. , De Backer, O. , Faraoni, I. , Lurquin, C. , Brasseur, F. , Boon, T. , 1996. The activation of human gene MAGE-1 in tumor cells is correlated with genome-wide demethylation. Proc. Natl. Acad. Sci. U.S.A. 93, 7149–7153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dews, M. , Fox, J.L. , Hultine, S. , Sundaram, P. , Wang, W. , Liu, Y.Y. , Furth, E. , Enders, G.H. , El-Deiry, W. , Schelter, J.M. , 2010. The myc-miR-17∼92 axis blunts TGF{beta} signaling and production of multiple TGF{beta}-dependent antiangiogenic factors. Cancer Res. 70, 8233–8246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohadwala, M. , Yang, S.C. , Luo, J. , Sharma, S. , Batra, R.K. , Huang, M. , Lin, Y. , Goodglick, L. , Krysan, K. , Fishbein, M.C. , 2006. Cyclooxygenase-2-dependent regulation of E-cadherin: prostaglandin E(2) induces transcriptional repressors ZEB1 and snail in non-small cell lung cancer. Cancer Res. 66, 5338–5345. [DOI] [PubMed] [Google Scholar]
- Driessens, G. , Beck, B. , Caauwe, A. , Simons, B.D. , Blanpain, C. , 2012. Defining the mode of tumour growth by clonal analysis. Nature. 488, 527–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, J. , Yang, S. , An, D. , Hu, F. , Yuan, W. , Zhai, C. , Zhu, T. , 2009. BMP-6 inhibits microRNA-21 expression in breast cancer through repressing deltaEF1 and AP-1. Cell. Res. 19, 487–496. [DOI] [PubMed] [Google Scholar]
- Dumont, N. , Wilson, M.B. , Crawford, Y.G. , Reynolds, P.A. , Sigaroudinia, M. , Tlsty, T.D. , 2008. Sustained induction of epithelial to mesenchymal transition activates DNA methylation of genes silenced in basal-like breast cancers. Proc. Natl. Acad. Sci. U.S.A. 105, 14867–14872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunbier, A. , Guilford, P. , 2001. Hereditary diffuse gastric cancer. Adv. Cancer Res. 83, 55–65. [DOI] [PubMed] [Google Scholar]
- Dylla, S.J. , Beviglia, L. , Park, I.K. , Chartier, C. , Raval, J. , Ngan, L. , Pickell, K. , Aguilar, J. , Lazetic, S. , Smith-Berdan, S. , 2008. Colorectal cancer stem cells are enriched in xenogeneic tumors following chemotherapy. PLoS One. 3, e2428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberle, I. , Pless, B. , Braun, M. , Dingermann, T. , Marschalek, R. , 2010. Transcriptional properties of human NANOG1 and NANOG2 in acute leukemic cells. Nucleic Acids Res. 38, 5384–5395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden, A. , Gaudet, F. , Waghmare, A. , Jaenisch, R. , 2003. Chromosomal instability and tumors promoted by DNA hypomethylation. Science. 300, 455 [DOI] [PubMed] [Google Scholar]
- Eger, A. , Aigner, K. , Sonderegger, S. , Dampier, B. , Oehler, S. , Schreiber, M. , Berx, G. , Cano, A. , Beug, H. , Foisner, R. , 2005. DeltaEF1 is a transcriptional repressor of E-cadherin and regulates epithelial plasticity in breast cancer cells. Oncogene. 24, 2375–2385. [DOI] [PubMed] [Google Scholar]
- Eramo, A. , Ricci-Vitiani, L. , Zeuner, A. , Pallini, R. , Lotti, F. , Sette, G. , Pilozzi, E. , Larocca, L.M. , Peschle, C. , De Maria, R. , 2006. Chemotherapy resistance of glioblastoma stem cells. Cell. Death Differ. 13, 1238–1241. [DOI] [PubMed] [Google Scholar]
- Ernst, A. , Campos, B. , Meier, J. , Devens, F. , Liesenberg, F. , Wolter, M. , Reifenberger, G. , Herold-Mende, C. , Lichter, P. , Radlwimmer, B. , 2010. De-repression of CTGF via the miR-17-92 cluster upon differentiation of human glioblastoma spheroid cultures. Oncogene. 29, 3411–3422. [DOI] [PubMed] [Google Scholar]
- Esquela-Kerscher, A. , Slack, F.J. , 2006. Oncomirs – microRNAs with a role in cancer. Nat. Rev. Cancer. 6, 259–269. [DOI] [PubMed] [Google Scholar]
- Esteller, M. , 2008. Epigenetics in cancer. N. Engl. J. Med. 358, 1148–1159. [DOI] [PubMed] [Google Scholar]
- Fearon, E.R. , Vogelstein, B. , 1990. A genetic model for colorectal tumorigenesis. Cell. 61, 759–767. [DOI] [PubMed] [Google Scholar]
- Feinberg, A.P. , Ohlsson, R. , Henikoff, S. , 2006. The epigenetic progenitor origin of human cancer. Nat. Rev. Genet. 7, 21–33. [DOI] [PubMed] [Google Scholar]
- Ferretti, E. , De Smaele, E. , Miele, E. , Laneve, P. , Po, A. , Pelloni, M. , Paganelli, A. , Di Marcotullio, L. , Caffarelli, E. , Screpanti, I. , 2008. Concerted microRNA control of Hedgehog signalling in cerebellar neuronal progenitor and tumour cells. Embo J. 27, 2616–2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fodde, R. , Brabletz, T. , 2007. Wnt/beta-catenin signaling in cancer stemness and malignant behavior. Curr. Opin. Cell. Biol. 19, 150–158. [DOI] [PubMed] [Google Scholar]
- Fraga, M.F. , Ballestar, E. , Villar-Garea, A. , Boix-Chornet, M. , Espada, J. , Schotta, G. , Bonaldi, T. , Haydon, C. , Ropero, S. , Petrie, K. , 2005. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat. Genet. 37, 391–400. [DOI] [PubMed] [Google Scholar]
- Fukuda, S. , Taga, T. , 2005. Cell fate determination regulated by a transcriptional signal network in the developing mouse brain. Anat. Sci. Int. 80, 12–18. [DOI] [PubMed] [Google Scholar]
- Gangaraju, V.K. , Lin, H. , 2009. MicroRNAs: key regulators of stem cell. Nat. Rev. Mol. Cell. Biol. 10, (2) 116–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzia, L. , Andolfo, I. , Cusanelli, E. , Marino, N. , Petrosino, G. , De Martino, D. , Esposito, V. , Galeone, A. , Navas, L. , Esposito, S. , 2009. MicroRNA-199b-5p impairs cancer stem cells through negative regulation of HES1 in medulloblastoma. PLoS One. 4, e4998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghoshal, P. , Nganga, A.J. , Moran-Giuati, J. , Szafranek, A. , Johnson, T.R. , Bigelow, A.J. , Houde, C.M. , Avet-Loiseau, H. , Smiraglia, D.J. , Ersing, N. , 2009. Loss of the SMRT/NCoR2 corepressor correlates with JAG2 overexpression in multiple myeloma. Cancer Res. 69, 4380–4387. [DOI] [PubMed] [Google Scholar]
- Gibbons, D.L. , Lin, W. , Creighton, C.J. , Rizvi, Z.H. , Gregory, P.A. , Goodall, G.J. , Thilaganathan, N. , Du, L. , Zhang, Y. , Pertsemlidis, A. , Kurie, J.M. , 2009. Contextual extracellular cues promote tumor cell EMT and metastasis by regulating miR-200 family expression. Genes Dev. 23, 2140–2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannetti, E. , Funel, N. , Peters, G.J. , Del Chiaro, M. , Erozenci, L.A. , Vasile, E. , Leon, L.G. , Pollina, L.E. , Groen, A. , Falcone, A. , 2010. MicroRNA-21 in pancreatic cancer: correlation with clinical outcome and pharmacologic aspects underlying its role in the modulation of gemcitabine activity. Cancer Res. 70, 4528–4538. [DOI] [PubMed] [Google Scholar]
- Gopisetty, G., Xu, J., Sampath, D., Colman, H., Puduvalli, V.K. Epigenetic regulation of CD133/PROM1 expression in glioma stem cells by Sp1/myc and promoter methylation. Oncogene, in press. [DOI] [PMC free article] [PubMed]
- Grand, F. , Kulkarni, S. , Chase, A. , Goldman, J.M. , Gordon, M. , Cross, N.C. , 1999. Frequent deletion of hSNF5/INI1, a component of the SWI/SNF complex, in chronic myeloid leukemia. Cancer Res. 59, 3870–3874. [PubMed] [Google Scholar]
- Gregory, P.A. , Bert, A.G. , Paterson, E.L. , Barry, S.C. , Tsykin, A. , Farshid, G. , Vadas, M.A. , Khew-Goodall, Y. , Goodall, G.J. , 2008. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat. Cell. Biol. 10, 593–601. [DOI] [PubMed] [Google Scholar]
- Guarino, M. , Rubino, B. , Ballabio, G. , 2007. The role of epithelial-mesenchymal transition in cancer pathology. Pathology. 39, 305–318. [DOI] [PubMed] [Google Scholar]
- Guo, Y. , Pakneshan, P. , Gladu, J. , Slack, A. , Szyf, M. , Rabbani, S.A. , 2002. Regulation of DNA methylation in human breast cancer. Effect on the urokinase-type plasminogen activator gene production and tumor invasion. J. Biol. Chem. 277, 41571–41579. [DOI] [PubMed] [Google Scholar]
- Gupta, P.B. , Chaffer, C.L. , Weinberg, R.A. , 2009. Cancer stem cells: mirage or reality?. Nat. Med. 15, 1010–1012. [DOI] [PubMed] [Google Scholar]
- Halkidou, K. , Gaughan, L. , Cook, S. , Leung, H.Y. , Neal, D.E. , Robson, C.N. , 2004. Upregulation and nuclear recruitment of HDAC1 in hormone refractory prostate cancer. Prostate. 59, 177–189. [DOI] [PubMed] [Google Scholar]
- Han, M. , Liu, M. , Wang, Y. , Chen, X. , Xu, J. , Sun, Y. , Zhao, L. , Qu, H. , Fan, Y. , Wu, C. , 2012. Antagonism of miR-21 reverses epithelial-mesenchymal transition and cancer stem cell phenotype through AKT/ERK1/2 inactivation by targeting PTEN. PLoS One. 7, e39520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan, D. , Coussens, L.M. , 2012. Accessories to the crime: functions of cells recruited to the tumor microenvironment. Cancer Cell. 21, 309–322. [DOI] [PubMed] [Google Scholar]
- Hashimoto, Y. , Akiyama, Y. , Otsubo, T. , Shimada, S. , Yuasa, Y. , 2010. Involvement of epigenetically silenced microRNA-181c in gastric carcinogenesis. Carcinogenesis. 31, 777–784. [DOI] [PubMed] [Google Scholar]
- He, L. , He, X. , Lim, L.P. , de Stanchina, E. , Xuan, Z. , Liang, Y. , Xue, W. , Zender, L. , Magnus, J. , Ridzon, D. , 2007. A microRNA component of the p53 tumour suppressor network. Nature. 447, 1130–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heddleston, J.M. , Li, Z. , McLendon, R.E. , Hjelmeland, A.B. , Rich, J.N. , 2009. The hypoxic microenvironment maintains glioblastoma stem cells and promotes reprogramming towards a cancer stem cell phenotype. Cell Cycle. 8, 3274–3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heddleston, J.M. , Wu, Q. , Rivera, M. , Minhas, S. , Lathia, J.D. , Sloan, A.E. , Iliopoulos, O. , Hjelmeland, A.B. , Rich, J.N. , 2012. Hypoxia-induced mixed-lineage leukemia 1 regulates glioma stem cell tumorigenic potential. Cell. Death Differ. 19, 428–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann, P.C. , Huber, S.L. , Herrler, T. , Aicher, A. , Ellwart, J.W. , Guba, M. , Bruns, C.J. , Heeschen, C. , 2007. Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell. 1, 313–323. [DOI] [PubMed] [Google Scholar]
- Herranz, N. , Pasini, D. , Diaz, V.M. , Franci, C. , Gutierrez, A. , Dave, N. , Escriva, M. , Hernandez-Munoz, I. , Di Croce, L. , Helin, K. , 2008. Polycomb complex 2 is required for E-cadherin repression by the Snail1 transcription factor. Mol. Cell. Biol. 28, 4772–4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera-Merchan, A. , Arranz, L. , Ligos, J.M. , de Molina, A. , Dominguez, O. , Gonzalez, S. , 2012. Ectopic expression of the histone methyltransferase Ezh2 in haematopoietic stem cells causes myeloproliferative disease. Nat. Commun. 3, 623 [DOI] [PubMed] [Google Scholar]
- Hiltunen, M.O. , Alhonen, L. , Koistinaho, J. , Myohanen, S. , Paakkonen, M. , Marin, S. , Kosma, V.M. , Janne, J. , 1997. Hypermethylation of the APC (adenomatous polyposis coli) gene promoter region in human colorectal carcinoma. Int. J. Cancer. 70, 644–648. [DOI] [PubMed] [Google Scholar]
- Hirata, H. , Hinoda, Y. , Ueno, K. , Nakajima, K. , Ishii, N. , Dahiya, R. , 2012. MicroRNA-1826 directly targets beta-catenin (CTNNB1) and MEK1 (MAP2K1) in VHL-inactivated renal cancer. Carcinogenesis. 33, 501–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho, L. , Crabtree, G.R. , 2010. Chromatin remodelling during development. Nature. 463, (7280) 474–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm, T.M. , Jackson-Grusby, L. , Brambrink, T. , Yamada, Y. , Rideout, W.M. , Jaenisch, R. , 2005. Global loss of imprinting leads to widespread tumorigenesis in adult mice. Cancer Cell. 8, 275–285. [DOI] [PubMed] [Google Scholar]
- Huang, S. , He, X. , Ding, J. , Liang, L. , Zhao, Y. , Zhang, Z. , Yao, X. , Pan, Z. , Zhang, P. , Li, J. , 2008. Upregulation of miR-23a approximately 27a approximately 24 decreases transforming growth factor-beta-induced tumor-suppressive activities in human hepatocellular carcinoma cells. Int. J. Cancer. 123, 972–978. [DOI] [PubMed] [Google Scholar]
- Hughes, D.P. , 2009. How the NOTCH pathway contributes to the ability of osteosarcoma cells to metastasize. Cancer Treat. Res. 152, 479–496. [DOI] [PubMed] [Google Scholar]
- Hussain, M. , Rao, M. , Humphries, A.E. , Hong, J.A. , Liu, F. , Yang, M. , Caragacianu, D. , Schrump, D.S. , 2009. Tobacco smoke induces polycomb-mediated repression of Dickkopf-1 in lung cancer cells. Cancer Res. 69, 3570–3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang-Verslues, W.W. , Chang, P.H. , Wei, P.C. , Yang, C.Y. , Huang, C.K. , Kuo, W.H. , Shew, J.Y. , Chang, K.J. , Lee, E.Y. , Lee, W.H. , 2011. miR-495 is upregulated by E12/E47 in breast cancer stem cells, and promotes oncogenesis and hypoxia resistance via downregulation of E-cadherin and REDD1. Oncogene. 30, (21) 2463–2474. [DOI] [PubMed] [Google Scholar]
- Hwang, J.H. , Voortman, J. , Giovannetti, E. , Steinberg, S.M. , Leon, L.G. , Kim, Y.T. , Funel, N. , Park, J.K. , Kim, M.A. , Kang, G.H. , 2010. Identification of microRNA-21 as a biomarker for chemoresistance and clinical outcome following adjuvant therapy in resectable pancreatic cancer. PLoS One. 5, e10630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliopoulos, D. , Lindahl-Allen, M. , Polytarchou, C. , Hirsch, H.A. , Tsichlis, P.N. , Struhl, K. , 2010. Loss of miR-200 inhibition of Suz12 leads to polycomb-mediated repression required for the formation and maintenance of cancer stem cells. Mol. Cell. 39, 761–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham, P.W. , Placzek, M. , 2006. Orchestrating ontogenesis: variations on a theme by sonic hedgehog. Nat. Rev. Genet. 7, 841–850. [DOI] [PubMed] [Google Scholar]
- Inui, M. , Martello, G. , Piccolo, S. , 2010. MicroRNA control of signal transduction. Nat. Rev. Mol. Cell. Biol. 11, 252–263. [DOI] [PubMed] [Google Scholar]
- Jagani, Z. , Mora-Blanco, E.L. , Sansam, C.G. , McKenna, E.S. , Wilson, B. , Chen, D. , Klekota, J. , Tamayo, P. , Nguyen, P.T. , Tolstorukov, M. , 2010. Loss of the tumor suppressor Snf5 leads to aberrant activation of the Hedgehog-Gli pathway. Nat. Med. 16, 1429–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji, J. , Yamashita, T. , Budhu, A. , Forgues, M. , Jia, H.L. , Li, C. , Deng, C. , Wauthier, E. , Reid, L.M. , Ye, Q.H. , 2009. Identification of microRNA-181 by genome-wide screening as a critical player in EpCAM-positive hepatic cancer stem cells. Hepatology. 50, 472–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji, Q. , Hao, X. , Meng, Y. , Zhang, M. , Desano, J. , Fan, D. , Xu, L. , 2008. Restoration of tumor suppressor miR-34 inhibits human p53-mutant gastric cancer tumorspheres. BMC Cancer. 8, 266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji, Q. , Hao, X. , Zhang, M. , Tang, W. , Yang, M. , Li, L. , Xiang, D. , Desano, J.T. , Bommer, G.T. , Fan, D. , 2009. MicroRNA miR-34 inhibits human pancreatic cancer tumor-initiating cells. PLoS One. 4, e6816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, J. , Hui, C.C. , 2008. Hedgehog signaling in development and cancer. Dev. Cell. 15, 801–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, X. , Tan, J. , Li, J. , Kivimae, S. , Yang, X. , Zhuang, L. , Lee, P.L. , Chan, M.T. , Stanton, L.W. , Liu, E.T. , 2008. DACT3 is an epigenetic regulator of Wnt/beta-catenin signaling in colorectal cancer and is a therapeutic target of histone modifications. Cancer Cell. 13, 529–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, X. , Jeon, H.Y. , Joo, K.M. , Kim, J.K. , Jin, J. , Kim, S.H. , Kang, B.G. , Beck, S. , Lee, S.J. , Kim, J.K. , 2011. Frizzled 4 regulates stemness and invasiveness of migrating glioma cells established by serial intracranial transplantation. Cancer Res. 71, (8) 3066–3075. [DOI] [PubMed] [Google Scholar]
- Johnson, S.M. , Grosshans, H. , Shingara, J. , Byrom, M. , Jarvis, R. , Cheng, A. , Labourier, E. , Reinert, K.L. , Brown, D. , Slack, F.J. , 2005. RAS is regulated by the let-7 microRNA family. Cell. 120, 635–647. [DOI] [PubMed] [Google Scholar]
- Jones, P.A. , Baylin, S.B. , 2007. The epigenomics of cancer. Cell. 128, 683–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, S. , Wang, T.L. , Shih Ie, M. , Mao, T.L. , Nakayama, K. , Roden, R. , Glas, R. , Slamon, D. , Diaz, L.A. , Vogelstein, B. , 2010. Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science. 330, 228–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagara, N. , Huynh, K.T. , Kuo, C. , Okano, H. , Sim, M.S. , Elashoff, D. , Chong, K. , Giuliano, A.E. , Hoon, D.S. , 2012. Epigenetic regulation of cancer stem cell genes in triple-negative breast cancer. Am. J. Pathol. 181, 257–267. [DOI] [PubMed] [Google Scholar]
- Katoh, M. , 2007. Networking of WNT, FGF, Notch, BMP, and Hedgehog signaling pathways during carcinogenesis. Stem Cell. Rev. 3, 30–38. [DOI] [PubMed] [Google Scholar]
- Kefas, B. , Comeau, L. , Floyd, D.H. , Seleverstov, O. , Godlewski, J. , Schmittgen, T. , Jiang, J. , diPierro, C.G. , Li, Y. , Chiocca, E.A. , 2009. The neuronal microRNA miR-326 acts in a feedback loop with notch and has therapeutic potential against brain tumors. J. Neurosci. 29, 15161–15168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirmizis, A. , Bartley, S.M. , Kuzmichev, A. , Margueron, R. , Reinberg, D. , Green, R. , Farnham, P.J. , 2004. Silencing of human polycomb target genes is associated with methylation of histone H3 Lys 27. Genes Dev. 18, 1592–1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klarmann, G.J. , Decker, A. , Farrar, W.L. , 2008. Epigenetic gene silencing in the Wnt pathway in breast cancer. Epigenetics. 3, 59–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose, R.J. , Bird, A.P. , 2006. Genomic DNA methylation: the mark and its mediators. Trends Biochem. Sci. 31, (2) 89–97. [DOI] [PubMed] [Google Scholar]
- Koizume, S. , Tachibana, K. , Sekiya, T. , Hirohashi, S. , Shiraishi, M. , 2002. Heterogeneity in the modification and involvement of chromatin components of the CpG island of the silenced human CDH1 gene in cancer cells. Nucleic Acids Res. 30, 4770–4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong, D. , Banerjee, S. , Ahmad, A. , Li, Y. , Wang, Z. , Sethi, S. , Sarkar, F.H. , 2010. Epithelial to mesenchymal transition is mechanistically linked with stem cell signatures in prostate cancer cells. PLoS One. 5, e12445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong, D. , Heath, E. , Chen, W. , Cher, M.L. , Powell, I. , Heilbrun, L. , Li, Y. , Ali, S. , Sethi, S. , Hassan, O. , 2012. Loss of let-7 up-regulates EZH2 in prostate cancer consistent with the acquisition of cancer stem cell signatures that are attenuated by BR-DIM. PLoS One. 7, e33729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong, W. , Yang, H. , He, L. , Zhao, J.J. , Coppola, D. , Dalton, W.S. , Cheng, J.Q. , 2008. MicroRNA-155 is regulated by the transforming growth factor beta/Smad pathway and contributes to epithelial cell plasticity by targeting RhoA. Mol. Cell. Biol. 28, 6773–6784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korpal, M. , Lee, E.S. , Hu, G. , Kang, Y. , 2008. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J. Biol. Chem. 283, 14910–14914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krivtsov, A.V. , Armstrong, S.A. , 2007. MLL translocations, histone modifications and leukaemia stem-cell development. Nat. Rev. Cancer. 7, 823–833. [DOI] [PubMed] [Google Scholar]
- Krivtsov, A.V. , Twomey, D. , Feng, Z. , Stubbs, M.C. , Wang, Y. , Faber, J. , Levine, J.E. , Wang, J. , Hahn, W.C. , Gilliland, D.G. , 2006. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature. 442, 818–822. [DOI] [PubMed] [Google Scholar]
- Kuang, W. , Tan, J. , Duan, Y. , Duan, J. , Wang, W. , Jin, F. , Jin, Z. , Yuan, X. , Liu, Y. , 2009. Cyclic stretch induced miR-146a upregulation delays C2C12 myogenic differentiation through inhibition of Numb. Biochem. Biophys. Res. Commun. 378, 259–263. [DOI] [PubMed] [Google Scholar]
- Lapidot, T. , Sirard, C. , Vormoor, J. , Murdoch, B. , Hoang, T. , Caceres-Cortes, J. , Minden, M. , Paterson, B. , Caligiuri, M.A. , Dick, J.E. , 1994. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 367, 645–648. [DOI] [PubMed] [Google Scholar]
- Lapouge, G. , Youssef, K.K. , Vokaer, B. , Achouri, Y. , Michaux, C. , Sotiropoulou, P.A. , Blanpain, C. , 2011. Identifying the cellular origin of squamous skin tumors. Proc. Natl. Acad. Sci. U.S.A. 108, (18) 7431–7436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, K.K. , Workman, J.L. , 2007. Hisotne acetyltransferase complex: one size doesn't fit all. Nat. Rev. Mol. Cell. Biol. 8, (4) 284–295. [DOI] [PubMed] [Google Scholar]
- Lee, J. , Son, M.J. , Woolard, K. , Donin, N.M. , Li, A. , Cheng, C.H. , Kotliarova, S. , Kotliarov, Y. , Walling, J. , Ahn, S. , 2008. Epigenetic-mediated dysfunction of the bone morphogenetic protein pathway inhibits differentiation of glioblastoma-initiating cells. Cancer Cell. 13, 69–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, T.I. , Jenner, R.G. , Boyer, L.A. , Guenther, M.G. , Levine, S.S. , Kumar, R.M. , Chevalier, B. , Johnstone, S.E. , Cole, M.F. , Isono, K. , 2006. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 125, 301–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X. , Gonzalez, M.E. , Toy, K. , Filzen, T. , Merajver, S.D. , Kleer, C.G. , 2009. Targeted overexpression of EZH2 in the mammary gland disrupts ductal morphogenesis and causes epithelial hyperplasia. Am. J. Pathol. 175, 1246–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X. , Pan, Y.Z. , Seigel, G.M. , Hu, Z.H. , Huang, M. , Yu, A.M. , 2011. Breast cancer resistance protein BCRP/ABCG2 regulatory microRNAs (hsa-miR-328, -519c and -520h) and their differential expression in stem-like ABCG2+ cancer cells. Biochem. Pharmacol. 81, 783–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y. , Guessous, F. , Zhang, Y. , Dipierro, C. , Kefas, B. , Johnson, E. , Marcinkiewicz, L. , Jiang, J. , Yang, Y. , Schmittgen, T.D. , 2009. MicroRNA-34a inhibits glioblastoma growth by targeting multiple oncogenes. Cancer Res. 69, 7569–7576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y. , VandenBoom, T.G. , Kong, D. , Wang, Z. , Ali, S. , Philip, P.A. , Sarkar, F.H. , 2009. Up-regulation of miR-200 and let-7 by natural agents leads to the reversal of epithelial-to-mesenchymal transition in gemcitabine-resistant pancreatic cancer cells. Cancer Res. 69, 6704–6712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Z. , Rich, J.N. , 2010. Hypoxia and hypoxia inducible factors in cancer stem cell maintenance. Curr. Top. Microbiol. Immunol. 345, 21–30. [DOI] [PubMed] [Google Scholar]
- Lindvall, C. , Bu, W. , Williams, B.O. , Li, Y. , 2007. Wnt signaling, stem cells, and the cellular origin of breast cancer. Stem Cell. Rev. 3, 157–168. [DOI] [PubMed] [Google Scholar]
- Liu, C. , Kelnar, K. , Liu, B. , Chen, X. , Calhoun-Davis, T. , Li, H. , Patrawala, L. , Yan, H. , Jeter, C. , Honorio, S. , 2011. The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nat. Med. 17, 211–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, S. , Tetzlaff, M.T. , Cai, R. , Xu, X. , 2012. miR-200c inhibits melanoma progression and drug resistance through down-regulation of Bmi-1. Am. J. Pathol. 181, (5) 1823–1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Z.L., Wang, H., Liu, J., Wang, Z.X. MicroRNA-21 (miR-21) expression promotes growth, metastasis, and chemo- or radioresistance in non-small cell lung cancer cells by targeting PTEN. Mol Cell Biochem., in press-b. (PMID: 22956424). [DOI] [PubMed]
- Lo, W.L. , Yu, C.C. , Chiou, G.Y. , Chen, Y.W. , Huang, P.I. , Chien, C.S. , Tseng, L.M. , Chu, P.Y. , Lu, K.H. , Chang, K.W. , 2011. MicroRNA-200c attenuates tumour growth and metastasis of presumptive head and neck squamous cell carcinoma stem cells. J. Pathol. 223, 482–495. [DOI] [PubMed] [Google Scholar]
- Lombaerts, M. , van Wezel, T. , Philippo, K. , Dierssen, J.W. , Zimmerman, R.M. , Oosting, J. , van Eijk, R. , Eilers, P.H. , van de Water, B. , Cornelisse, C.J. , Cleton-Jansen, A.M. , 2006. E-cadherin transcriptional downregulation by promoter methylation but not mutation is related to epithelial-to-mesenchymal transition in breast cancer cell lines. Br. J. Cancer. 94, 661–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lujambio, A. , Ropero, S. , Ballestar, E. , Fraga, M.F. , Cerrato, C. , Setien, F. , Casado, S. , Suarez-Gauthier, A. , Sanchez-Cespedes, M. , Git, A. , 2007. Genetic unmasking of an epigenetically silenced microRNA in human cancer cells. Cancer Res. 67, 1424–1429. [DOI] [PubMed] [Google Scholar]
- Ma, S. , Lee, T.K. , Zheng, B.J. , Chan, K.W. , Guan, X.Y. , 2008. CD133+ HCC cancer stem cells confer chemoresistance by preferential expression of the Akt/PKB survival pathway. Oncogene. 27, 1749–1758. [DOI] [PubMed] [Google Scholar]
- Ma, S. , Tang, K.H. , Chan, Y.P. , Lee, T.K. , Kwan, P.S. , Castilho, A. , Ng, I. , Man, K. , Wong, N. , To, K.F. , 2010. miR-130b Promotes CD133(+) liver tumor-initiating cell growth and self-renewal via tumor protein 53-induced nuclear protein 1. Cell Stem Cell. 7, 694–707. [DOI] [PubMed] [Google Scholar]
- Majumdar, A. , Curley, S.A. , Wu, X. , Brown, P. , Hwang, J.P. , Shetty, K. , Yao, Z.X. , He, A.R. , Li, S. , Katz, L. , 2012. Hepatic stem cells and transforming growth factor beta in hepatocellular carcinoma. Nat. Rev. Gastroenterol. Hepatol. 9, 530–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani, S.A. , Guo, W. , Liao, M.J. , Eaton, E.N. , Ayyanan, A. , Zhou, A.Y. , Brooks, M. , Reinhard, F. , Zhang, C.C. , Shipitsin, M. , 2008. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 133, 704–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massague, J. , 2008. TGFbeta in Cancer. Cell. 134, 215–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May, R. , Riehl, T.E. , Hunt, C. , Sureban, S.M. , Anant, S. , Houchen, C.W. , 2008. Identification of a novel putative gastrointestinal stem cell and adenoma stem cell marker, doublecortin and CaM kinase-like-1, following radiation injury and in adenomatous polyposis coli/multiple intestinal neoplasia mice. Stem Cells. 26, 630–637. [DOI] [PubMed] [Google Scholar]
- May, R. , Sureban, S.M. , Lightfoot, S.A. , Hoskins, A.B. , Brackett, D.J. , Postier, R.G. , Ramanujam, R. , Rao, C.V. , Wyche, J.H. , Anant, S. , Houchen, C.W. , 2010. Identification of a novel putative pancreatic stem/progenitor cell marker DCAMKL-1 in normal mouse pancreas. Am. J. Physiol. Gastrointest. Liver Physiol. 299, G303–G310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald, O.G. , Wu, H. , Timp, W. , Doi, A. , Feinberg, A.P. , 2012. Genome-scale epigenetic reprogramming during epithelial-to-mesenchymal transition. Nat. Struct. Mol. Biol. 18, 867–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlos-Suarez, A. , Barriga, F.M. , Jung, P. , Iglesias, M. , Cespedes, M.V. , Rossell, D. , Sevillano, M. , Hernando-Momblona, X. , da Silva-Diz, V. , Munoz, P. , 2012. The intestinal stem cell signature identifies colorectal cancer stem cells and predicts disease relapse. Cell Stem Cell. 8, 511–524. [DOI] [PubMed] [Google Scholar]
- Mestdagh, P. , Bostrom, A.K. , Impens, F. , Fredlund, E. , Van Peer, G. , De Antonellis, P. , von Stedingk, K. , Ghesquiere, B. , Schulte, S. , Dews, M. , 2010. The miR-17-92 microRNA cluster regulates multiple components of the TGF-beta pathway in neuroblastoma. Mol. Cell. 40, 762–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miele, L. , Miao, H. , Nickoloff, B.J. , 2006. NOTCH signaling as a novel cancer therapeutic target. Curr. Cancer Drug Targets. 6, 313–323. [DOI] [PubMed] [Google Scholar]
- Misawa, A. , Katayama, R. , Koike, S. , Tomida, A. , Watanabe, T. , Fujita, N. , 2010. AP-1-Dependent miR-21 expression contributes to chemoresistance in cancer stem cell-like SP cells. Oncol. Res. 19, 23–33. [DOI] [PubMed] [Google Scholar]
- Mohyeldin, A. , Garzon-Muvdi, T. , Quinones-Hinojosa, A. , 2010. Oxygen in stem cell biology: a critical component of the stem cell niche. Cell Stem Cell. 7, 150–161. [DOI] [PubMed] [Google Scholar]
- Moriyama, T. , Ohuchida, K. , Mizumoto, K. , Yu, J. , Sato, N. , Nabae, T. , Takahata, S. , Toma, H. , Nagai, E. , Tanaka, M. , 2009. MicroRNA-21 modulates biological functions of pancreatic cancer cells including their proliferation, invasion, and chemoresistance. Mol. Cancer Ther. 8, 1067–1074. [DOI] [PubMed] [Google Scholar]
- Nagel, R. , le Sage, C. , Diosdado, B. , van der Waal, M. , Oude Vrielink, J.A. , Bolijn, A. , Meijer, G.A. , Agami, R. , 2008. Regulation of the adenomatous polyposis coli gene by the miR-135 family in colorectal cancer. Cancer Res. 68, 5795–5802. [DOI] [PubMed] [Google Scholar]
- Nakamura, N. , Takenaga, K. , 1998. Hypomethylation of the metastasis-associated S100A4 gene correlates with gene activation in human colon adenocarcinoma cell lines. Clin. Exp. Metastasis. 16, 471–479. [DOI] [PubMed] [Google Scholar]
- Nishigaki, M. , Aoyagi, K. , Danjoh, I. , Fukaya, M. , Yanagihara, K. , Sakamoto, H. , Yoshida, T. , Sasaki, H. , 2005. Discovery of aberrant expression of R-RAS by cancer-linked DNA hypomethylation in gastric cancer using microarrays. Cancer Res. 65, 2115–2124. [DOI] [PubMed] [Google Scholar]
- Northcott, P.A. , Fernandez, L.A. , Hagan, J.P. , Ellison, D.W. , Grajkowska, W. , Gillespie, Y. , Grundy, R. , Van Meter, T. , Rutka, J.T. , Croce, C.M. , 2009. The miR-17/92 polycistron is up-regulated in sonic hedgehog-driven medulloblastomas and induced by N-myc in sonic hedgehog-treated cerebellar neural precursors. Cancer Res. 69, 3249–3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa, O. , Eccles, M.R. , Szeto, J. , McNoe, L.A. , Yun, K. , Maw, M.A. , Smith, P.J. , Reeve, A.E. , 1993. Relaxation of insulin-like growth factor II gene imprinting implicated in Wilms' tumour. Nature. 362, 749–751. [DOI] [PubMed] [Google Scholar]
- Ohm, J.E. , McGarvey, K.M. , Yu, X. , Cheng, L. , Schuebel, K.E. , Cope, L. , Mohammad, H.P. , Chen, W. , Daniel, V.C. , Yu, W. , 2007. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat. Genet. 39, 237–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano, M. , Bell, D.W. , Hber, D.A. , Li, E. , 1999. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 99, (3) 247–257. [DOI] [PubMed] [Google Scholar]
- Otte, A.P. , Kwaks, T.H. , 2003. Gene expression by Polycomb group of protein complexes: a distinct complex for every occasion?. Curr. Opin. Genet. Dev. 13, (5) 448–454. [DOI] [PubMed] [Google Scholar]
- Pang, R.T. , Leung, C.O. , Ye, T.M. , Liu, W. , Chiu, P.C. , Lam, K.K. , Lee, K.F. , Yeung, W.S. , 2010. MicroRNA-34a suppresses invasion through downregulation of Notch1 and Jagged1 in cervical carcinoma and choriocarcinoma cells. Carcinogenesis. 31, 1037–1044. [DOI] [PubMed] [Google Scholar]
- Park, J.K. , Lee, E.J. , Esau, C. , Schmittgen, T.D. , 2009. Antisense inhibition of microRNA-21 or -221 arrests cell cycle, induces apoptosis, and sensitizes the effects of gemcitabine in pancreatic adenocarcinoma. Pancreas. 38, e190–199. [DOI] [PubMed] [Google Scholar]
- Park, S.M. , Gaur, A.B. , Lengyel, E. , Peter, M.E. , 2008. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 22, 894–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasca di Magliano, M. , Hebrok, M. , 2003. Hedgehog signalling in cancer formation and maintenance. Nat. Rev. Cancer. 3, 903–911. [DOI] [PubMed] [Google Scholar]
- Pece, S. , Serresi, M. , Santolini, E. , Capra, M. , Hulleman, E. , Galimberti, V. , Zurrida, S. , Maisonneuve, P. , Viale, G. , Di Fiore, P.P. , 2004. Loss of negative regulation by Numb over Notch is relevant to human breast carcinogenesis. J. Cell. Biol. 167, 215–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peinado, H. , Ballestar, E. , Esteller, M. , Cano, A. , 2004. Snail mediates E-cadherin repression by the recruitment of the Sin3A/histone deacetylase 1 (HDAC1)/HDAC2 complex. Mol. Cell. Biol. 24, 306–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Pomares, J.M. , Munoz-Chapuli, R. , 2002. Epithelial-mesenchymal transitions: a mesodermal cell strategy for evolutive innovation in Metazoans. Anat. Rec. 268, 343–351. [DOI] [PubMed] [Google Scholar]
- Petrocca, F. , Vecchione, A. , Croce, C.M. , 2008. Emerging role of miR-106b-25/miR-17-92 clusters in the control of transforming growth factor beta signaling. Cancer Res. 68, 8191–8194. [DOI] [PubMed] [Google Scholar]
- Pierce, G.B. , Speers, W.C. , 1988. Tumors as caricatures of the process of tissue renewal: prospects for therapy by directing differentiation. Cancer Res. 48, 1996–2004. [PubMed] [Google Scholar]
- Plikus, M.V. , Mayer, J.A. , de la Cruz, D. , Baker, R.E. , Maini, P.K. , Maxson, R. , Chuong, C.M. , 2008. Cyclic dermal BMP signalling regulates stem cell activation during hair regeneration. Nature. 451, 340–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polakis, P. , 2000. Wnt signaling and cancer. Genes Dev. 14, 1837–1851. [PubMed] [Google Scholar]
- Polyak, K. , Weinberg, R.A. , 2009. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat. Rev. Cancer. 9, 265–273. [DOI] [PubMed] [Google Scholar]
- Qian, B. , Katsaros, D. , Lu, L. , Preti, M. , Durando, A. , Arisio, R. , Mu, L. , Yu, H. , 2009. High miR-21 expression in breast cancer associated with poor disease-free survival in early stage disease and high TGF-beta1. Breast Cancer Res. Treat. 117, 131–140. [DOI] [PubMed] [Google Scholar]
- Quintana, E. , Shackleton, M. , Sabel, M.S. , Fullen, D.R. , Johnson, T.M. , Morrison, S.J. , 2008. Efficient tumour formation by single human melanoma cells. Nature. 456, 593–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao, X. , Di Leva, G. , Li, M. , Fang, F. , Devlin, C. , Hartman-Frey, C. , Burow, M.E. , Ivan, M. , Croce, C.M. , Nephew, K.P. , 2011. MicroRNA-221/222 confers breast cancer fulvestrant resistance by regulating multiple signaling pathways. Oncogene. 30, 1082–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raver-Shapira, N. , Marciano, E. , Meiri, E. , Spector, Y. , Rosenfeld, N. , Moskovits, N. , Bentwich, Z. , Oren, M. , 2007. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol. Cell. 26, 731–743. [DOI] [PubMed] [Google Scholar]
- Reisman, D. , Glaros, S. , Thompson, E.A. , 2009. The SWI/SNF complex and cancer. Oncogene. 28, 1653–1668. [DOI] [PubMed] [Google Scholar]
- Reisman, D.N. , Sciarrotta, J. , Wang, W. , Funkhouser, W.K. , Weissman, B.E. , 2003. Loss of BRG1/BRM in human lung cancer cell lines and primary lung cancers: correlation with poor prognosis. Cancer Res. 63, 560–566. [PubMed] [Google Scholar]
- Rendl, M. , Polak, L. , Fuchs, E. , 2008. BMP signaling in dermal papilla cells is required for their hair follicle-inductive properties. Genes Dev. 22, 543–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringrose, L. , Paro, R. , 2004. Epigenetic regulation of cellular memory by Polycomb and THritorax group proteins. Annu. Rev. Genet. 38, 413–443. [DOI] [PubMed] [Google Scholar]
- Rizzo, S. , Hersey, J.M. , Mellor, P. , Dai, W. , Santos-Silva, A. , Liber, D. , Luk, L. , Titley, I. , Carden, C.P. , Box, G. , 2011. Ovarian cancer stem cell-like side populations are enriched following chemotherapy and overexpress EZH2. Mol. Cancer Ther. 10, 325–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts, C.W. , Orkin, S.H. , 2004. The SWI/SNF complex–chromatin and cancer. Nat. Rev. Cancer. 4, 133–142. [DOI] [PubMed] [Google Scholar]
- Santos-Rosa, H. , Schneider, R. , Bannister, A.J. , Sherriff, J. , Bernstein, B.E. , Emre, N.C. , 2002. Active genes are tri-methylated at K4 of histone H3. Nature. 419, (6905) 407–411. [DOI] [PubMed] [Google Scholar]
- Saydam, O. , Shen, Y. , Wurdinger, T. , Senol, O. , Boke, E. , James, M.F. , Tannous, B.A. , Stemmer-Rachamimov, A.O. , Yi, M. , Stephens, R.M. , 2009. Downregulated microRNA-200a in meningiomas promotes tumor growth by reducing E-cadherin and activating the Wnt/beta-catenin signaling pathway. Mol. Cell. Biol. 29, 5923–5940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepers, A.G. , Snippert, H.J. , Stange, D.E. , van den Born, M. , van Es, J.H. , van de Wetering, M. , Clevers, H. , 2012. Lineage tracing reveals Lgr5+ stem cell activity in mouse intestinal adenomas. Science. 337, 730–735. [DOI] [PubMed] [Google Scholar]
- Schlesinger, Y. , Straussman, R. , Keshet, I. , Farkash, S. , Hecht, M. , Zimmerman, J. , Eden, E. , Yakhini, Z. , Ben-Shushan, E. , Reubinoff, B.E. , 2007. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat. Genet. 39, 232–236. [DOI] [PubMed] [Google Scholar]
- Schoenhals, M. , Kassambara, A. , De Vos, J. , Hose, D. , Moreaux, J. , Klein, B. , 2009. Embryonic stem cell markers expression in cancers. Biochem. Biophys. Res. Commun. 383, 157–162. [DOI] [PubMed] [Google Scholar]
- Seidel, S. , Garvalov, B.K. , Wirta, V. , von Stechow, L. , Schanzer, A. , Meletis, K. , Wolter, M. , Sommerlad, D. , Henze, A.T. , Nister, M. , 2010. A hypoxic niche regulates glioblastoma stem cells through hypoxia inducible factor 2 alpha. Brain. 133, 983–995. [DOI] [PubMed] [Google Scholar]
- Seligson, D.B. , Horvath, S. , Shi, T. , Yu, H. , Tze, S. , Grunstein, M. , Kurdistani, S.K. , 2005. Global histone modification patterns predict risk of prostate cancer recurrence. Nature. 435, 1262–1266. [DOI] [PubMed] [Google Scholar]
- Shah, A.N. , Summy, J.M. , Zhang, J. , Park, S.I. , Parikh, N.U. , Gallick, G.E. , 2007. Development and characterization of gemcitabine-resistant pancreatic tumor cells. Ann. Surg. Oncol. 14, 3629–3637. [DOI] [PubMed] [Google Scholar]
- Shiao, S.L. , Ganesan, A.P. , Rugo, H.S. , Coussens, L.M. , 2011. Immune microenvironments in solid tumors: new targets for therapy. Genes Dev. 25, 2559–2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimono, Y. , Zabala, M. , Cho, R.W. , Lobo, N. , Dalerba, P. , Qian, D. , Diehn, M. , Liu, H. , Panula, S.P. , Chiao, E. , 2009. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell. 138, 592–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu, M.K., Wong, E.S., Kong, D.S., Chan, H.Y., Jiang, L., Wong, O.G., Lam, E.W., Chan, K.K., Ngan, H.Y., Le, X.F., Cheung, A. N. Stem cell transcription factor NANOG controls cell migration and invasion via dysregulation of E-cadherin and FoxJ1 and contributes to adverse clinical outcome in ovarian cancers. Oncogene, in press. [DOI] [PubMed]
- Somervaille, T.C. , Cleary, M.L. , 2006. Identification and characterization of leukemia stem cells in murine MLL-AF9 acute myeloid leukemia. Cancer Cell. 10, 257–268. [DOI] [PubMed] [Google Scholar]
- Song, J. , Noh, J.H. , Lee, J.H. , Eun, J.W. , Ahn, Y.M. , Kim, S.Y. , Lee, S.H. , Park, W.S. , Yoo, N.J. , Lee, J.Y. , Nam, S.W. , 2005. Increased expression of histone deacetylase 2 is found in human gastric cancer. APMIS. 113, 264–268. [DOI] [PubMed] [Google Scholar]
- Sparmann, A. , van Lohuizen, M. , 2006. Polycomb silencers control cell fate, development and cancer. Nat. Rev. Cancer. 6, 846–856. [DOI] [PubMed] [Google Scholar]
- Stylianou, S. , Clarke, R.B. , Brennan, K. , 2006. Aberrant activation of notch signaling in human breast cancer. Cancer Res. 66, 1517–1525. [DOI] [PubMed] [Google Scholar]
- Sun, J.F. , Sui, J.L. , Zhou, P.K. , Geng, Y. , Hu, Y.C. , Cao, Z.S. , Ge, S.L. , Lou, T.Z. , Wu, D.C. , 2002. Decreased efficiency of gamma-ray-induced DNA double-strand break rejoining in malignant transformants of human bronchial epithelial cells generated by alpha-particle exposure. Int. J. Radiat. Biol. 78, 773–780. [DOI] [PubMed] [Google Scholar]
- Sun, J.Y. , Huang, Y. , Li, J.P. , Zhang, X. , Wang, L. , Meng, Y.L. , Yan, B. , Bian, Y.Q. , Zhao, J. , Wang, W.Z. , 2012. MicroRNA-320a suppresses human colon cancer cell proliferation by directly targeting beta-catenin. Biochem. Biophys. Res. Commun. 420, 787–792. [DOI] [PubMed] [Google Scholar]
- Sureban, S.M. , May, R. , Lightfoot, S.A. , Hoskins, A.B. , Lerner, M. , Brackett, D.J. , Postier, R.G. , Ramanujam, R. , Mohammed, A. , Rao, C.V. , 2011. DCAMKL-1 regulates epithelial-mesenchymal transition in human pancreatic cells through a miR-200a-dependent mechanism. Cancer Res. 71, 2328–2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suva, M.L. , Riggi, N. , Janiszewska, M. , Radovanovic, I. , Provero, P. , Stehle, J.C. , Baumer, K. , Le Bitoux, M.A. , Marino, D. , Cironi, L. , 2009. EZH2 is essential for glioblastoma cancer stem cell maintenance. Cancer Res. 69, 9211–9218. [DOI] [PubMed] [Google Scholar]
- Suzuki, H. , Watkins, D.N. , Jair, K.W. , Schuebel, K.E. , Markowitz, S.D. , Chen, W.D. , Pretlow, T.P. , Yang, B. , Akiyama, Y. , Van Engeland, M. , 2004. Epigenetic inactivation of SFRP genes allows constitutive WNT signaling in colorectal cancer. Nat. Genet. 36, 417–422. [DOI] [PubMed] [Google Scholar]
- Swarbrick, A. , Woods, S.L. , Shaw, A. , Balakrishnan, A. , Phua, Y. , Nguyen, A. , Chanthery, Y. , Lim, L. , Ashton, L.J. , Judson, R.L. , 2010. miR-380-5p represses p53 to control cellular survival and is associated with poor outcome in MYCN-amplified neuroblastoma. Nat. Med. 16, 1134–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarasov, V. , Jung, P. , Verdoodt, B. , Lodygin, D. , Epanchintsev, A. , Menssen, A. , Meister, G. , Hermeking, H. , 2007. Differential regulation of microRNAs by p53 revealed by massively parallel sequencing: miR-34a is a p53 target that induces apoptosis and G1-arrest. Cell Cycle. 6, 1586–1593. [DOI] [PubMed] [Google Scholar]
- Tellez, C.S. , Juri, D.E. , Do, K. , Bernauer, A.M. , Thomas, C.L. , Damiani, L.A. , Tessema, M. , Leng, S. , Belinsky, S.A. , 2011. EMT and stem cell-like properties associated with miR-205 and miR-200 epigenetic silencing are early manifestations during carcinogen-induced transformation of human lung epithelial cells. Cancer Res. 71, (8) 3087–3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiery, J.P. , Sleeman, J.P. , 2006. Complex networks orchestrate epithelial-mesenchymal transitions. Nat. Rev. Mol. Cell. Biol. 7, 131–142. [DOI] [PubMed] [Google Scholar]
- Tili, E. , Michaille, J.J. , Liu, C.G. , Alder, H. , Taccioli, C. , Volinia, S. , Calin, G.A. , Croce, C.M. , 2010. GAM/ZFp/ZNF512B is central to a gene sensor circuitry involving cell-cycle regulators, TGF{beta} effectors, Drosha and microRNAs with opposite oncogenic potentials. Nucleic Acids Res. 38, 7673–7688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To, K.K. , Polgar, O. , Huff, L.M. , Morisaki, K. , Bates, S.E. , 2008. Histone modifications at the ABCG2 promoter following treatment with histone deacetylase inhibitor mirror those in multidrug-resistant cells. Mol. Cancer Res. 6, 151–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todaro, M. , Alea, M.P. , Di Stefano, A.B. , Cammareri, P. , Vermeulen, L. , Iovino, F. , Tripodo, C. , Russo, A. , Gulotta, G. , Medema, J.P. , Stassi, G. , 2007. Colon cancer stem cells dictate tumor growth and resist cell death by production of interleukin-4. Cell Stem Cell. 1, 389–402. [DOI] [PubMed] [Google Scholar]
- Torres-Padilla, M.E. , Parfitt, D.E. , Kouzarides, T. , Zernicka-Goetz, M. , 2007. Histone arginine methylation regulates pluripotency in the early mouse embryo. Nature. 445, 214–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trowbridge, J.J. , Sinha, A.U. , Zhu, N. , Li, M. , Armstrong, S.A. , Orkin, S.H. , 2012. Haploinsufficiency of Dnmt1 impairs leukemia stem cell function through derepression of bivalent chromatin domains. Genes Dev. 26, 344–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai, H.C. , Baylin, S.B. , 2011. Cancer epigenetics: linking basic biology to clinical medicine. Cell. Res. 21, 502–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang, D.P. , Cheng, A.S. , 2011. Epigenetic regulation of signaling pathways in cancer: role of the histone methyltransferase EZH2. J. Gastroenterol. Hepatol. 26, 19–27. [DOI] [PubMed] [Google Scholar]
- Uziel, T. , Karginov, F.V. , Xie, S. , Parker, J.S. , Wang, Y.D. , Gajjar, A. , He, L. , Ellison, D. , Gilbertson, R.J. , Hannon, G. , Roussel, M.F. , 2009. The miR-17∼92 cluster collaborates with the Sonic Hedgehog pathway in medulloblastoma. Proc. Natl. Acad. Sci. U.S.A. 106, 2812–2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallejo, D.M. , Caparros, E. , Dominguez, M. , 2011. Targeting Notch signalling by the conserved miR-8/200 microRNA family in development and cancer cells. Embo J. 30, 756–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen, L. , De Sousa, E.M.F. , van der Heijden, M. , Cameron, K. , de Jong, J.H. , Borovski, T. , Tuynman, J.B. , Todaro, M. , Merz, C. , Rodermond, H. , 2010. Wnt activity defines colon cancer stem cells and is regulated by the microenvironment. Nat. Cell. Biol. 12, 468–476. [DOI] [PubMed] [Google Scholar]
- Vincan, E. , Barker, N. , 2008. The upstream components of the Wnt signalling pathway in the dynamic EMT and MET associated with colorectal cancer progression. Clin. Exp. Metastasis. 25, 657–663. [DOI] [PubMed] [Google Scholar]
- Vire, E. , Brenner, C. , Deplus, R. , Blanchon, L. , Fraga, M. , Didelot, C. , Morey, L. , Van Eynde, A. , Bernard, D. , Vanderwinden, J.M. , 2006. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 439, 871–874. [DOI] [PubMed] [Google Scholar]
- Visvader, J.E. , 2011. Cells of origin in cancer. Nature. 469, 314–322. [DOI] [PubMed] [Google Scholar]
- Viswanathan, S.R. , Powers, J.T. , Einhorn, W. , Hoshida, Y. , Ng, T.L. , Toffanin, S. , O'Sullivan, M. , Lu, J. , Phillips, L.A. , Lockhart, V.L. , 2009. Lin28 promotes transformation and is associated with advanced human malignancies. Nat. Genet. 41, 843–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein, B. , Kinzler, K.W. , 2004. Cancer genes and the pathways they control. Nat. Med. 10, 789–799. [DOI] [PubMed] [Google Scholar]
- Volinia, S. , Calin, G.A. , Liu, C.G. , Ambs, S. , Cimmino, A. , Petrocca, F. , Visone, R. , Iorio, M. , Roldo, C. , Ferracin, M. , 2006. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. U.S.A. 103, 2257–2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, B. , Hsu, S.H. , Majumder, S. , Kutay, H. , Huang, W. , Jacob, S.T. , Ghoshal, K. , 2010. TGFbeta-mediated upregulation of hepatic miR-181b promotes hepatocarcinogenesis by targeting TIMP3. Oncogene. 29, 1787–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J. , Lu, F. , Ren, Q. , Sun, H. , Xu, Z. , Lan, R. , Liu, Y. , Ward, D. , Quan, J. , Ye, T. , Zhang, H. , 2011. Novel histone demethylase LSD1 inhibitors selectively target cancer cells with pluripotent stem cell properties. Cancer Res. 71, 7238–7249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X. , Lam, E.K. , Zhang, J. , Jin, H. , Sung, J.J. , 2009. MicroRNA-122a functions as a novel tumor suppressor downstream of adenomatous polyposis coli in gastrointestinal cancers. Biochem. Biophys. Res. Commun. 387, 376–380. [DOI] [PubMed] [Google Scholar]
- Wang, Y., Shang, Y. Epigenetic control of epithelial-to-mesenchymal transition and cancer metastasis. Exp Cell Res., in press. (pii: S0014-4827(12)00361-8). [DOI] [PubMed]
- Wang, Z. , Li, Y. , Kong, D. , Banerjee, S. , Ahmad, A. , Azmi, A.S. , Ali, S. , Abbruzzese, J.L. , Gallick, G.E. , Sarkar, F.H. , 2009. Acquisition of epithelial-mesenchymal transition phenotype of gemcitabine-resistant pancreatic cancer cells is linked with activation of the notch signaling pathway. Cancer Res. 69, 2400–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellner, U. , Schubert, J. , Burk, U.C. , Schmalhofer, O. , Zhu, F. , Sonntag, A. , Waldvogel, B. , Vannier, C. , Darling, D. , zur Hausen, A. , 2009. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat. Cell. Biol. 11, 1487–1495. [DOI] [PubMed] [Google Scholar]
- Weng, A.P. , Millholland, J.M. , Yashiro-Ohtani, Y. , Arcangeli, M.L. , Lau, A. , Wai, C. , Del Bianco, C. , Rodriguez, C.G. , Sai, H. , Tobias, J. , 2006. c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes Dev. 20, 2096–2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, A.C. , Tran, K. , Khuu, J. , Dang, C. , Cui, Y. , Binder, S.W. , Lowry, W.E. , 2011. Defining the origins of Ras/p53-mediated squamous cell carcinoma. Proc. Natl. Acad. Sci. U.S.A. 108, (18) 7425–7430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widschwendter, M. , Fiegl, H. , Egle, D. , Mueller-Holzner, E. , Spizzo, G. , Marth, C. , Weisenberger, D.J. , Campan, M. , Young, J. , Jacobs, I. , Laird, P.W. , 2007. Epigenetic stem cell signature in cancer. Nat. Genet. 39, 157–158. [DOI] [PubMed] [Google Scholar]
- Wong, D.J. , Liu, H. , Ridky, T.W. , Cassarino, D. , Segal, E. , Chang, H.Y. , 2008. Module map of stem cell genes guides creation of epithelial cancer stem cells. Cell Stem Cell. 2, 333–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Y. , Sun, J. , Xu, J. , Li, Q. , Guo, Y. , Zhang, Q. , 2012. miR-21 is a promising novel biomarker for lymph node metastasis in patients with gastric cancer. Gastroenterol. Res. Pract. 2012, 640168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, J. , Chai, L. , Liu, F. , Fink, L.M. , Lin, P. , Silberstein, L.E. , Amin, H.M. , Ward, D.C. , Ma, Y. , 2007. Bmi-1 is a target gene for SALL4 in hematopoietic and leukemic cells. Proc. Natl. Acad. Sci. U.S.A. 104, 10494–10499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, J. , Mani, S.A. , Donaher, J.L. , Ramaswamy, S. , Itzykson, R.A. , Come, C. , Savagner, P. , Gitelman, I. , Richardson, A. , Weinberg, R.A. , 2004. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 117, 927–939. [DOI] [PubMed] [Google Scholar]
- Yang, J. , Weinberg, R.A. , 2008. Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev. Cell. 14, 818–829. [DOI] [PubMed] [Google Scholar]
- Yang, S., Li, Y., Gao, J., Zhang, T., Li, S., Luo, A., Chen, H., Ding, F., Wang, X., Liu, Z. MicroRNA-34 suppresses breast cancer invasion and metastasis by directly targeting Fra-1. Oncogene, in press. [DOI] [PubMed]
- Yang, X. , Lin, X. , Zhong, X. , Kaur, S. , Li, N. , Liang, S. , Lassus, H. , Wang, L. , Katsaros, D. , Montone, K. , 2010. Double-negative feedback loop between reprogramming factor LIN28 and microRNA let-7 regulates aldehyde dehydrogenase 1-positive cancer stem cells. Cancer Res. 70, 9463–9472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, X.J. , 2004. The diverse superfamily of lysine acetyltransferases and their roles in leukemia and other diseases. Nucleic Acids Res. 32, 959–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi, J.M. , Tsai, H.C. , Glockner, S.C. , Lin, S. , Ohm, J.E. , Easwaran, H. , James, C.D. , Costello, J.F. , Riggins, G. , Eberhart, C.G. , 2008. Abnormal DNA methylation of CD133 in colorectal and glioblastoma tumors. Cancer Res. 68, 8094–8103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, F. , Jiao, Y. , Zhu, Y. , Wang, Y. , Zhu, J. , Cui, X. , Liu, Y. , He, Y. , Park, E.Y. , Zhang, H. , 2012. MicroRNA 34c gene down-regulation via DNA methylation promotes self-renewal and epithelial-mesenchymal transition in breast tumor-initiating cells. J. Biol. Chem. 287, 465–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, F. , Yao, H. , Zhu, P. , Zhang, X. , Pan, Q. , Gong, C. , Huang, Y. , Hu, X. , Su, F. , Lieberman, J. , Song, E. , 2007. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 131, 1109–1123. [DOI] [PubMed] [Google Scholar]
- Zhang, H. , Li, W. , Nan, F. , Ren, F. , Wang, H. , Xu, Y. , Zhang, F. , 2011. MicroRNA expression profile of colon cancer stem-like cells in HT29 adenocarcinoma cell line. Biochem. Biophys. Res. Commun. 404, 273–278. [DOI] [PubMed] [Google Scholar]
- Zhang, W. , Glockner, S.C. , Guo, M. , Machida, E.O. , Wang, D.H. , Easwaran, H. , Van Neste, L. , Herman, J.G. , Schuebel, K.E. , Watkins, D.N. , 2008. Epigenetic inactivation of the canonical Wnt antagonist SRY-box containing gene 17 in colorectal cancer. Cancer Res. 68, 2764–2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Z. , Tavoosidana, G. , Sjolinder, M. , Gondor, A. , Mariano, P. , Wang, S. , Kanduri, C. , Lezcano, M. , Sandhu, K.S. , Singh, U. , 2006. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat. Genet. 38, 1341–1347. [DOI] [PubMed] [Google Scholar]
- Zhou, A.D. , Diao, L.T. , Xu, H. , Xiao, Z.D. , Li, J.H. , Zhou, H. , Qu, L.H. , 2012. beta-Catenin/LEF1 transactivates the microRNA-371-373 cluster that modulates the Wnt/beta-catenin-signaling pathway. Oncogene. 31, 2968–2978. [DOI] [PubMed] [Google Scholar]


