Abstract
Lysophosphatidylcholine acyltransferase 1 (LPCAT1) has been suggested to play a role in cancer. To assess its role in prostate cancer, LPCAT1 expression was analyzed on a tissue microarray containing samples from 11,152 prostate cancer patients. In benign prostate glands, LPCAT1 immunostaining was absent or weak. In prostate cancer, LPCAT1 positivity was found in 73.8% of 8786 interpretable tumors including 29.2% with strong expression. Increased LPCAT1 expression was associated with advanced tumor stage (pT3b/T4) (p < 0.0001), high Gleason score (≥4 + 4) (p < 0.0001), positive nodal involvement (p = 0.0002), positive surgical margin (p = 0.0005), and early PSA recurrence (p < 0.0001). High LPCAT1 expression was strongly linked to ERG‐fusion type prostate cancer. Strong LPCAT1 staining was detected in 45.3% of ERG positive but in only 16.7% of ERG negative tumors (p < 0.0001). Within ERG negative cancers, LPCAT1 staining was strongly increased within the subgroup of PTEN deleted cancers (p < 0.0001). Further subgroup analyses revealed that associations of high LPCAT1 expression with PSA recurrence and unfavorable tumor phenotype were largely driven by ERG negative cancers (p < 0.0001) while these effects were substantially mitigated in ERG positive cancers (p = 0.0073). The prognostic impact of LPCAT1 expression was independent of histological and clinical parameters. It is concluded, that LPCAT1 measurement, either alone or in combination, may be utilized for better clinical decision‐making. These data also highlight the potentially important role of lipid metabolism in prostate cancer biology.
Keywords: LPCAT1, Tissue microarray, Prostate cancer
Highlights
Increased LPCAT1 expression was associated with unfavorable tumor phenotype and early PSA recurrence in prostate cancers.
High LPCAT1 expression was strongly linked to ERG‐fusion type prostate cancer.
Within ERG negative cancers, LPCAT1 staining was strongly increased within the subgroup of PTEN deleted cancers.
LPCAT1 measurement, either alone or in combination, may be utilized for better clinical decision‐making.
1. Introduction
Prostate cancer represents a major cause of cancer‐related mortality and morbidity (Siegel et al., 2013). In consequence of the clinical and histological heterogeneity of prostate cancer, possible treatment options vary from active surveillance to surgical or radiation therapy. As the common pre‐operative prognostic parameters Gleason grade, tumor extent on biopsies, preoperative PSA and clinical parameters are not satisfactory for optimal individual treatment decisions, the identification of novel biomarkers reflecting the tumor aggressiveness are desperately needed.
Alterations in lipid metabolism affect numerous cellular processes that are relevant for cancer biology, including cell growth, proliferation, differentiation and motility (Santos and Schulze, 2012). Lysophosphatidylcholine acyltransferase 1 (LPCAT1) is a key enzyme of the Lands cycle that contributes to the synthesis of diverse individual phosphatidylcholine specimens, which are prominent constituents of eukaryotic membranes and major structural features of serum lipoproteins (Kent, 2005). LPCAT1 has physiological roles in the lung where it generates the component dipalmitoyl phosphatidylcholine of pulmonary surfactant (Bridges et al., 2010; Chen et al., 2006; Nakanishi et al., 2006), in non‐inflammatory platelet‐activation factor remodeling pathway (Harayama et al., 2008) and in retinal photoreceptor homeostasis (Cheng et al., 2009).
Only recently, overexpression of LPCAT1 has also been described in human malignancies, such as human colorectal cancer (Mansilla et al., 2009). Functional analyses in colon cancer cell lines had also demonstrated that overexpressed LPCAT1 results in a significant growth advantage (Mansilla et al., 2009).
In one recent study elevated LPCAT1 expression was also found in prostate cancer and an association of LPCAT1 expression levels with PSA recurrence was suggested in a cohort of 148 patients (Zhou et al., 2012).
To further evaluate the potential of LPCAT1 as a clinically relevant prognostic prostate cancer biomarker, and to search for possible associations with molecularly defined cancer subgroups, a preexisting tissue microarray (TMA) containing 11,152 prostate cancer specimens with follow‐up data and attached molecular information was analyzed for LPCAT1 expression. Our data identify high LPCAT1 expression as a strong and independent prognostic biomarker of early PSA recurrence in prostate cancer.
2. Materials and methods
2.1. Patients
Radical prostatectomy specimens were available from 11,152 patients, undergoing surgery between 1992 and 2011 at the Department of Urology, and the Martini Clinics at the University Medical Center Hamburg‐Eppendorf. Follow‐up data were available of 9695 patients with a median follow‐up of 36.8 months (range: 1–228 months; Table 1). Prostate specific antigen values were measured following surgery and recurrence was defined as a postoperative PSA of 0.2 ng/ml and rising. All prostate specimens were analyzed according to a standard procedure, including complete embedding of the entire prostate for histological analysis (Erbersdobler et al., 2002). The Gleason score had been diagnosed in the prostatectomy specimens. The TMA manufacturing process was described earlier in detail (Mirlacher and Simon, 2010). In short, one 0.6 mm core was taken from a representative tissue block from each patient. The tissues were distributed among 24 TMA blocks, each containing 144 to 522 tumor samples. Presence or absence of cancer tissue was validated by immunohistochemical AMACR and 34BE12 analysis on adjacent TMA sections. For internal controls, each TMA block also contained various control tissues, including normal prostate tissue. The molecular database attached to this TMA contained results on ERG expression in 9,628, ERG break apart fluorescence in‐situ hybridization (FISH) analysis in 6106 (expanded from Minner et al. (2011a)), and deletion status of 5q21 in 3037 (Burkhardt et al., 2013), 6q15 in 3528 (extended from Kluth et al. (2013)), PTEN in 6130 (Krohn et al., 2012), and 3p13 in 1290 (unpublished data) tumors.
Table 1.
Composition of the prognostic tissue microarray containing 11,152 prostate cancer specimens.
| No. of patients | ||
|---|---|---|
| Study cohort on TMA (n = 11,152) | Biochemical relapse among categories (i=1824) | |
| Follow‐up (mo) | ||
| Mean | 53.4 | – |
| Median | 36.8 | – |
| Age (y) | ||
| <50 | 318 | 49 |
| 50–60 | 2.768 | 460 |
| 60–70 | 6.548 | 1.081 |
| >70 | 1.439 | 232 |
| Pretreatment PSA (ng/ml) | ||
| <4 | 1.407 | 142 |
| 4–10 | 6735 | 827 |
| 10–20 | 2159 | 521 |
| >20 | 720 | 309 |
| pT category (AJCC, 2002) | ||
| pT2 | 7.370 | 570 |
| pT3a | 2.409 | 587 |
| pT3b | 1.262 | 618 |
| pT4 | 63 | 49 |
| Gleason grade | ||
| ≤3 + 3 | 2.859 | 193 |
| 3 + 4 | 1.565 | 573 |
| 4 + 3 | 6.183 | 849 |
| ≥4 + 4 | 482 | 208 |
| pN category | ||
| pN0 | 6.117 | 1.126 |
| pN+ | 561 | 291 |
| Surgical margin | ||
| Negative | 8.984 | 1.146 |
| Positive | 1.970 | 642 |
Note: Numbers do not always add up to 11,152 in the different categories because of cases with missing data. Abbreviation: AJCC, American Joint Committee on Cancer.
2.2. Immunohistochemistry
Freshly cut TMA sections were immunostained in one day and in one experiment. Primary antibody specific for LPCAT1 (rabbit, ProteinTech; at 1/1350 dilution) was applied, slides were deparaffinized and exposed to heat‐induced antigen retrieval for 5 min in an autoclave at 121 °C in pH 7.8 Tris‐EDTA‐Citrate buffer. Bound antibody was then visualized using the EnVision Kit (Dako). LPCAT1 staining was analyzed by one person (KG) experienced in immunohistochemisty. LPCAT1 staining was evaluated according to the following scoring system: The staining intensity (0, 1+, 2+, and 3+) and the fraction of positive tumor cells were recorded for each tissue spot. A final score was built from these two parameters according to the following established score, as previously described (Grupp et al., 2013, 2011, 2011, 2013) Negative scores had absence of LPCAT1 staining, weak scores had staining intensity of 1+ in ≤70% of tumor cells or staining intensity of 2+ in ≤30% of tumor cells; moderate scores had staining intensity of 1+ in ≥70% of tumor cells, staining intensity of 2+ in >30% but in ≤70% of tumor cells or staining intensity of 3+ in ≤30% of tumor cells; strong scores had staining intensity of 2+ in >70% of tumor cells or staining intensity of 3+ in >30% of tumor cells.
2.3. Statistics
Statistical calculations were performed with JPM 9 software (SAS Institute Inc., NC, USA). Contingency tables and the chi2‐test were performed to search for associations between molecular parameters and tumor phenotype. Survival curves were calculated according to Kaplan–Meier. The Log–Rank test was applied to detect significant survival differences between groups. COX proportional hazards regression analysis was performed to test the statistical independence and significance between pathological, molecular and clinical variables.
3. Results
3.1. Technical issues
A total of 2366 of 11,152 (21.2%) tissue cores were non‐informative for LPCAT1 immunohistochemistry due to the complete lack of tissue or absence of unequivocal cancer cells on individual TMA spots.
3.2. Immunohistochemistry of LPCAT1
LPCAT1 expression in epithelial cells was ‐ if present ‐ generally cytoplasmic and occasionally showed a granular pattern. In prostate cancers, 6482 of our 8786 interpretable tumors (73.8%) showed positive LPCAT1 immunostaining, which was considered weak in 17.7%, moderate in 26.9% and strong in 29.2% of cases. Representative images of LPCAT1 expression in benign and cancerous prostate tissue are given in Figure 1. Increased LPCAT1 expression was significantly linked to advanced pathological tumor stage (pT3b/T4) (p < 0.0001), high Gleason score (≥4 + 4) (p < 0.0001), positive nodal involvement (p = 0.0002) and positive surgical margin (p = 0.0005) if all tumors were analyzed (data not shown).
Figure 1.
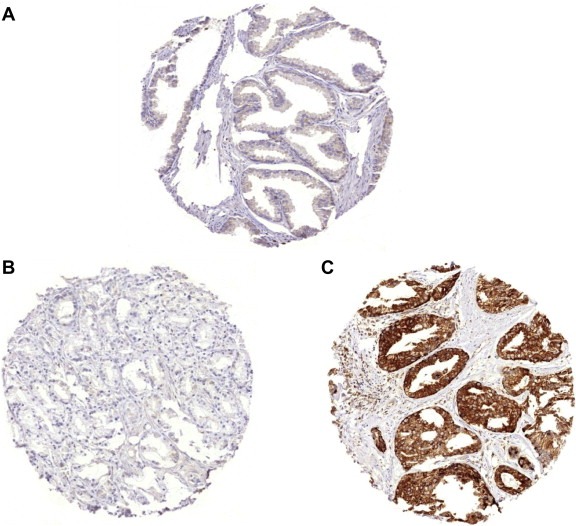
Representative immunohistochemical pictures of (A) weak LPCAT1 staining in benign prostate, (B) negative LPCAT1 expression in prostate cancer and (C) strong LPCAT1 staining in prostate cancer.
3.3. Relationship with fusion type in prostate cancer and ERG protein expression
To evaluate whether LPCAT1 expression is linked to fusion type in prostate cancer, our pre‐existing database including data on ERG‐fusion status obtained by FISH in 5414 patients and by IHC in 8526 tumors with available LPCAT1 expression data was used. Strong LPCAT1 expression was significantly associated with TMPRSS2‐ERG fusion type (p < 0.0001). Strong LPCAT1 expression was seen in 1104 of 2494 (44.3%) cancers with ERG rearrangement detected by FISH but in only 471 of 2920 (16.1%) cancers without ERG rearrangement (p < 0.0001, Figure 2). Accordingly, strong LPCAT1 staining was detected in 1707 of 3765 (45.3%) tumors with ERG expression positivity but in only 794 of 4761 (16.7%) cancers with ERG expression negativity by IHC (p < 0.0001, Figure 2). The associations retained its significance in subset analysis of tumors with a Gleason grade ≤3 + 3, 3 + 4, 4 + 3, and ≥4 + 4. Furthermore, associations with tumor phenotype and clinical cancer features were separately analyzed in the subsets of ERG positive and negative prostate cancers (Tables 2 and 3). In 4761 ERG negative cancers, high LPCAT1 expression was significantly associated with advanced tumor stage, high Gleason grade, positive nodal involvement (p < 0.0001 each) and positive surgical margin (p = 0.006). LPCAT1 immunostaining was only significantly linked to advanced Gleason grade (p < 0.0001) in 3765 ERG positive prostate cancers, however.
Figure 2.
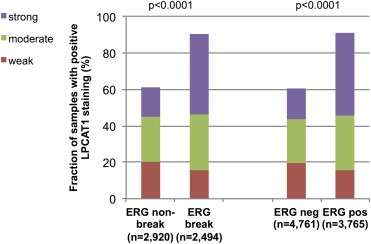
Relationship of LPCAT1 expression with ERG‐fusion probed by fluorescence in‐situ hybridization analysis and immunohistochemistry in all prostate cancers.
Table 2.
Associations between LPCAT1 expression results and ERG negative prostate cancer phenotype.
| Parameter | n Evaluable | LPCAT1 IHC result | p value | |||
|---|---|---|---|---|---|---|
| Negative (%) | Weak (%) | Moderate (%) | Strong (%) | |||
| All cancers | 4.761 | 39.36 | 19.3 | 24.66 | 16.68 | |
| Tumor stage | ||||||
| pT2 | 3.225 | 42.73 | 20.12 | 23.41 | 13.74 | <0.0001 |
| pT3a | 958 | 36.12 | 18.16 | 25.47 | 20.25 | |
| pT3b | 532 | 25.56 | 16.17 | 30.45 | 27.82 | |
| pT4 | 28 | 25 | 25 | 21.43 | 28.57 | |
| Gleason grade | ||||||
| ≤3 + 3 | 1.112 | 52.16 | 19.15 | 17.9 | 10.79 | <0.0001 |
| 3 + 4 | 2.646 | 38.7 | 20.26 | 24.87 | 16.18 | |
| 4 + 3 | 727 | 28.89 | 17.47 | 31.22 | 22.42 | |
| ≥4 + 4 | 252 | 20.24 | 15.48 | 32.94 | 31.35 | |
| Lymph node metastasis | ||||||
| N0 | 2.691 | 36.94 | 19.73 | 25.12 | 18.21 | <0.0001 |
| N+ | 236 | 20.76 | 17.37 | 36.44 | 25.42 | |
| Surgical margin | ||||||
| negative | 3.819 | 40.35 | 19.46 | 24.38 | 15.82 | 0.006 |
| positive | 856 | 35.51 | 18.57 | 25.82 | 20.29 | |
Table 3.
Associations between LPCAT1 expression results and ERG positive prostate cancer phenotype.
| Parameter | n Evaluable | LPCAT1 IHC result | p value | |||
|---|---|---|---|---|---|---|
| Negative (%) | Weak (%) | Moderate (%) | Strong (%) | |||
| All cancers | 3.765 | 9.0 | 15.62 | 30.04 | 45.34 | |
| Tumor stage | ||||||
| pT2 | 2.260 | 9.42 | 15.58 | 29.78 | 45.22 | 0.5767 |
| pT3a | 1.006 | 8.85 | 15.81 | 30.02 | 45.33 | |
| pT3b | 460 | 7.39 | 14.78 | 31.52 | 46.3 | |
| pT4 | 21 | 0 | 9.52 | 33.33 | 57.14 | |
| Gleason grade | ||||||
| ≤3 + 3 | 854 | 11.94 | 18.27 | 32.67 | 37.12 | <0.0001 |
| 3 + 4 | 2.249 | 8.49 | 15.07 | 29.57 | 46.87 | |
| 4 + 3 | 521 | 6.91 | 13.44 | 28.6 | 51.06 | |
| ≥4 + 4 | 116 | 6.03 | 12.93 | 27.59 | 53.45 | |
| Lymph node metastasis | ||||||
| N0 | 2.092 | 7.17 | 15.58 | 29.54 | 47.71 | 0.57 |
| N+ | 205 | 9.76 | 14.15 | 27.8 | 48.29 | |
| Surgical margin | ||||||
| Negative | 2.975 | 9.31 | 15.73 | 30.29 | 44.67 | 0.2687 |
| Positive | 724 | 7.73 | 14.5 | 29.56 | 48.2 | |
3.4. Relationship with key genomic deletions associated with distinct subgroups of prostate cancers
Earlier studies had provided evidence for distinct molecular subgroups of prostate cancers defined by TMPRSS2‐ERG fusions and several genomic deletions. We and others had described a strong link of PTEN and 3p13 deletion to ERG positivity and of 5q21 and 6q15 deletions to ERG negativity (Berger et al., 2011; Burkhardt et al., 2013; Kluth et al., 2013; Krohn et al., 2012; Lapointe et al., 2007; Taylor et al., 2010). To study, whether LPCAT1 expression might be associated with one of these genomic deletions, LPCAT1 IHC results were compared with preexisting findings on deletions of PTEN, 3p13, 6q15 and 5q21. In all cancers, deletions at PTEN and 3q13 were significantly linked to increase LPCAT1 expression (p < 0.0001, p = 0.0028) while deletions at 6q15 were associated with slightly reduced LPCAT1 expression (p = 0.0395, Figure 3). Based on the strong associations of both LPCAT1 and these deletions with ERG status, such associations were expected in mixed cohorts. It was therefore not surprising, that none of these associations was retained within ERG positive cancers (Figure 4). However, a strong positive association between LPCAT1 expression and PTEN deletions was retained in ERG negative cancers (p < 0.0001, Figure 5). In the same subgroup, statistically significant associations were also observed between LPCAT1 and 6q15 as well as 5q21 deletions (p = 0.0005 and p = 0.0065), although the differences in absolute numbers were small for these deletions. Additional analysis revealed that these associations mostly retained its significance in subset analysis of tumors with a Gleason grade ≤3 + 3, 3 + 4, 4 + 3, and ≥4 + 4 although the absolute number of analyzable tumors was rather small.
Figure 3.
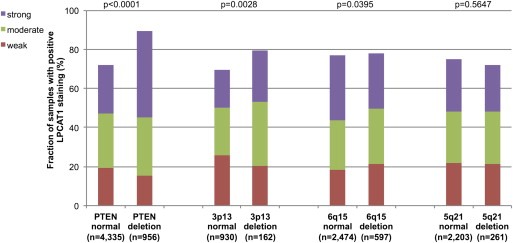
LPCAT1 expression versus deletions at PTEN, 3p13, 6q15 and 5q21 probed by fluorescence in‐situ hybridization analysis in all prostate cancers.
Figure 4.
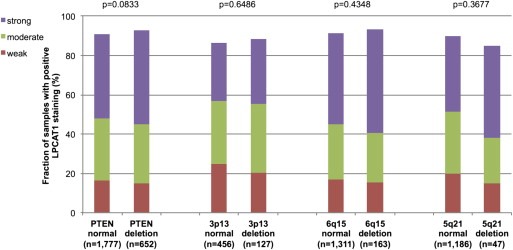
LPCAT1 expression versus deletions at PTEN, 3p13, 6q15 and 5q21 probed by fluorescence in‐situ hybridization analysis in ERG positive prostate cancers.
Figure 5.
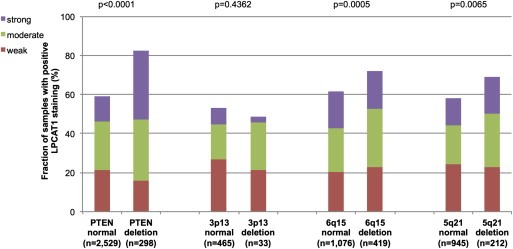
LPCAT1 expression versus deletions at PTEN, 3p13, 6q15 and 5q21 probed by fluorescence in‐situ hybridization analysis in ERG negative prostate cancers.
3.5. Prognostic role of LPCAT1 expression
Follow‐up data were available for 7620 patients with informative LPCAT1 data. The prognostic role of Gleason grade is given for this patient subset in order to demonstrate the overall validity of our follow‐up data (p < 0.0001, Figure 6A). LPCAT1 was related to early biochemical recurrence in the analysis of all cancers (p < 0.0001, Figure 6B). This association was particularly evident in 4102 ERG negative cancers (p < 0.0001, Figure 6C) but was only marginal in 3283 ERG positive cancers (p = 0.0073, Figure 6D). Another analysis stratifying according to the PTEN deletion status revealed that LPCAT1 expression was significantly associated with prognosis in the subgroup of 3627 PTEN non‐deleted cancers (p = 0.0045, Figure 6E) but not in the smaller subset of 846 PTEN deleted cancers (p = 0.6603, Figure 6F).
Figure 6.
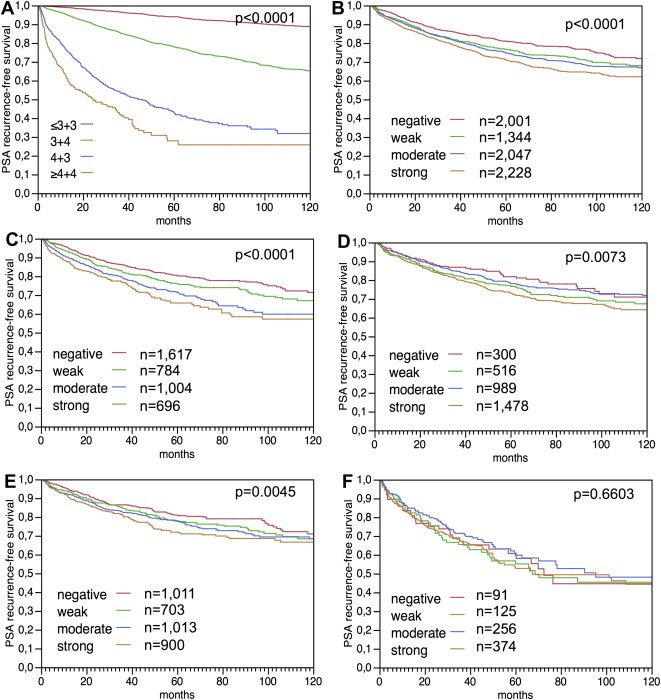
Relationship of Gleason grade with biochemical recurrence (A). Association of LPCAT1 immunostaining intensity with biochemical recurrence (PSA) in the analysis of (B) all prostate cancers (n = 7620), (C) in the subset of ERG negative cancers (n = 4102) and (D) in the subset of ERG positive cancers (n = 3283). Figure E and F include the PTEN deletions status and demonstrate the relationship of LPCAT1 immunostaining intensity with biochemical recurrence in (E) prostate cancers lacking PTEN deletions (n = 3627), and (F) prostate cancers harboring PTEN deletions (n = 846).
3.6. Multivariate analysis
Four multivariate analyses were performed evaluating the clinical relevance of LPCAT1 expression in different scenarios (Table 4). No 1 was utilizing all post‐operatively available parameters including pT, pN, margin status, pre‐operative PSA value and Gleason grade obtained on the resected prostate. Scenario 2 included all postoperatively available parameters with the exception of the lymph node status. The rational for this approach was that lymphadenectomy is not a routine procedure in the surgical therapy of prostate cancer and that excluding the nodal status in multivariate analysis increases case numbers. The next two scenarios tried to better model the pre‐operative situation. Scenario 3 included the LPCAT1 expression, pre‐operative PSA, clinical stage (cT) and the Gleason grade obtained on the prostatectomy specimen. However, the pre‐operative determination of a tumors Gleason grade is subject to sampling errors and therefore results in under grading in more than one third of cases (Epstein et al., 2012). Because the post‐operative Gleason grade thus varies from the pre‐operative Gleason grade, another multivariate analysis was added as scenario 4. In this scenario, the pre‐operative Gleason grade obtained on the original biopsy was combined with pre‐operative PSA, clinical stage and LPCAT1 expression. If all tumors are analyzed, all four scenarios suggest a tendency towards LPCAT1 representing an independent predictor of prognosis, especially in scenarios 3 and 4 using preoperative parameters. Subgroup analyses further revealed that independent prognostic value was found in both ERG negative and positive cancers if preoperative parameters were used (Table 4b and c). In multivariate analysis including parameters that are available after surgery, independent prognostic relevance of LPCAT1 was found in ERG positive prostate cancers only (Table 4c).
Table 4.
Multivariate analysis including LPCAT1 expression status in a) all cancers, b) ERG negative, and c) ERG positive prostate cancers.
| Scenario (n) | p value | |||||||
|---|---|---|---|---|---|---|---|---|
| Preoperative PSA‐level | pT stage | cT stage | Gleason grade prostatectomy | Biopsy Gleason grade | N status | R status | LPCAT1 | |
| a) | ||||||||
| 1 (n = 4499) | <0.0001 | <0.0001 | – | <0.0001 | – | <0.0001 | <0.0001 | 0.1493 |
| 2 (n = 7445) | <0.0001 | <0.0001 | – | <0.0001 | – | – | <0.0001 | 0.0983 |
| 3 (n = 7308) | <0.0001 | – | <0.0001 | <0.0001 | – | – | – | 0.0324 |
| 4 (n = 7185) | <0.0001 | – | <0.0001 | – | <0.0001 | – | – | 0.0001 |
| b) | ||||||||
| 1 (n = 2437) | 0.0002 | <0.0001 | – | <0.0001 | – | <0.0001 | 0.0191 | 0.4607 |
| 2 (n = 4008) | <0.0001 | <0.0001 | – | <0.0001 | – | – | 0.0003 | 0.214 |
| 3 (n = 3965) | <0.0001 | – | <0.0001 | <0.0001 | – | – | – | 0.0381 |
| 4 (n = 3912) | <0.0001 | – | <0.0001 | – | <0.0001 | – | – | 0.0001 |
| c) | ||||||||
| 1 (n = 1934) | 0.0266 | <0.0001 | – | <0.0001 | – | 0.083 | 0.0004 | 0.0218 |
| 2 (n = 3205) | 0.0009 | <0.0001 | – | <0.0001 | – | – | <0.0001 | 0.0157 |
| 3 (n = 3120) | <0.0001 | – | <0.0001 | <0.0001 | – | – | – | 0.052 |
| 4 (n = 3068) | <0.0001 | – | <0.0001 | – | <0.0001 | – | – | 0.0218 |
4. Discussion
The results of our study identify LPCAT1 immunostaining as a strong predictor of an increased risk for biochemical recurrence in prostate cancer.
Our immunohistochemical analysis showed positive LPACT1 staining in 73.8% of the interpretable prostate cancers. The intensity of the immunohistochemical signal was typically stronger in malignant than in benign prostate epithelium. These data are generally in line with one earlier study analyzing LPCAT1 expression on a TMA containing 251 samples from 148 patients who had undergone prostatectomy, or transurethral resection of the prostate (Zhou et al., 2012). In this study, LPCAT1 expression was assessed by a score defined by intensity and extent of LPCAT1 immunostaining with a maximum score of 9 (Zhou et al., 2012). In accordance with our study, the authors found that LPCAT1 staining was predominantly localized in the cytoplasm of the cells (Zhou et al., 2012). Increasing levels of LPCAT1 positivity were then described from benign prostatic changes (mean IHC score: 2.68) to high‐grade prostatic intraepithelial neoplasia (mean IHC score: 2.72), non‐metastatic (mean IHC score: 4.63) and metastatic (mean IHC score: 8.00) prostate cancers (Zhou et al., 2012).
The most striking finding in our study is the strong association of LPCAT1 expression with unfavorable histological phenotype and clinical outcome. The strong and independent prognostic value of LPCAT1 overexpression suggests a biologically relevant role in prostate cancer cells. This assumption is consistent with numerous previous studies implicating tumor relevant functional consequences of alterations in the lipid metabolism (Furuta et al., 2010; Menendez, 2010; Menendez and Lupu, 2007; Santos and Schulze, 2012; Suburu and Chen, 2012). For example, several studies have presented data suggesting that activation of lipid biosynthesis and lipid remodeling is a common feature of cancer cells, and that high expression of proteins involved in lipid metabolism, such as fatty acid‐binding protein (C‐FABP) (Morgan et al., 2008), fatty acid synthase (FASN) (Flavin et al., 2010; Shah et al., 2006), Caveolin‐1 (Freeman et al., 2012) or fatty acid elongase 7 (ELOVL7) (Tamura et al., 2009) may be linked to progression and metastatic growth in prostate cancer. Our data, identifying LPCAT1 as a strong prognostic factor for prostate cancer, further support the emerging concept of an important role of lipid metabolism for the behavior of prostate cancer cells (Faas et al., 1996, 2001).
It was one major aim of this project to investigate for possible interactions of LPCAT1 with key genomic alterations in prostate cancer. About 50% of prostate cancers are characterized by gene fusions linking the androgen‐regulated gene TMPRSS2 with the transcription factor ERG of the ETS family (Tomlins et al., 2005). In consequence of this rearrangement, the ERG expression becomes massively overexpressed. Our data demonstrate that high level LPCAT1 expression is significantly associated with ERG fusion type prostate cancer. As this link was found by IHC and FISH, two independent approaches for ERG fusion detection, a false positive association due to inefficient immunostaining for both LPCAT1 and ERG in a subset of damaged non‐reactive tissues can be largely excluded. Our finding of high LPCAT1 expression in fusion positive prostate cancers is coherent with earlier data showing that ERG dependent pathways target the lipid metabolism of cancer cells (Iljin et al., 2006; Vainio et al., 2011a). For example, ERG induces phospholipase PLA2G7, an important enzyme for cell growth, signaling and maintenance of membrane phospholipids (Dong et al., 2006, 2006, 2010, 2011, 2011) required for the growth of ERG positive, but not of ERG negative prostate cancer cells (Vainio et al., 2011a). PLA2G7 releases lysophosphatidylcholine from the cell membrane, which in turn is a substrate for LPACT1. Upregulation of LPCAT1 in ERG positive cancer cells may thus be a result of PLA2G7 activation in these cells. It remains to be shown whether ERG is also directly involved in LPCAT1 regulation. Of note, ETS1, another member of the ETS family of transcription factors, induces LPCAT2, an isoform of LPCAT1, in ovarian cancer cells (Verschoor et al., 2010). Based on these data, it could be possible, that ERG increases the cellular phosphatidylcholine content by a dual action on both PLA2G7 and LPCAT1.
While LPCAT1 was generally high in ERG positive prostate cancer and independent of any of the key chromosomal deletions (5q, 6q, 3p, PTEN) in this subgroup, a comparable level of LPCAT1 expression could be observed in the PTEN deleted subset of ERG negative cancers. This observation may possibly be explained by the known role of PTEN to upregulate the LPCAT1‐antagoists phospholipase D and C (Horie et al., 2004). PTEN inactivation may thus – through reduced activity of LPCAT1 antagonists ‐ induce increased LPCAT1 expression to a similar level as achieved by ERG activation. At the same time our findings also demonstrate, that such a PTEN induced effect on LPCAT1 expression is no longer operational in ERG positive cancers where other mechanisms are obviously in place to induce high LPCAT1 expression. Alternatively, it cannot be completely excluded, that the striking association of PTEN deletions with LPCAT1 overexpression in ERG negative cancers is caused by the known link of both alterations to fusion type prostate cancer. As our classifier for fusion type prostate cancer only includes the most frequent ETS family member (ERG), it must be assumed that a small fraction of fusion type positive prostate cancers are missed by our approach. It is estimated, that about 5–10% of all fusion type prostate cancers do not have ERG activation (Rubin et al., 2011).
The large number of cancers analyzed in this study enabled us to determine, that associations of LPCAT1 expression with unfavorable tumor phenotype and PSA recurrence were largely driven by the subset of ERG negative cancers, while the impact of LPCAT1 expression on tumor phenotype and PSA recurrence was substantially less obvious in ERG positive cancers. These observations again, suggest a markedly different role of LPCAT1 in fusion type and non‐fusion type prostate cancer. It is remarkable, that the prognostic effect of LPCAT1 expression predominantly occurs in the subset of ERG negative cancers with a markedly lower basic LPCAT1 expression than in ERG positive cancers. It is tempting to speculate, that the marked attenuation of the prognostic relevance of LPCAT1 in ERG positive cancers could be due to interference of ERG with other molecules involved in LPCAT1 action, thus rendering effects of LPCAT1 expression on tumor aggressiveness inactive. Candidate genes for such a role include the lipoxygenase ALOX15, the lipid transporter SORL1, the lipoprotein receptor VLDLR, and the phospholipases PLA1A and PLA2G2A, all of which have been reported to be differentially expressed in ERG negative and ERG positive prostate cancers (Brase et al., 2011; Jhavar et al., 2009) and all of which may have a potential impact on LPCAT function by either involvement in lipoprotein uptake (Klinger et al., 2011; Shen et al., 2012), production of lipid signaling mediators (Il Lee et al., 2011), glycerophospholipids and lysophosholipids (Kudo and Murakami, 2002). The mechanism by which LPCAT1 exerts a direct impact on cancer cell aggressiveness remains unclear. It appears possible that LPCAT1‐mediated phospholipid accumulation increases the membrane potential and membrane fluidity, important factors for cell proliferation, adhesion and motility that have been linked to tumor progression and metastasis before (Dobrzynska et al., 2005; Kohno et al., 1998; Monet et al., 2009; Zeisig et al., 2007).
Our TMA containing more than 10,000 prostate cancer specimens represents a suitable system for assessing potential prognostic markers. In earlier studies we had successfully validated all established prognostic biomarkers in prostate cancer such as nuclear p53 accumulation (Schlomm et al., 2008), PTEN inactivation (Krohn et al., 2012), and Ki67 labeling index (Zellweger et al., 2009) on even smaller prostate cancer TMAs, and identified several other prognostic biomarkers such as CRISP3 overexpression (Grupp et al., 2013) and deletions at 8p (El Gammal et al., 2010), 6q15 (Kluth et al., 2013), 5q21 (Burkhardt et al., 2013). It is noteworthy, that our approach of analyzing molecular features on one‐minute tissue specimen per patient on a TMA measuring 0.6 mm in diameter represents a close model of molecularly analyzing core needle biopsies. Core needle biopsies enable the molecular analysis of comparable amounts of tissue as on a TMA. The optimal biomarker evaluation strategy would include the molecular analysis of the original needle biopsy of a patient and compare its prognostic value with preoperative Gleason grade obtained on the same biopsy as well as the preoperative PSA value. For practical purposes, this approach is not feasible because preoperative biopsies are typically distributed over many different centers and not available for studies. Moreover, even if available, these precious core needle biopsies would be exhausted after only few studies. A convoluted approach evaluating multiple different scenarios was thus utilized in this study and scenario 4 including the pre‐operative Gleason grade obtained on the original biopsy, the pre‐operative PSA, clinical stage and LPCAT1 expression was used. Overall, these data suggest a strong independent prognostic relevance of LPCAT1 expression in the subset of ERG positive prostate cancer. The statistical significance becomes particularly high in scenario 4 where the biopsy Gleason grade is considered instead of the more representative Gleason grade obtained from the radical prostatectomy specimen. Although this scenario is somewhat biased since it overestimates the true prognostic value of LPCAT1, it strongly suggests that LPCAT1 might be a powerful marker in this clinically relevant scenario.
In summary, our data identify LPCAT1 expression as a potential biomarker with clinical utility in prostate cancer. It appears well possible, that LPCAT1 measurement, either alone or in combination, may be utilized for better clinical decision‐making. The findings also highlight the potentially important role of lipid metabolism in prostate cancer biology.
Disclosure/Conflict of interest
The authors declare no conflicts of interest.
Supporting information
The following are the supplementary data related to this article.
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Acknowledgments
We thank Christina Koop, Julia Schumann, Sünje Seekamp, and Inge Brandt for excellent technical assistance.
Supplementary data 1.
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.molonc.2013.07.009.
Grupp Katharina, Sanader Stella, Sirma Hüseyin, Simon Ronald, Koop Christina, Prien Kristina, Hube-Magg Claudia, Salomon Georg, Graefen Markus, Heinzer Hans, Minner Sarah, Izbicki Jakob R., Sauter Guido, Schlomm Thorsten, Tsourlakis Maria Christina, (2013), High lysophosphatidylcholine acyltransferase 1 expression independently predicts high risk for biochemical recurrence in prostate cancers, Molecular Oncology, 7, doi: 10.1016/j.molonc.2013.07.009.
Contributor Information
Katharina Grupp, Email: k.grupp@uke.de.
Stella Sanader, Email: stella.sanader@hotmail.de.
Hüseyin Sirma, Email: h.sirma@uke.de.
Ronald Simon, Email: r.simon@uke.de.
Christina Koop, Email: c.koop@uke.de.
Kristina Prien, Email: k.prien@uke.de.
Claudia Hube-Magg, Email: c.hube@uke.de.
Georg Salomon, Email: g.salomon@uke.de.
Markus Graefen, Email: graefen@uke.de.
Hans Heinzer, Email: heinzer@uke.de.
Sarah Minner, Email: s.minner@uke.de.
Jakob R. Izbicki, Email: izbicki@uke.de
Guido Sauter, Email: g.sauter@uke.de.
Thorsten Schlomm, Email: tschlomm@uke.de.
Maria Christina Tsourlakis, Email: mtsourlakis@uke.de.
References
- Berger, M.F. , Lawrence, M.S. , Demichelis, F. , Drier, Y. , Cibulskis, K. , Sivachenko, A.Y. , Sboner, A. , Esgueva, R. , Pflueger, D. , Sougnez, C. , Onofrio, R. , Carter, S.L. , Park, K. , Habegger, L. , Ambrogio, L. , Fennell, T. , Parkin, M. , Saksena, G. , Voet, D. , Ramos, A.H. , Pugh, T.J. , Wilkinson, J. , Fisher, S. , Winckler, W. , Mahan, S. , Ardlie, K. , Baldwin, J. , Simons, J.W. , Kitabayashi, N. , MacDonald, T.Y. , Kantoff, P.W. , Chin, L. , Gabriel, S.B. , Gerstein, M.B. , Golub, T.R. , Meyerson, M. , Tewari, A. , Lander, E.S. , Getz, G. , Rubin, M.A. , Garraway, L.A. , 2011. The genomic complexity of primary human prostate cancer. Nature. 470, 214–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brase, J.C. , Johannes, M. , Mannsperger, H. , Falth, M. , Metzger, J. , Kacprzyk, L.A. , Andrasiuk, T. , Gade, S. , Meister, M. , Sirma, H. , Sauter, G. , Simon, R. , Schlomm, T. , Beissbarth, T. , Korf, U. , Kuner, R. , Sultmann, H. , 2011. TMPRSS2-ERG -specific transcriptional modulation is associated with prostate cancer biomarkers and TGF-beta signaling. BMC Cancer. 11, 507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges, J.P. , Ikegami, M. , Brilli, L.L. , Chen, X. , Mason, R.J. , Shannon, J.M. , 2010. LPCAT1 regulates surfactant phospholipid synthesis and is required for transitioning to air breathing in mice. J. Clin. Invest.. 120, 1736–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhardt, L. , Fuchs, S. , Krohn, A. , Masser, S. , Mader, M. , Kluth, M. , Bachmann, F. , Huland, H. , Steuber, T. , Graefen, M. , Schlomm, T. , Minner, S. , Sauter, G. , Sirma, H. , Simon, R. , 2013. CHD1 is a 5q21 tumor suppressor required for ERG rearrangement in prostate cancer. Cancer Res.. 73, 2795–2805. [DOI] [PubMed] [Google Scholar]
- Chen, X. , Hyatt, B.A. , Mucenski, M.L. , Mason, R.J. , Shannon, J.M. , 2006. Identification and characterization of a lysophosphatidylcholine acyltransferase in alveolar type II cells. Proc. Natl. Acad. Sci. U. S. A.. 103, 11724–11729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, L. , Han, X. , Shi, Y. , 2009. A regulatory role of LPCAT1 in the synthesis of inflammatory lipids, PAF and LPC, in the retina of diabetic mice. Am. J. Physiol. Endocrinol. Metab.. 297, E1276–E1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrzynska, I. , Szachowicz-Petelska, B. , Sulkowski, S. , Figaszewski, Z. , 2005. Changes in electric charge and phospholipids composition in human colorectal cancer cells. Mol. Cell Biochem.. 276, 113–119. [DOI] [PubMed] [Google Scholar]
- Dong, Q. , Patel, M. , Scott, K.F. , Graham, G.G. , Russell, P.J. , Sved, P. , 2006. Oncogenic action of phospholipase A2 in prostate cancer. Cancer Lett.. 240, 9–16. [DOI] [PubMed] [Google Scholar]
- El Gammal, A.T. , Bruchmann, M. , Zustin, J. , Isbarn, H. , Hellwinkel, O.J. , Kollermann, J. , Sauter, G. , Simon, R. , Wilczak, W. , Schwarz, J. , Bokemeyer, C. , Brummendorf, T.H. , Izbicki, J.R. , Yekebas, E. , Fisch, M. , Huland, H. , Graefen, M. , Schlomm, T. , 2010. Chromosome 8p deletions and 8q gains are associated with tumor progression and poor prognosis in prostate cancer. Clin. Cancer Res.. 16, 56–64. [DOI] [PubMed] [Google Scholar]
- Epstein, J.I. , Feng, Z. , Trock, B.J. , Pierorazio, P.M. , 2012. Upgrading and downgrading of prostate cancer from biopsy to radical prostatectomy: incidence and predictive factors using the modified Gleason grading system and factoring in tertiary grades. Eur. Urol.. 61, 1019–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erbersdobler, A. , Fritz, H. , Schnoger, S. , Graefen, M. , Hammerer, P. , Huland, H. , Henke, R.P. , 2002. Tumour grade, proliferation, apoptosis, microvessel density, p53, and bcl-2 in prostate cancers: differences between tumours located in the transition zone and in the peripheral zone. Eur. Urol.. 41, 40–46. [DOI] [PubMed] [Google Scholar]
- Faas, F.H. , Dang, A.Q. , Pollard, M. , Hong, X.M. , Fan, K. , Luckert, P.H. , Schutz, M. , 1996. Increased phospholipid fatty acid remodeling in human and rat prostatic adenocarcinoma tissues. J. Urol.. 156, 243–248. [PubMed] [Google Scholar]
- Faas, F.H. , Dang, A.Q. , White, J. , Schaefer, R. , Johnson, D. , 2001. Increased prostatic lysophosphatidylcholine acyltransferase activity in human prostate cancer: a marker for malignancy. J. Urol.. 165, 463–468. [DOI] [PubMed] [Google Scholar]
- Flavin, R. , Peluso, S. , Nguyen, P.L. , Loda, M. , 2010. Fatty acid synthase as a potential therapeutic target in cancer. Future Oncol.. 6, 551–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman, M.R. , Yang, W. , Di Vizio, D. , 2012. Caveolin-1 and prostate cancer progression. Adv. Exp. Med. Biol.. 729, 95–110. [DOI] [PubMed] [Google Scholar]
- Furuta, E. , Okuda, H. , Kobayashi, A. , Watabe, K. , 2010. Metabolic genes in cancer: their roles in tumor progression and clinical implications. Biochim. Biophys. Acta. 1805, 141–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grupp, K. , Kohl, S. , Sirma, H. , Simon, R. , Steurer, S. , Becker, A. , Adam, M. , Izbicki, J. , Sauter, G. , Minner, S. , Schlomm, T. , Tsourlakis, M.C. , 2013. Cysteine-rich secretory protein 3 overexpression is linked to a subset of PTEN-deleted ERG fusion-positive prostate cancers with early biochemical recurrence. Mod. Pathol.. 26, 733–742. [DOI] [PubMed] [Google Scholar]
- Harayama, T. , Shindou, H. , Ogasawara, R. , Suwabe, A. , Shimizu, T. , 2008. Identification of a novel noninflammatory biosynthetic pathway of platelet-activating factor. J. Biol. Chem.. 283, 11097–11106. [DOI] [PubMed] [Google Scholar]
- Horie, Y. , Suzuki, A. , Kataoka, E. , Sasaki, T. , Hamada, K. , Sasaki, J. , Mizuno, K. , Hasegawa, G. , Kishimoto, H. , Iizuka, M. , Naito, M. , Enomoto, K. , Watanabe, S. , Mak, T.W. , Nakano, T. , 2004. Hepatocyte-specific Pten deficiency results in steatohepatitis and hepatocellular carcinomas. J. Clin. Invest.. 113, 1774–1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Il Lee, S. , Zuo, X. , Shureiqi, I. , 2011. 15-Lipoxygenase-1 as a tumor suppressor gene in colon cancer: is the verdict in?. Cancer Metastasis Rev.. 30, 481–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iljin, K. , Wolf, M. , Edgren, H. , Gupta, S. , Kilpinen, S. , Skotheim, R.I. , Peltola, M. , Smit, F. , Verhaegh, G. , Schalken, J. , Nees, M. , Kallioniemi, O. , 2006. TMPRSS2 fusions with oncogenic ETS factors in prostate cancer involve unbalanced genomic rearrangements and are associated with HDAC1 and epigenetic reprogramming. Cancer Res.. 66, 10242–10246. [DOI] [PubMed] [Google Scholar]
- Jhavar, S. , Brewer, D. , Edwards, S. , Kote-Jarai, Z. , Attard, G. , Clark, J. , Flohr, P. , Christmas, T. , Thompson, A. , Parker, M. , Shepherd, C. , Stenman, U.H. , Marchbank, T. , Playford, R.J. , Woodhouse, C. , Ogden, C. , Fisher, C. , Kovacs, G. , Corbishley, C. , Jameson, C. , Norman, A. , De-Bono, J. , Bjartell, A. , Eeles, R. , Cooper, C.S. , 2009. Integration of ERG gene mapping and gene-expression profiling identifies distinct categories of human prostate cancer. BJU. Int.. 103, 1256–1269. [DOI] [PubMed] [Google Scholar]
- Kent, C. , 2005. Regulatory enzymes of phosphatidylcholine biosynthesis: a personal perspective. Biochim. Biophys. Acta. 1733, 53–66. [DOI] [PubMed] [Google Scholar]
- Klinger, S.C. , Glerup, S. , Raarup, M.K. , Mari, M.C. , Nyegaard, M. , Koster, G. , Prabakaran, T. , Nilsson, S.K. , Kjaergaard, M.M. , Bakke, O. , Nykjaer, A. , Olivecrona, G. , Petersen, C.M. , Nielsen, M.S. , 2011. SorLA regulates the activity of lipoprotein lipase by intracellular trafficking. J. Cell. Sci.. 124, 1095–1105. [DOI] [PubMed] [Google Scholar]
- Kluth, M. , Hesse, J. , Heinl, A. , Krohn, A. , Steurer, S. , Sirma, H. , Simon, R. , Mayer, P.S. , Schumacher, U. , Grupp, K. , Izbicki, J.R. , Pantel, K. , Dikomey, E. , Korbel, J.O. , Plass, C. , Sauter, G. , Schlomm, T. , Minner, S. , 2013. Genomic deletion of MAP3K7 at 6q12-22 is associated with early PSA recurrence in prostate cancer and absence of TMPRSS2:ERG fusions. Mod. Pathol.. 26, 975–983. [DOI] [PubMed] [Google Scholar]
- Kohno, M. , Yokokawa, K. , Yasunari, K. , Minami, M. , Kano, H. , Hanehira, T. , Yoshikawa, J. , 1998. Induction by lysophosphatidylcholine, a major phospholipid component of atherogenic lipoproteins, of human coronary artery smooth muscle cell migration. Circulation. 98, 353–359. [DOI] [PubMed] [Google Scholar]
- Krohn, A. , Diedler, T. , Burkhardt, L. , Mayer, P.S. , De Silva, C. , Meyer-Kornblum, M. , Kotschau, D. , Tennstedt, P. , Huang, J. , Gerhauser, C. , Mader, M. , Kurtz, S. , Sirma, H. , Saad, F. , Steuber, T. , Graefen, M. , Plass, C. , Sauter, G. , Simon, R. , Minner, S. , Schlomm, T. , 2012. Genomic deletion of PTEN is associated with tumor progression and early PSA recurrence in ERG fusion-positive and fusion-negative prostate cancer. Am. J. Pathol.. 181, 401–412. [DOI] [PubMed] [Google Scholar]
- Kudo, I. , Murakami, M. , 2002. Phospholipase A2 enzymes. Prostaglandins Other Lipid Mediat.. 68-69, 3–58. [DOI] [PubMed] [Google Scholar]
- Lapointe, J. , Li, C. , Giacomini, C.P. , Salari, K. , Huang, S. , Wang, P. , Ferrari, M. , Hernandez-Boussard, T. , Brooks, J.D. , Pollack, J.R. , 2007. Genomic profiling reveals alternative genetic pathways of prostate tumorigenesis. Cancer Res.. 67, 8504–8510. [DOI] [PubMed] [Google Scholar]
- Mansilla, F. , da Costa, K.A. , Wang, S. , Kruhoffer, M. , Lewin, T.M. , Orntoft, T.F. , Coleman, R.A. , Birkenkamp-Demtroder, K. , 2009. Lysophosphatidylcholine acyltransferase 1 (LPCAT1) overexpression in human colorectal cancer. J. Mol. Med. (Berl). 87, 85–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menendez, J.A. , 2010. Fine-tuning the lipogenic/lipolytic balance to optimize the metabolic requirements of cancer cell growth: molecular mechanisms and therapeutic perspectives. Biochim. Biophys. Acta. 1801, 381–391. [DOI] [PubMed] [Google Scholar]
- Menendez, J.A. , Lupu, R. , 2007. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat. Rev. Cancer. 7, 763–777. [DOI] [PubMed] [Google Scholar]
- Minner, S. , Enodien, M. , Sirma, H. , Luebke, A.M. , Krohn, A. , Mayer, P.S. , Simon, R. , Tennstedt, P. , Muller, J. , Scholz, L. , Brase, J.C. , Liu, A.Y. , Schluter, H. , Pantel, K. , Schumacher, U. , Bokemeyer, C. , Steuber, T. , Graefen, M. , Sauter, G. , Schlomm, T. , 2011. ERG status is unrelated to PSA recurrence in radically operated prostate cancer in the absence of antihormonal therapy. Clin. Cancer Res.. 17, 5878–5888. [DOI] [PubMed] [Google Scholar]
- Minner, S. , Kraetzig, F. , Tachezy, M. , Kilic, E. , Graefen, M. , Wilczak, W. , Bokemeyer, C. , Huland, H. , Sauter, G. , Schlomm, T. , 2011. Low activated leukocyte cell adhesion molecule expression is associated with advanced tumor stage and early prostate-specific antigen relapse in prostate cancer. Hum. Pathol.. 42, 1946–1952. [DOI] [PubMed] [Google Scholar]
- Minner, S. , Wittmer, C. , Graefen, M. , Salomon, G. , Steuber, T. , Haese, A. , Huland, H. , Bokemeyer, C. , Yekebas, E. , Dierlamm, J. , Balabanov, S. , Kilic, E. , Wilczak, W. , Simon, R. , Sauter, G. , Schlomm, T. , 2011. High level PSMA expression is associated with early PSA recurrence in surgically treated prostate cancer. Prostate. 71, 281–288. [DOI] [PubMed] [Google Scholar]
- Mirlacher, M. , Simon, R. , 2010. Recipient block TMA technique. Methods Mol. Biol.. 664, 37–44. [DOI] [PubMed] [Google Scholar]
- Monet, M. , Gkika, D. , Lehen'kyi, V. , Pourtier, A. , Vanden Abeele, F. , Bidaux, G. , Juvin, V. , Rassendren, F. , Humez, S. , Prevarsakaya, N. , 2009. Lysophospholipids stimulate prostate cancer cell migration via TRPV2 channel activation. Biochim. Biophys. Acta. 1793, 528–539. [DOI] [PubMed] [Google Scholar]
- Morgan, E.A. , Forootan, S.S. , Adamson, J. , Foster, C.S. , Fujii, H. , Igarashi, M. , Beesley, C. , Smith, P.H. , Ke, Y. , 2008. Expression of cutaneous fatty acid-binding protein (C-FABP) in prostate cancer: potential prognostic marker and target for tumourigenicity-suppression. Int. J. Oncol.. 32, 767–775. [PubMed] [Google Scholar]
- Muller, J. , Ehlers, A. , Burkhardt, L. , Sirma, H. , Steuber, T. , Graefen, M. , Sauter, G. , Minner, S. , Simon, R. , Schlomm, T. , Michl, U. , 2013. Loss of pSer2448-mTOR expression is linked to adverse prognosis and tumor progression in ERG-fusion-positive cancers. Int. J. Cancer. 132, 1333–1340. [DOI] [PubMed] [Google Scholar]
- Nakanishi, H. , Shindou, H. , Hishikawa, D. , Harayama, T. , Ogasawara, R. , Suwabe, A. , Taguchi, R. , Shimizu, T. , 2006. Cloning and characterization of mouse lung-type acyl-CoA:lysophosphatidylcholine acyltransferase 1 (LPCAT1). Expression in alveolar type II cells and possible involvement in surfactant production. J. Biol. Chem.. 281, 20140–20147. [DOI] [PubMed] [Google Scholar]
- Rubin, M.A. , Maher, C.A. , Chinnaiyan, A.M. , 2011. Common gene rearrangements in prostate cancer. J. Clin. Oncol.. 29, 3659–3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos, C.R. , Schulze, A. , 2012. Lipid metabolism in cancer. FEBS J.. 279, 2610–2623. [DOI] [PubMed] [Google Scholar]
- Schlomm, T. , Iwers, L. , Kirstein, P. , Jessen, B. , Kollermann, J. , Minner, S. , Passow-Drolet, A. , Mirlacher, M. , Milde-Langosch, K. , Graefen, M. , Haese, A. , Steuber, T. , Simon, R. , Huland, H. , Sauter, G. , Erbersdobler, A. , 2008. Clinical significance of p53 alterations in surgically treated prostate cancers. Mod. Pathol.. 21, 1371–1378. [DOI] [PubMed] [Google Scholar]
- Scott, K.F. , Sajinovic, M. , Hein, J. , Nixdorf, S. , Galettis, P. , Liauw, W. , de Souza, P. , Dong, Q. , Graham, G.G. , Russell, P.J. , 2010. Emerging roles for phospholipase A2 enzymes in cancer. Biochimie. 92, 601–610. [DOI] [PubMed] [Google Scholar]
- Shah, U.S. , Dhir, R. , Gollin, S.M. , Chandran, U.R. , Lewis, D. , Acquafondata, M. , Pflug, B.R. , 2006. Fatty acid synthase gene overexpression and copy number gain in prostate adenocarcinoma. Hum. Pathol.. 37, 401–409. [DOI] [PubMed] [Google Scholar]
- Shen, G.M. , Zhao, Y.Z. , Chen, M.T. , Zhang, F.L. , Liu, X.L. , Wang, Y. , Liu, C.Z. , Yu, J. , Zhang, J.W. , 2012. Hypoxia-inducible factor-1 (HIF-1) promotes LDL and VLDL uptake through inducing VLDLR under hypoxia. Biochem. J.. 441, 675–683. [DOI] [PubMed] [Google Scholar]
- Siegel, R. , Naishadham, D. , Jemal, A. , 2013. Cancer statistics, 2013. CA. Cancer J. Clin.. 63, 11–30. [DOI] [PubMed] [Google Scholar]
- Suburu, J. , Chen, Y.Q. , 2012. Lipids and prostate cancer. Prostaglandins Other Lipid Mediat.. 98, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura, K. , Makino, A. , Hullin-Matsuda, F. , Kobayashi, T. , Furihata, M. , Chung, S. , Ashida, S. , Miki, T. , Fujioka, T. , Shuin, T. , Nakamura, Y. , Nakagawa, H. , 2009. Novel lipogenic enzyme ELOVL7 is involved in prostate cancer growth through saturated long-chain fatty acid metabolism. Cancer Res.. 69, 8133–8140. [DOI] [PubMed] [Google Scholar]
- Taylor, B.S. , Schultz, N. , Hieronymus, H. , Gopalan, A. , Xiao, Y. , Carver, B.S. , Arora, V.K. , Kaushik, P. , Cerami, E. , Reva, B. , Antipin, Y. , Mitsiades, N. , Landers, T. , Dolgalev, I. , Major, J.E. , Wilson, M. , Socci, N.D. , Lash, A.E. , Heguy, A. , Eastham, J.A. , Scher, H.I. , Reuter, V.E. , Scardino, P.T. , Sander, C. , Sawyers, C.L. , Gerald, W.L. , 2010. Integrative genomic profiling of human prostate cancer. Cancer Cell. 18, 11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlins, S.A. , Rhodes, D.R. , Perner, S. , Dhanasekaran, S.M. , Mehra, R. , Sun, X.W. , Varambally, S. , Cao, X. , Tchinda, J. , Kuefer, R. , Lee, C. , Montie, J.E. , Shah, R.B. , Pienta, K.J. , Rubin, M.A. , Chinnaiyan, A.M. , 2005. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 310, 644–648. [DOI] [PubMed] [Google Scholar]
- Vainio, P. , Gupta, S. , Ketola, K. , Mirtti, T. , Mpindi, J.P. , Kohonen, P. , Fey, V. , Perala, M. , Smit, F. , Verhaegh, G. , Schalken, J. , Alanen, K.A. , Kallioniemi, O. , Iljin, K. , 2011. Arachidonic acid pathway members PLA2G7, HPGD, EPHX2, and CYP4F8 identified as putative novel therapeutic targets in prostate cancer. Am. J. Pathol.. 178, 525–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vainio, P. , Lehtinen, L. , Mirtti, T. , Hilvo, M. , Seppanen-Laakso, T. , Virtanen, J. , Sankila, A. , Nordling, S. , Lundin, J. , Rannikko, A. , Oresic, M. , Kallioniemi, O. , Iljin, K. , 2011. Phospholipase PLA2G7, associated with aggressive prostate cancer, promotes prostate cancer cell migration and invasion and is inhibited by statins. Oncotarget. 2, 1176–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verschoor, M.L. , Wilson, L.A. , Verschoor, C.P. , Singh, G. , 2010. Ets-1 regulates energy metabolism in cancer cells. PloS One. 5, e13565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeisig, R. , Koklic, T. , Wiesner, B. , Fichtner, I. , Sentjurc, M. , 2007. Increase in fluidity in the membrane of MT3 breast cancer cells correlates with enhanced cell adhesion in vitro and increased lung metastasis in NOD/SCID mice. Arch. Biochem. Biophys.. 459, 98–106. [DOI] [PubMed] [Google Scholar]
- Zellweger, T. , Gunther, S. , Zlobec, I. , Savic, S. , Sauter, G. , Moch, H. , Mattarelli, G. , Eichenberger, T. , Curschellas, E. , Rufenacht, H. , Bachmann, A. , Gasser, T.C. , Mihatsch, M.J. , Bubendorf, L. , 2009. Tumour growth fraction measured by immunohistochemical staining of Ki67 is an independent prognostic factor in preoperative prostate biopsies with small-volume or low-grade prostate cancer. Int. J. Cancer. 124, 2116–2123. [DOI] [PubMed] [Google Scholar]
- Zhou, X. , Lawrence, T.J. , He, Z. , Pound, C.R. , Mao, J. , Bigler, S.A. , 2012. The expression level of lysophosphatidylcholine acyltransferase 1 (LPCAT1) correlates to the progression of prostate cancer. Exp. Mol. Pathol.. 92, 105–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The following are the supplementary data related to this article.
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data


