Abstract
The Estrogen Receptor (ER) is an established predictive marker for the selection of adjuvant endocrine treatment in early breast cancer. During the 1990s Immunohistochemistry (IHC) replaced cytosol based assays for determination of ER status. This study examined the association between ER protein level determined by two different methods and ESR1 gene copy number. From 289 primary high‐risk breast cancer patients, randomized in the Danish Breast Cancer Cooperative Group (DBCG) 77C trial, results from cytosolic ER levels were available from ligand binding assays. Archival tumor tissue was retrieved from 257 patients. ESR1/CEN‐6 ratio was analyzed successfully by Fluorescence In Situ Hybridization (FISH) in 220 (86%) patients. ESR1 amplification (ESR1/CEN‐6 ≥ 2.00) was observed in 23% of the patients and ESR1 deletion (ESR1/CEN‐6 < 0.80) was observed in 32%. Further, we identified ESR1 gain (ratio ESR1/CEN‐6 from 1.30 to 1.99) in 19% of the patients. A positive correlation of ESR1 FISH with both ER‐cytosol and ER IHC was found (p < 0.0001). Amplification and gain of the ESR1 gene are associated with higher ER protein content measured by ligand binding assay and a more intense nuclear staining by IHC compared to tumors with normal ESR1 gene status. Major variations in ER measured by ligand binding assay and IHC are observed within all ESR1 copy number subgroups and other mechanisms than gene copy number seem to contribute to the ER protein content in the tumors.
Keywords: Estrogen receptor, ESR1, ER, FISH, Immunohistochemistry
Highlights
The Estrogen Receptor (ER) is an established predictive marker for adjuvant endocrine treatment in early breast cancer.
We examined the association between ER protein level determined by two different methods and ESR1 gene copy number.
ESR1/CEN‐6 ratio was analyzed successfully by Fluorescence In Situ Hybridization (FISH).
ESR1 amplification (ESR1/CEN‐6 ≥ 2.00) was observed in 23% of the cases.
A positive correlation of ESR1 FISH with both ER‐cytosol and ER IHC was found (p < 0.0001).
1. Introduction
Estrogen Receptor (ER) positivity is a predictor for response to adjuvant endocrine therapy in breast cancer and ER determination should be performed in all invasive breast tumors. Originally, from the 1970s to early 1990s, very sensitive biochemical ligand binding assays were used. Initially analyses were based on the dextran‐coated charcoal (DCC) method which was used for ER analysis in this study. In brief, cytosols of homogenized fresh frozen tumor tissue were incubated with radioactively labeled estradiol and the unbound estradiol was removed after absorption to dextran‐coated charcoal (DeSombre et al., 1979). Being less methodological demanding, requiring less tissue and yielding concurrent histological information, immunohistochemical (IHC) evaluation of ER replaced the ligand binding assays during the 1990s (Grabau et al., 2000). Retrospective studies comparing ER values measured by IHC and ligand binding assays have shown good concordance ranging between 86 and 88% (Harvey et al., 1999; Khoshnoud et al., 2011; Regan et al., 2006).
Quantitatively, ER level determined by ligand binding assay spans a larger range, from zero to several thousands fmol/mg and expression of ER in 80–100% of tumors cells may correspond to ER content of 31–2126 fmol/mg (Grabau et al., 2000). Interestingly, IHC has been unable to detect the negative prognostic impact demonstrated from a high ER content measured by ligand binding assay (Thorpe et al., 1993). Presently, IHC is the standard method for determining ER (Hammond et al., 2010) with approximately 80% of malignant breast tumors being ER positive (Allred et al., 2009; Talman et al., 2008) and ER positive patients benefit the most from endocrine treatment (Viale et al., 2007). Nevertheless, at 10‐years follow up 26% of the patients receiving adjuvant endocrine treatment experience relapse (Davies et al., 2011) and consequently research has focused on both identification of other potential important biomarkers reflecting active ER signaling (Drury et al., 2011; Henriksen et al., 2009) and on the impact of the ESR1 gene level (Schuur and Weigel, 2000).
Recently, the existence of amplifications (Ejlertsen et al., 2011; Holst et al., 2007; Nielsen et al., 2011b; Tomita et al., 2009) and deletions (Ejlertsen et al., 2011; Nielsen et al., 2011b) in the ESR1 gene encoding ER has been documented by Fluorescence in Situ Hybridization (FISH) and several other methods as qPCR, Southern analysis, Comparative Genome Hybridization, and Chromogen In Situ Hybridization. By FISH the frequency of ESR1 gene amplifications in invasive breast tumors range from 13 to 23.7% (Ejlertsen et al., 2011; Holst et al., 2007; Nielsen et al., 2011b; Tomita et al., 2009), and has also been reported in carcinoma in situ (Burkhardt et al., 2010). In a previous study (Nielsen et al., 2011b) we observed ESR1 deletions with a very low frequency (5%) among ER positive samples. Thus ESR1 gene copy number is mirrored by ER expression level, but not in complete agreement (Holst et al., 2007; Tomita et al., 2009). The present study was designed to investigate the correlation of ESR1 gene status with the ER protein level measured by both ligand binding assay and IHC in a high‐risk breast cancer cohort and to further explore the existence of deletions in this patient cohort which also includes ER negative patients as well. Finally, we wanted to investigate whether tumors with very high ER‐cytosol level constituted a separate group of patients with respect to ESR1 gene alterations. We hypothesize that ESR1 copy number alterations may assist explaining discordant results when comparing ER expression by IHC with ER content determined by ligand binding assay, especially in ER poor and highly ER positive tumors.
2. Material and methods
2.1. Patients
Patients in this study represent a subset of 289 participants in the DBCG 77C trial. Postmenopausal patients were eligible for the 77C trial following mastectomy and axillary sampling if they met at least one of the following criteria: node positive, tumor size above 50 mm, or involvement of skin or deep fascia, and were without signs of distant metastasis. All patients received radiotherapy. Between August 1977 and November 1982, 1716 eligible patients were randomly allocated to radiotherapy or to radiotherapy and additional 1 year of tamoxifen (Knoop et al., 2001). At the time ER analysis was not standard procedure but for 289 of the 1716 patients ER was assessed by ligand binding assay and for the present study archival paraffin embedded tumor tissue was identified from 257 of the 289 patients with known results from the ER ligand binding assay (Table 1).
Table 1.
Tumor samples available for the study
| Tumor samples from the DBCG 77‐C available for the presented study | Number of samples |
|---|---|
| Number of tumor samples with ER protein values (DCC) | 289 |
| Number of tumor samples available for ESR1 FISH analysis | 257 |
| Number of tumor samples with successful ESR1 FISH analysis | 220 |
| Number of tumor samples with analyses for both ER protein values (DCC), ER (IHC) and ESR1 FISH | 220 |
2.2. Dextran‐coated charcoal assay (DCC)
The DCC steroid binding assay on tumor cytosols was introduced in 1977 by the DBCG and performed as described in the EORTC publication from 1979 (EORTC, 1973). The total binding capacity of high affinity receptors as well as the dissociation constant were determined. Fresh frozen tumor tissue was homogenized and cytosol prepared as described previously (EORTC, 1973). The cytosol was incubated overnight at 2–4 °C with 3H‐17β‐estradiol and unbound ligand removed by adding a dextran‐coated charcoal suspension (EORTC, 1973). Finally, the radioactivity in an aliquot of the supernatant was determined by liquid scintillation counting. The biochemical assays were performed at three different laboratories in Denmark (Talman et al., 2008). Values above or equal to 10 fmol ER/mg protein were considered positive. Values ≥108 fmol/mg protein were considered very high positive (Thorpe et al., 1993).
2.3. Immunohistochemical staining
Thin tissue sections (5 μm) were mounted on Dako Chemmate Capillary Gap© microscope slides and immunostained using Techmate 1000© (Dako). Heat induced antigen retrieval was performed in citrate buffer in microwave oven for 25 min. Slides with primary antibody ER1D5 (1:200) were incubated over night and the Chemmate labeled streptavidin biotin kit (K5001; Dako) was applied as detection system. Staining was performed according to the Dako streptavidin‐peroxidase protocol. Both positive and negative controls were implemented. In samples from 149 patients, the percentage of ER positive tumor cells was scored semiquantitatively by bright field microscopy as 10% intervals, and the remaining were classified as negative (0–9%), low positive (10–74%), or high positive (≥75%). Only nuclear staining was considered positive. ER IHC was retrospectively performed for patients enrolled in DBCG 77C (Knoop et al., 2001).
2.4. Fluorescence in situ hybridization (FISH) analysis
ESR1 copy number was analyzed with Dako Histology FISH Accessory Kit (K5599, Dako A/S, Glostrup, Denmark) using a custom‐made probe covering the ESR1 gene at 6q25 and a centromere 6 reference probe (Nielsen et al., 2011, 2004). The FISH procedure was performed according to the manufacturer's instructions and described in an earlier study (Nielsen et al., 2011b). Briefly the method implies pretreatment for 10 min at 95–99 °C in pretreatment solution followed by pepsin treatment for 10 min at room temperature. After denaturation at 82 °C for 5 min and overnight hybridization at 45 °C, excess of probe is removed by stringent wash at 65 °C for 10 min. After dehydration in ethanol, the slides are mounted with DAPI. Leica DM microscope equipped with filters for Texas Red, FITC, and DAPI was used for gene signal evaluation.
2.5. FISH scoring
To allow detection of both ESR1 gene amplification and deletion, the slide evaluation was performed according to the TOP2A FISH scoring guidelines (Dako K5333 USA package insert, 1st edition 2008.01.18, approved by the American Food and Drug Administration). These guidelines recommend quantification of at least 60 red gene signals along with the green reference probe signals in the same nuclei producing a final ESR1/CEN‐6 ratio score. The recommendations are based on a previously published pilot study (Olsen et al., 2004). In contrast to the HER2 guidelines that merely focus on identifying HER2 gene amplification (Wolff et al., 2007), the guidelines for TOP2A enables identification of gene deletion, as it is accepted to count signals in tumor cells with only green reference probe signals present. In accordance with the TOP2A scoring guidelines a ratio below 0.80 was defined as ESR1 gene deletion and ratios above or equal to 2.00 as ESR1 gene amplification. A previous assay validation (Nielsen et al., 2011b) showed that the distribution of ESR1/CEN‐6 ratios in 120 samples from normal breast tissue varied between 0.96 and 1.29. Consequently, in this study the ESR1/CEN‐6 ratios between 1.30 and 1.99 were defined as ESR1 gene gains, covering the ratios above the normal range and below clear amplifications. A successful FISH analysis is characterized by distinguishable point‐shaped red and green signals of quite balanced size. Samples with fuzzy appearance of the signals were classified unsuccessful.
2.6. Statistics
Spearman rank correlation was used to compare quantitative values for two methods (ER DCC vs. ER IHC, ER DCC vs. ESR1 and ER IHC vs. ESR1). Agreement for ordered grouping of hormone receptor status was measured by the weighted K statistic.
Box plots were designed to visualize the correlations between methods and ESR1 gene subgroups. The associations between ESR1 gene status and clinicopathological variables were assessed by Fishers exact test. P‐values are two‐tailed. Statistical analyses were done with SAS v9.2 (SAS Institute, Inc., Cary, USA).
3. Results
ER values measured by DCC were available for 289 patients participating in the DBCG 77C trial. Paraffin embedded tissue sections were retrieved from 257 patients and ESR1 FISH analysis was performed successfully in 220 (86%) cases (Table 1). Only samples with bright, point‐shaped and clearly separated signals were considered successful. For 149 of these patients the values for ER IHC were available as percentage positive tumor cells. The remaining patients samples were classified by IHC as negative (0–9%), low positive (10–74%), or high positive (≥75%).
3.1. Comparison of ER expression by IHC and ligand binding assay
Of the 220 patients where ER IHC, ER DCC and ESR1 results were all available a total of 45 cases were ER negative measured by both ligand binding assay (<10 fmol ER/mg protein) and IHC (<10%) and 140 cases were ER positive for both methods (Table 2) corresponding to Spearman correlation value of 0.74 (p < 0.0001) for quantitative values, with Kappa value of 0.57, p < 0.0001 when classified into groups. Only two ER IHC positive patients had negative ER DCC value in contrast to 33 cases with positive ER DCC values whom were ER IHC negative.
Table 2.
ER status by IHC compared with ER DCC
| DCC (fmol/mg protein) | ER‐IHC negative (0–9%) | ER‐IHC positive (10–74%) | ER‐IHC positive (75–100%) | Total |
|---|---|---|---|---|
| DCC negative (<10) | 45 (58%) | 2 (3%) | 0 (0%) | 47 |
| DCC positive (≥10–107) | 26 (33%) | 32 (46%) | 9 (13%) | 67 |
| DCC positive (≥108) | 7 (9%) | 36 (51%) | 63 (88%) | 106 |
| Total | 78 | 70 | 72 | 220 |
3.2. ESR1 by FISH
A total of 220 tumors were assessable by FISH and ESR1 gene amplification (ratio ESR1/CEN‐6 ≥ 2.00) was observed in 50 (23%) tumors in the range of 2.11–16.43 with an average ESR1/CEN‐6 ratio of 3.96 (Table 3). ESR1 deletion (ratio ESR1/CEN‐6 < 0.80) was observed in 70 (32%) tumors within the range of 0.30–0.79 and with an average ESR1/CEN‐6 ratio of 0.60. ESR1 gain (ratio ESR1/CEN‐6 from 1.30 to 1.94) was observed in 41 (19%) of the cases with an average ratio of 1.57. An average ratio of 1.10 was found within the group of patients (n = 59, 27%) with normal ESR1/CEN‐6 ratio. Examples of ESR1 gene amplification and deletion are illustrated in Figures 1A and B.
Table 3.
Patient and tumor characteristics by ESR1 gene status including ESR1 gene status in accordance to average values of ESR1/CEN‐6 ratio, ESR1 copy number, CEN‐6 copy number, ER DCC and ER IHC
| Patient and tumor characteristics by ESR1 copy number status | ||||
|---|---|---|---|---|
| Variable | ESR1 Deletion (<0.80) | ESR1 Normal (0.80–<1.30) | ESR1 Gain (1.30–<2.00) | ESR1 Amplification (≥2.00) |
| ESR1 gene status: primary tumor N = 220 (%) | 70 (32%) | 59 (27%) | 41 (19%) | 50 (23%) |
| Average ESR1/CEN‐6 Ratio | 0.60 | 1.10 | 1.57 | 3.96 |
| Average number of ESR1/nucleus | 1.63 | 2.23 | 2.70 | 5.87 |
| Average number of CEN‐6/nucleus | 2.76 | 2.04 | 1.73 | 1.55 |
| Average ER DCC value (fmol/mg protein) | 128 | 158 | 232 | 377 |
| Average percentage of ER IHC positive cells (N = 149) | 25% (51) | 40% (35) | 52% (29) | 58% (34) |
| Lymph node status: (n pos. nodes) | ||||
| 0 | 6 | 9 | 4 | 4 |
| 1–3 | 33 | 37 | 23 | 23 |
| >3 | 28 | 10 | 14 | 22 |
| Unknown | 3 | 3 | 0 | 1 |
| Tumor size (mm) | ||||
| 0–20 | 20 | 7 | 9 | 14 |
| 21–50 | 35 | 38 | 24 | 29 |
| >50 | 15 | 14 | 8 | 7 |
| Histology | ||||
| Ductal carcinoma | 63 | 53 | 39 | 47 |
| Lobular carcinoma | 2 | 4 | 2 | 0 |
| Other | 5 | 2 | 0 | 3 |
| Malignancy grade | ||||
| Grade 1 | 13 | 15 | 6 | 10 |
| Grade 2 | 39 | 30 | 29 | 29 |
| Grade 3 | 15 | 13 | 6 | 9 |
| Unknown | 3 | 1 | 0 | 2 |
Figure 1.
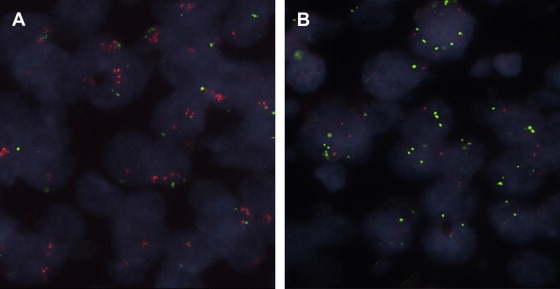
Examples of ESR1 gene status in invasive breast tumors (60× objective, oil emulsion). The red ESR1 probe is labeled with Texas Red fluorochrome and the green reference probe with fluorescein (FITC). In (A) the ESR1 gene is amplified (FISH ratio ≥ 2,00) and in (B) ESR1 gene deletion is present (FISH ratio < 0.80).
The distribution of deletions, gains and amplifications according to ploidy level is seen in Table 4. The background for the presented distribution of ploidy levels is described elsewhere (Nielsen et al., 2011a). Overall aneuploidy (also referred to as polyploidy) defined as more than three CEN‐6 copies, was present in 19 (9%) tumors, very close to the frequency of 8% described by Wolff et al. (25). Five (2%) tumors had loss of CEN‐6 indicating monosomy (haploid CEN‐6 copy number), all being ESR1 amplified tumors. ESR1 gain was solely observed in diploid tumors whereas ESR1 normal copy numbers were seen in diploid, triploid and tetraploid tumors. ESR1 deletions were restricted to triploid, tetraploid and higher ploidy levels while amplifications were only observed in haploid and diploid tumors.
Table 4.
ESR1 gene status in accordance to ploidy levels
| Ploidy level | CEN‐6 copies | Deletion | Normal | Gain | Amplification | All patients | Percentage |
|---|---|---|---|---|---|---|---|
| Haploid | <1.25 | 0 | 0 | 0 | 5 | 5 | 2% |
| Diploid | 1.26–<2.10 | 0 | 41 | 41 | 45 | 127 | 58% |
| Triploid | 2.10–2.93 | 52 | 17 | 0 | 0 | 69 | 31% |
| Tetraploidy | 2.94–3.77 | 15 | 1 | 0 | 0 | 16 | 7% |
| High ploidy | >3.78 | 3 | 0 | 0 | 0 | 3 | 1% |
| Total | 70 | 59 | 41 | 50 | 220 | 100% |
3.3. ESR1 FISH compared to ER expression by IHC and ER content by DCC
Table 5 shows the association between FISH, DCC and IHC. Among the ESR1 amplified tumors, 94% were ER DCC positive and 86% were ER IHC positive using cut‐off values for DCC positivity of ≥10 fmol/mg protein and for IHC positivity of ≥10% stained cells. Among the samples with ESR1 gain 88% were ER DCC positive and 78% were ER‐IHC positive, thus resembling ESR1 amplified tumors. Among samples with ESR1 normal status, 76% were ER DCC positive and 68% were ER‐IHC positive. The frequency of ER positive samples (DCC/IHC) were markedly lower (64% and 39%, respectively) in the groups of ESR1 deleted tumors.
Table 5.
ER DCC and ER IHC according to ESR1 gene status
| ESR1 Deletion (<0.80) | ESR1 Normal (0.80–<1.30) | ESR1 Gain (1.30–<2.00) | ESR1 Amplification (≥2.00) | Total | |
|---|---|---|---|---|---|
| ER DCC negative (<10 fmol/mg protein) | 25 (36%) | 14 (24%) | 5 (12%) | 3 (6%) | 47 |
| ER DCC low positive (10–107 fmol/mg protein) | 23 (33%) | 22 (37%) | 10 (24%) | 12 (24%) | 67 |
| ER DCC high positive (≥108 fmol/mg protein) | 22 (31%) | 23 (39%) | 26 (63%) | 35 (70%) | 106 |
| ER‐IHC negative (<10% stained cells) | 43 (61%) | 19 (32%) | 9 (22%) | 7 (14%) | 78 |
| ER IHC low positive (10–74% stained cells) | 13 (19%) | 19 (32%) | 20 (49%) | 18 (36%) | 70 |
| ER IHC high positive (≥75% stained cells) | 14 (20%) | 21 (36%) | 12 (29%) | 25 (50%) | 72 |
| Total (N = 220) | 70 | 59 | 41 | 50 | |
| ER IHC and ER DCC negative | 24 (56%) | 13 (68%) | 5 (56%) | 3 (43%) | 45 |
| ER DCC positive and ER IHC negative | 19 (44%) | 6 (32%) | 4 (44%) | 4 (57%) | 33 |
| Total (N = 78) | 43 | 19 | 9 | 7 |
The quantitative values of the two different ER assays were both significantly (p < 0.0001) correlated with ESR1 copy number. The ESR1 amplified tumors had higher average ER values compared to ESR1 normal tumors and tumors with ESR1 gain, while ESR1 deleted tumors had lower ER values (Table 3). This is illustrated by Box Plots in Figures 2 and 3, which also show that although a positive correlation exists there is a wide range in ER protein values in each group. A broad variation of ER DCC values was especially found in the ESR1 amplified group ranging from 0 to 1510 fmol/mg protein with a mean value of 377 fmol/mg protein (Table 3). The clinicopathological characteristics according to ESR1 gene status are presented in Table 3. No significant correlations with known prognostic factors were observed.
Figure 2.
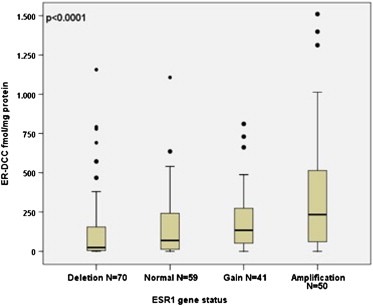
Boxplot illustrating ER DCC (fmol ER/mg protein) according to ESR1/CEN‐6 ratio in 220 malignant breast tumors. The boxplot visualizes the significant positive correlation (p < 0.0001) between the amount of ER protein measured by DCC and ESR1 ratio measured by FISH.
Figure 3.
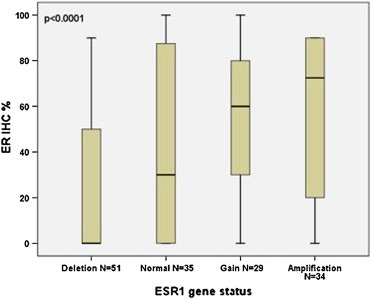
Boxplot illustrating ER IHC (% positive tumors cells) according to ESR1 ratio in 149 patients. The boxplot visualizes the significant positive correlation (p < 0.0001) between the amount of ER protein measured by IHC and ESR1 ratio measured by FISH.
3.4. ESR1 status in discordant cases between IHC and DCC
Among 78 ER IHC negative tumors, 45 cases were ER DCC negative and 33 ER DCC positive. No significant difference was identified with respect to ESR1 gene status (p = 0.66).
4. Discussion
In the present study we could confirm the existence of amplifications, gains and deletions of the ESR1 gene. In addition we found a strong positive correlation between ESR1 gene level and ER content by DCC and between ESR1 gene level and ER expression by IHC (p < 0.0001). By IHC, one third of the patients (35%) had ER negative tumors and a large fraction of these tumors had ESR1 deletion (55%). Similarly, 53% of the tumors, which were classified as ER negative after DCC analysis, had ESR1 deletion. Also, ESR1 gain or amplification was observed in tumors classified as ER negative by IHC or by DCC analysis but only in 21% and 17% of the cases, respectively. ESR1 amplification was most frequent in tumors with more than 75% ER positive cells or high ER level determined by the DCC method, 35% and 33% of the cases, respectively. Our data suggest a close association between genetic alterations in the ESR1 gene and ER protein content as measured by DCC or IHC.
The strength of the study is the availability of ER protein measurements from two different methods. Although the samples were collected more than 15 years ago, ESR1 FISH analysis was successful in 86% of the samples available. The limitations concerns the fact that the results of ER IHC were reported with the percentage positive cells for only 149 of the patients analyzed, but fortunately we could show that the classification in ER positive and negative tumors yielded similar results.
ESR1 was amplified in 23% of the tumor samples in the current study, and this frequency is in line with previous reports using FISH (Holst et al., 2007; Nielsen et al., 2011b; Tomita et al., 2009). The frequency of ESR1 deletions was considerable higher (32% versus 5%) than in our previous study of ER‐IHC positive patients (Nielsen et al., 2011b). One possible explanation is that one third of the patients in the current study were ER negative. We have previously reported elevated number of deletions in a patient population with high number of ER negative tumors (Ejlertsen et al., 2007). Thus ESR1 deleted tumors might constitute a unique group with respect to protein content.
The FISH technique seems to be particular suited for detection of low level amplifications and deletions, while other techniques (Albertson, 2008; Brown et al., 2008; Drury et al., 2007; Horlings et al., 2008; Moelans et al., 2010b; Reis‐Filho et al., 2008; Vincent‐Salomon et al., 2008) seem to underestimate the frequency of amplifications and miss the deletions (Albertson, 2008). Only two studies have made direct comparison between ESR1 FISH and other techniques (Moelans et al., 2010b; Tomita et al., 2009). In one study the number of amplifications was 23% by FISH analysis and 1% by quantitative PCR analysis (Tomita et al., 2009) while the other showed a decrease from 12.5% amplifications by FISH to only 2% by the multiplex ligation‐dependent probe method (Moelans et al., 2010b). Thus, the FISH analysis appears to be the most sensitive technique, particularly for detection of low level amplifications. Assay comparison is often based on the high level amplified HER2 gene (Moelans et al., 2010a), but results from HER2 assays seem not easily transferred to other genes. ERS1 FISH signals have recently been described as homogeneously staining regions with fuzzy and cloudy appearance (Ooi et al., 2011). We cannot recognize this description of FISH signals in our series of samples where fuzzy and cloudy signals were considered as unsuccessful FISH analysis. An explanation to this difference in appearance of signals could either be the difference in probe length or the tissue fixation time.
Our study demonstrates a statistically significant correlation between ESR1 gene level and ER protein content measured by both DCC and IHC. Although ESR1 amplification and gain are associated with higher ER IHC expression and ER DCC content in the majority of the ESR1 enriched tumors, while only approximately one third of the IHC or DCC positive tumors have increased ESR1 gene copy number. This relation between gene copy numbers and protein content corresponds to the one found for the TOP2A gene and protein (Callagy et al., 2005). In contrast, seven (14%) of the ESR1 amplified tumors were ER IHC negative, but four of these tumors had positive ER DCC values. Only three (6%) of the ESR1 amplified tumors were DCC negative. Apart from the differences in ESR1 copy number the identified discrepant tumor values might be explained by variation in sensitivity between methods or by tumor heterogeneity. Finally, the cut‐off value for ER IHC in this study was ≥10% positive tumor cells. This might explain some of the discrepancy as well since a few of the tumors with low DCC values and negative ER IHC might have positive reaction in 1–9% of the tumor cells. However, only a minor fraction (5%) of patients samples show positive staining in 1–9% of the tumor cells (Iwamoto et al., 2012).
Disagreement on the clinical impact of ERS1 amplifications has been reported as two studies find correlation between ESR1 amplification and improved survival (Holst et al., 2007; Tomita et al., 2009) while a third study (Nielsen et al., 2011b) reports that ESR1 amplification was significantly associated with poor survival. Recently Ejlertsen et al. (2011) found that ESR1 amplification was correlated to poor disease free survival in a sub‐group of patients enrolled in the BIG‐98 trial. However in multivariate analysis ESR1 gene status did not provide significant prognostic nor predictive information for this specific population of ER positive patients randomized to endocrine treatment with either tamoxifen or letrozole.
The highest values of ER DCC (≥108 fmol/mg protein) have been associated with the worst prognosis but also with the greatest benefit from adjuvant treatment with tamoxifen (Thorpe et al., 1993). Since the average value of both ER IHC and ER DCC in the present study is increased according to ESR1 gene status one could speculate that the patients with both ESR1 gene amplification and high protein expression/content benefit most from tamoxifen treatment. However, since we observed major variations in ER content and immunoreactivity within every ESR1 group, high protein expression is not always the result of amplification and other mechanisms than gene aberrations seem to contribute (Dunbier et al., 2011).
In conclusion, we observed a close association between ESR1 gene copy number and ER protein content measured by ER DCC as well as by IHC. Both amplification and gain of ESR1 was associated with highly elevated ER protein levels and a large fraction of ER negative tumors had ESR1 deletion.
Conflict of interest
Bent Ejlertsen, Sven Müller and Kirsten Vang Nielsen are inventors on a patent application on ESR1. Bent Ejlertsen has a relative employment by Dako A/S and Birgitte Bruun Rasmussen has received a minor fee from Dako A/S.
Acknowledgments
This study is supported by the Danish Ministry of Science, Innovation and Higher Education (Grant No. 2101‐07‐0022)
Laenkholm Anne-Vibeke, Knoop Ann, Ejlertsen Bent, Rudbeck Tine, Jensen Maj-Britt, Müller Sven, Lykkesfeldt Anne Elisabeth, Rasmussen Birgitte Bruun, Nielsen Kirsten Vang, (2012), ESR1 gene status correlates with estrogen receptor protein levels measured by ligand binding assay and immunohistochemistry, Molecular Oncology, 6, doi: 10.1016/j.molonc.2012.04.003.
References
- Albertson, D.G. , 2008. Conflicting evidence on the frequency of ESR1 amplification in breast cancer. Nat. Genet.. 40, 821–822. [DOI] [PubMed] [Google Scholar]
- Allred, D.C. , Carlson, R.W. , Berry, D.A. , Burstein, H.J. , Edge, S.B. , Goldstein, L.J. , Gown, A. , Hammond, M.E. , Iglehart, J.D. , Moench, S. , Pierce, L.J. , Ravdin, P. , Schnitt, S.J. , Wolff, A.C. , 2009. NCCN task force report: estrogen receptor and progesterone receptor testing in breast cancer by immunohistochemistry. J. Natl. Compr. Canc Netw.. 7, (Suppl. 6) S1–S21. quiz S22–23 [DOI] [PubMed] [Google Scholar]
- Brown, L.A. , Hoog, J. , Chin, S.F. , Tao, Y. , Zayed, A.A. , Chin, K. , Teschendorff, A.E. , Quackenbush, J.F. , Marioni, J.C. , Leung, S. , Perou, C.M. , Neilsen, T.O. , Ellis, M. , Gray, J.W. , Bernard, P.S. , Huntsman, D.G. , Caldas, C. , 2008. ESR1 gene amplification in breast cancer: a common phenomenon?. Nat. Genet.. 40, 806–807. author reply 810–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhardt, L. , Grob, T.J. , Hermann, I. , Burandt, E. , Choschzick, M. , Janicke, F. , Muller, V. , Bokemeyer, C. , Simon, R. , Sauter, G. , Wilczak, W. , Lebeau, A. , 2010. Gene amplification in ductal carcinoma in situ of the breast. Breast Cancer Res. Treat.. 123, 757–765. [DOI] [PubMed] [Google Scholar]
- Callagy, G. , Pharoah, P. , Chin, S.F. , Sangan, T. , Daigo, Y. , Jackson, L. , Caldas, C. , 2005. Identification and validation of prognostic markers in breast cancer with the complementary use of array-CGH and tissue microarrays. J. Pathol.. 205, 388–396. [DOI] [PubMed] [Google Scholar]
- Davies, C. , Godwin, J. , Gray, R. , Clarke, M. , Cutter, D. , Darby, S. , McGale, P. , Pan, H.C. , Taylor, C. , Wang, Y.C. , Dowsett, M. , Ingle, J. , Peto, R. , 2011. Relevance of breast cancer hormone receptors and other factors to the efficacy of adjuvant tamoxifen: patient-level meta-analysis of randomised trials. Lancet. 378, 771–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSombre, E.R. , Carbone, P.P. , Jensen, E.V. , McGuire, W.L. , Wells, S.A. , Wittliff, J.L. , Lipsett, M.B. , 1979. Special report. Steriod receptors in breast cancer. N. Engl. J. Med.. 301, 1011–1012. [DOI] [PubMed] [Google Scholar]
- Drury, S. , Detre, S. , Leary, A. , Salter, J. , Reis-Filho, J. , Barbashina, V. , Marchio, C. , Lopez-Knowles, E. , Ghazoui, Z. , Habben, K. , Arbogast, S. , Johnston, S.R. , Dowsett, M. , 2011. Changes in breast cancer biomarkers in the IGF1R/PI3K pathway in recurrent breast cancer after tamoxifen treatment. Endocr. Relat. Cancer. 18, 565–577. [DOI] [PubMed] [Google Scholar]
- Drury, S., Lambros, M., Marchio, C., Johnson, N., Salter, J., Levey, P., Fletcher, O., Peto, J., Reis-Filho, J., Dowsett, M., 2007. Gene copy number variability of oestrogen receptor alpha in breast cancer, San Antonio Breast Cancer Symposium. Breast Cancer Res Treat 106, p. (abstr 404).
- Dunbier, A.K. , Anderson, H. , Ghazoui, Z. , Lopez-Knowles, E. , Pancholi, S. , Ribas, R. , Drury, S. , Sidhu, K. , Leary, A. , Martin, L.A. , Dowsett, M. , 2011. ESR1 is co-expressed with closely adjacent uncharacterised genes spanning a breast cancer susceptibility locus at 6q25.1. PLoS Genet.. 7, e1001382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ejlertsen, B. , Aldridge, J. , Nielsen, K.V. , Regan, M.M. , Henriksen, K.L. , Lykkesfeldt, A.E. , Muller, S. , Gelber, R.D. , Price, K.N. , Rasmussen, B.B. , Viale, G. , Mouridsen, H. , 2011. Prognostic and predictive role of ESR1 status for postmenopausal patients with endocrine-responsive early breast cancer in the Danish cohort of the BIG 1-98 trial. Ann. Oncol. Epub ahead of print. Oct 12. 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ejlertsen, B. , Nielsen, K. , Rasmussen, B. , Jensen, M.-B. , Møller, S. , Balslev, E. , Müller, S. , Mouridsen, H. , 2007. Relationship between ESR1 copy number and ER expression in the DBCG 89D trial. Breast Cancer Res. Treat.. 106, abstr 403 [Google Scholar]
- EORTC, B.C.C.G. 1973. Standards for the assessment of estrogen receptors in human breast cancer. Report of a workshop on September 29, 1972, at the Antoni van Leeuwenhoek-Huis, Amsterdam Eur. J. Cancer. 9, 379–381. [DOI] [PubMed]
- Grabau, D.A. , Thorpe, S.M. , Knoop, A. , Vach, W. , Schroder, H.D. , Blichert-Toft, M. , Al-Suliman, N.N. , Graversen, H.P. , Rose, C. , 2000. Immunohistochemical assessment of oestrogen and progesterone receptors: correlations with the DCC method and clinical outcome in primary breast cancer patients. Breast. 9, 208–217. [DOI] [PubMed] [Google Scholar]
- Hammond, M.E. , Hayes, D.F. , Dowsett, M. , Allred, D.C. , Hagerty, K.L. , Badve, S. , Fitzgibbons, P.L. , Francis, G. , Goldstein, N.S. , Hayes, M. , Hicks, D.G. , Lester, S. , Love, R. , Mangu, P.B. , McShane, L. , Miller, K. , Osborne, C.K. , Paik, S. , Perlmutter, J. , Rhodes, A. , Sasano, H. , Schwartz, J.N. , Sweep, F.C. , Taube, S. , Torlakovic, E.E. , Valenstein, P. , Viale, G. , Visscher, D. , Wheeler, T. , Williams, R.B. , Wittliff, J.L. , Wolff, A.C. , 2010. American Society of Clinical Oncology/College Of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. J. Clin. Oncol.. 28, 2784–2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey, J.M. , Clark, G.M. , Osborne, C.K. , Allred, D.C. , 1999. Estrogen receptor status by immunohistochemistry is superior to the ligand-binding assay for predicting response to adjuvant endocrine therapy in breast cancer. J. Clin. Oncol.. 17, 1474–1481. [DOI] [PubMed] [Google Scholar]
- Henriksen, K.L. , Rasmussen, B.B. , Lykkesfeldt, A.E. , Moller, S. , Ejlertsen, B. , Mouridsen, H.T. , 2009. An ER activity profile including ER, PR, Bcl-2 and IGF-IR may have potential as selection criterion for letrozole or tamoxifen treatment of patients with advanced breast cancer. Acta Oncol.. 48, 522–531. [DOI] [PubMed] [Google Scholar]
- Holst, F. , Stahl, P.R. , Ruiz, C. , Hellwinkel, O. , Jehan, Z. , Wendland, M. , Lebeau, A. , Terracciano, L. , Al-Kuraya, K. , Janicke, F. , Sauter, G. , Simon, R. , 2007. Estrogen receptor alpha (ESR1) gene amplification is frequent in breast cancer. Nat. Genet.. 39, 655–660. [DOI] [PubMed] [Google Scholar]
- Horlings, H.M. , Bergamaschi, A. , Nordgard, S.H. , Kim, Y.H. , Han, W. , Noh, D.Y. , Salari, K. , Joosse, S.A. , Reyal, F. , Lingjaerde, O.C. , Kristensen, V.N. , Borresen-Dale, A.L. , Pollack, J. , van de Vijver, M.J. , 2008. ESR1 gene amplification in breast cancer: a common phenomenon?. Nat. Genet.. 40, 807–808. author reply 810–802 [DOI] [PubMed] [Google Scholar]
- Iwamoto, T. , Booser, D. , Valero, V. , Murray, J.L. , Koenig, K. , Esteva, F.J. , Ueno, N.T. , Zhang, J. , Shi, W. , Qi, Y. , Matsuoka, J. , Yang, E.J. , Hortobagyi, G.N. , Hatzis, C. , Symmans, W.F. , Pusztai, L. , 2012. Estrogen receptor (ER) mRNA and ER-related gene expression in breast cancers that are 1% to 10% ER-positive by immunohistochemistry. J. Clin. Oncol.. 30, 729–734. [DOI] [PubMed] [Google Scholar]
- Khoshnoud, M.R. , Lofdahl, B. , Fohlin, H. , Fornander, T. , Stal, O. , Skoog, L. , Bergh, J. , Nordenskjold, B. , 2011. Immunohistochemistry compared to cytosol assays for determination of estrogen receptor and prediction of the long-term effect of adjuvant tamoxifen. Breast Cancer Res. Treat.. 126, 421–430. [DOI] [PubMed] [Google Scholar]
- Knoop, A.S. , Bentzen, S.M. , Nielsen, M.M. , Rasmussen, B.B. , Rose, C. , 2001. Value of epidermal growth factor receptor, HER2, p53, and steroid receptors in predicting the efficacy of tamoxifen in high-risk postmenopausal breast cancer patients. J. Clin. Oncol.. 19, 3376–3384. [DOI] [PubMed] [Google Scholar]
- Moelans, C.B. , de Weger, R.A. , van Blokland, M.T. , van der Wall, E. , van Diest, P.J. , 2010. Simultaneous detection of TOP2A and HER2 gene amplification by multiplex ligation-dependent probe amplification in breast cancer. Mod. Pathol.. 23, 62–70. [DOI] [PubMed] [Google Scholar]
- Moelans, C.B. , Monsuur, H.N. , de Pinth, J.H. , Radersma, R.D. , de Weger, R.A. , van Diest, P.J. , 2010. ESR1 amplification is rare in breast cancer and is associated with high grade and high proliferation: a multiplex ligation-dependent probe amplification study. Anal. Cell Pathol. (Amst). 33, 13–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen, K.V. , Ejlertsen, B. , Moller, S. , Jensen, M.B. , Balslev, E. , Muller, S. , Knoop, A. , Mouridsen, H.T. , 2011. Lack of independent prognostic and predictive value of centromere 17 copy number changes in breast cancer patients with known HER2 and TOP2A status. Mol. Oncol.. 6, 88–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen, K.V. , Ejlertsen, B. , Muller, S. , Moller, S. , Rasmussen, B.B. , Balslev, E. , Laenkholm, A.V. , Christiansen, P. , Mouridsen, H.T. , 2011. Amplification of ESR1 may predict resistance to adjuvant tamoxifen in postmenopausal patients with hormone receptor positive breast cancer. Breast Cancer Res. Treat.. 127, 345–355. [DOI] [PubMed] [Google Scholar]
- Nielsen, K.V. , Müller, S. , Poulsen, T.S. , Gabs, S. , Schonau, A. , 2004. Combined use of PNA and DNA for fluorescence in situ hybridization (FISH). In Nielsen P.E., Peptide Nucleic Acids: Protocols and Applications. second ed. Horizon Bioscience; Norfolk: 227–260. [Google Scholar]
- Olsen, K.E. , Knudsen, H. , Rasmussen, B.B. , Balslev, E. , Knoop, A. , Ejlertsen, B. , Nielsen, K.V. , Schonau, A. , Overgaard, J. , 2004. Amplification of HER2 and TOP2A and deletion of TOP2A genes in breast cancer investigated by new FISH probes. Acta Oncol.. 43, 35–42. [DOI] [PubMed] [Google Scholar]
- Ooi, A. , Inokuchi, M. , Harada, S. , Inazawa, J. , Tajiri, R. , Kitamura, S.S. , Ikeda, H. , Kawashima, H. , Dobashi, Y. , 2011. Gene amplification of ESR1 in breast cancers-fact or fiction? A fluorescence in situ hybridization and multiplex ligation-dependent probe amplification study. J. Pathol.. Epub ahead of print. Dec 13, 2011 [DOI] [PubMed] [Google Scholar]
- Regan, M.M. , Viale, G. , Mastropasqua, M.G. , Maiorano, E. , Golouh, R. , Carbone, A. , Brown, B. , Suurkula, M. , Langman, G. , Mazzucchelli, L. , Braye, S. , Grigolato, P. , Gelber, R.D. , Castiglione-Gertsch, M. , Price, K.N. , Coates, A.S. , Goldhirsch, A. , Gusterson, B. , 2006. Re-evaluating adjuvant breast cancer trials: assessing hormone receptor status by immunohistochemical versus extraction assays. J. Natl. Cancer Inst.. 98, 1571–1581. [DOI] [PubMed] [Google Scholar]
- Reis-Filho, J.S. , Drury, S. , Lambros, M.B. , Marchio, C. , Johnson, N. , Natrajan, R. , Salter, J. , Levey, P. , Fletcher, O. , Peto, J. , Ashworth, A. , Dowsett, M. , 2008. ESR1 gene amplification in breast cancer: a common phenomenon?. Nat. Genet.. 40, 809–810. author reply 810–802 [DOI] [PubMed] [Google Scholar]
- Schuur, E.R. , Weigel, R.J. , 2000. Monoallelic amplification of estrogen receptor-alpha expression in breast cancer. Cancer Res.. 60, 2598–2601. [PubMed] [Google Scholar]
- Talman, M.L. , Rasmussen, B.B. , Andersen, J. , Christensen, I.J. , 2008. Estrogen receptor analyses in the Danish Breast Cancer Cooperative Group. History, methods, prognosis and clinical implications. Acta Oncol.. 47, 789–794. [DOI] [PubMed] [Google Scholar]
- Thorpe, S.M. , Christensen, I.J. , Rasmussen, B.B. , Rose, C. , 1993. Short recurrence-free survival associated with high oestrogen receptor levels in the natural history of postmenopausal, primary breast cancer. Eur. J. Cancer. 29A, 971–977. [DOI] [PubMed] [Google Scholar]
- Tomita, S. , Zhang, Z. , Nakano, M. , Ibusuki, M. , Kawazoe, T. , Yamamoto, Y. , Iwase, H. , 2009. Estrogen receptor alpha gene ESR1 amplification may predict endocrine therapy responsiveness in breast cancer patients. Cancer Sci.. 100, 1012–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viale, G. , Regan, M.M. , Maiorano, E. , Mastropasqua, M.G. , Dell'Orto, P. , Rasmussen, B.B. , Raffoul, J. , Neven, P. , Orosz, Z. , Braye, S. , Ohlschlegel, C. , Thurlimann, B. , Gelber, R.D. , Castiglione-Gertsch, M. , Price, K.N. , Goldhirsch, A. , Gusterson, B.A. , Coates, A.S. , 2007. Prognostic and predictive value of centrally reviewed expression of estrogen and progesterone receptors in a randomized trial comparing letrozole and tamoxifen adjuvant therapy for postmenopausal early breast cancer: BIG 1-98. J. Clin. Oncol.. 25, 3846–3852. [DOI] [PubMed] [Google Scholar]
- Vincent-Salomon, A. , Raynal, V. , Lucchesi, C. , Gruel, N. , Delattre, O. , 2008. ESR1 gene amplification in breast cancer: a common phenomenon?. Nat. Genet.. 40, 809 author reply 810–802 [DOI] [PubMed] [Google Scholar]
- Wolff, A.C. , Hammond, M.E. , Schwartz, J.N. , Hagerty, K.L. , Allred, D.C. , Cote, R.J. , Dowsett, M. , Fitzgibbons, P.L. , Hanna, W.M. , Langer, A. , McShane, L.M. , Paik, S. , Pegram, M.D. , Perez, E.A. , Press, M.F. , Rhodes, A. , Sturgeon, C. , Taube, S.E. , Tubbs, R. , Vance, G.H. , van de Vijver, M. , Wheeler, T.M. , Hayes, D.F. , 2007. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. J. Clin. Oncol.. 25, 118–145. [DOI] [PubMed] [Google Scholar]


