Abstract
Medulloblastoma cells exhibit varied responses to therapy by all‐trans retinoic acid (RA). The underlying mechanism for such diverse effects however remains largely unclear. In this study, we attempted to elucidate the molecular basis of RA resistance through the study of RA signaling components in both RA‐sensitive (Med‐3) and RA‐resistant (UW228‐2 and UW228‐3) medulloblastoma cells. The results revealed that RARα/β/γ and RXRα/β/γ were found in the three cell lines. Expression of CRABP‐I and CRABP‐II was seen in Med‐3 cells, up‐regulated when treated with RA, but was absent in UW228‐2 and UW228‐3 cells regardless of RA treatment. Bisulfite sequencing revealed 8 methylated CG sites at the promoter region of CRABP‐II in UW228‐2 and UW228‐3 but not in Med‐3 cells. Demethylation by 5‐aza‐2′‐deoxycytidine recovered CRABP‐II expression. Upon restoration of CRABP‐II expression, both UW228‐2 and UW228‐3 cells responded to RA treatment by forming neuronal‐like differentiation, synaptophysin expression, β‐III tubulin upregulation, and apoptosis. Furthermore, CRABP‐II specific siRNA reduced RA sensitivity in Med‐3 cells. Tissue microarray‐based immunohistochemical staining showed variable CRABP‐II expression patterns among 104 medulloblastoma cases, ranging from negative (42.3%), partly positive (14.4%) to positive (43.3%). CRABP‐II expression was positively correlated with synaptophysin (rs = 0.317; p = 0.001) but not with CRABP‐I expression (p > 0.05). In conclusion, aberrant methylation in CRABP‐II reduces the expression of CRABP‐II that in turn confers RA resistance in medulloblastoma cells. Determination of CRABP‐II expression or methylation status may enable a personalized RA therapy in patients with medulloblastomas and other types of cancers.
Keywords: Medulloblastoma, Retinoic acid, RA signaling, CRABP‐II, DNA methylation, Epigenetic modulation: Gene transfection
Highlights
CRABP‐II is silenced in RA‐resistant medulloblastoma/MB cells due to promoter methylation.
Restoration of CRABP‐II expression overcomes RA‐resistant property of MB cells.
Inhibition of CRABP‐II expression reduces RA sensitivity of MB cells.
CRABP‐II expression patterns are highly variable in medulloblastoma tissues.
Evaluation of CRABP‐II expression would be of values in personalized RA therapy for MB patients.
1. Introduction
Retinoic acid/RA has been commonly‐used in cancer chemotherapy because it can induce differentiation and apoptosis (Freemantle, 2003). The anticancer signal of RA is transmitted from cytoplasm to the nucleus through stepwise processes. RA first binds to protein/CRABP‐II and then transports lipophilic RA to the nucleus where RA regulates its target gene expression through binding with the heterodimers of its nuclear receptors, RARα/β/γ and RXRα/β/γ (Delva et al., 1999; Dong et al., 1999; Budhu and Noy, 2002; Sessler and Noy, 2005). Nevertheless, RA resistance often occurs at first‐time use or during the treatment (Warrell et al., 1993; Freemantle et al., 2003). Although it is known that aberrant expression of nuclear RA receptors, increased rate of cytoplasmic RA metabolism, differential BMP‐2 expression, disrupted expression of CRABP‐II and the fatty acid binding protein/FABP‐5‐mediated RA signaling pathway may influence the RA sensitivity of cancer cells (van der Burg et al., 1993; Hallahan et al., 2003; Schug et al., 2007; Schug et al., 2008; Corlazzoli et al., 2009), the detailed mechanism(s) resulting in RA‐resistance remains to be further elucidated. Since a normal operation of CRABP‐II mediated RA signaling is essential for RA to exert anticancer effects, it is worthwhile to evaluate the status of this signaling in RA‐sensitive and RA‐resistant cancer cells with the same tissue origin.
Medulloblastoma is the most frequent primary brain malignancy in childhood and characterized with rapid growth, earlier intracranial dissemination and high recurrence incidence (Chatty and Earle, 1971; Ho et al., 2000; Fossati et al., 2009). Although the combination of operation with craniospinal radiation and/or multi‐agent chemotherapy have been adapted in clinical settings (Mueller and Chang, 2009; Ohta et al., 2011), the outcome of medulloblastomas remains poor due to the difficulty in removing the highly invasive tumor radically and the long‐term side effects of conventional adjuvant therapies (Gururangan et al., 2008). As a kind of neuroectodermal tumors, medulloblastomas maintain the potential for further differentiation. However, the medulloblastoma cells respond to RA differently by showing both differentiation and apoptosis (Gumireddy et al., 2003) or growth arrest only (Chang et al., 2007) or almost no response (Kitamura et al., 2004). According to our previous observations, human medulloblastoma cell line Med‐3 is relatively sensitive to RA treatment (Liu et al., 2000), while UW228‐1, UW228‐2 and UW228‐3 cells are not [unpublished data]. We therefore selected Med‐3, UW228‐2 and UW228‐3 cells as an experimental model to investigate the underlying molecular mechanism for diverse sensitivities of medulloblastomas to the treatment by RA.
2. Materials and methods
2.1. Cell culture and treatment
The Med‐3 medulloblastoma cell line was kindly provided by the doctors in the Department of Neurosurgery, Kobe University School of Medicine (Matsumoto, 1991). UW228‐2 and UW228‐3 cell lines were established and provided by the Department of Neurological Surgery, University of Washington at Seattle (Keles et al., 1995). Med‐3 cells were cultured in MEM (Invitrogen, California, USA) containing 10% fetal bovine serum/FBS (Gibco Life Science, Grand Island, NY) and UW228‐2 and UW228‐3 cells in DMEM (Invitrogen) supplemented with 10% FBS. 5 × 104/ml of the cells were plated to 100 mm dishes (Nunc A/S, Roskilde, Denmark) and incubated for 24 h before further experiments. The gelatin‐coated coverslips were put onto the dishes before cell seeding and collected regularly during drug treatments for immunocytochemical staining and terminal deoxynucleotidyl transferase dUTP nick end labeling/TUNEL assay. All‐trans retinoic acid (RA; Sigma–Aldrich, St. Louis, USA) was dissolved in dimethylsulfoxide/DMSO (Sigma–Aldrich) and diluted with culture medium to optimal working concentration of 10 μM. The treatments lasted for 3 days and the cells were observed in 6‐h intervals. The cells cultured in conventional medium with and without 0.2% DMSO supplementation were cited as controls. To determine the effects of RA on cell growth, the treated cells were collected after 72‐h 10 μM RA exposure, re‐suspended in 1 ml propidium iodide solution containing RNase, incubated at 37 °C for 30 min and subjected to flow cytometry analysis (Becton Dickinson, San Jose, CA). Meanwhile, total cell numbers, cell viability and the feature(s) of cell death were determined in 24‐h intervals by staining the suspended single cells with 0.25% trypan blue, MTT (3‐[4,5‐dimethylthiazol‐ 2‐yl]‐2,5‐diphenyl‐tetrazolium bromide) assay and TUNEL. Each of the experiments was repeated at least three times to establish a confident conclusion and the data were statistically analyzed by ANOVA (analysis of variance).
2.2. RT‐PCR for major components of CRABP‐II mediated RA signaling
After 3 days of RA treatment, total RNA samples were isolated from the cells using Trizol solution (Life Tech, Texas, USA) and subjected to RT‐PCR for RARα, RARβ, RARγ, RXRα, RXRβ, RXRγ, CRABP‐I, CRABP‐II and CYP26A1 according to producer's protocols (Takara, Dalian Branch, Dalian, China). NGN1 was amplified to check the impact of RA in neuronal differentiation. The sequences of PCR primers for each of the gene transcripts were listed in Table 1. The PCR products were resolved on ethidium bromide‐stained 1.5% agarose gel and photographed under ultraviolet illumination (UVP, LLC Upland, CA, USA). β‐actin was used as the quantitative and qualitative control.
Table 1.
Primer sequences for conventional RT‐PCR and methylation‐specific PCR
| Gene | Primers | Amplicon size (bp) | Annealing temperature (°C) |
|---|---|---|---|
| RARα | F:5′‐CTGCCAGTACTGCCGACTGC‐3′ | 325 | 66 |
| R:5′‐ACGTTGTTCTGAGCTGTTGTTCGTA‐3′ | |||
| β | F:5′‐GAATTGAAACACAGAGCACC‐3′ | 1180 | 54 |
| R:5′‐GCAGGAGTGGTGACTGACTG‐3′ | |||
| γ | F:5′‐CCACCAATAAGGAGCGACTCTTTG‐3′ | 358 | 55 |
| R:5′‐TTCTTCTGGATGCTTCGGCG‐3′ | |||
| RXRα | F:5′‐CCCTGTCACCAACATTTGC‐3′ | 90 | 60 |
| R:5′‐AGAAGTGTGGGATCCGCTTG‐3′ | |||
| β | F:5′‐CTCTGGATGATCAGGTCATATTGCT‐3′ | 92 | 60 |
| R:5′‐GCCATCTCG AACATCAATGGA‐3′ | |||
| γ | F:5′‐GGGAAGCTGTGCAAGAAGAAA‐3′ | 69 | 60 |
| R:5′‐TGGTAGCACATTCTGCCTCACT‐3′ | |||
| CYP26A1 | F:5′‐GAGACCCTTCGACTGAATCC‐3′ | 332 | 56 |
| R:5′‐GGAGGTCCATTTAGAAGCTGC‐3′ | |||
| CRABP‐I | F: 5′‐GCCATGCTGAGGAAAGTG‐3′ | 132 | 65 |
| R: 5′‐TTCTCCGACCTTGAAGTTG‐3′ | |||
| II | F: 5′‐ATGCCCAACTTCTCTGGCAA‐3′ | 375 | 59 |
| R: 5′‐CGTCATGGTCAGGATCAGTT‐3′ | |||
| NGN1 | F: 5′‐TCAGCAGGCAATAGATTGGG ‐3′ | 200 | 58 |
| R: 5′‐AAAGGAAAGGCCGTCTAGGG‐3′ | |||
| β‐III tubulin | F: 5′‐CTCAGGGGCCTTTGGACATC‐3′ | 160 | 60 |
| R: 5′‐CAGGCAGTCGCAGTTTTCAC‐3′ | |||
| Primers for bisulfite sequencing PCR | |||
| CRABP‐I | F:5′‐ AGGGAGGTGGAGGTTTTTTAGT ‐3′ | 359 | 53 |
| R:5′‐ ACCAACTTACCCAATACCTTAAAC‐3′ | |||
| CRABP‐II | F:5′‐GGGTTTTTGTTTAATTTTTTAATGTT‐3′ | 214 | 58 |
| R:3′‐AACTAAATCCAATAAAACCACTCC‐5′ | |||
2.3. Immunocytochemical and immunohistochemical staining
Based on RT‐PCR findings, CRABP‐I, CRABP‐II and synaptophysin were selected for further immunocytochemical (ICC) and tissue microarray‐based immunohistochemical (IHC) analyses (Wang et al., 2008), because of their distinct transcription patterns between Med‐3 and UW228‐2 or UW228‐3 cells. The antibodies used were the rabbit anti‐human CRABP‐I and CRABP‐II antibodies (Protein Tech, Chicago, USA) and anti‐human synaptophysin antibody (Dako, Glostrup, Denmark). The experiment was performed on the cell‐bearing coverslips and the paraffin sections of tissue microarray constructed in duplicate with the representative tumor and, where possible, tumor‐surrounding non‐cancerous regions of 104 medulloblastoma cases ranging from ages 1‐year to 58‐years‐old. The sections without the first antibody incubation were used as background control. The staining results were scored as “positive (+)” when the target proteins were evenly distributed in the tissues, “negative (−)” if no immunolabeling was observed, and “mixed pattern (+/−)” when only part (>20% and <80%) of the cells showed positive staining (Malanchi et al., 2008). Spearman rank‐correlation test was used to analyze the distribution of three CRABP‐I and CRABP‐II staining patterns in the three medulloblastoma subtypes and their relevance with synaptophysin expression. Bivariate correlation analysis was applied to evaluate the relationship between CRABP‐I expression and CRABP‐II silencing in the three medulloblastoma subtypes.
2.4. Protein preparation and Western blotting for CRABP‐I and CRABP‐II
The sample proteins (50μg/lane) were prepared conventionally (Wang et al., 2008) and separated by electrophoresis in 10% sodium dodecylsulfate–polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride membrane (Amersham, Buckinghamshire, UK). The membrane was blocked with 5% nonfat milk in TBS‐T (10 mM Tris–Cl, pH 8.0, 150 mM NaCl, and 0.5% Tween 20) at 4 °C overnight, rinsed three times with TBS‐T, followed by 3‐h incubation at room temperature with the first antibodies (1:1000) and 1‐h incubation with HRP‐conjugated anti‐rabbit IgG (Zymed). The bound rabbit anti‐CRABP‐I was detected using the enhanced chemiluminescence system (Roche, Penzberg, Germany). After removing the labeling signal by incubation with stripping buffer (62.5 mM Tris–HCl, pH 6.7, 100 mM 2‐mercaptoethanol, 2% SDS) at 55 °C for 30 min, the membrane was reprobed one‐by‐one with rabbit anti‐human CRABP‐II antibody and with anti‐β‐actin antibody.
2.5. Bisulfite sequencing PCR
DNA samples were extracted from Med‐3, UW228‐2 and UW228‐3 cells, respectively with DNAzol (Invitrogen). They were modified with sodium bisulfite by the method described previously (Zhang et al., 2008) and then purified by the Wizard DNA Clean‐Up System (Promega, Madison, USA). Parts of CRABP‐I and CRABP‐II promoter regions (Figure 3) were amplified using the forward and reverse primers for modified CRABP‐I and CRABP‐II (Table 1). DNA fragments were connected according to the protocol provided with the DNA ligation kit (D6022, TaKaRa). Altogether, 9 ampicillin‐resistance clones were collected separately for DNA isolation and sequencing (ABI PRISMTM 3730XL DNA Analyzer, ABI, CA, USA). The sample DNA isolated from CRABP‐I and CRABP‐II expressing Med‐3 cells was used as negative control for promoter methylation of the two genes. The BSP generated sequences were compared with the sequence of the corresponding region in the normal human genome (http://www.ncbi.nlm.nih.gov/nuccore/NC_000001.10report=genbank&from=156669398&to=156675608&strand=true).
Figure 3.

BSP sequencing‐based DNA methylation analyses of CRABP‐II promoter region encompassing the position of −734 to −521 and 8 CG sites. The sample DNAs isolated from UW228‐2 and UW228‐3 cells were positive in methylation. The methylation status in Med‐3 cells was determined as the negative control. a. Scheme of CRABP‐II gene and the location of BSP amplified promoter region. b. The nucleotide sequence of BSP product and the locations of 8 CG sites. c. The sequencing result of the underlined region (b) of CRABP‐II BSP product generated from UW228‐2 and Med‐3 sample DNAs, respectively. The sequencing result of UW228‐3 BSP product is the same as that of UW228‐2 product and therefore not shown.
2.6. 5‐aza‐2′‐deoxycytidine demethylation and RA treatment
5‐aza‐2′‐deoxycytidine (Sigma–Aldrich) was dissolved in water and diluted with culture medium to the working concentrations. UW228‐2 and UW228‐3 cells were cultured in the presence of 5 μM and 10 μM 5‐aza‐2′‐deoxycytidine for 4‐days by daily renewing 5‐aza‐2′‐deoxycytidine containing medium (Lusher et al., 2002). The demethylated cells were collected directly or co‐incubated with 10 μM RA for another 2 days, followed by total RNA isolation and RT‐PCR for CRABP‐I, CRABP‐II, NGN1, β‐III tubulin, RARα and RARβ, respectively. Meanwhile, cell‐bearing coverslips were collected for H/E staining and CRABP‐I and CRABP‐II immunocytochemistry. Total number and viability of the cells treated by 5‐aza‐2′‐deoxycytidine and RA combination were determined in 12‐h intervals by 0.25% trypan blue staining. The cells cultured normally and treated by 0.2% DMSO, 10 μM RA or 10 μM 5‐aza‐2′‐deoxycytidine were used as normal and background controls.
2.7. Transfection of CRABP‐II expressing plasmid to UW228‐2 cells
Because UW228‐2 and UW228‐3 cells were resistant to RA treatment and shared similar expression pattern of major RA signaling components, the former was selected for transfection assay. Plasmid harboring full‐length CRABP‐II cDNA was purchased from GeneCopoeia, Rockvillie, MD, USA and purified using the PureYieldTM Plasmid Midiprep System (Promega, Madison, USA). After estimating DNA concentration, plasmid DNA was introduced into UW228‐2 cells using Lipofectamine™2000 reagent (Invitrogen, California, USA). Briefly, the cells were conventionally cultured in 6‐well plates until about 90% confluence. 2 μg plasmid DNA was suspended in 50 μl of DMEM and mixed with 2 μl Lipofectamine 2000 in 50 μl DMEM. The mixture was incubated for 20 min at room temperature to allow the formation of DNA‐liposome complexes and added drop‐by‐drop to the culture dishes containing serum‐free DMEM. The cells were incubated at 37 °C in 5% CO2 for 4–6 h. After incubation, the transfection medium was replaced by DMEM supplemented with 10% fetal bovine serum (Gibco Life Science, Grand Island, NY, USA). The transfection efficiency was confirmed by checking the expression of the enhanced green fluorescent EGFP reporter protein at 24‐h time‐point under fluorescence microscope. The transfected cells were then treated by 10 μM RA for 2 days. The morphology, feature of cell death, ratios of viable/unviable cells and CRABP‐II expression of the treated cells were checked by conventional approaches. The results were compared with that obtained from the cells cultured in normal medium without and with RA treatment, treated by Lipo2000 without and with 10 μM RA supplementation and transfected with CRABP‐II expressing plasmids.
2.8. RNA interference and RA treatments
The influence of CRABP‐II down‐regulation in RA sensitivity of Med‐3 cells was elucidated by using the RNA interference/siRNA approach. The CRABP‐II siRNA sequences were sense‐5′‐GAGACAUUUCUACAUCAATT‐3′ and antisense‐5′‐UUGAUGUAGAAAGUGUCUCCC‐3′. The mock oligonucleotides (sense‐5′‐UUCUCCGAACGUG UCACGUTT‐3′ and antisense‐5′‐ACGUGACACGUUCGGAGA) and β‐actin siRNAs (sense‐5′‐UGAAGAUCAAGAUCAUUGCdTdT‐3′ and antisense‐5′‐GCAAUGAUCUUGAUCUUCAdTdT‐3′) were used as negative and positive controls of transfection efficiency (Bergamaschi et al., 2003). Those siRNAs were synthesized by Genepharma Company, Shanghai, China. The RA‐sensitive Med‐3 cells were conventionally cultured in 6‐well plates to 70%–80% confluence and then transfected with 0.3 nmol siRNA/well for 2 or 3 days using 4 μl X‐tremeGENE siRNA transfection reagent according to manufacturer's manual (Roche, Penzberg, Germany). After confirming the efficiency of CRABP‐II inhibition, Med‐3 cells were further incubated with 10 μM RA for 3 days after 2‐day siRNA treatment; afterward, the cells were examined by morphological staining, viable and unviable cell counting, flow cytometry and MTT assay and the results were compared with those obtained from the normally cultured cells and the cells treated by CRABP‐II siRNA or mock oligonucleotides.
3. Results
3.1. Differential response of Med‐3 and UW228 cells to RA treatment
By comparing Med‐3 with UW228‐2 and UW228‐3 cells, we observed that the treatment by 10 μM RA induced elongated changes in cell morphology, growth suppression, and the signs of apoptosis in Med‐3 cells. In contrast, no distinct morphologic changes were seen in RA‐treated UW228‐2 and UW228‐3 cells (Figure 1a). Flow cytometry analysis demonstrated increased fraction of apoptotic cells in RA‐treated Med‐3 but not in UW228‐2 and UW228‐3 cells (Figure 1b). In accordance, TUNEL (Figure 1b), MTT (Figure 1c) and cell viability tests (data not shown) showed significant growth inhibition and apoptosis induction only in RA‐treated Med‐3 cell population. NGN1 was undetectable in normally cultured Med‐3, UW228‐2 and UW228‐3 cells but became expressed only in RA‐treated Med‐3 cells (Figure 2a). The above results suggest that Med‐3 is in fact RA‐sensitive while UW228‐2 and UW228‐3 are RA‐resistant or ‐insensitive medulloblastoma cells.
Figure 1.
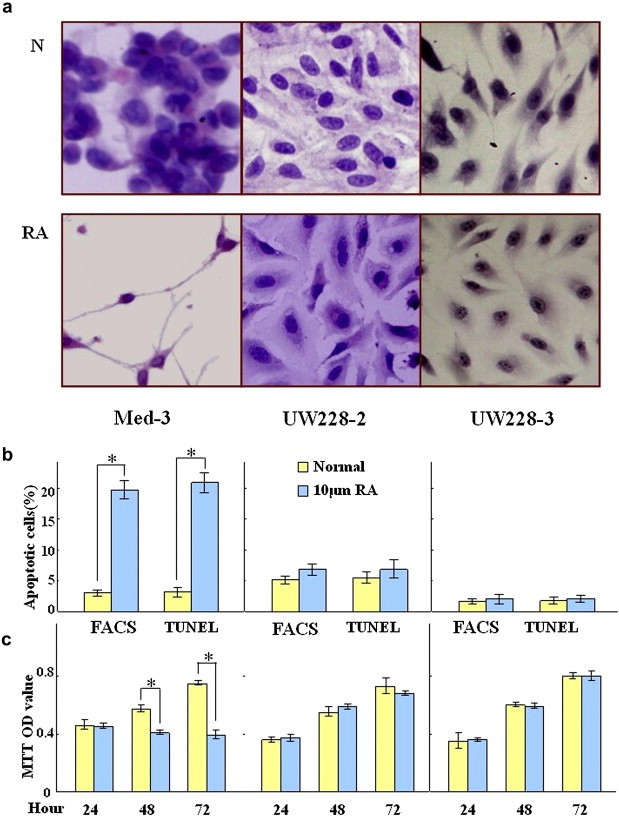
H&E morphological staining (a), flow cytometry (b), TUNEL (b) and MTT assay (c) performed on Med‐3, UW228‐2 and UW228‐3 cells under normal culture condition (N) and incubated with 10 μM RA for 3 days (RA). * indicates p < 0.01.
Figure 2.
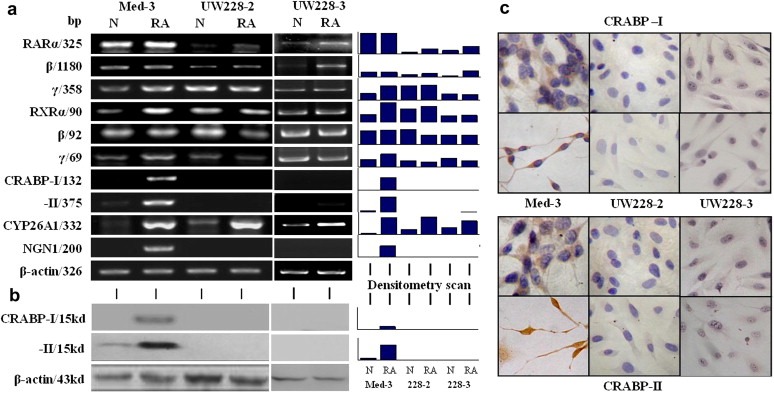
Evaluation of the expression of RA signaling components (RARα, RARβ, RARγ, RXRα, RXRβ, RXRγ, CRABP‐I, CRABP‐II and CYP26A1) and neuronal differentiation biomarker NGN1 in Med‐3, UW228‐2 and UW228‐3 cells by RT‐PCR (a), followed by Western blotting (b) and ICC staining (c) for CRABP‐I and ‐II. Grayscale quantitative analysis was performed on the RT‐PCR and Western blot results. N, normal culture; RA, treatment with 10 μM RA for 72 h.
3.2. RA‐induced CYP26A1 expression
CYP26A1, as a major retinoic acid catabolic enzyme and a supposed target of RA signaling, promotes RA degradation and extracellular elimination (Niederreither et al., 2002; Osanai et al., 2010). The results of RT‐PCR and grayscale analysis clearly showed that under normal culture condition, CYP26A1 was expressed in low levels in UW228‐2, UW228‐3 and Med‐3 cells and its expression was remarkably enhanced in the three cell lines after RA treatment (Figure 2a). This suggests that the expression of CYP26A1 can be enhanced by exogenous RA and this process is independent of RA sensitivity of the medulloblastoma cells.
3.3. Expression of nuclear RA receptors
Because of the importance of nuclear RA receptors in mediating RA target gene transcription, their expression patterns in the three medulloblastoma cell lines were examined before and after RA treatment. The results of RT‐PCR revealed that the three members of RAR and RXR families were expressed in those cells although the level of RARα and RARβ were lower in UW228‐3 and especially in UW228‐2 cells (Figure 2a). In RA‐treated Med‐3 cells, RARα, RARβ and RXRβ expression remained stable, while RXRα, RARγ and RXRγ were increased. In RA‐treated UW228‐2 cells, the expression of RARγ, RXRβ and RXRγ was more or less reduced, RARβ was elevated, and RARα and RXRα remained almost unchanged. In the case of UW228‐3 cells, RARα and RARβ were clearly up‐regulated, while the expression levels of other RA nuclear receptors were almost unchanged.
3.4. Absence of CRABP‐I and CRABP‐II in UW228 cells
The results of RT‐PCR revealed that CRABP‐I was almost undetectable and CRABP‐II was expressed weakly in Med‐3 cells; the expression of these two genes was apparently enhanced upon RA treatment (Figure 2a and b). In contrast, neither control nor the RA‐treated UW228‐2 and UW228‐3 cells showed CRABP‐I and CRABP‐II expression. In agreement with RT‐PCR findings, immunocytochemical staining and Western blot analysis showed a RA‐enhanced expression of CRABP‐I and CRABP‐II in Med‐3 but neither proteins were present in UW228‐2 and UW228‐3 cells without and with RA treatment (Figure 2b and c). Moreover, distinct nuclear translocation of CRABP‐II was observed in RA‐treated Med‐3 cells (Figure 2c).
3.5. CRABP‐II promoter methylation in UW228 cells
To understand the molecular mechanism underlying the loss of CRABP‐I and CRABP‐II expression in UW228‐2 and UW228‐3 cells, the methylation statuses of promoter regions in the genes for CRABP‐I and CRABP‐II were examined by bisulfite sequencing PCR (BSP). The PCR products for CRABP‐I (359bp) and for CRABP‐II (214bp) were sequenced, which demonstrated the presence of methylation at 8 CG sites in the CRABP‐II promoter region, while no methylation was found in the BSP product of Med‐3 cells (Figure 3). In contrast, no promoter methylation of CRABP‐I could be detected in UW228‐2, UW228‐3 and Med‐3 cells (data not shown).
3.6. 5‐aza‐2′‐deoxycytidine recovered CRABP‐II but not CRABP‐I expression
In order to further evaluate the signficance of DNA methylation in regulation of CRABP‐II expression, UW228‐2 and UW228‐3 cells were treated with two different doses (5 μM and 10 μM) of the demethylation agent 5‐aza‐2′‐deoxycytidine for 4 days. The study by RT‐PCR and ICC demonstrated that CRABP‐II expression in the two cell lines was successfully recovered by 5‐aza‐2′‐deoxycytidine, whereas CRABP‐I remained undetectable in both RNA and protein levels (Figure 4a). These data thus provide evidence for the role of DNA methylation to control expression of CRABP‐II but not CRABP‐I in UW228‐2 and UW228‐3 cells. In a separate control study, we showed that the expressions of RARα and RARβ expression were unchanged in both UW228‐2 and UW228‐3 cells by 5‐aza‐2′‐deoxycytidine. The data suggests that DNA methylation may be a specific regulatory mechanism that controls the expression of CRABP‐II in UW228‐2 cells.
Figure 4.
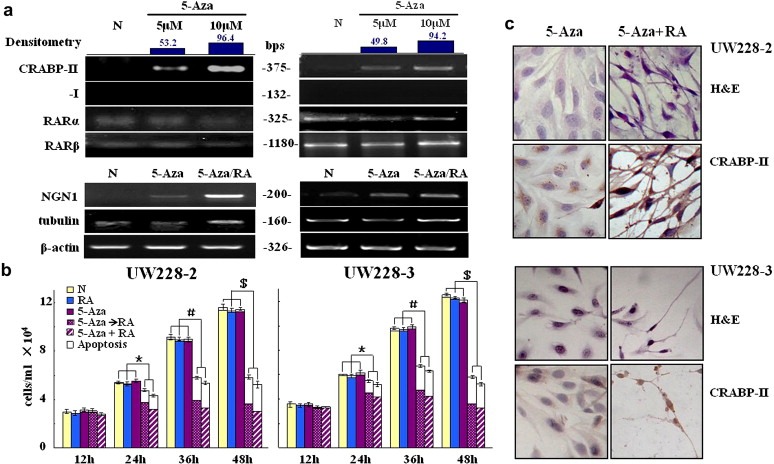
Effects of 5‐aza‐2′‐deoxycytidine induced demethylation on gene expression and RA sensitivity of UW228‐2 and −3 cells. a. RT‐PCR examination of CRABP‐II, CRABP‐I, RARα and RARβ transcription after 4‐day demethylation treatments with 5 μM and 10 μM 5‐aza‐2′‐deoxycytidine. b. Total cell numbers and viable fractions of UW228‐2 and −3 cells cultured for 2 days under the following conditions: 1, normal culture; 2, 10 μM RA; 3, 10 μM 5‐aza‐2′‐deoxycytidine; 4, 10 μM RA after 10 μM 5‐aza‐2′‐deoxycytidine pre‐treatment and 5, co‐treatment with 10 μM 5‐aza‐2′‐deoxycytidine and 10 μM RA. White block, nonviable cells; gray block, viable cells. *, # and $ indicate p < 0.01, respectively. c. Morphological features (H&E staining; X40) and immunocytochemical illustration of CRABP‐II expression of UW228‐2 and −3 cells treated by 10 μM 5‐aza‐2′‐deoxycytidine and by 10 μM 5‐aza‐2′‐deoxycytidine and 10 μM RA combination. RT‐PCR was performed to examine NGN1 transcription in normally cultured cells (N), 3 day 10 μM 5‐aza‐2′‐deoxycytidine treated cells (5‐aza) and the cells pre‐treated by 10 μM 5‐aza‐2′‐deoxycytidine for 3 days, followed by 2 day 10 μM RA treatment (5‐aza+RA).
3.7. Overcome RA resistance by 5‐aza‐2′‐deoxycytidine and CRABP‐II transfection
To elucidate the importance of CRABP‐II expression in determining RA sensitivity of medulloblastoma cells, CRABP‐II expression in UW228‐2 and UW228‐3 cells was recovered by 5‐aza treatment and by CRABP‐II expressing plasmid transfection, respectively. In the first option, CRABP‐II+/CRABP‐I− cells were incubated with 10 μM RA for 2 days after 4‐day 5‐aza‐2′‐deoxycytidine treatment. H/E morphologic examination, trypan blue cell discrimination, ICC and RT‐PCR analyses demonstrated that 10 μM 5‐aza‐2′‐deoxycytidine treatment alone brought about little cellular change, while when 10 μM RA was used, the cells showed growth arrest, apparent neuron‐like morphology and cell death (Figure 4b and c). Furthermore, NGN1, a pivotal bHLH transcription factor in neurogenesis, became expressed in low levels in 5‐aza‐2′‐deoxycytidine treated UW228‐2 and UW228‐3 cells and apparently up‐regulated in the cells treated with both 5‐aza‐2′‐deoxycytidine (10 μM) and RA (10 μM). β‐III tubulin, the early neuronal differentiation biomarker, was expressed in UW228‐2 and UW228‐3 cells and became up‐regulated by 5‐aza‐2′‐deoxycytidine (10 μM) and RA (10 μM) combination (Figure 4a). These phenomena indicate the possible pre‐determination of neuron‐oriented differentiation in UW228 cells and, meanwhile, suggest RA promoted differentiation of 5‐aza‐2′‐deoxycytidine treated cells. More meaningfully, CRABP‐II expression in UW228‐2 cells was specifically recovered by transfecting the plasmids harboring full‐length CRABP‐II cDNA (Figure 5a) and the transfectants became sensitive to RA in the forms as similar as that of 5‐aza‐2′‐deoxycytidine/RA‐treated cells (Figure 5a and b).
Figure 5.

a. H/E morphological staining, CRABP‐II oriented immunocytochemical staining and TUNEL assay performed on UW228‐2 cells transfected with CRABP‐II expressing plasmids and cultured without (Transfection only) or with 10 μM RA supplementation (Transfection/RA). b. Trypan blue viable/nonviable cell discrimination performed on UW228‐2 cells under six experimental conditions: N. normal culture; RA, 10 mM RA treatment; Lipo2000, treatment with transfection reagent only; Lipo2000/RA, co‐treatment with transfection reagent and 10 μM RA; Transfection, CRABP‐II siRNA transfection; Transfection/RA, co‐treatment with CRABP‐II siRNA and 10 μM RA. *, indicates p < 0.01.
3.8. Reduced RA sensitivity of Med‐3 cells by CRABP‐II siRNA
The importance of CRABP‐II in determining RA sensitivity of medulloblastoma cells was confirmed by transfecting Med‐3 cells with CRABP‐II siRNA candidates. Among three siRNA sequences tested, the siRNA construct containing the following sequences, sense‐5′‐GAGACAUUUCUACAUCAATT‐3′ and antisense‐5′‐UUGAUGUAGAAAGUGUCUCCC‐3′, showed an optimal inhibitory effect (∼83%) on CRABP‐II expression (Figure 6a and b). We then used this siRNA in the subsequent experiments. MTT assay and morphological examination revealed that a 3‐day treatment with 10 μM RA failed to bring about growth arrest and neuron‐like differentiation of the Med‐3 cells transfected by CRABP‐II siRNA (Figure 6b and c), and the apoptosis fractions were similar between the Med‐3 cells treated by siRNA only and by the siRNA/RA combination (Figure 6d). In contrast, the RA‐sensitive features remained unchanged when Med‐3 cells were treated by 10 μM mock oligonucleotides and 10 μM RA.
Figure 6.
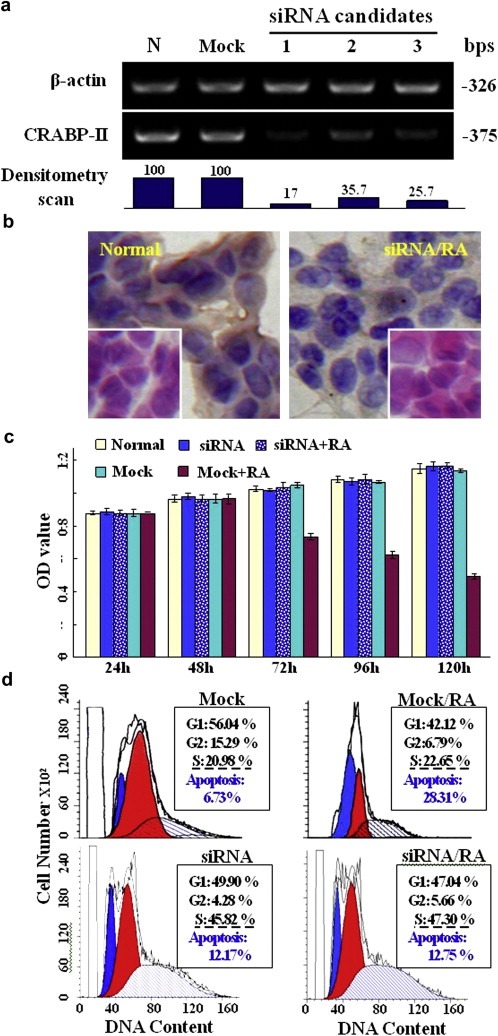
siRNA suppressed CRABP‐II expression and its influence in RA sensitivity of Med‐3 cells. a. Down‐regulation of CRABP‐II expression by siRNA and subsequent reduction of RA sensitivity of Med‐3 cells. Densitometry scan values of the RT‐PCR products showed that the three siRNA candidates exerted different inhibitory effects on CRABP‐II transcripts. b. H&E staining C and RABP‐II oriented immunocytochemical staining performed on normally cultured Med‐3 cells and the cells treated by siRNA and RA combination. c. MTT‐based proliferation assay of Med‐3 cells under normal culture condition (normal), transfected with mock siRNA (mock) or with CRABP‐II specific siRNA (siRNA) and pre‐treated by CRABP‐II specific siRNA for 48 h, followed by 10 μM RA supplementation (siRNA + RA). No statistical significance of OD values among the four experimental groups (p > 0.05). d. Flow cytometry analysis of Med‐3 cells under four culture conditions: mock, transfected with mock siRNA; siRNA, transfected with CRABP‐II specific siRNA; mock + RA, treated by mock siRNA for 48 h and then 10 μM RA for 72 h; siRNA + RA, treated by CRABP‐II specific siRNA for 48 h and then 10 μM RA for 72 h.
3.9. Differential in vivo expression of CRABP‐I, CRABP‐II and synaptophysin
Since no comprehensive report is available concerning CRABP‐I and CRABP‐II expression in human medulloblastoma tumors, tissue microarray‐based IHC staining was performed to address this issue. The results revealed that all of the 12 non‐cancerous cerebella were positive in CRABP‐I and CRABP‐II production together with synaptophysin expression in the neurons (Figure 7a). In contrast, 45 (43.3%) of the 104 tumor samples were positive (+), 44 (42.3%) were negative (−) and 15 (14.4%) were partly positive in CRABP‐II expression (Figure 7b and Table 2). According to the criterion reported by Eberhart et al. 2002, 58 samples were diagnosed as the classic subtype, 40 as anaplastic/large cell subtype, and 6 as nodular subtype. Three CRABP‐II staining patterns were observed in the corresponding three histological groups as mentioned above. As shown in Figure 7c and Table 2, the expression of CRABP‐II and synaptophysin was strongly correlated (Spearman rank‐correlation, rs = 0.317; P = 0.001) among the 104 medulloblastoma specimens examined and this correlation was more remarkable (91.7%; 22/24) in the classic subtype (rs = 0.576; P = 0.000). Paralleled analysis demonstrated that 60 (57.7%) of 104 tumor cases were negative in CRABP‐I expression and distributed in 36/58 (62.1%) of the classic, 18/40 (45%) of the anaplastic/large cell and 6/6 (100%) of the nodular/desmoplastic medulloblastomas. The overlapped CRABP‐I and CRABP‐II silencing was only found in 18/58 (31.0%) of the classic, 10/40 (25%) of the anaplastic/large cell and 3/6 (50%) of the anaplastic/desmoplastic tumors (p > 0.05; Figure 7c and d).
Figure 7.

a. Immunohistochemical illustration of CRABP‐I, CRABP‐II and synaptophysin expression in medulloblastoma‐surrounding non‐cancerous tissues. b. Immunohistochemical profiling of CRABP‐II expression patterns in the classic and the anaplastic subtypes of medulloblastomas. The staining patterns were scored as “Negative (−)” if no immunolabeling was observed in the tumor cells, “Positive (+)” if distinct staining was generally observed and “Mixed (+/−)” when CRABP‐II positive and negative tumor cells co‐existed in the same observed region. c. The incidences of three CRABP‐I IHC staining patterns in the medulloblastoma subtypes. d. Fractionation of CRABP‐I and CRABP‐II expression patterns in the three medulloblastoma subtypes. Chi‐square analysis showed no statistic difference between the expression of these two genes in any of the histological groups (p > 0.05).
Table 2.
Tissue microarray‐based immunohistochemical staining of CRABP‐I, CRABP‐II and synaptophysin in the three histological subtypes of medulloblastomas
| CRABP‐I (%) | Synaptophysin (%) | CRABP‐II (%) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | – | + | +/− | – | + | +/− | – | + | +/− | |||
| Medulloblastomas | 104 | 60 (57.7) | 39 (37.5) | 5 (4.8) | $ | 24 (23.1) | 74 (71.1) | 6 (5.8) | # | 44 (42.3) | 45 (43.3) | 15 (14.4) |
| Classic | 58 | 36 (62.1) | 20 (34.5) | 2 (3.4) | $$ | 15 (25.9) | 38 (65.5) | 5 (8.6) | ∗ | 28 (48.3) | 24 (41.4) | 6 (10.3) |
| Anaplastic/large cell | 40 | 18 (45) | 19 (47.5) | 3 (7.5) | $$$ | 6 (15) | 33 (82.5) | 1 (2.5) | ∗∗ | 13 (32.5) | 19 (47.5) | 8 (20) |
| Nodular/desmoplastic | 6 | 6 (100) | 0 | 0 | 3 (50) | 3 (50) | 0 | 3 (50) | 2 (33.3) | 1 (16.7) | ||
The marked correlations between CRABP‐I and Synaptophysin expression levels: $rs = −0.109, p>0.05; $$rs = 0.036, p>0.05; $$$rs = −0.252, p>0.05.
The marked correlations between CRABP‐II and Synaptophysin expression levels: # rs = 0.317, p<0.01; * rs = 0.576, p<0.01; ** rs = 0.09, p = 0.579.
4. Discussion
RA has been widely used in cancer chemotherapy because of its property to suppress growth and to induce differentiation and apoptosis (Meyskens and Manetta, 1995; Zusi et al., 2002; Ravandi, 2009). Several cytosolic elements can influence the biological effects of RA. For example, the significances of CRABP‐II in RA resistance had been reported using a mouse model of breast cancer MMTV‐neu in which the expression of CRABP‐II is disrupted (Schug et al., 2008). CYP26A1 can metabolize RA to less bioactive forms and eliminate metabolites to the extracellular space (Abu‐Abed et al., 2002). CRABP‐I influences the biological effects of RA in cell‐selective manners by enhancing the physiological function of RA in keratinocytes (Aström et al., 1991; Tang et al., 2008) or inhibiting RA‐induced differentiation of neuroblastoma cells (Uhrig et al., 2008). Because any imbalanced expression of RA associated proteins may lead to different cellular outcomes (Warrell et al., 1993; Zhu et al., 2009), a full understanding of the mechanism for RA‐resistant mechanism(s) will potentially help clinicians in the selection of personalized cancer therapy for their patients.
Our study revealed that CRABP‐I and ‐II were up‐regulated in Med‐3 cells treated with RA but absent in both UW228‐2 and UW228‐3 cells regardless of RA treatment. Although CYP26A1 was expressed in low levels in the three medulloblastoma cell lines, it could be remarkably up‐regulated by RA in those cells. As for the nuclear RA receptors, the three subtypes of RARs and RXRs were found either in Med‐3 or in UW228‐2 and UW228‐3 cells. Given the above findings, we considered that CYP26A1 seems not a central player in determining RA therapeutic effects because it is inducible in both RA‐sensitive and RA‐resistant medulloblastoma cells. However, the fundamental difference of CRABP‐I and ‐II expression patterns between Med‐3 and UW228 cells suggests that these proteins likely contribute to the differences in RA sensitivity. Tissue microarray‐based immunohistochemistry revealed that CRABP‐I and CRABP‐II were constitutively expressed in the tumor‐surrounding non‐cancerous cerebella but absent in 57.7% and 42.3% of medulloblastoma tumors, respectively. Interestingly, 15 out of 104 (14.4%) medulloblastoma cases were partly positive in CRABP‐II expression and 5 (4.8%) in CRABP‐I expression. This further suggests the presence of molecular heterogeneity (Pfister et al., 2010), the multiclonal origins of medulloblastomas and independent expression of these two genes (Eberhart et al., 2002).
Synaptophysin is regarded as a reliable biomarker of neuron and an indicator of neuron‐like differentiation of brain tumors (Kawashima et al., 2000; Kido et al., 2009). In order to evaluate potential relevance of synaptophysin production with CRABP‐II expression, a correlative analysis of these two parameters was conducted using the data from tissue microarray‐based immunohistochemistry. Although CRABP‐II and synaptophysin expression was closely related not only in non‐cancerous cerebella but also among the three subtypes of medulloblastoma tissues, there are still some cases with synaptophysin but without CRABP‐II expression (31.2%; 23/74) and vice versa (13.3%; 6/45). These phenomena suggest that the concurrent expression of these two genes in medulloblastoma cells may not be directly linked. Therefore, CRABP‐II expression is unnecessarily restricted to a particular medulloblastoma subtype and can not be inferred from the commonly‐used synaptophysin staining.
DNA methylation is known to regulate gain or loss of gene expression, which also happens to the promoter region of CRABP‐I (Wei and Lee, 1994; Tanaka et al., 2007) and CRABP‐II (Gupta et al., 2008; Calmon et al., 2009). Although differential responses of medulloblastoma cells to RA treatment have been documented, there has been no report yet concerning the effect of CRABP‐I and ‐II expression patterns on RA sensitivity. Our study thus demonstrated for the first‐time the fundamental difference of CRABP‐I and CRABP‐II expression in RA‐sensitive Med‐3 and RA‐resistant UW228‐2 and ‐3 cells. Furthermore, more than half of medulloblastoma cases checked so far were either totally or partly negative in CRABP‐I and CRABP‐II expression. These findings prompted us to investigate the underlying reason(s) leading to the silencing of these two genes. The presence of CG methylation at the amplified CRABP‐II promoter region (−521 to −734) and 5‐aza‐2′‐deoxycytidine recovered CRABP‐II expression of UW228‐2 and UW228‐3 cells suggest a close correlation of DNA methylation with CRABP‐II silencing. The distinct CRABP‐II expression patterns in RA‐sensitive and RA‐resistant medulloblastoma cells thus provide an ideal experimental model to evaluate the importance of CRABP‐II in determining the RA sensitivities of medulloblastoma cells. In the case of CRABP‐I, the parallel sequencing analysis showed no methylation in the BSP product and 5‐aza‐2′‐deoxycytidine failed to restore its expression in both UW228‐2 and UW228‐3 cells. These findings indicate the presence of an alternative regulatory pathway for CRABP‐I expression. CYP26A1 is supposed to be the target gene of CRABP‐II mediated RA signaling (Abu‐Abed et al., 2002), while this gene can be up‐regulated in RA‐treated UW228‐2 cells in CRABP‐II independent fashion. Since CYP26A1 expression is also under complex feed‐forward control by FGF signaling that blocks the proliferation of mouse medulloblastoma cells via inhibiting Sonic hedgehog signaling (White et al., 2007), the potential influence of RA in FGF‐regulated CYP26A1 expression in human medulloblastoma cells should be taken into account.
So far, the influence of altered CRABP‐I and ‐II expression in the RA sensitivities of human cancers, especially medulloblastomas, remains lesser known. This issue has been addressed in the current study by modulating CRABP‐II expression in UW228‐2, UW228‐3 and Med‐3 cells. An increased RA sensitivity accompanied with appearance of NGN1 expression could be observed in 5‐aza‐2′‐deoxycytidine treated UW228‐2 and −3 cells in which CRABP‐II but not CRABP‐I expression was recovered. More importantly, UW228‐2 cells transfected with CRABP‐II expressing element became sensitive to RA and, conversely, Med‐3 cells showed declined RA sensitivities when CRABP‐II expression was largely inhibited by siRNA approach. It would be therefore considered that the presence or absence of CRABP‐II but not CRABP‐I would be more critical in determining RA sensitivities of human medulloblastoma cells. In this context, CRABP‐II can be regarded as a potential epigenetic marker for personalized RA therapy of medulloblastomas and other applicable cancers.
Acknowledgment
This work is supported by the grants from National Natural Science Foundation of China (30670946, 30971038, 81072063 and 81071971) and by the special grants of Liaoning Department of Education for creative research team (2007‐7‐26 and 2008T028) and for the key laboratory (20060193). We thank Dr. Hai‐Feng Wu and his colleagues at Ohio State University for their critical reading.
Fu Yuan-Shan, Wang Qian, Ma Jing-Xin, Yang Xiang-Hong, Wu Mo-Li, Zhang Kai-Li, Kong Qing-You, Chen Xiao-Yan, Sun Yuan, Chen Nan-Nan, Shu Xiao-Hong, Li Hong, Liu Jia, (2012), CRABP‐II methylation: A critical determinant of retinoic acid resistance of medulloblastoma cells, Molecular Oncology, 6, doi: 10.1016/j.molonc.2011.11.004.
Contributor Information
Hong Li, Email: lihongmcn@dlmedu.edu.cn, Email: lihongmcn@yahoo.com.cn.
Jia Liu, Email: jialiudl@dlmedu.edu.cn.
References
- Abu-Abed, S. , MacLean, G. , Fraulob, V. , Chambon, P. , Petkovich, M. , Dollé, P. , 2002. Differential expression of the retinoic acid-metabolizing enzymes CYP26A1 and CYP26B1 during murine organogenesis. Mech. Dev.. 110, 173–177. [DOI] [PubMed] [Google Scholar]
- Aström, A. , Tavakkol, A. , Pettersson, U. , Cromie, M. , Elder, J.T. , Voorhees, J.J. , 1991. Molecular cloning of two human cellular retinoic acid-binding proteins (CRABP). Retinoic acid-induced expression of CRABP-II but not CRABP-I in adult human skin in vivo and in skin fibroblasts in vitro. J. Biol. Chem.. 266, 17662–17666. [PubMed] [Google Scholar]
- Bergamaschi, D. , Gasco, M. , Hiller, L. , Sullivan, A. , Syed, N. , Trigiante, G. , Yulug, I. , Merlano, M. , Numico, G. , Comino, A. , Attard, M. , Reelfs, O. , 2003. P53 polymorphism influences response in cancer chemotherapy via modulation of p73-dependent apoptosis. Cancer Cell.. 3, 387–402. [DOI] [PubMed] [Google Scholar]
- Budhu, A.S. , Noy, N. , 2002. Direct channeling of retinoic acid between cellular retinoic acid-binding protein II and retinoic acid receptor sensitizes mammary carcinoma cells to retinoic acid-induced growth arrest. Mol. Cell Biol.. 22, 2632–2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calmon, M.F. , Rodrigues, R.V. , Kaneto, C.M. , Moura, R.P. , Silva, S.D. , Mota, L.D. , Pinheiro, D.G. , Torres, C. , de Carvalho, A.F. , Cury, P.M. , Nunes, F.D. , Nishimoto, I.N. , 2009. Epigenetic silencing of CRABP2 and MX1 in Head and Neck tumors. Neoplasia. 11, 1329–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, Q. , Chen, Z. , You, J. , McNutt, M.A. , Zhang, T. , Han, Z. , Zhang, X. , Gong, E. , Gu, J. , 2007. All-trans-retinoic acid induces cell growth arrest in a human medulloblastoma cell line. J. Neurooncol.. 84, 263–267. [DOI] [PubMed] [Google Scholar]
- Chatty, E.M. , Earle, K.M. , 1971. Medulloblastoma. A report of 201 cases with emphasis on the relationship of histologic variants to survival. Cancer. 28, 977–983. [DOI] [PubMed] [Google Scholar]
- Corlazzoli, F. , Rossetti, S. , Bistulfi, G. , Ren, M. , Sacchi, N. , 2009. Derangement of a factor upstream of RARalpha triggers the repression of a pleiotropic epigenetic network. PLoS One. 4, e4305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delva, L. , Bastie, J.N. , Rochette-Egly, C. , Kraïba, R. , Balitrand, N. , Despouy, G. , Chambon, P. , Chomienne, C. , 1999. Physical and functional interactions between cellular retinoic acid binding protein II and the retinoic acid-dependent nuclear complex. Mol. Cell Biol.. 19, 7158–7167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, D. , Ruuska, S.E. , Levinthal, D.J. , Noy, N. , 1999. Distinct roles for cellular retinoic acid-binding proteins I and II in regulating signaling by retinoic acid. J. Biol. Chem.. 274, 23695–23698. [DOI] [PubMed] [Google Scholar]
- Eberhart, C.G. , Kepner, J.L. , Goldthwaite, P.T. , Kun, L.E. , Duffner, P.K. , Friedman, H.S. , Strother, D.R. , Burger, P.C. , 2002. Histopathologic grading of medulloblastomas: a Pediatric Oncology Group study. Cancer. 94, 552–560. [DOI] [PubMed] [Google Scholar]
- Fossati, P. , Ricardi, U. , Orecchia, R. , 2009. Pediatric medulloblastoma: toxicity of current treatment and potential role of protontherapy. Cancer Treat. Rev.. 35, 79–96. [DOI] [PubMed] [Google Scholar]
- Freemantle, S.J. , Spinella, M.J. , Dmitrovsky, E. , 2003. Retinoids in cancer therapy and chemoprevention: promise meets resistance. Oncogene. 22, 7305–7315. [DOI] [PubMed] [Google Scholar]
- Gumireddy, K. , Sutton, L.N. , Phillips, P.C. , Reddy, C.D. , 2003. All-trans-retinoic acid-induced apoptosis in human medulloblastoma: activation of caspase-3/poly(ADP-ribose) polymerase 1 pathway. Clin. Cancer Res.. 9, 4052–4059. [PubMed] [Google Scholar]
- Gururangan, S. , Krauser, J. , Watral, M.A. , Driscoll, T. , Larrier, N. , Reardon, D.A. , Rich, J.N. , Quinn, J.A. , Vredenburgh, J.J. , Desjardins, A. , McLendon, R.E. , Fuchs, H. , 2008. Efficacy of high-dose chemotherapy or standard salvage therapy in patients with recurrent medulloblastoma. Neuro Oncol.. 10, 745–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta, A. , Kessler, P. , Rawwas, J. , Williams, B.R. , 2008. Regulation of CRABP-II expression by MycN in Wilms tumor. Exp. Cell Res.. 314, 3663–3668. [DOI] [PubMed] [Google Scholar]
- Hallahan, A.R. , Pritchard, J.I. , Chandraratna, R.A. , Ellenbogen, R.G. , Geyer, J.R. , Overland, R.P. , Strand, A.D. , Tapscott, S.J. , Olson, J.M. , 2003. BMP-2 mediates retinoid-induced apoptosis in medulloblastoma cells through a paracrine effect. Nat. Med.. 9, 1033–1038. [DOI] [PubMed] [Google Scholar]
- Ho, D.M. , Hsu, C.Y. , Wong, T.T. , Ting, L.T. , Chiang, H. , 2000. Atypical teratoid/rhabdoid tumor of the central nervous system: a comparative study with primitive neuroectodermal tumor/medulloblastoma. Acta Neuropathol.. 99, 482–488. [DOI] [PubMed] [Google Scholar]
- Kawashima, M. , Suzuki, S.O. , Doh-ura, K. , Iwaki, T. , 2000. alpha-Synuclein is expressed in a variety of brain tumors showing neuronal differentiation. Acta Neuropathol.. 99, 154–160. [DOI] [PubMed] [Google Scholar]
- Keles, G.E. , Berger, M.S. , Srinivasan, J. , Kolstoe, D.D. , Bobola, M.S. , Silber, J.R. , 1995. Establishment and characterization of four human medulloblastoma-derived cell lines. Oncol. Res.. 7, 493–503. [PubMed] [Google Scholar]
- Kido, M. , Ueno, M. , Onodera, M. , Matsumoto, K. , Imai, T. , Haba, R. , Tamiya, T. , Huang, C.L. , Sakamoto, H. , 2009. Medulloblastoma with myogenic differentiation showing double immunopositivity for synaptophysin and myoglobin. Pathol. Int.. 59, 255–260. [DOI] [PubMed] [Google Scholar]
- Kitamura, K. , Kiyoi, H. , Yoshida, H. , Tobita, T. , Takeshita, A. , Ohno, R. , Naoe, T. , 2004. New retinoids and arsenic compounds for the treatment of refractory acute promyelocytic leukemia: clinical and basic studies for the next generation. Cancer Chemother. Pharmacol.. 40, 36S–41S. [DOI] [PubMed] [Google Scholar]
- Liu, J. , Guo, L. , Li, J.W. , Liu, N. , Li, H. , 2000. All-trans retinoic acid modulates fas expression and enhances chemosensitivity of human medulloblastoma cells. Int. J. Mol. Med.. 5, 145–149. [DOI] [PubMed] [Google Scholar]
- Lusher, M.E. , Lindsey, J.C. , Latif, F. , Pearson, A.D. , Ellison, D.W. , Clifford, S.C. , 2002. Biallelic epigenetic inactivation of the RASSF1A tumor suppressor gene in medulloblastoma development. Cancer Res.. 62, 5906–5911. [PubMed] [Google Scholar]
- Malanchi, I. , Peinado, H. , Kassen, D. , Hussenet, T. , Metzger, D. , Chambon, P. , Huber, M. , Hohl, D. , Cano, A. , Birchmeier, W. , Huelsken, J. , 2008. Cutaneous cancer stem cell maintenance is dependent on β-catenin signalling. Nature. 452, 650–653. [DOI] [PubMed] [Google Scholar]
- Matsumoto, M. , 1991. Inhibitors for protein tyrosine kinases, erbstatin, genistein and herbimycin A, induce differentiation of human neural tumor cell lines. Nippon Geka Hokan. 60, 113–121. [PubMed] [Google Scholar]
- Meyskens, F.L. , Manetta, A. , 1995. Prevention of cervical intraepithelial neoplasia and cancer. Am. J. Clin. Nutr.. 62, 1417S–1419S. [DOI] [PubMed] [Google Scholar]
- Mueller, S. , Chang, S. , 2009. Pediatric brain tumors: current treatment strategies and future therapeutic approaches. Neurotherapeutics. 6, 570–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niederreither, K. , Abu-Abed, S. , Schuhbaur, B. , Petkovich, M. , Chambon, P. , Dollé, P. , 2002. Genetic evidence that oxidative derivatives of retinoic acid are not involved in retinoid signaling during mouse development. Nat. Genet.. 31, 84–88. [DOI] [PubMed] [Google Scholar]
- Ohta, M. , Taga, T. , Nomura, A. , Kato, H. , Takano, T. , Maruo, Y. , Takeuchi, Y. , Ishida, M. , Ohta, S. , 2011. Epstein-Barr virus-related lymphoproliferative disorder, cytomegalovirus reactivation, and varicella zoster virus encephalitis during treatment of medulloblastoma. J. Med. Virol.. 83, 1582–1584. [DOI] [PubMed] [Google Scholar]
- Osanai, M. , Sawada, N. , Lee, G.H. , 2010. Oncogenic and cell survival properties of the retinoic acid metabolizing enzyme, CYP26A1. Oncogene. 29, 1135–1144. [DOI] [PubMed] [Google Scholar]
- Pfister, S.M. , Korshunov, A. , Kool, M. , Hasselblatt, M. , Eberhart, C. , Taylor, M.D. , 2010. Molecular diagnostics of CNS embryonal tumors. Acta Neuropathol.. 120, 553–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravandi, F. , 2009. Therapy-related acute promyelocytic leukemia: further insights into the molecular basis of the disease and showing the way forward in therapy. Leuk. Lymphoma. 50, 1073–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schug, T.T. , Berry, D.C. , Shaw, N.S. , Travis, S.N. , Noy, N. , 2007. Opposing effects of retinoic acid on cell growth result from alternate activation of two different nuclear receptors. Cell. 129, 723–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schug, T.T. , Berry, D.C. , Toshkov, I.A. , Cheng, L. , Nikitin, A.Y. , Noy, N. , 2008. Overcoming retinoic acid-resistance of mammary carcinomas by diverting retinoic acid from PPARbeta/delta to RAR. Proc. Natl. Acad. Sci. USA. 105, 7546–7551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessler, R.J. , Noy, N. , 2005. A ligand-activated nuclear localization signal in cellular retinoic acid binding protein-II. Mol. Cell.. 18, 343–353. [DOI] [PubMed] [Google Scholar]
- Tanaka, K. , Imoto, I. , Inoue, J. , Kozaki, K. , Tsuda, H. , Shimada, Y. , Aiko, S. , Yoshizumi, Y. , Iwai, T. , Kawano, T. , Inazawa, J. , 2007. Frequent methylation-associated silencing of a candidate tumor-suppressor, CRABP1, in esophageal squamous-cell carcinoma. Oncogene. 26, 6456–6468. [DOI] [PubMed] [Google Scholar]
- Tang, X.H. , Vivero, M. , Gudas, L.J. , 2008. Overexpression of CRABPI in suprabasal keratinocytes enhances the proliferation of epidermal basal keratinocytes in mouse skin topically treated with all-trans retinoic acid. Exp. Cell Res.. 314, 38–51. [DOI] [PubMed] [Google Scholar]
- Uhrig, M. , Brechlin, P. , Jahn, O. , Knyazev, Y. , Weninger, A. , Busia, L. , Honarnejad, K. , Otto, M. , Hartmann, T. , 2008. Upregulation of CRABP1 in human neuroblastoma cells overproducing the Alzheimer-typical Abeta42 reduces their differentiation potential. BMC Med.. 6, 38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Burg, B. , van der Leede, B.M. , Kwakkenbos-Isbrücker, L. , Salverda, S. , de Laat, S.W. , van der Saag, P.T. , 1993. Retinoic acid resistance of estradiol-independent breast cancer cells coincides with diminished retinoic acid receptor function. Mol. Cell Endocrinol.. 91, 149–157. [DOI] [PubMed] [Google Scholar]
- Wang, Q. , Li, H. , Liu, N. , Chen, X.Y. , Wu, M.L. , Zhang, K.L. , Kong, Q.Y. , Liu, J. , 2008. Correlative analyses of notch signaling with resveratrol-induced differentiation and apoptosis of human medulloblastoma cells. Neurosci. Lett.. 438, 168–173. [DOI] [PubMed] [Google Scholar]
- Warrell, R.P. , de Thé, H. , Wang, Z.Y. , Degos, L. , 1993. Acute promyelocytic leukemia. N. Engl. J. Med.. 329, 177–189. [DOI] [PubMed] [Google Scholar]
- Wei, L.N. , Lee, C.H. , 1994. Demethylation in the 5'-flanking region of mouse cellular retinoic acid binding protein-I gene is associated with its high level of expression in mouse embryos and facilitates its induction by retinoic acid in P19 embryonal carcinoma cells. Dev. Dyn.. 201, 1–10. [DOI] [PubMed] [Google Scholar]
- White, R.J. , Nie, Q. , Lander, A.D. , Schilling, T.F. , 2007. Complex regulation of cyp26a1 Creates a Robust retinoic acid gradient in the Zebrafish Embryo. PLoS Biol.. 5, e304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, K.L. , Sun, Y. , Li, Y. , Liu, M. , Qu, B. , Cui, S.H. , Kong, Q.Y. , Chen, X.Y. , Li, H. , Liu, J. , 2008. Increased frequency of CpG island methylator phenotype and CDH1 methylation in a gastric cancer high-risk region of China. Transl Oncol.. 1, 28–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, G.H. , Huang, J. , Bi, Y. , Su, Y. , Tang, Y. , He, B.C. , He, Y. , Luo, J. , Wang, Y. , Chen, L. , Zuo, G.W. , Jiang, W. , Luo, Q. , Shen, J. , Liu, B. , Zhang, W.L. , Shi, Q. , Zhang, B.Q. , Kang, Q. , Zhu, J. , Tian, J. , Luu, H.H. , Haydon, R.C. , Chen, Y. , He, T.C. , 2009. Activation of RXR and RAR signaling promotes myogenic differentiation of myoblastic C2C12 cells. Differentiation. 78, 195–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zusi, F.C. , Lorenzi, M.V. , Vivat-Hannah, V. , 2002. Selective retinoids and rexinoids in cancer therapy and chemoprevention. Drug Discov. Today. 7, 1165–1174. [DOI] [PubMed] [Google Scholar]


